Abstract
Transcription factors NF-κB1 and c-Rel, individually dispensable during embryogenesis, serve similar, yet distinct, roles in the function of mature hemopoietic cells. Redundancy among Rel/NF-κB family members prompted an examination of the combined roles of c-Rel and NF-κB1 by using mice that lack both proteins. Embryonic development and the maturation of hemopoietic progenitors were unaffected in nfkb1−/−c-rel−/− mice. Peripheral T cell populations developed normally, but follicular, marginal zone, and CD5+ peritoneal B cell populations all were reduced. In culture, a failure of mitogen-stimulated nfkb1−/−c-rel−/− B cells to proliferate was caused by a cell cycle defect in early G1 that prevented growth. In vivo, defects in humoral immunity and splenic architecture seen in nfkb1−/− and c-rel−/− mice were exacerbated in the double mutant mice. These findings demonstrate that in the B lineage overlapping roles for NF-κB1 and c-Rel appear to be restricted to regulating the activation and function of mature cells.
Rel/NF-κB transcription factors comprise a group of dimeric proteins assembled from a family of related subunits. The mammalian polypeptides (NF-κB1, NF-κB2, RelA, RelB, and c-Rel) share a conserved amino terminus (Rel homology domain, RHD) that encompasses sequences essential for DNA binding, dimerization, and nuclear localization (1). c-Rel, RelA, and RelB possess carboxyl-terminal transcriptional transactivation domains, whereas the cleaved forms of NF-κB1 and NF-κB2, which only comprise the RHD, function as homodimeric transcriptional repressors or modulators of transactivating dimer partners (1, 2). Rel/NF-κB factors exist mainly in the cytoplasm as inactive complexes with IκB proteins (1, 2) and in response to diverse signals are translocated to the nucleus as a result of IKK phosphorylation-induced IκB degradation (3). In the nucleus, Rel/NF-κB factors control transcription by binding specific sequences (κB elements) found within the regulatory regions of many cellular genes (1, 2).
In hemopoietic cells, essential roles served by individual Rel/NF-κB proteins have been revealed in mutant mice generated by gene targeting (4). In the absence of c-Rel, RelA, NF-κB1, or NF-κB2, progenitor differentiation appears normal (4), but the activation and function of mature cells are impaired. For lymphocytes, this includes proliferative defects associated with division and survival and impaired isotype switching and cytokine expression (4), which collectively affect cell-mediated and humoral immunity (2, 4, 5). In single mutant mice, overlapping activities among Rel/NF-κB proteins can lead to phenotypic masking or diminished severity of the defects (4, 5). This process is best illustrated in the B cell lineage, where different combinations of Rel/NF-κB null mutations result in novel defects at distinct developmental junctures. In the absence of NF-κB1 and RelA, B220+ precursors are absent (6). In nfkb1−/−nfkb2−/− mice, B cell development is blocked at the immature IgMhiIgDlo stage (7), whereas a loss of c-Rel and RelA results in differentiation being stalled at the transitional stage (IgMhiIgDhi), a point preceding entry into the mature peripheral B cell pool (8). Here we examined what effects the combined loss of c-Rel and NF-κB1 has on hemopoiesis, in particular B cell development and function. Whereas nfkb1−/−c-rel−/− hemopoietic stem cell differentiation appeared normal, humoral immunity was severely impaired, probably in part a result of profound B cell activation defects that include a failure of B cells to undergo normal growth.
Materials and Methods
Mice.
Mice used in this study were aged between 6 and 10 weeks. nfkb1−/−c-rel−/− mice were generated by intercrossing nfkb1−/− (9) and c-rel−/− (10) mice previously backcrossed eight and 10 times, respectively with the C57BL/6 strain. Wild-type and null alleles for c-rel and nfkb1 were discriminated by PCR of tail biopsy DNA samples (8).
Bone Marrow Cultures.
Bone marrow agar cultures were performed as described (10) by using murine growth factors granulocyte-macrophage colony-stimulating factor, IL-3 (each at 10 ng/ml), and stem cell factor (100 ng/ml). Cultures were incubated at 37°C for 7 days, fixed, and stained, and colonies were identified and enumerated.
Immunofluorescence Staining And Flow Cytometry.
Dispersed cells from the thymus, spleen, bone marrow, lymph nodes, or peritoneum were used for two- and three-color immunofluorescent staining. For two-color stains, T cells were visualized with FITC-conjugated anti-CD4 and R-phycoerythrin (PE)-conjugated anti-CD8 (Caltag, South San Francisco, CA), and B cells were stained with FITC-conjugated B220 and biotinylated anti-IgM or anti-CD5 as described (11, 12). For three-color stains of splenic B lymphocytes, cells were incubated with PE-conjugated anti-IgM, FITC-conjugated anti-CD23, and biotinylated anti-CD21. Biotinylated antibodies were revealed by secondary staining with Streptavidin-Tricolor (Caltag). Between 5,000 and 10,000 viable cells were analyzed by using a FACScan flow cytometer (Becton Dickinson).
Immunization and ELISA Assays.
Mice were immunized with the T-dependent antigen nitrophenyl (NP) coupled to keyhole limpet hemocyanin (KLH) (NP/KLH conjugation ratio of 17:1) by i.p. injection with 100 μg of immunogen precipitated in alum. Serum samples were collected before and 21 days after immunization. NP-specific Ig levels were determined by ELISA (10) using NP17-BSA as a capture agent and goat anti-mouse isotype-specific sera conjugated to horseradish peroxidase as the revealing reagent. Absolute levels of each isotype were determined by using purified myeloma protein standards.
Lymphocyte Activation in Tissue Culture.
Small resting B cells purified to >95% purity by negative sorting (11) were stimulated in culture at an input of 3 × 105/ml in 100 μl with lipoplysaccharide (LPS) (20 μg/ml), goat anti-mouse IgM (Fab′)2 fragments (Jackson ImmunoResearch, 20 μg/ml), rat anti-mouse RP mAb (1 μg/ml), a mitogenic antibody specific for the B cell surface protein RP (10), or recombinant IL-4 (100 units/ml). Proliferation was measured by pulsing cultures with 0.5 μCi [3H]thymidine (Amersham Pharmacia) for 6 h and measuring incorporated radioactivity by scintillation counting.
Apoptosis, Cell Cycle, and Cell Turnover Analysis.
Viability and cell division in mitogen-stimulated B cell cultures were determined by monitoring cellular DNA content with propidium iodide (PI) staining (11). For cell cycle analysis, cells were fixed (>8 h at 4°C) in 70% ethanol, treated with 0.5 μg/ml of DNase-free RNase A for 20 min at room temperature, and stained with 69 μM PI in 0.1 M sodium citrate (pH 7.4) for 30 min at 4°C. Flow cytometric analysis (10,000 cells/sample) was performed on a FACScan and cell cycle distributions were determined with the cellfit program (Becton Dickinson). Cell turnover in mice was determined by labeling proliferating cells with BrdUrd as described (11). Incorporated BrdUrd was detected in immature (sIgMhiIgDlo) and mature (sIgMloIgDhi) splenic B cells by flow cytometry using the FITC-labeled BU-1 mAb (Becton Dickinson) in conjunction with anti-IgM and anti-IgD-specific antibodies.
PCR Detection of Germ-Line Ig CH Transcripts.
Total RNA isolated from B cell cultures was used as a template for cDNA synthesis followed by 30 cycles of PCR amplification as described (8). Oligonucleotides used for PCR amplification of germ-line γ3, γ1, ɛ, α, and β actin transcripts have been described (13). PCR samples were fractionated on 1.5% agarose gels, with the identity of PCR products confirmed by Southern blotting.
Staining of Germinal Centers in Tissue Sections.
Splenic sections were stained with horseradish peroxidase-labeled anti-B220 (6B2) and biotinylated PNA (Vector Laboratories). Bound PNA was visualized by using streptavidin-alkaline phosphatase followed by enzymatic detection with naphthol AS-MX phosphate (Sigma), Fast blue BB Salt (Sigma), and levamisole (Sigma) at 0.125, 0.25, and 0.25 mg/ml, respectively in 0.1 M Tris⋅HCl, pH 8.5. Bound anti-B220 was visualized by staining with anti-rat Ig antibodies coupled to horseradish peroxidase followed by incubation with the horseradish peroxidase substrate.
Results
Embryonic Development and Hemopoiesis Are Normal in nfkb1−/− c-rel−/− Mice.
To determine whether the combined activity of NF-κB1 and c-Rel was essential for mouse development, nfkb1+/−c-rel−/− mice were intercrossed. nfkb1−/−c-rel−/− offspring, born at a frequency of ≈25%, reached maturity without an increased incidence of death, indicating that both transcription factors were dispensable for fetal and neonatal development. Adults displayed no morphological or histological abnormalities, were fertile, and exhibited normal behavior.
Because the loss of NF-κB1 or c-Rel specifically afflicted the function of mature hemopoietic cells (4, 5, 9, 10), we focused our analysis on these lineages in nfkb1−/−c-rel−/− mice. Mature hemopoietic cells from all lineages were observed in the bone marrow, spleen, and blood of naïve nfkb1−/−c-rel−/− mice, with the number of erythrocytes, platelets, granulocytes, eosinophils, and macrophages falling within normal ranges (results not shown). The finding that hemopoiesis in nfkb1−/−c-rel−/− mice appeared to be unaffected was supported by bone marrow colony-forming cell assays, which demonstrated that the frequency of nfkb1−/−c-rel−/− hemopoietic progenitors and their differentiative capacity in culture was normal (see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org).
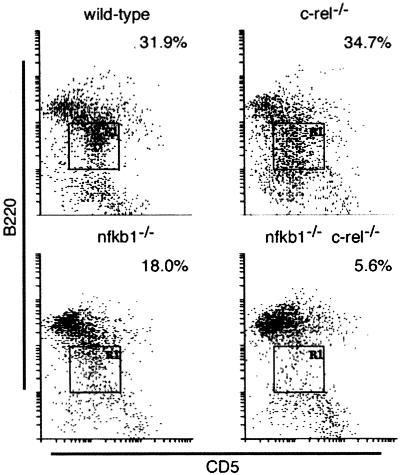
A survey of thymic and splenic lymphoid populations in nfkb1−/−c-rel−/− mice by using flow cytometric analysis of cells stained with labeled mAbs specific for cell surface markers is summarized in Table 1. All major thymocyte subpopulations (CD4−CD8−, CD4+CD8+, CD4+CD8−, CD4−CD8+) were present in normal numbers, as were the CD4+ and CD8+ splenic T cell populations. Thus, T cell development and selection did not appear to be affected. In contrast, various mature nfkb1−/−c-rel−/− B cell populations were reduced. In the spleen, follicular B cell numbers were marginally diminished compared with that of wild-type and single mutant mice. Although follicular B cells comprise a lower proportion of the cellular population in nfkb1−/− spleens compared with wild-type and c-rel−/− mice, the increased splenic cellularity in nfkb1−/− mice (mainly caused by elevated granulocyte levels, unpublished results) meant that absolute follicular B cell numbers were normal. Marginal zone B cells, reduced in nfkb1−/− mice (14), were reduced to a similar extent in nfkb1−/−c-rel−/− animals. The number of CD5+ peritoneal B cells also varied in the different Rel/NF-κB mutants (Fig. 1), with 2- and 6-fold reductions, respectively, in nfkb1−/− and nfkb1−/−c-rel−/− mice.
Table 1.
The proportion of the different T and B cell populations within the thymus and spleen of wild-type, nfkb1−/−, c-rel−/−, and nfkb1−/− c-rel−/− mice of 8–10 weeks of age were determined by flow cytometry using the B- and T cell-specific mAbs as described in Materials and Methods
| Genotype | Spleen
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Thymus
|
T cells, %
|
B cells, %
|
||||||||
| Cellularity, ×107 | CD4−CD8−, % | CD4+CD8−, % | CD4−CD8+, % | CD4+CD8+, % | Cellularity, ×106 | CD4+CD8−, % | CD4−CD8+, % | Marginal zone | Follicular | |
| Wild type | 16 ± 2 | 3 ± 0.4 | 7 ± 1 | 2 ± 0.3 | 88 ± 4 | 70 ± 21 | 21 ± 2 | 16 ± 2 | 4.1 ± 0.3 | 46 ± 7 |
| nfkb1−/− | 17 ± 1 | 2 ± 0.3 | 8 ± 2 | 2 ± 0.3 | 89 ± 4 | 92 ± 17 | 13 ± 3 | 11 ± 3 | 1.7 ± 0.3 | 37 ± 6 |
| c-rel−/− | 15 ± 2 | 2 ± 0.4 | 6 ± 1 | 3 ± 0.2 | 86 ± 4 | 76 ± 18 | 19 ± 2 | 11 ± 1 | 3.7 ± 0.7 | 46 ± 9 |
| nfkb1−/− c-rel−/− | 17 ± 2 | 3 ± 0.2 | 6 ± 1 | 2 ± 0.3 | 87 ± 5 | 77 ± 28 | 18 ± 3 | 13 ± 2 | 1.7 ± 0.6 | 29 ± 7 |
The mean ± SD were based upon the analysis of six mice of each genotype.
Figure 1.
The peritoneal CD5+ B cell population is diminished in nfkb1−/− c-rel−/− mice. Cells isolated from the peritoneal cavity of wild-type, c-rel−/−, nfkb1−/−, and nfkb1−/− c-rel−/− mice were stained with mAbs for B220 and CD5, then examined by flow cytometry. A typical series of staining profiles is shown, with the proportion of CD5+ B cells (boxed region) observed in mice of each genotype expressed as a percentage of the total peritoneal cell population.
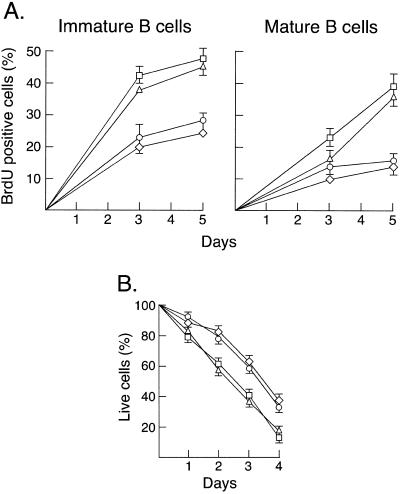
Various studies have highlighted crucial antiapoptotic roles for Rel/NF-κB in mature B cells (15). NF-κB1 promotes the survival of resting splenic B cells (11), whereas c-Rel prevents apoptosis in activated B cells by up-regulating bcl-2-like prosurvival gene expression (16, 17). Because the death of quiescent nfkb1−/− follicular B cells is abnormally high in culture and their turnover is increased in vivo (11), these parameters were examined in nfkb1−/−c-rel−/− mice to determine whether the diminished follicular B cell population reflected levels of apoptosis greater than that observed in nfkb1−/− animals. These results (Fig. 2) show that in the combined absence of NF-κB1 and c-Rel, B cell turnover in vivo measured by BrdUrd incorporation (Fig. 2A) and spontaneous cell death in culture (Fig. 2B) were equivalent to that observed for nfkb1−/− mice. This finding suggests that the reduction in follicular B cells seen in nfkb1−/−c-rel−/− mice is not caused by a survival defect.
Figure 2.
Absence of c-Rel does not enhance spontaneous death in culture or turnover of nfkb1−/− B cells in vivo. (A) Splenic B cell turnover in vivo. Splenocytes isolated from wild-type and mutant mice fed BrdUrd over 5 days were subjected to three-color immunofluorescent staining and flow cytometry to determine the fraction of immature (sIgMhiIgDlo) and mature (sIgMloIgDhi) B cells incorporating BrdUrd. The kinetics of BrdUrd incorporation by immature and mature wild-type (○), nfkb1−/− (▵), c-rel−/− (◊), and nfkb1−/−c-rel−/− (□) B cells over a 5-day time course are summarized. Values represent the mean ± SD from four mice per genotype at each time point; the data are representative of two experiments. (B) Survival of quiescent B cells in culture. Resting splenic B cells from wild-type (○), nfkb1−/− (▵), c-rel−/− (◊), and nfkb1−/−c-rel−/− (□) mice were cultured in DME/10% FCS without mitogens or cytokines for 72 h. At 24-h intervals, the frequency of dead cells was determined by flow cytometric analysis of PI-stained cells. At the start of the experiments, >99% of all cells were viable. The data represent the mean ± SD of five experiments.
Mitogens Fail to Induce nfkb1−/−c-rel−/− B Cell Proliferation.
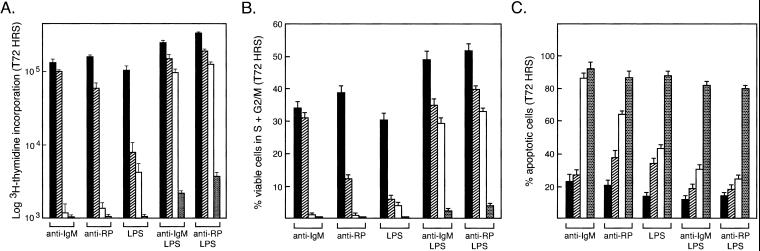
NF-κB1 and c-Rel are critical for B cell proliferation (5, 15), with nfkb1−/− and c-rel−/− B cells exhibiting similar, yet distinct, proliferative defects that arise from impaired cell cycle progression and increased apoptosis (5, 11). nfkb1−/− B cells are hyporesponsive to LPS but respond normally to anti-IgM antibodies (9), whereas c-rel−/− cells fail to divide in response to a variety of single mitogenic signals (10, 11). Because the c-rel−/− B cell proliferative defect is overcome by certain combined stimuli (11), which individually are ineffective, the ability to bypass the defect in c-rel−/− B cells may reflect complementary roles served by other family members such as NF-κB1. This model was tested by examining the proliferative response of nfkb1−/−c-rel−/− B cells (Fig. 3A). Whereas nfkb1−/− and c-rel−/− B cell responses to single and multiple mitogens were as expected (9–11), dual stimuli, such as anti-IgM/LPS or anti-RP/LPS that overcame the defect in c-rel−/− cells, induced only a weak response by nfkb1−/−c-rel−/− B cells. The failure of nfkb1−/−c-rel−/− B cells to proliferate in response to these and other mitogenic signals, including CD40 ligand and IL-4 (results not shown), coincided with a low frequency of cells in the S and G2/M phases of the cell cycle after 72 h stimulation (Fig. 3B), plus a marked increase in apoptosis compared with single mutant B cells (Fig. 3C). The increased severity of the nfkb1−/−c-rel−/− proliferative defect was not caused by reduced survival, as bcl-2 transgene expression inhibited activation-induced apoptosis, but failed to enhance nfkb1−/−c-rel−/− B cell division (results not shown).
Figure 3.
nfkb1−/−c-rel−/− B cells fail to proliferate in response to mitogenic stimulation. (A) B cell proliferative responses. Purified resting wild-type (filled bars), nfkb1−/− (hatched bars), c-rel−/− (empty bars), or c-rel−/−nfkb1−/− (stippled bars) splenic B cells were stimulated for 72 h with optimal concentrations of anti-IgM (Fab)2 antibodies, anti-RP antibodies, LPS, anti-IgM antibodies plus LPS, or anti-RP antibodies plus LPS. Proliferation was measured by [3H]thymidine incorporation. (B) Cell cycle analysis. The fraction of viable cells of each genotype (identification code as above) in the S, G2, or M phases of the cell cycle after stimulation for 72 h with the indicated mitogens was determined by flow cytometric analysis of permeabilized cells stained with PI. Before stimulation, more than 99% of resting B cells were viable and <1% of cells had entered the cell cycle. (C) Cell death. The frequency of apoptotic cells (genotypic code as for A) expressed as a proportion of the total cell number after 72 h of mitogen stimulation was determined by flow cytometric analysis of permeablized cells stained with PI. Cells with a DNA content <2n were deemed to be apoptotic. All results shown represent the mean ± SD from five experiments.
NF-κB1 and c-Rel Cooperate to Regulate B Cell Growth.
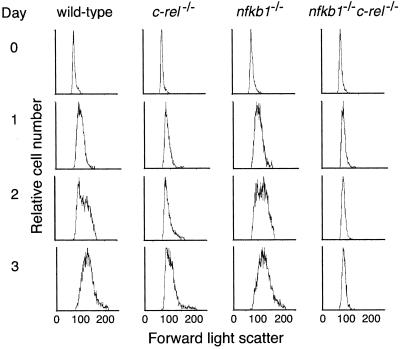
The failure of nfkb1−/−c-rel−/− B cells to respond to any mitogens indicated that the proliferative defect may be distinct from that afflicting nfkb1−/− and c-rel−/− B cells. In the single mutant B cells, the cell cycle block occurs late in G1, after an increase in cell size (11). To test whether the nfkb1−/−c-rel−/− defect occurs earlier in G1, entry of stimulated cells into the cell cycle, as measured by cell size was assessed by using flow cytometry. Forward scatter profiles of untreated and stimulated B cells are shown in Fig. 4. Stimulated nfkb1−/− B cells increase in size normally, whereas c-rel−/− B cells form blasts with slightly delayed kinetics. Activated nfkb1−/−c-rel−/− B cells, however, fail to undergo enlargement, even after 72 h. The inability to form blasts was accompanied by a failure to up-regulate expression of the cell surface markers CD25, B7.1, and B7.2 (results not shown), which normally increase within 24 h of mitogenic activation (18). These results indicate that nfkb1−/−c-rel−/− B cell proliferation is blocked in early G1 at a point preceding growth.
Figure 4.
A combined absence of NF-κB1 and c-Rel prevents mitogen-induced B cell growth. Normal, c-rel−/−, nfkb1−/−, and nfkb1−/−c-rel−/− splenocytes stimulated in culture with anti-RP antibodies plus LPS for 72 h were stained at 24-h intervals with biotinylated B220, then examined by flow cytometry. Forward light scatter profiles (x axis: linear scale) are a measure of cell size. The data shown were electronically gated to exclude dead cells and are representative of three experiments. Similar results were obtained by using different stimuli (results not shown).
The Combined Loss of NF-κB1 and c-Rel Causes Severe Humoral Immune Defects.
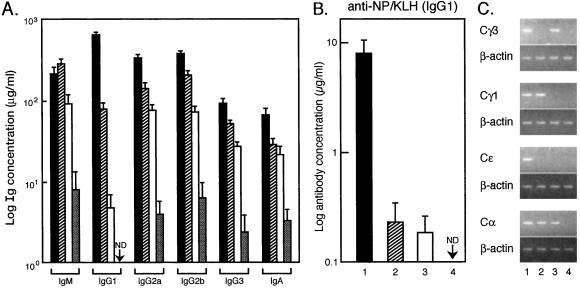
To determine whether the varying severity of mitogen-associated activation defects for nfkb1−/−, c-rel−/−, and nfkb1−/−c-rel−/− B cells in culture were manifest to the same degree in vivo, humoral immune responses in the mutant mice were compared. In naïve animals, most serum Igs, which are lower than normal in the single mutants (9, 10), were further reduced in nfkb1−/−c-rel−/− mice (Fig. 5A), a 30-fold drop in serum IgM compared with relatively normal IgM levels in c-rel−/− and nfkb1−/− mice being the most striking change. In response to immunization with NP-KLH, for which an IgG1 response normally predominates (19), antigen-specific IgG1, although reduced in the single mutants as reported (9, 10), was undetectable in nfkb1−/−c-rel−/− mice 21 days postimmunization (Fig. 5B) and at later times (data not shown).
Figure 5.
Antibody production and germ-line CH gene expression in nfkb1−/−c-rel−/− mice. (A) Serum Ig levels in naive mice. Levels of serum Igs (μg/ml) in naive 8- to 10-week-old litter-matched mice was determined by isotype-specific ELISA. Filled, hatched, empty, and stippled bars correspond to wild-type, nfkb1−/−, c-rel−/−, and nfkb1−/c-rel−/− mice, respectively. Five mice of each genotype were examined. IgG1 levels were below the level of detection (ND) in nfkb1−/c-rel−/− mice. (B) Humoral response to NP-KLH. Five age-matched mice of each genotype were immunized with NP-KLH and serum samples were collected after 21 days. The concentration (μg/ml) of NP-specific IgG1 is indicated for wild-type (filled bars), nfkb1−/− (hatched bars), and c-rel−/− (empty bars) mice. Antigen-specific IgG1 was below the levels of detection (ND) in nfkb1−/c-rel−/− mice. (C) Germ-line CH transcript expression. RNA isolated from equal numbers of wild-type (lane 1), nfkb1−/− (lane 2), c-rel−/− (lane 3), and nfkb1−/−c-rel−/− (lane 4), B cells stimulated in culture for 6 h with various mitogen plus cytokine combinations (13) was subjected to semiquantitative reverse transcription–PCR using primers specific for Cγ3, Cγ1, Cɛ, and Cα germ-line RNAs and β-actin mRNA as a control. The PCR products were then fractionated on agarose gels. B cells were stimulated with LPS alone (Cγ3 RNA), LPS plus IL-4 (Cγ1,Cɛ RNA), and LPS plus transforming growth factor β (Cα RNA).
Poor antibody production by nfkb1−/−c-rel−/− mice could be caused in part by compound defects in isotype switching that result from impaired antigen and cytokine-induced germ-line CH gene transcription in the absence of NF-κB1 (13) or c-Rel (15). This was examined in culture by measuring mitogen plus cytokine-induced germ-line CH gene expression using semiquantitative reverse transcription–PCR (Fig. 5C). Because CH RNA levels were measured within 6 h of stimulation, impaired proliferation and survival should not impact RNA levels. In contrast to restricted defects in CH gene expression seen in nfkb1−/− (Cγ3 and Cɛ RNAs) and c-rel−/− (Cγ1and Cɛ RNAs) B cells, all germ-line CH RNAs examined were undetectable in activated nfkb1−/−c-rel−/− B cells.
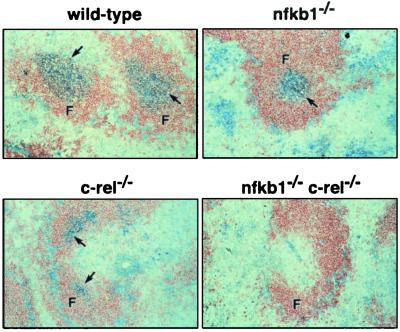
The effect an absence of NF-κB1 and c-Rel had on germinal centers, areas within secondary lymphoid organs where intense B cell proliferation and antigen receptor affinity maturation occurs during an immune response, was also examined. Splenic cryosections from normal and mutant mice immunized with NP-KLH were stained with biotinylated peanut agglutanin (PNA) (Fig. 6), with high-level binding corresponding to germinal center B cells. Whereas normal spleens display large strongly staining PNA+ regions within B cell follicles, germinal center formation was impaired or absent in the mutant mice. Least affected were the nfkb1−/− mice; despite a reduction in the number and size of the germinal centers, PNAhi staining cells were present. In the absence of c-Rel, even fewer clearly demarcated PNA+ germinal centers were present, with those found being small in size and irregular in shape. In nfkb1−/− c-rel−/− mice, no PNA+ cells were observed in the B cell follicles. These findings indicate that either the generation or maintenance of germinal center B cells depends on the activity of NF-κB1 and c-Rel.
Figure 6.
Germinal centers do not form in nfkb1−/−c-rel−/− mice. Sections of spleens from wild-type, nfkb1−/−, c-rel−/−, and nfkb1−/−c-rel−/− mice 14 days postimmunization with NP-KLH were stained with horseradish peroxidase-labeled B220 and biotinylated PNA. Follicles (F) and germinal centers (arrows) are indicated (×100 magnification). The stained sections shown are representative of mice of each genotype.
Discussion
In addition to unique roles ascribed to each Rel/NF-κB subunit, some functional overlap among these transcription factors has meant mutant mice lacking different combinations of proteins are necessary to determine the complete spectrum of functions each family member serves. Here we show that NF-κB1 and c-Rel, despite being coexpressed in various embryonic tissues as early as embryonic day 12.5 (20), are not essential for fetal or neonatal development. Relatively normal development of all hemopoietic lineages in nfkb1−/−c-rel−/− mice was unexpected given the coexpression of NF-κB1 and c-Rel in hemopoietic progenitors (20) and the finding that different aspects of hemopoietic differentiation are disrupted in mice lacking other combinations of Rel/NF-κB proteins (4).
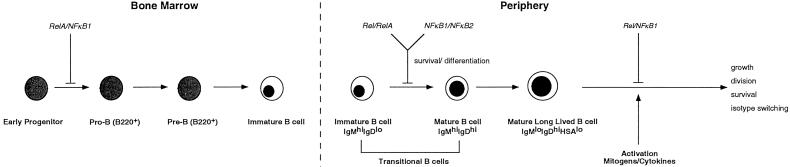
In the B lineage, NF-κB1/RelA and RelA/c-Rel are the predominant heterodimeric complexes in progenitors (21, 22). During the pre-B to B cell transition, up-regulation of NF-κB1 and c-Rel (21, 23, 24) leads to a shift in the relative abundance of different Rel/NF-κB proteins, with NF-κB1/c-Rel and NF-κB1 homodimers emerging as the major complexes found in mature B cells. The importance of NF-κB1/RelA and NF-κB1/c-Rel in normal immature and mature B cells, respectively, is supported by the phenotypes resulting from the loss of these Rel/NF-κB combinations. Whereas NF-κB1 and RelA are essential for the development or survival of B cell progenitors (6), loss of NF-κB1 and c-Rel does not disrupt lymphopoiesis, but instead affects activation-associated functions in mature B cells. Together with roles for NF-κB1/NF-κB2 and c-Rel/RelA in promoting the development and survival of immature and transitional B cells, respectively (7, 8), these findings indicate that different Rel/NF-κB complexes serve essential, nonredundant functions at distinct checkpoints throughout B cell ontogeny (Fig. 7).
Figure 7.
Roles of Rel/NF-κB proteins during B cell development and activation. The functions of different Rel/NF-κB proteins within the B cell lineage were deduced from the phenotype of single and multiple combinations of null mutations for these proteins.
The basis of the selective reduction of various mature B cell populations in nfkb1−/−c-rel−/− mice remains to be determined. Whereas NF-κB1 promotes the survival of quiescent follicular B cells in culture, steady-state numbers are normal in vivo (11). Because both survival in culture and turnover in vivo is equivalent for nfkb1−/− and nfkb1−/−c-rel−/− follicular B cells, the reduction in nfkb1−/−c-rel−/− follicular B cell numbers does not appear to involve increased apoptosis. One possibility is impaired homing of this population in nfkb1−/−c-rel−/− mice. This notion is given credence by the finding that B cell homing is abnormal in nfkb1−/−nfkb2−/− mice caused by reduced expression of the BLR1 B cell chemokine receptor (25). Although survival defects cannot be ruled out as an explanation for the reduction in nfkb1−/−c-rel−/− CD5+ B cells, the observation that CD5+ B cells often are diminished in mice that exhibit B cell proliferative defects (26) is consistent with the findings presented here on the combined roles of NF-κB1 and c-Rel in B cell activation.
Whereas the proliferative defects that afflict nfkb1−/− and c-rel−/− B cells result in part from a cell cycle block in late G1 that occurs after the mitogen-induced increase in cell size, mitogen-treated nfkb1−/−c-rel−/− B cells fail to undergo enlargement. This finding implicates the Rel/NF-κB signaling pathway in the control of mitogen-induced cell growth. Together with previous work (15), it also establishes that in B cells, NF-κB1 and c-Rel regulate a number of distinct steps within the G1 phase of the cell cycle. This combined role for NF-κB1 and c-Rel dimers in growth control within the B lineage appears to be restricted to mature cells, as the growth of nfkb1−/−c-rel−/− B cell progenitors is normal (unpublished results). Whereas growth is intimately linked with the increase in RNA and protein synthesis that precedes DNA replication, little is known about the key molecular regulators of growth in mammalian cells. Additional studies are needed to identify the target genes regulated by Rel/NF-κB that are important for B cell growth.
The results presented here establish that the functions of c-Rel and NF-κB1 in the control of humoral immunity partially overlap. In naïve nfkb1−/−c-rel−/− mice, all serum Igs are further reduced compared with already abnormally low levels seen in the nfkb1−/− or c-rel−/− mutants. The marked drop in serum IgM is particularly interesting, given the finding that CD5+ B cells, which are thought to be a major source of secreted IgM (27, 28), are greatly reduced in nfkb1−/−c-rel−/− mice. Similarly, in response to antigenic challenge, NP-specific antibodies, which were reduced in nfkb1−/− and c-rel−/− mice, were undetectable in the double mutants. Coupled with the drop in serum Igs, the absence of germinal center B cells in nfkb1−/−c-rel−/− mice indicates that affinity maturation of antigen-specific humoral responses is also unlikely. Impaired humoral immunity observed in nfkb1−/−c-rel−/− mice most likely reflects defects in both the B cell and non-B cell compartments of the immune system. Certainly B cell intrinsic defects such as cell division (29) and CH germ-line gene transcription (30) will impact isotype switching. However, the extent to which the disrupted architecture of secondary lymphoid organs, coupled with defects in the function of nfkb1−/−c-rel−/− T cells (31) and antigen-presenting cells (32), accounts for impaired humoral immunity is unclear and will need to be addressed by reconstitution experiments using various combinations of different wild-type and mutant cells. Finally, the generation and characterization of mice lacking multiple members of the Rel/NF-κB family, in addition to revealing interactive roles served by these transcription factors, offers an opportunity to identify relevant target genes whose impaired expression can account for the increasingly complex phenotypes observed in these mutant mice.
Supplementary Material
Acknowledgments
This work was supported by the National Health and Medical Research Council (Australia), the Anti-Cancer Council of Victoria, and the Leukemia and Lymphoma Society of America.
Abbreviations
- NP
nitrophenyl
- KLH
keyhole limpet hemocyanin
- LPS
lipoplysaccharide
- PNA
peanut agglutanin
- PI
propidium iodide
References
- 1.Gilmore T D, Koedood M, Piffat K A, White D W. Oncogene. 1996;13:1367–1378. [PubMed] [Google Scholar]
- 2.Ghosh S, May M J, Kopp E B. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 3.Karin M. Oncogene. 1999;18:6867–6874. doi: 10.1038/sj.onc.1203219. [DOI] [PubMed] [Google Scholar]
- 4.Gerondakis S, Grossmann M, Nakamura Y, Pohl T, Grumont R J. Oncogene. 1999;18:6888–6895. doi: 10.1038/sj.onc.1203236. [DOI] [PubMed] [Google Scholar]
- 5.Gerondakis S, Grumont R, Rourke I, Grossmann M. Curr Opin Immunol. 1998;10:353–359. doi: 10.1016/s0952-7915(98)80175-1. [DOI] [PubMed] [Google Scholar]
- 6.Horwitz B H, Scott M L, Cherry S R, Bronson R T, Baltimore D. Immunity. 1997;6:765–772. doi: 10.1016/s1074-7613(00)80451-3. [DOI] [PubMed] [Google Scholar]
- 7.Franzoso G, Carlson L, Xing L, Poljak L, Shores E W, Brown K D, Leonardi A, Tran T, Boyce B F, Siebenlist U. Genes Dev. 1997;11:3482–3496. doi: 10.1101/gad.11.24.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grossmann M, O'Reilly L A, Gugasyan R, Strasser A, Adams J M, Gerondakis S. EMBO J. 2000;19:6351–6360. doi: 10.1093/emboj/19.23.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sha W C, Liou H C, Tuomanen E I, Baltimore D. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 10.Kontgen F, Grumont R J, Strasser A, Metcalf D, Li R, Tarlinton D, Gerondakis S. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 11.Grumont R J, Rourke I J, O'Reilly L A, Strasser A, Miyake K, Sha W, Gerondakis S. J Exp Med. 1998;187:663–674. doi: 10.1084/jem.187.5.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerondakis S, Strasser A, Metcalf D, Grigoriadis G, Scheerlinck J-P Y, Grumont R J. Proc Natl Acad Sci USA. 1996;93:3405–3409. doi: 10.1073/pnas.93.8.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snapper C M, Zelazowski P, Rosas F R, Kehry M R, Tian M, Baltimore D, Sha W C. J Immunol. 1996;156:183–191. [PubMed] [Google Scholar]
- 14.Cariappa A, Liou H C, Horwitz B H, Pillai S. J Exp Med. 2000;192:1175–1182. doi: 10.1084/jem.192.8.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gugasyan R, Gumont R, Grossmann M, Nakamura Y, Pohl T, Nesic D, Gerondakis S. Immunol Rev. 2000;176:134–140. doi: 10.1034/j.1600-065x.2000.00615.x. [DOI] [PubMed] [Google Scholar]
- 16.Grumont R J, Rourke I J, Gerondakis S. Genes Dev. 1999;13:400–411. doi: 10.1101/gad.13.4.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee H H, Dadgoster H, Cheng Q, Shu J, Cheng J. Proc Natl Acad Sci USA. 1999;96:9136–9141. doi: 10.1073/pnas.96.16.9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gold M R, DeFranco A L. Adv Immunol. 1994;55:221–295. doi: 10.1016/s0065-2776(08)60511-8. [DOI] [PubMed] [Google Scholar]
- 19.Lalor P A, Nossal G J V, Sanderson R D, McHeyzer-Williams M G. J Immuol. 1992;22:3001–3011. doi: 10.1002/eji.1830221136. [DOI] [PubMed] [Google Scholar]
- 20.Carrasco D, Weih F, Bravo R. Development (Cambridge, UK) 1994;120:2991–3004. doi: 10.1242/dev.120.10.2991. [DOI] [PubMed] [Google Scholar]
- 21.Grumont R J, Gerondakis S. Cell Growth Differ. 1994;5:1321–1331. [PubMed] [Google Scholar]
- 22.Klug C A, Gerety S J, Shah P C, Chen Y-Y, Rice N R, Rosenberg N, Singh H. Genes Dev. 1994;8:678–687. doi: 10.1101/gad.8.6.678. [DOI] [PubMed] [Google Scholar]
- 23.Liou H C, Sha W C, Scott M L, Baltimore D. Mol Cell Biol. 1994;14:5349–5359. doi: 10.1128/mcb.14.8.5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyamoto S, Schmitt M J, Verma I M. Proc Natl Acad Sci USA. 1994;91:5056–5060. doi: 10.1073/pnas.91.11.5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolf I, Pevzner V, Kaiser E, Bernhardt G, Claudio E, Siebenlist U, Forster R, Lipp M. J Biol Chem. 1998;273:28831–28836. doi: 10.1074/jbc.273.44.28831. [DOI] [PubMed] [Google Scholar]
- 26.Hayakawa K, Hardy R R. Curr Opin Immunol. 2000;12:346–353. doi: 10.1016/s0952-7915(00)00098-4. [DOI] [PubMed] [Google Scholar]
- 27.Haughton G, Arnold L W, Whitmore A C, Clarke S H. Immunol Today. 1993;14:84–87. doi: 10.1016/0167-5699(93)90064-R. [DOI] [PubMed] [Google Scholar]
- 28.Kantor A B, Herzenberg L A. Annu Rev Immunol. 1993;11:501–538. doi: 10.1146/annurev.iy.11.040193.002441. [DOI] [PubMed] [Google Scholar]
- 29.Hodgkin P D, Lee J H, Lyons A B. J Exp Med. 1996;184:277–281. doi: 10.1084/jem.184.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snapper C M, Marcu K B, Zelazowski P. Immunity. 1997;6:217–223. doi: 10.1016/s1074-7613(00)80324-6. [DOI] [PubMed] [Google Scholar]
- 31.Zheng Y, Ouaaz F, Bruzzo P, Singh V, Gerondakis S, Beg A A. J Immunol. 2001;166:4949–4957. doi: 10.4049/jimmunol.166.8.4949. [DOI] [PubMed] [Google Scholar]
- 32.Grumont R, Hochrein H, O'Keeffe M, Gugasyan R, White C, Caminschi I, Cook W, Gerondakis S. J Exp Med. 2001;194:1021–1032. doi: 10.1084/jem.194.8.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.