Abstract
Shc, a prototypic adapter molecule, has been implicated in T cell receptor (TCR) signal transduction, but its role has not been identified clearly. Here we report that Shc is essential for TCR-induced IL-2 production but is dispensable for CD69 or CD25 expression. Engagement of TCR in mutant Jurkat T cells lacking Shc fails to produce IL-2 because of impaired mitogen-activated protein kinase activation. Activation of c-Rel, a transcription factor essential for IL-2 expression, was impaired also. In contrast, activation of nuclear factor of activated T cell and expression of CD69/CD25 were comparable between the mutant and wild-type Jurkat cells. These defects were rescued by expression of exogenous Shc. Activation of c-Rel using the estrogen receptor fusion protein restored the activation of the IL-2 promoter in an estrogen-dependent manner. These results show that Shc plays an essential role in the TCR-induced activation of c-Rel and the IL-2 promoter.
The T cell receptor (TCR) initiates signal transduction through the intracellular regions of the CD3 and ζ molecules. Each of these molecules contains a sequence of (D/E)XXYXX(I/L)X6–8YXX(I/L), referred to as an immunoreceptor tyrosine-based activation motif (ITAM; refs. 1–3). Antigenic stimulation leads to phosphorylation of both tyrosines in ITAMs. A Src family tyrosine kinase, Lck, plays a major role in this phosphorylation process (4, 5). When both tyrosines in a single motif are phosphorylated, ITAM forms a binding site for ZAP-70 and recruits this kinase from the cytoplasm (4, 6, 7). After this recruitment, ZAP-70 gets activated and initiates the downstream signaling cascade (8, 9).
LAT, a prominent 36–38-kDa tyrosine-phosphorylated protein present after TCR engagement, is a ZAP-70 substrate and plays critical roles in recruitment and activation of downstream signaling molecules such as phospholipase Cγ-1 (PLCγ-1) and Gads/Grb2 (10–13). LAT contains a transmembrane domain and palmitoylation sites (10, 14). Palmitoylation is required for LAT localization into the glycolipid-enriched microdomains and for efficient tyrosine phosphorylation. Based on these results, a model was proposed whereby activated ZAP-70 phosphorylates LAT, leading to the subsequent recruitment of Grb2, PLCγ-1, and other signaling molecules to the glycolipid-enriched microdomains of the plasma membrane (15). Such recruitment could lead to an increase in intracellular Ca2+ and activation of Ras (16, 17).
Shc, a prototypic adapter molecule (18, 19), also was implicated in TCR-induced T cell activation (20–26). Shc is a Zap-70 substrate, is phosphorylated at tyrosine residues, and forms a complex with Grb2 after TCR stimulation (22, 24). This Shc–Grb2 complex formation leads to enhanced association of Grb2 and SOS, a GDP-GTP exchanger for Ras (23). Expression of mutant forms of Shc blocks TCR-induced signal transduction (20, 21, 26), and glycolipid-enriched microdomain-targeted expression of Shc leads to the enhancement of TCR signals (25). However, the specific function of Shc in TCR activation is yet to be determined.
IL-2 is an essential lymphokine for T cell growth and immune regulation (27). Its expression mechanism has been studied extensively, and several transcription factors including NF-AT, AP-1, and NF-κB have been identified as major regulators (28). NF-AT is activated mainly by a Ca2+-dependent protein phosphatase, calcinuerin (29). The increase of intracellular calcium activates this phosphatase and induces nuclear translocation of NF-AT. AP-1, which consists of two transcription factors, Fos and Jun, is another key component for activation of the IL-2 promoter. AP-1 binding sites are found in juxtaposition to NF-AT and NF-κB binding sites, and it has been shown that NF-AT and AP-1 function in a cooperative manner (30). Binding sites for NF-κB, a group of transcription factors involved in the regulation of many genes (31), are found in the IL-2 promoter region, and members of the NF-κB family, RelA and c-Rel, are activated by TCR stimulation (27, 31). Transgenic mice expressing the dominant active form of the inhibitor of NF-κB, IκBα, have been established, and it was demonstrated that inhibition of NF-κB results in severe suppression of lymphokine production (32). Moreover, c-Rel knockout mice have been generated, and it has been shown that the absence of c-Rel expression impairs TCR-induced production of IL-2 and other cytokines (33–36). In this report, we present data from experiments using J.SL1 cells, a Shc loss mutant of Jurkat cells, and demonstrate that Shc is required for activation of c-Rel and plays an essential role for IL-2 production.
Materials and Methods
Cells, Antibodies, and Chemicals.
Jurkat cells were a gift from A. Weiss (University of California, San Francisco). The J.SL1 cell line was isolated from Jurkat cells transfected with an expression construct for the Shc (239, 240F) mutant (37). J.MAK14 was established by transfecting J.SL1 with the expression construct for human Shc cDNA using pEneo-Mos vector (a gift from Y. Takagaki, Kitasato University, Kanagawa, Japan). Anti-Jurkat TCR (C305) was obtained from A. Weiss, anti-Lck (MOL171) from Y. Koga (Tokai University, Kanagawa, Japan), and anti-Slp76 from G. Koretzky (University of Pennsylvania, Philadelphia). Anti-phosphotyrosine (4G10), anti-Vav, and anti-PLCγ-1 were purchased from Upstate Biotechnology (Lake Placid, NY), anti-Shc, anti-Ras, and anti-Grb2 from Transduction Laboratories (San Diego), anti-Cbl, anti-SOS, anti-mitogen-activated protein kinase kinase (MEK), anti-extracellular response kinase (ERK), anti-estrogen receptor (ER), and anti-c-Rel from Santa Cruz Biotechnology, and anti-CD69 and anti-CD25 from PharMingen. Anti-ZAP-70 (2F3.2) has been described (4). PD98059, anti-phospho-ERK, anti-phospho-p38, and anti-phospho-c-Jun N-terminal kinase were from New England Biolabs), and phorbol 12-myristate 13-acetate (PMA) and ionomycin from Calbiochem.
Cell Stimulation, Immunoprecipitation, Western Blot, and Gel Shift Assay.
For Western blot and Ca2+ assays, cells were stimulated with soluble C305 (1 mg/ml) purified from ascites fluid. PMA was used at 10 ng/ml. Ionomycin was used at 1.0 μM. For IL-2 production and fluorescence-activated cell sorter (CD69 and CD25 expression) and Northern blot analyses, cells were stimulated with plate-coated C305 (coating conditions: C305 in PBS at 0.3 μg/ml for 3 h at room temperature or overnight at 4°C). The conditions of lysis buffer, immunoprecipitation, and Western blot were described elsewhere (38). For nuclear extracts, samples were prepared according to the procedure reported previously (39). The oligonucleotide corresponding to the NF-IL-2B site (5′-gtttaaagaaattccaaagagtcatcagaagagg-3′; ref. 40) was used as the probe for the gel shift assay. The binding reaction with the antibody was carried out according to the protocol suggested by the manufacturer.
Reporter and Expression Constructs.
NF-AT luciferase was obtained from G. Crabtree (Stanford University, Stanford, CA). AP-1 luciferase was from Takara (Shiga, Japan) and modified by inserting four sites of 12-O-tetradecanoylphorbol-13-acetate response elements (TGAGTC) into the original construct. NF-κB luciferase was from J. Fujisawa (Kansai Medical University, Osaka). IL-2 luciferase was constructed by inserting luciferase cDNA into the first exon of the IL-2 gene at the translation initiation site. The construct contains the region spanning 2 kb 5′ from exon 1 to the 3′ end of exon 3. For the transient expression construct of MEK and active MEK (MEKDSESE; ref. 41), Xenopus cDNA (from E. Nishida and Y. Gotoh, Kyoto University, Kyoto) were inserted into pME18S vector (a gift from K. Maruyama, Tokyo Medical and Dental College, Tokyo). c-Rel–ER fusion construct from M. Zurobech (University of Prague, Prague); ref. 42) was cloned into pMKIT-neo vector (from K. Maruyama, Tokyo Medical and Dental College, Tokyo).
Transfection, Luciferase Assay, Fluorescence-Activated Cell Sorter Analysis, and IL-2 Assay.
Luciferase assays were carried out by using assay kits from Promega using the conditions described (9). Each reporter construct was transfected with a cytomegalovirus promoter-driven expression construct for Renilla luciferase or a simian virus 40 promoter-driven β-galactosidase gene. Transfection efficiency was adjusted by the activity of β-galactosidase or Renilla luciferase. The fold induction for each sample was normalized to an unstimulated Jurkat sample. Fluorescence-activated cell sorter analysis was carried out by using a FACScan (Becton Dickinson). The IL-2 assay was carried out by using kits from R & D Systems.
Northern Blotting.
Northern blotting was carried out by using 10 μg of total RNA as described (43). Total RNA for c-fos blots was isolated from cells cultured for 2 h with or without anti-TCR stimulation. For IL-2 blots, total RNA was isolated from cells cultured for 16 h. The cDNA for shc was from N. Gotoh (Institute of Medical Science, University of Tokyo, Tokyo). IL-2 cDNA was from T. Taniguchi (University of Tokyo, Tokyo), and c-fos cDNA was from Health Science Research Resources Bank (Osaka). The cDNA fragment for G3PDH (glyceraldehyde-3-phosphate dehydrogenase) was purchased from CLONTECH.
Ca2+ Assay.
Cells were loaded with Fura-2 acetoxymethyl ester (Dojindo, Kumamoto, Japan), and the level of Ca2+ was determined on the basis of the ratio between fluorescence from excitation at 340 nm to that from excitation at 380 nm by using a spectrofluorometer RF1500 (Shimadzu).
Results
Expression of Shc Is Required for Induction of IL-2 but Not CD69/CD25.
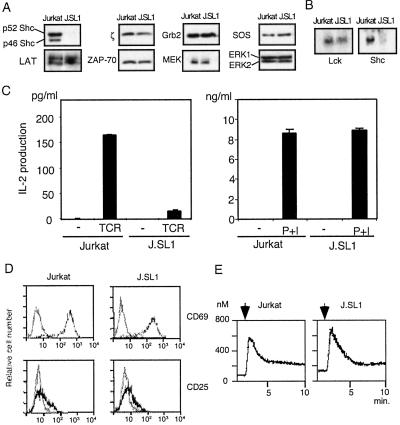
While screening Jurkat cells transfected with the Shc mutant expression construct, we found a cell line that no longer expressed Shc protein (J.SL1; Fig. 1A). The p52 and p46 forms of Shc, derived from the same gene but with different translation initiation sites (18), both were undetectable by Western blotting of J.SL1 lysates. Other molecules involved in the TCR signaling pathway were expressed at the same level between Jurkat and J.SL1 cells (Fig. 1A). J.SL1 and Jurkat cells expressed the surface TCR complex at the same level (data not shown). Northern blot analysis showed that the lack of Shc protein expression is caused by the absence of Shc transcripts (Fig. 1B).
Figure 1.
J.SL1 cells lack expression of Shc and are defective in IL-2 production. (A) Cell lysates of Jurkat and J.SL1 cells were analyzed by Western blots with the antibodies shown on the side of each panel. (B) Northern blot for lck and shc mRNA. Total RNA from Jurkat and J.SL1 cells was analyzed by using the cDNA fragment for lck (Left) or shc (Right). (C) IL-2 production by Jurkat and J.SL1 cells. Cells were cultured 16 h with (TCR) or without (−) stimulation by anti-TCR (Left) or with PMA and ionomycin (P+I, Right). The amount of IL-2 in the culture supernatant from each sample was determined by ELISA. A representative result of several experiments is shown. (D) Surface expression of CD69 and CD25 on Jurkat and J.SL1 cells. Jurkat and J.SL1 cells were treated as described for C, and expression of CD69 and CD25 were examined by using FITC-conjugated antibodies. The dark lines show fluorescence levels detected from activated cells, and gray lines show that from unstimulated cells. (E) Induction of intracellular Ca2+ in Jurkat and J.SL1 cells. Jurkat (Left) and J.SL1 (Right) cells were loaded with Fura-2 acetoxymethyl ester, and the cells were stimulated with anti-TCR at the times shown by the arrow above each panel. The level of Ca2+ was determined on the basis of the ratio between fluorescence from excitation at 340 nm to that from excitation at 380 nm.
To test the functional consequence of the loss of Shc, IL-2 production induced by TCR stimulation was compared between Jurkat and J.SL1 cells. Treatment of Jurkat cells with the plate-coated anti-TCR antibody (C305) induced a significant level of IL-2 secretion, whereas the same treatment of J.SL1 cells induced a minimum amount of IL-2 (Fig. 1C Left). Treatment of cells with a T cell mitogen, phytohemagglutinin, induced IL-2 production only in Jurkat cells and not in J.SL1 (data not shown). However, Jurkat and J.SL1 cells treated with PMA plus calcium ionophore (ionomycin) produced equal amounts of IL-2 (Fig. 1C Right), indicating that the defect of J.SL1 cells lies in the proximal signaling events. In contrast, activation-induced surface antigens such as CD25 (IL-2 receptor α chain) or CD69 were expressed to equivalent levels in activated Jurkat and J.SL1 cells (Fig. 1D). Additionally, engagement of TCR increased intracellular Ca2+ levels in J.SL.1 cells in a manner indistinguishable from Jurkat cells (Fig. 1E).
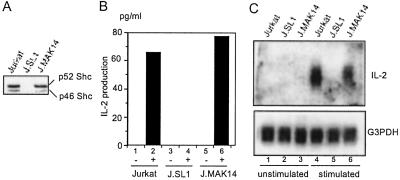
To test whether loss of IL-2 production by J.SL1 cells is caused by a loss of Shc expression, we transfected J.SL1 cells with the expression construct for wild-type Shc and established stable cell lines. J.MAK14, one of these transfectants, expressed Shc protein at an equivalent level to Jurkat cells (Fig. 2A). As shown in Fig. 2B, J.MAK14 responded to anti-TCR stimulation and produced IL-2 at nearly equivalent levels to Jurkat cells. We then measured IL-2 mRNA levels in the three cell lines by using Northern blotting (Fig. 2C). mRNA for IL-2 was not detectable from TCR- stimulated J.SL1 cells, whereas stimulated Jurkat and J.MAK14 cells expressed IL-2 mRNA at equivalent levels.
Figure 2.
The loss of IL-2 production by J.SL1 cells can be reconstituted by the expression of exogenous Shc. (A) Total cell lysates from Jurkat, J.SL1, and Shc-reconstituted J.MAK14 cells were analyzed by Western blot by using anti-Shc antibody. (B) The culture supernatants from unstimulated (lanes 1, 3, and 5) and anti-TCR stimulated (lanes 2, 4, and 6) Jurkat, J.SL1, and J.MAK14 cells were assayed for IL-2 secretion. (C) Total RNA samples from unstimulated (lanes 1, 2, and 3) and stimulated (lanes 4, 5, and 6) Jurkat, J.SL1, and J.MAK14 cells were analyzed by Northern blot with human IL-2 cDNA (Upper). The same filter was rehybridized with glyceraldehyde-3-phosphate dehydrogenase (G3PDH) cDNA (Lower) to show that equivalent amounts of RNA were loaded.
Reduced Activation of Mitogen-Activated Protein Kinase (MAPK) in J.SL1 Cells Is Responsible for Loss of IL-2 Promoter Activity.
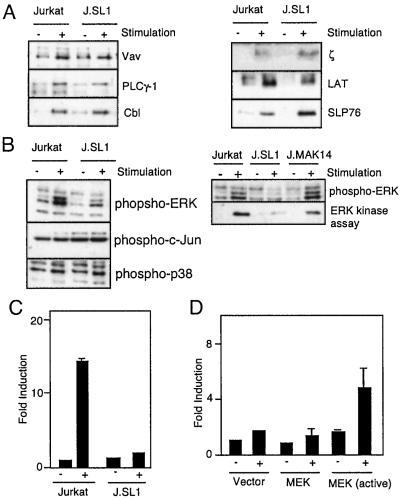
To determine the cause of loss of IL-2 production by J.SL1, we first examined tyrosine phosphorylation of signaling molecules. Jurkat and J.SL1 cells were stimulated with anti-TCR antibody (C305) for 5 min, and signaling molecules were isolated from cell lysates. As shown in Fig. 3A, equivalent amounts of tyrosine phosphorylation were observed in Jurkat and J.SL1 cells for Vav, PLCγ-1, Cbl, LAT, and SLP76. Each immunoprecipitate contained equivalent amounts of protein as confirmed by Western blot analysis (data not shown).
Figure 3.
MAPK activation is impaired in J.SL1 cells and is responsible for the loss of IL-2 production. (A) Tyrosine phosphorylation of signaling molecules in Jurkat and J.SL1 cells. Jurkat and J.SL1 cells were stimulated with anti-TCR antibody. Tyrosine phosphorylation of immunoprecipitated signaling molecules was determined by Western blot with anti-phosphotyrosine antibody. (B) Activation of MAPK (ERK1 and ERK2), c-Jun N-terminal kinase, and p38 by TCR in Jurkat, J.SL1, and J.MAK14 cells. Each cell line was stimulated by anti-TCR antibody. Total cell lysates were prepared at 5 min after stimulation. The lysates were analyzed (Left) by using Western blots with the antibody against phospho-MAPK (Top: ERK1, Upper; ERK2, Lower), phospho-c-Jun, and phospho-p38 (Bottom Left). J.MAK14 cells were also characterized (Right) for MAPK phosphorylation (Upper) and mitogen-activated protein kinase activity (Lower) by using Elk1 as the substrate for in vitro kinase assay of immunoprecipitated MAPK from each cell lysate. (C) Impaired IL-2 promoter activity in J.SL1 cells. Jurkat and J.SL1 cells were transfected with an IL-2 luciferase reporter construct. Each transfectant was stimulated (+) or nonstimulated (−) with anti-TCR antibody, and luciferase activity was determined 24 h later. Transfection efficiency was adjusted by using a dual reporter system (Promega). (D) Reconstitution of IL-2 promoter activity by expression of active MAPK in J.SL1 cells. J.SL1 cells were transfected with the expression construct for the activated MEK [MEK (active)], wild-type MEK (MEK), or the vector alone (Vector) with the IL-2 reporter construct. Sixteen hours after transfection, cells were not stimulated (−) or stimulated with anti-TCR antibody (+), and luciferase activity was determined 24 h after stimulation. Transfection efficiency was adjusted by using the dual reporter system.
We next tested activation of MAPK (ERK1 and ERK2). Shc is involved in activation of MAPK via Ras activation in the signaling pathway for the receptor type tyrosine kinases (44). In T cells, MAPK is also known to play a critical role in the production of IL-2 (45). Thus, one potential cause of loss of IL-2 production by J.SL1 cells is impairment of MAPK activation. To test this hypothesis, phosphorylation and activation of MAPK (ERK1 and ERK2) were examined in TCR-stimulated Jurkat, J.SL1, and J.MAK14 cells (Fig. 3B, Left). TCR stimulation of Jurkat cells induced rapid activation of MAPK. In contrast, the level of phosphorylation and activation of ERK1 and ERK2 were reduced significantly in stimulated J.SL1 cells. Activation of other members of the MAPK family, p38 and c-Jun N-terminal kinase, appeared normal in J.SL1 cells as indicated by the levels of phosphorylation of p38 and c-Jun (Fig. 3B, Left, Bottom). In vitro kinase assays of MAPK using Elk1 as substrate also confirmed that activation of ERK1 and ERK2 were impaired in J.SL1 cells (Fig. 3B Right). Activation of MAPK was restored in J.MAK14 cells to the level observed in Jurkat cells (Fig. 3B Right).
We next tested whether loss of IL-2 mRNA production in activated J.SL1 cells is caused by impaired activation of MAPK. First, the promoter activity of the IL-2 gene in J.SL1 cells was examined. An IL-2-luciferase reporter construct was transfected into Jurkat and J.SL1 cells. As shown in Fig. 3C, TCR stimulation of Jurkat cells induced more than a 10-fold increase of luciferase activity, whereas only a minimal increase was observed in stimulated J.SL1 cells. Next, we cotransfected J.SL1 cells with the reporter gene and an expression construct for wild-type MEK, a mitogen-activated protein kinase kinase, or the activated form of MEK (MEKΔSESE; ref. 41). Cells were stimulated with anti-TCR antibody 16 h after transfection, and the luciferase activity was compared. As shown in Fig. 3D, TCR engagement on J.SL1 cells transfected with the vector alone or with the construct for wild-type MEK showed no induction of promoter activity over the unstimulated samples. In contrast, when the active form of MEK was transfected, anti-TCR stimulation of J.SL1 cells greatly enhanced IL-2 promoter activity. This result demonstrates that the active form of MEK in J.SL1 cells is able to partially restore the signal required for IL-2 production and suggests that MAPK is involved in the downstream signaling process of Shc.
Loss of Shc Caused Impairment of NF-κB Activation but Not AP-1/NF-AT Activation.
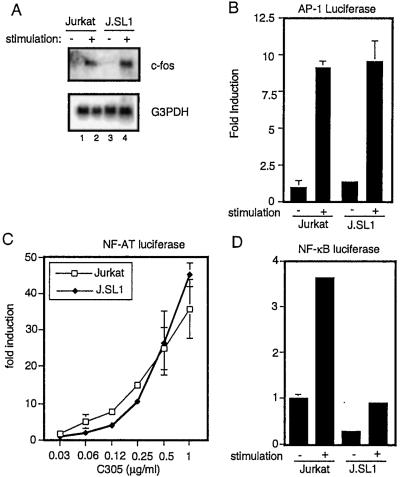
Next we examined the activity of transcription factors that are regulated by the presence of Shc. It has been shown that activation of MAPK induces c-Fos expression (46). c-Fos binds to Jun to form the AP-1 transcription complex. There are several binding sites for AP-1 in the IL-2 promoter including composite binding sites with NF-AT. These sites are indispensable for NF-AT activity (28). Thus, we compared c-fos induction by TCR between Jurkat and J.SL1 cells. Surprisingly, TCR stimulation of J.SL1 cells induced c-fos mRNA expression equally as well as the wild-type Jurkat cells (Fig. 4A). The activity of AP-1-dependent transcription also was equivalent between Jurkat and J.SL1 cells (Fig. 4B). Furthermore, NF-AT-dependent transcription in the two cell lines was induced to almost identical levels by stimulation with a limiting amount of anti-TCR antibody (Fig. 4C). These data indicate that loss of Shc caused the defect in IL-2 production by impairment of elements other than c-Fos or NF-AT.
Figure 4.
Loss of Shc impairs NF-κB-dependent transcription. (A) Total RNA samples from unstimulated (lanes 1 and 3) and anti-TCR-stimulated (lanes 2 and 4) Jurkat and J.SL1 cells were analyzed by Northern blots using c-Fos (Top) and glyceraldehyde-3-phosphate dehydrogenase (G3PDH, Bottom) cDNA. (B) Jurkat and J.SL1 cells were transfected with the reporter construct AP-1-luciferase. Each transfectant was cultured with (+) or without (−) stimulation by the plate-coated anti-TCR antibody. The luciferase activity of each sample was measured in duplicate and is shown as a fold induction over unstimulated Jurkat cells. Transfection efficiency was determined by using a simian virus 40 promoter-driven β-galactosidase expression vector. (C) Jurkat and J.SL1 cells were transfected with the reporter construct NF-AT-luciferase. Each transfectant was cultured with the plate-coated anti-TCR antibody. Concentrations of the coating antibody are shown on the x axis. The luciferase activity of each sample was measured in duplicate and is shown as a fold induction over the unstimulated sample. (D) Jurkat and J.SL1 cells were transfected with the reporter construct NF-κB-luciferase. Each transfectant was cultured with (+) or without (−) stimulation by the plate-bound anti-TCR antibody. The luciferase activity of each sample was measured in duplicate and is shown as a fold induction over the unstimulated Jurkat cells. Transfection efficiency was adjusted by using a cytomegalovirus promoter-driven Renilla luciferase expression vector.
One of the known transcription factors involved in IL-2 gene expression is the NF-κB transcription factors (28). To test whether NF-κB function is impaired in J.SL1, transcriptional activity from the NF-κB family consensus binding sequence was tested for stimulated and unstimulated J.SL1 cells. As shown in Fig. 4D, NF-κB activity was induced in activated Jurkat T cells, whereas the induction in J.SL1 cells was reduced significantly.
c-Rel Activation Is Impaired in J.SL1 Cells.
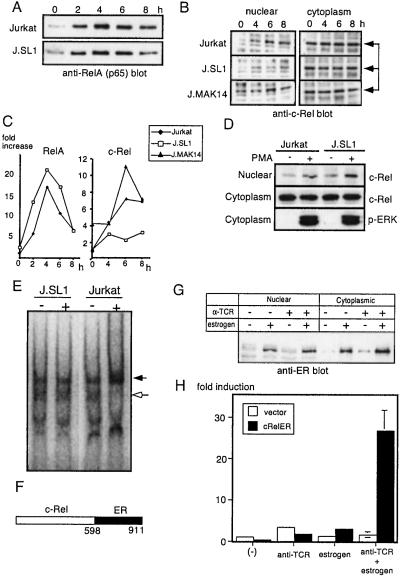
Among the members of the NF-κB family, RelA and c-Rel have been shown to play critical roles in IL-2 gene activation (28). To test whether one of these factors is impaired in J.SL1 cells, nuclear translocation of RelA and c-Rel were examined by using Western blotting. As shown in Fig. 5 A and C, nuclear translocation of RelA was observed at a comparable level between Jurkat and J.SL1 cells after stimulation. In contrast, TCR stimulation induced almost no increase in nuclear localization of c-Rel in J.SL1 cells (Fig. 5 B and C). In a clear correlation with IL-2 expression, stimulation of J.MAK14 cells led to an increase of c-Rel in the nucleus at a level comparable to Jurkat cells and with the same kinetics (Fig. 5 B and C). When Jurkat and J.SL1 cells were treated with PMA to bypass proximal signaling events, c-Rel nuclear localization was induced to the same levels in both cell lines (Fig. 5D). Loss of c-Rel activation in J.SL1 cells also was confirmed by using a gel supershift assay. As shown in Fig. 5E, nuclear extracts mixed with the anti-c-Rel antibody showed a band (indicated by the darkened arrow) induced in activated Jurkat cell extracts but not in unstimulated Jurkat or stimulated/unstimulated J.SL1 cell extracts.
Figure 5.
Reconstitution of IL-2 promoter activity by J.SL1 cells with activation of the c-Rel–ER fusion protein. (A) Nuclear localization of RelA in Jurkat and J.SL1 cells. The time course of nuclear localization of RelA was examined for extracts from Jurkat and J.SL1 cells by Western blot. (B) Nuclear localization of c-Rel is reduced in J.SL1 cells but restored in J.MAK14 cells. The nuclear extract (Left) and cytoplasmic protein (Right) were isolated 4, 6, and 8 h after TCR stimulation. Samples were analyzed by Western blotting using anti-c-Rel antibody. (C) Relative amount of RelA and c-Rel in the nucleus. Relative intensities of bands from the data shown in A and B were determined by using NIH image software. The amount of RelA or c-Rel in the nucleus of Jurkat cells at unstimulated stage (0 h) was taken as 1. (D) Nuclear localization of c-Rel induced by PMA. Jurkat and J.SL1 cells were stimulated with PMA for 30 min. Nuclear and cytoplasmic fractions were analyzed by Western blotting using anti-c-Rel antibody (Top and Middle). An equivalent level of ERK activation was confirmed with anti-phospho-ERK blot (Bottom). (E) Gel supershift assay for c-Rel. Nuclear extracts from stimulated (+) and unstimulated (−) Jurkat and J.SL1 cells were mixed with the 32P-labeled NF-IL-2B site containing oligonucleotide and anti-c-Rel antibody. The shifted band (shown by the darkened arrow) was observed only in activated Jurkat and not in J.SL1 cell nuclear extracts. Note the loss of unshifted band in the activated Jurkat extracts (open arrow). (F) A schematic presentation of the c-Rel–ER fusion protein used in this study with amino acid numbers indicated. (G) Nuclear localization of c-Rel–ER. J.SL1 cells were transfected with the expression construct for c-Rel–ER. Twenty-four hours after transfection, cells were treated with anti-TCR antibody or estrogen as indicated by + above each lane. Nuclear extracts and cytoplasmic fractions of transfectants were analyzed by Western blot using anti-ER antibody. (H) Transient IL-2 promoter assay for J.SL1 transfectants with c-Rel–ER. J.SL1 cells were transfected with the expression vector or the c-Rel–ER expression construct along with the IL-2–luciferase reporter construct as indicated. Four hours after transfection, cells were stimulated with anti-TCR antibody alone, estrogen alone, or anti-TCR antibody and estrogen. Luciferase activity was assayed 18 h after stimulation.
Expression of c-Rel Fusion Protein with ER Restores the Ability of IL-2 Production by J.SL1 Cells.
To test whether loss of c-Rel activation causes impairment of IL-2 production by J.SL1 cells, a c-Rel–ER fusion protein (ref. 42; Fig. 5F) was expressed transiently in J.SL1 cells. This fusion protein stabilizes and translocates into the nucleus when estrogen is present in the culture medium as shown in Fig. 5G. When J.SL1 cells were transfected with the expression construct and stimulated with anti-TCR antibody, a significant level of IL-2 promoter activity was observed only in the presence of estrogen (Fig. 5H). Activation of the IL-2 promoter was not observed when transfectants were treated with anti-TCR alone or with estrogen alone. These results show that c-Rel nuclear translocation can rescue the impaired IL-2 gene expression in J.SL1 cells and that c-Rel is a downstream transcription factor in the signaling pathway of Shc.
Discussion
This study shows that Shc plays a limited but essential role in IL-2 production. Loss of IL-2 production by J.SL1 cells is caused, at least in part, by impairment of MAPK and c-Rel activation. In these cells, TCR-induced activation of MAPK and c-Rel was impaired severely, and expression of the active form of MEK and activation of c-Rel–ER fusion protein restored the activity of the IL-2 promoter.
Although ERK activation is reduced, there is sufficient activity to induce activation of transcription factors (NF-AT and AP-1) and expression of surface antigens (CD25 and CD69). This activation is likely caused by a signal that goes through LAT. However, for the production of IL-2, another signaling pathway that involves Shc apparently is required, and ERK and c-Rel are in the downstream of the pathway. The phenotypic resemblance between J.SL1 and T cells from c-rel knockout mice also supports this model (33–36). Moreover, nuclear localization of c-Rel was blocked completely by treatment of wild-type Jurkat cells with an MEK inhibitor, PD98059 (T.K., H.Y., Y.H., A. Fukushima, and M.I., unpublished data). The amount of PD98059 required for blocking c-Rel translocation was equivalent to that required for inhibition of IL-2 production. These data indicate that the combination of LAT- and Shc-dependent ERK activation is required for a sufficient signal to induce IL-2 expression.
The question remains as to what the differences are between LAT- and Shc-mediated ERK activation. In the thymus, negative- and positive-selecting peptides were shown to induce different patterns of ERK activation (47–51). Negatively selecting peptide induces a strong but short ERK activation, whereas positively selecting peptide induces a long-lasting but less prominent ERK activation, which could be caused by two different ERK activation pathways involving LAT and Shc. In the LAT signaling pathway, it has been shown that the PLCγ-1 binding site is critical for Ras activation (52, 53), which strongly suggests that RasGRP plays a major role in Ras activation downstream of LAT (54). In contrast, the Shc–Grb2 complex enhances binding between Grb2 and SOS, indicating that Shc can activate Ras via SOS (23). In the absence of LAT, the Shc–Grb2 complex is induced normally by TCR stimulation, and sustained Ras activation also is observed at a reduced level (M.T., Y.H., A. Fukushima, J.-W. Chang, and M.I., unpublished data). These two Ras activators could act differently in terms of their kinetics as well as their targets via different scaffold proteins (55, 56). Alternatively, quantitatively stronger activation obtained by a combination of the two pathways with LAT and Shc may lead to phosphorylation of rare or minor substrates that are required for IL-2 production.
Similar requirements of ERK activation were observed previously in neuroblastoma PC12 cells (57). With epidermal growth factor (EGF) stimulation, PC12 cells proliferate without differentiation. In contrast, stimulation of PC12 cells with nerve growth factor (NGF) leads to differentiation of this cell line. Both NGF and EGF induced ERK activation, but activation of ERK by NGF was much stronger and more sustained when compared with EGF stimulation. Thus, ERK activation seems to be partly responsible for the functional difference between the two receptors.
It has been demonstrated that TCR-induced activation of NF-κB requires the presence of PKCθ (58, 59). Although c-Rel and RelA both are controlled by similar mechanisms involving Iκ-B, they behave differently with TCR stimulation (28). RelA nuclear translocation can be observed as early as 15 min after stimulation, whereas it takes 3–4 h for c-Rel to translocate into the nucleus. Here we demonstrate that in the absence of Shc, nuclear translocation of RelA but not of c-Rel is induced by TCR stimulation. These data indicate that Shc provides the signal required for c-Rel activation in addition to that transduced by PKCθ.
Unlike previous reports (20), the data presented here show that Shc is dispensable for AP-1 and NF-AT activation by TCR. This contradiction could be because of different experimental systems. Engagement of TCR induces LAT tyrosine phosphorylation, and the tyrosine-phosphorylated form of LAT forms a complex with Grb2 and Gads (10–13). Binding of these molecules is critical for recruitment of SLP-76, which is essential for the activation of PLCγ-1 (60, 61). Overexpression of Shc mutants potentially competes with LAT for Grb2–Gads binding and inhibits the pathway leading to activation of AP-1 and NF-AT. It also could compete to inhibit phosphorylation of ZAP-70 substrates such as LAT and SLP-76, because Shc is a direct substrate of ZAP-70 (ref. 24; M.T., Y.H., A. Fukushima, J.-W. Chang, and M.I., unpublished data).
A controversial issue regarding Shc involvement in TCR signaling is how it is recruited to the activated TCR complex (62). It has been shown that the affinity of the Src homology 2 domain of Shc to the phosphorylated ITAM is much lower that that of ZAP-70. In this regard, our recent finding that Lck and Shc form a stable complex provides a compelling explanation (M.T., Y.H., A. Fukushima, J.-W. Chang, and M.I., unpublished data). The complex can provide a direct connection between TCR and Shc at the site of activated TCR.
It has been reported that c-Rel is capable of up-regulating AP-1 activity in T cells (63). In J.SL1 cells, although c-Rel activation is impaired greatly, loss of Shc did not affect AP-1 activation. One explanation is that c-Rel and RelA have a redundant function in activating AP-1, and thus loss of c-Rel activation in J.SL1 cells did not affect the AP-1 activity, but overexpression of c-Rel can enhance it. This function of c-Rel may play a critical role in certain T cell activation processes, although it may not be critical for IL-2 gene activation.
In summary, this study demonstrates that Shc is an essential molecule that connects TCR stimulation and IL-2 gene expression by activation of ERK and nuclear localization of c-Rel.
Acknowledgments
We are very grateful to Drs. A. Weiss, G. Crabtree, G. Koretzky, E. Nishida, Y. Gotoh, K. Maruyama, and Y. Takagaki for reagents, A. Asada, J. Goldenring, A. Mellor, and R. B. Markowitz for critical reading of the manuscript, and M. Shibuya, K. Okumura, H. Nakauchi, M. Iwata, S. Sakaguchi, and M. Muramatsu for support. This work was supported in part by grants from the Medical College of Georgia Research Institute and Kanagawa Academy of Science and Technology and National Institutes of Health Grant 1RO1AI47266-01 (to M.I.). This paper is dedicated to the memory of Joel E. Adams, Masao Fukushima, and Toru and Satoshi Iwashima.
Abbreviations
- TCR
T cell receptor
- ITAM
immunoreceptor tyrosine-based activation motif
- PLCγ-1
phospholipase Cγ-1
- ERK
extracellular response kinase
- MEK
mitogen-activated protein kinase kinase
- PMA
phorbol 12-myristate 13-acetate
- ER
estrogen receptor
- MAPK
mitogen-activated protein kinase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Weiss A, Littman D R. Cell. 1994;76:263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 2.Wange R L, Samelson L E. Immunity. 1996;5:197–205. doi: 10.1016/s1074-7613(00)80315-5. [DOI] [PubMed] [Google Scholar]
- 3.Reth M. Nature (London) 1989;338:383–384. [Google Scholar]
- 4.Iwashima M, Irving B A, van Oers N S, Chan A C, Weiss A. Science. 1994;263:1136–1139. [PubMed] [Google Scholar]
- 5.Straus D B, Weiss A. Cell. 1992;70:585–593. doi: 10.1016/0092-8674(92)90428-f. [DOI] [PubMed] [Google Scholar]
- 6.Wange R L, Malek S N, Desiderio S, Samelson L E. J Biol Chem. 1993;268:19797–19801. [PubMed] [Google Scholar]
- 7.Huby R D J, Iwashima M, Weiss A, Ley S C. J Cell Biol. 1997;137:1639–1649. doi: 10.1083/jcb.137.7.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neumeister E, Zhu Y, Richard S, Terhorst C, Chan A C, Shaw A S. Mol Cell Biol. 1995;15:3171–3178. doi: 10.1128/mcb.15.6.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamasaki S, Takamatsu M, Iwashima M. Mol Cell Biol. 1996;16:7151–7160. doi: 10.1128/mcb.16.12.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Sloan-Lancaster J, Kitchen J, Trible R P, Samelson L E. Cell. 1998;92:83–92. doi: 10.1016/s0092-8674(00)80901-0. [DOI] [PubMed] [Google Scholar]
- 11.Gilliland L K, Schieven G L, Norris N A, Kanner S B, Aruffo A, Ledbetter J A. J Biol Chem. 1992;267:13610–13616. [PubMed] [Google Scholar]
- 12.Sieh M, Batzer A, Schlessinger J, Weiss A. Mol Cell Biol. 1994;14:4435–4442. doi: 10.1128/mcb.14.7.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buday L, Egan S E, Viciana P R, Cantrell D A, Downward J. J Biol Chem. 1994;269:9019–9023. [PubMed] [Google Scholar]
- 14.Zhang W, Trible R P, Samelson L E. Immunity. 1998;9:239–246. doi: 10.1016/s1074-7613(00)80606-8. [DOI] [PubMed] [Google Scholar]
- 15.Rudd C E. Cell. 1999;96:5–8. doi: 10.1016/s0092-8674(00)80953-8. [DOI] [PubMed] [Google Scholar]
- 16.Finco T S, Kadlecek T, Zhang W, Samelson L E, Weiss A. Immunity. 1998;9:617–626. doi: 10.1016/s1074-7613(00)80659-7. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, Irvin B J, Trible R P, Abraham R T, Samelson L E. Int Immunol. 1999;11:943–950. doi: 10.1093/intimm/11.6.943. [DOI] [PubMed] [Google Scholar]
- 18.Pelicci G, Lanfrancone L, Grignani F, McGlade J, Cavallo F, Forni G, Nicoletti I, Grignani F, Pawson T, Pelicci P G. Cell. 1992;70:93–104. doi: 10.1016/0092-8674(92)90536-l. [DOI] [PubMed] [Google Scholar]
- 19.Bonfini L, Migliaccio E, Pelicci G, Lanfrancone L, Pelicci P. Trends Biochem Sci. 1996;21:257–261. [PubMed] [Google Scholar]
- 20.Milia E, Di Somma M M, Baldoni F, Chiari R, Lanfrancone L, Pelicci P G, Telford J L, Baldari C T. Oncogene. 1996;13:767–775. [PubMed] [Google Scholar]
- 21.Pratt J C, van den Brink M R, Igras V E, Walk S F, Ravichandran K S, Burakoff S J. J Immunol. 1999;163:2586–2591. [PubMed] [Google Scholar]
- 22.Ravichandran K S, Lee K K, Songyang Z, Cantley L C, Burn P, Burakoff S J. Science. 1993;262:902–905. doi: 10.1126/science.8235613. [DOI] [PubMed] [Google Scholar]
- 23.Ravichandran K S, Lorenz U, Shoelson S E, Burakoff S J. Mol Cell Biol. 1995;15:593–600. doi: 10.1128/mcb.15.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walk S F, March M E, Ravichandran K S. Eur J Immunol. 1998;28:2265–2275. doi: 10.1002/(SICI)1521-4141(199808)28:08<2265::AID-IMMU2265>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 25.Plyte S, Majolini M B, Pacini S, Scarpini F, Bianchini C, Lanfrancone L, Pelicci P, Baldari C T. Oncogene. 2000;19:1529–1537. doi: 10.1038/sj.onc.1203451. [DOI] [PubMed] [Google Scholar]
- 26.Pratt J C, van den Brink M R, Igras V E, Walk S F, Ravichandran K S, Burakoff S J. J Immunol. 1999;163:2586–2591. [PubMed] [Google Scholar]
- 27.Waldmann T A, Dubois S, Tagaya Y. Immunity. 2001;14:105–110. [PubMed] [Google Scholar]
- 28.Jain J, Loh C, Rao A. Curr Opin Immunol. 1995;7:333–342. doi: 10.1016/0952-7915(95)80107-3. [DOI] [PubMed] [Google Scholar]
- 29.Crabtree G R. Cell. 1999;96:611–614. doi: 10.1016/s0092-8674(00)80571-1. [DOI] [PubMed] [Google Scholar]
- 30.Macian F, Lopez-Rodriguez C, Rao A. Oncogene. 2001;20:2476–2489. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- 31.Ghosh S, May M J, Kopp E B. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 32.Boothby M R, Mora A L, Scherer D C, Brockman J A, Ballard D W. J Exp Med. 1997;185:1897–1907. doi: 10.1084/jem.185.11.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kontgen F, Grumont R J, Strasser A, Metcalf D, Li R, Tarlinton D, Gerondakis S. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 34.Gerondakis S, Strasser A, Metcalf D, Grigoriadis G, Scheerlinck J Y, Grumont R J. Proc Natl Acad Sci USA. 1996;93:3405–3409. doi: 10.1073/pnas.93.8.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liou H C, Jin Z, Tumang J, Andjelic S, Smith K A, Liou M L. Int Immunol. 1999;11:361–371. doi: 10.1093/intimm/11.3.361. [DOI] [PubMed] [Google Scholar]
- 36.Strasser A, Grumont R J, Stanley M L, Gerondakis S. Eur J Immunol. 1999;29:928–935. doi: 10.1002/(SICI)1521-4141(199903)29:03<928::AID-IMMU928>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 37.Gotoh N, Toyoda M, Shibuya M. Mol Cell Biol. 1997;17:1824–1831. doi: 10.1128/mcb.17.4.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irving B A, Weiss A. Cell. 1991;64:891–901. doi: 10.1016/0092-8674(91)90314-o. [DOI] [PubMed] [Google Scholar]
- 39.Schreiber E, Matthias P, Muller M M, Schaffner W. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durand D B, Shaw J P, Bush M R, Replogle R E, Belagaje R, Crabtree G R. Mol Cell Biol. 1988;8:1715–1724. doi: 10.1128/mcb.8.4.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gotoh Y, Matsuda S, Takenaka K, Hattori S, Iwamatsu A, Ishikawa M, Kosako H, Nishida E. Oncogene. 1994;9:1891–1898. [PubMed] [Google Scholar]
- 42.Zurovec M, Petrenko O, Roll R, Enrietto P J. Oncogene. 1998;16:3133–3142. doi: 10.1038/sj.onc.1201860. [DOI] [PubMed] [Google Scholar]
- 43.Chien Y H, Iwashima M, Kaplan K B, Elliott J F, Davis M M. Nature (London) 1987;327:677–682. doi: 10.1038/327677a0. [DOI] [PubMed] [Google Scholar]
- 44.Pawson T. Nature (London) 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 45.Pastor M I, Reif K, Cantrell D. Immunol Today. 1995;16:159–164. doi: 10.1016/0167-5699(95)80134-0. [DOI] [PubMed] [Google Scholar]
- 46.Hill C S, Treisman R. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- 47.Hogquist K A. Curr Opin Immunol. 2001;13:225–231. doi: 10.1016/s0952-7915(00)00208-9. [DOI] [PubMed] [Google Scholar]
- 48.Alberola-Ila J, Takaki S, Kerner J D, Perlmutter R M. Annu Rev Immunol. 1997;15:125–154. doi: 10.1146/annurev.immunol.15.1.125. [DOI] [PubMed] [Google Scholar]
- 49.Delgado P, Fernandez E, Dave V, Kappes D, Alarcon B. Nature (London) 2000;406:426–430. doi: 10.1038/35019102. [DOI] [PubMed] [Google Scholar]
- 50.Werlen G, Hausmann B, Palmer E. Nature (London) 2000;406:422–426. doi: 10.1038/35019094. [DOI] [PubMed] [Google Scholar]
- 51.Mariathasan S, Ho S S, Zakarian A, Ohashi P S. Eur J Immunol. 2000;30:1060–1068. doi: 10.1002/(SICI)1521-4141(200004)30:4<1060::AID-IMMU1060>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 52.Lin J, Weiss A. J Biol Chem. 2001;276:29588–29595. doi: 10.1074/jbc.M102221200. [DOI] [PubMed] [Google Scholar]
- 53.Zhang W, Trible R P, Zhu M, Liu S K, McGlade C J, Samelson L E. J Biol Chem. 2000;275:23355–23361. doi: 10.1074/jbc.M000404200. [DOI] [PubMed] [Google Scholar]
- 54.Dower N A, Stang S L, Bottorff D A, Ebinu J O, Dickie P, Ostergaard H L, Stone J C. Nat Immunol. 2000;1:317–321. doi: 10.1038/79766. [DOI] [PubMed] [Google Scholar]
- 55.Bardwell A J, Flatauer L J, Matsukuma K, Thorner J, Bardwell L. J Biol Chem. 2001;276:10374–10386. doi: 10.1074/jbc.M010271200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu W, Fantl W J, Harrowe G, Williams L T. Curr Biol. 1998;8:56–64. doi: 10.1016/s0960-9822(98)70020-x. [DOI] [PubMed] [Google Scholar]
- 57.Marshall C J. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 58.Sun Z, Arendt C W, Ellmeier W, Schaeffer E M, Sunshine M J, Gandhi L, Annes J, Petrzilka D, Kupfer A, Schwartzberg P L, Littman D R. Nature (London) 2000;404:402–407. doi: 10.1038/35006090. [DOI] [PubMed] [Google Scholar]
- 59.Altman A, Isakov N, Baier G. Immunol Today. 2000;21:567–573. doi: 10.1016/s0167-5699(00)01749-7. [DOI] [PubMed] [Google Scholar]
- 60.Tomlinson M G, Lin J, Weiss A. Immunol Today. 2000;21:584–591. doi: 10.1016/s0167-5699(00)01716-3. [DOI] [PubMed] [Google Scholar]
- 61.Clements J L, Boerth N J, Lee J R, Koretzky G A. Annu Rev Immunol. 1999;17:89–108. doi: 10.1146/annurev.immunol.17.1.89. [DOI] [PubMed] [Google Scholar]
- 62.Osman N, Lucas S C, Turner H, Cantrell D. J Biol Chem. 1995;270:13981–13986. doi: 10.1074/jbc.270.23.13981. [DOI] [PubMed] [Google Scholar]
- 63.Shapiro V S, Mollenauer M N, Greene W C, Weiss A. J Exp Med. 1996;184:1663–1669. doi: 10.1084/jem.184.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]