Abstract
Chromatin remodeling enzymes are increasingly implicated in a variety of important cellular functions. Various components of chromatin remodeling complexes, including several members of the SWI/SNF family, have been shown to be disrupted in cancer. In this study we identified as a target for gene inactivation in colon cancer the gene for helicase-like transcription factor (HLTF), a SWI/SNF family protein. Loss of HLTF expression accompanied by HLTF promoter methylation was noted in nine of 34 colon cancer cell lines. In these cell lines HLTF expression was restored by treatment with the demethylating agent 5-azacytidine. In further studies of primary colon cancer tissues, HLTF methylation was detected in 27 of 63 cases (43%). No methylation of HLTF was detected in breast or lung cancers, suggesting selection for HLTF methylation in colonic malignancies. Transfection of HLTF suppressed 75% of colony growth in each of three different HLTF-deficient cell lines, but showed no suppressive effect in any of three HLTF-proficient cell lines. These findings show that HLTF is a common target for methylation and epigenetic gene silencing in colon cancer and suggest HLTF is a candidate colon cancer suppressor gene.
Keywords: methylation‖SWI/SNF
The SWI/SNF family of genes, which encode a group of chromatin remodeling engines, have recently been implicated as playing a role in cancer (1–3). Membership in the SWI/SNF superfamily is defined by a structure of seven DNA-dependent ATPase motifs that is conserved in organisms from yeast to humans, and as members of multiprotein complexes SWI/SNF proteins use the energy of ATP hydrolysis to alter nucleosome position or spacing (4, 5). In addition to chromatin reorganization, SWI/SNF-related proteins have been implicated in many aspects of cellular function, including maintenance of chromosome stability and proper chromosome segregation (lodestar) (6), recombination (RAD54 and RAD54b), nucleotide excision repair (RAD4, ERCC6, and RAD16) (7), and transcriptional activation or repression (MOT1 and HLTF) (8–11). Although most SWI/SNF proteins lack specific DNA-binding motifs and are thought to contribute to transcriptional activation by perturbing chromatin structure to allow promoter access for sequence-specific activator proteins, select members of the SWI/SNF family, such as CHD1 and HLTF (helicase-like transcription factor, also called HIP116a, Zbu1, RUSH1a, and Smarca3) have sequence-specific DNA-binding domains and so can be targeted to specific promoters directly (9, 12, 13).
Recently, mutations of some SWI/SNF family genes were reported in cancers. Human hSNF5/INI1 gene was found to be inactivated frequently in pediatric malignant rhabdoid tumors (3), meningiomas (2), and lymphoid tumors (14). BRG1 inactivation mutations have been reported in human prostate, breast, pancreas, and lung tumor cell lines (15), whereas mutations of RAD54b were reported in human primary lymphoma and colon cancer (1), suggesting that SWI/SNF genes can function as tumor suppressor genes.
In this study, we evaluated HLTF, a SWI/SNF family member gene, as a candidate target for inactivation in cancers. We report here that the HLTF gene was specifically silenced by hypermethylation in 43% of colon cancers. In contrast, no gene silencing was observed in lung or breast samples tested. In addition, transfection of HLTF suppressed colony-forming ability of HLTF-deficient, but not HLTF-proficient, colon cancer cells. HLTF is thus a common target for methylation and epigenetic gene silencing in colon cancer and is a candidate colon cancer suppressor gene.
Materials and Methods
Reverse Transcription–PCR (RT-PCR) Detection and Sequencing of HLTF cDNA.
HLTF cDNA sequence was obtained from GenBank (accession no. Z46606), and primers were chosen by using macvector software (Oxford Molecular, Madison, WI). The HLTF transcript expression was detected by RT-PCR using either of two primer sets that amplify two overlapping fragments spanning the full coding region (base pairs 286-1381 and 1317–3456). Primer sequences were: HLTF-286F (5′-GCTCCTCTTGTCATCCCACTCA), HLTF-1381R (5′-CGTCTTTGCTTAGTCCATCTGCCTT), HLTF-1317F (5′′-CGATGGTCTATGAAACTTGGA), and HLTF-3456R (5′-GAAATTG- TGTCAGTAATACCTCTTCAC).
Methylation-Specific PCR (MS-PCR).
MS-PCR was performed as described (16). Primers for amplifying methylated HLTF promoter templates were P-HLTF1352MF (5′-TGGGGTTTCGTGGTTTTTTCGCGC-3′) and P-HLTF1606MR (5′-CCGCGAATCCAATCAAACGTCGACG-3′). Primers for amplifying unmethylated templates were P-HLTF 1347UF (5′-ATTTTTGGGGTTTTGTGGTTTTTTTGTGT-3′) and P-HLTF1610UR (5′-ATCACCACAAATCCAATCAAACATCAACA-3′). PCR was carried out by using a hot start at 95°C (9 min) and the following cycling parameters: 33 cycles of 95°C (45 s), 66°C (45 s), 72°C (45 s), 72°C (5 min), and 4°C to cool.
Cell Culture and 5-Azacytidine (5-azaC) Treatment.
The Vaco series of colon cancer cell lines was derived and grown as described (17). FET and RCA cells were the kind gift of Michael Brattain, Roswell Park Cancer Institute (18). The cultures were grown and treated with 5-azaC as described (19). The optimal tolerated 5-azaC doses were determined for each treated line, and two doses were used for some lines, ranging from 1 μg/ml to 3 μg/ml.
Clonogenic Assays and Transfections.
Cells were plated in 6-well dishes (12,000–20,00 cells/well) 24 h before transfection in regular growth medium and transfected with 0.4 μg DNA/well with effectene (Qiagen, Chatsworth, CA) according to the manufacturer's protocol. G418 was added to the wells 48 h after transfection, and cells were kept in G418 media (replaced biweekly) for 3 weeks, until tight colonies were observed. Colonies were stained with trypan blue and counted.
Statistical Methods.
Associations of HLTF methylation with sex, human MutL homolog1 (MLH1) methylation status, and CpG island methylator phenotype (CIMP) status were analyzed by using two-tailed Fisher's exact tests. Associations of HLTF methylation status with tumor site or stage were analyzed by using Pearson's χ2 statistics, and we tested for trends by using the Mantel χ2 test for ordered categorical data. Comparisons of age distributions based on HLTF methylation status were done by using Wilcoxon nonparametric tests. Comparisons of colony counts after transfection with different vectors were done by t tests and linear models.
Results
HLTF Is Not Mutated in Colon Cancers.
As several SWI/SNF family genes have been found to be altered in human cancers (1–3, 14, 15), we first determined the sequence of the HLTF cDNA amplified by RT-PCR from 34 colon cancer cell lines matched to primary patient samples in our colon cancer bank. Only one mutation was detected, a hemizygous nonsense mutation at codon 979. Thus HLTF is not a common target for gene mutation in colon cancer.
HLTF Is Frequently Methylated and Silenced in Colon Cancer Cell Lines.
In the process of HLTF sequence analysis in colon cancer cell lines we noted that nine of 34 cell lines did not express HLTF cDNA (Fig. 1A and data not shown). Southern analysis did not identify any alterations in the HLTF locus. Coincidentally, in some of the cell lines that had lost HLTF expression we previously had demonstrated silencing of the hMLH1 gene caused by promoter methylation (19). We therefore examined the genomic sequence upstream of the HLTF (GenBank accession no. NT_005616) where we identified a putative HLTF promoter that we noted indeed contained a CpG dense region that could potentially be methylated (Fig. 2A). No TATA box consensus sequence was found within the putative promoter. However, it did contain a consensus initiator element and two SP1 sites that are typical of TATA-less housekeeping gene promoters.
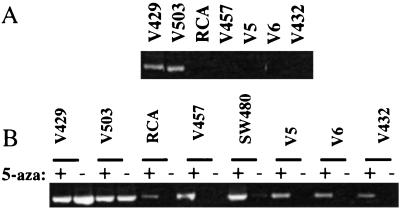
Figure 1.
HLTF silencing in colon cancer cell lines. (A) HLTF RNA expression. Shown is RT-PCR assay for HLTF expression in colon cancer cell lines. (B) HLTF expression reactivation. Shown is RT-PCR assay for HLTF expression in colon cancer cell lines treated (+) or untreated (−) with 5-azaC. Cell lines V429 and V503 are controls with constitutive HLTF expression. 5-AzaC treatment reactivates HLTF expression in cell lines RCA, V457, SW480, V5, V6, and V432.
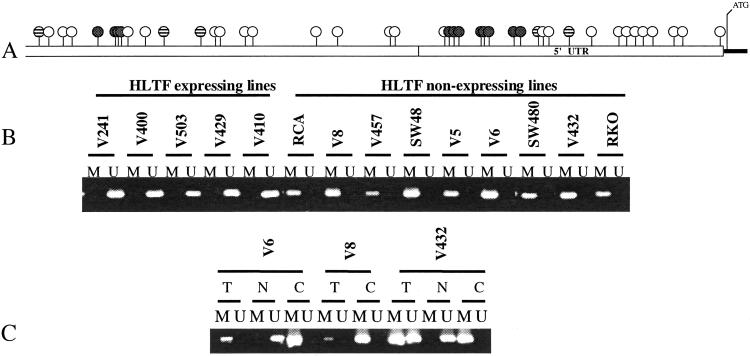
Figure 2.
HLTF promoter methylation. (A) Diagram of the putative HLTF promoter. CpG sites are shown with circles. Shaded circles represent CpG sites that are tested in MS-PCR assays. Hatched circles represent CpG sites that overlap HpaII restriction sites. (B) MS-PCR assay of the putative HLTF promoter. Shown are the results of MS-PCR assay of the putative HLTF promoter by using primers specific for amplification of either methylated (M) or unmethylated (U) templates. (C) HLTF MS-PCR of matched cell lines and tissues. Shown are the results of HLTF MS-PCR assay of colon cancer cell lines (C), matched antecedent tumor tissue (T), or matched normal colon mucosa (N).
To test for methylation of the CpG-rich region of the putative HLTF promoter, we used MS-PCR (16), using PCR primers specific for amplification of either methylated or unmethylated DNA templates (Fig. 2A). As shown in Fig. 2B, all colon cancer cell lines that lacked HLTF gene expression demonstrated methylation of CpG sites within the putative HLTF promoter; whereas, methylation was not detected in the HLTF-expressing cell lines. These results were confirmed by two independent MS-PCR assays that tested different HLTF promoter CpG sites, as well as by resistance of the HLTF promoter to digestion with a methylation-sensitive restriction enzyme, HpaII (data not shown). Thus, cell lines that had silenced the HLTF gene demonstrated methylation that spanned the entire HLTF promoter-associated CpG island, whereas HLTF-expressing cell lines assayed as free of methylated CpG sites.
For three of these HLTF-methylated cell lines (V6, V8, and V432), DNA from matched normal and antecedent tumor DNA was additionally available. In each of these cases, HLTF promoter methylation was detected in the primary tumors, but was absent in the matched normal tissues (Fig. 2C), verifying that HLTF methylation and silencing was a true somatic event and was not an artifact of cell line culture.
Reinduction of HLTF Expression.
To establish that methylation was responsible for silencing HLTF gene expression, cell lines with HLTF promoter methylation were treated with 5-azaC, a demethylating agent. In each of the seven cell lines tested, HLTF expression was strongly reactivated by 5-azaC treatment (Fig. 1B, plus data not shown). However, 5-azaC did not alter HLTF expression in control cell lines in which HLTF expression was constitutive and in which the basal HLTF promoter was unmethylated. In five of the cell lines in which 5-azaC treatment induced re-expression of HLTF, the re-expressed transcripts were PCR-amplified and sequenced. In all five cases, the sequencing confirmed that the re-expressed HLTF allele was wild type. However, in three of these cases the presence of common silent HLTF sequence polymorphisms enabled us to recognize that 5-azaC had reactivated expression of two distinguishable HLTF alleles, of presumptive maternal and paternal origin, both of which had been independently silenced by methylation. Thus aberrant methylation would appear to be an efficient and preferred mechanism for inactivation of the HLTF gene (which maps to cytogenetic position 3q25.1–26.1; ref. 20).
HLTF Methylation Is Widespread in Primary Colon Cancer.
To further establish the frequency of HLTF methylation in primary colon cancer tumors, we analyzed an additional 63 pairs of primary colon tumors along with matched normal tissues (Fig. 3 and data not shown). HLTF methylation was detected in 27 of 63 (43%) primary colon cancers. In contrast, no evidence of HLTF gene silencing was detected in 30 lung cancer cell lines or eight breast cancer samples tested (data not shown).
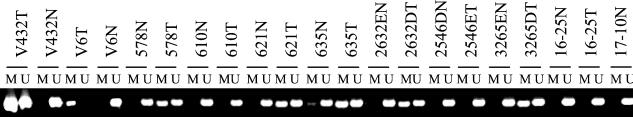
Figure 3.
Methylation status of the putative HLTF promoter in primary tumors and matched normal tissues. Shown are the results of MS-PCR assay of the putative HLTF promoter in matched paired tumor (T) and normal (N) colon tissue samples amplified with primers specific for methylated (M) or unmethylated (U) templates.
The finding of HLTF methylation in colon cancer tumors and cell lines was not significantly correlated with patients' sex (P = 0.31) or age (P = 0.14), with a median age of 72 in persons with HLTF-methylated cancers versus 68 in those with HLTF-unmethylated cancers. In the overwhelming majority of cases (84%), HLTF methylation was detected only in the colon cancers and was absent from the same individuals' normal colon tissues. HLTF methylation thus substantially arose in these individuals specifically during the neoplastic process. In the uncommon cases in which methylation was detected in normal colon tissue, the signal was very faint. Surveying 78 normal colon tissues recovered from cancer resections, we detected such faint HLTF methylation in 12% of cases (nine of 78), with individuals showing such methylation having a median age of 81 compared with 67 for those demonstrating unmethylated HLTF only (P = 0.02). Compared with HLTF-expressing cancers, cancers with methylated and silenced HLTF alleles showed a borderline significant trend (P = 0.06) to be more likely to arise in the proximal right colon and less likely to arise in the rectum, a trend similar to that previously observed for hereditary nonpolyposis colon cancer and sporadic microsatellite instability colon cancer (21) (Table 1). The distribution by tumor stage (adenoma; Dukes' stage B, C, or D cancer primary; or metastatic lesion) was also significantly different between HLTF-methylated and nonmethylated colon neoplasms (P = 0.02) (Table 2). An a postiori grouping of the tumors into metastatic (Dukes' D primary cancers plus cancers from distant metastatic sites) versus nonmetastatic subsets suggests the hypothesis that this difference is caused by a lesser likelihood of HLTF-methylated tumors being metastatic (nominal P value = 0.01).
Table 1.
Correlation of HLTF methylation and tumor site
| Tumor site | HLTF methylated | HLTF nonmethylated |
|---|---|---|
| Right colon | 19 (63%) | 32 (48%) |
| Left colon | 9 (30%) | 20 (30%) |
| Rectal | 2 (7%) | 15 (22%) |
Shown are numbers (and %) of colon neoplasms (tumors and cell lines) in each category defined by location of the tumor in the colon and HLTF methylation status.
Table 2.
Correlation of HLTF methylation and tumor stage
| Tumor stage | HLTF methylated | HLTF nonmethylated |
|---|---|---|
| Adenoma | 7 (18%) | 23 (23%) |
| Dukes' B | 14 (35%) | 31 (31%) |
| Dukes' C | 16 (40%) | 19 (19%) |
| Dukes' D | 2 (5%) | 13 (13%) |
| Metastatic lesion | 1 (3%) | 15 (15%) |
Shown are numbers (and %) of colon neoplasms (tumors and cell lines) in each category defined by clinical stage and HLTF methylation status.
To determine the timing of onset of HLTF silencing during colon carcinogenesis, we additionally analyzed a group of 28 early and late adenomas for HLTF CpG island methylation. HLTF methylation was detected in seven of the adenomas, all of which were larger than 1.5 cm in size and/or demonstrated villous histology, suggesting that HLTF methylation commences at the advanced adenoma stage of colon neoplasia.
HLTF Methylation Defines a Singular Group of Colon Cancers.
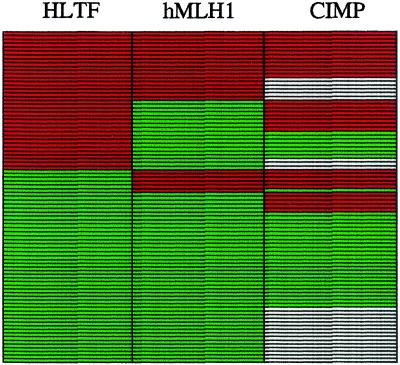
Recently, it has been suggested that certain colon cancers are typified by a high frequency of gene promoter methylation and represent a distinct pathway termed CIMP+ (22, 23). Tumors exhibiting this phenotype (CIMP+) show concordant CpG island methylation affecting multiple genes, including hMLH1, p16, and THBS1. To establish whether HLTF methylation correlates with hMLH1 methylation, and/or with CIMP+, 87 colon cancer cases examined for HLTF methylation were also examined for hMLH1 methylation, and 63 were further assayed for CIMP+ or CIMP− as determined by methylation status of two or more of MINT1, MINT2, MINT31, and MINT27 loci (22). HLTF methylation correlated with CIMP+ (P < 0.001) (Table 3) and as well with hMLH1 gene methylation (P < 0.0001) (Table 4). However, HLTF-methylated tumors essentially defined a distinct subclass of colon cancers that did not fall exclusively into either the hMLH1-methylated or CIMP+ groups (Fig. 4). This finding suggests that aberrant methylation of HLTF might involve at least in part a mechanism distinct from that leading to the CIMP+ or hMLH1 gene methylation, and/or that the current assay for CIMP+ may not be 100% sensitive for recognizing all cancers in which an underlying hypermethylation defect is present.
Table 3.
Correlation of HLTF methylation and CIMP status
| CIMP status | HLTF methylated | HLTF nonmethylated |
|---|---|---|
| CIMP+ | 20 | 11 |
| CIMP− | 7 | 25 |
Shown are the numbers of primary colon cancers in each of the categories defined by combined HLTF methylation and CIMP status.
Table 4.
Correlation of HLTF and hMLH1 methylation
| HLTF methylated | HLTF nonmethylated | |
|---|---|---|
| MLH1 methylated | 20 | 6 |
| MLH1 nonmethylated | 17 | 44 |
Shown are numbers of colon cancers (tumors and cell lines) in each category defined by combined hMLH1 and HLTF methylation status.
Figure 4.
Comparative analysis of methylation in colon cancer. Shown for 87 colon cancer cases are comparative analysis for HLTF methylation, hMLH1 methylation, and CIMP status. Red bars represent positive assays for methylation (HLTF methylated, hMLH1 methylated, or CIMP+ status). Green bars represent nonmethylated findings. White bars indicate data unavailable.
To further determine whether HLTF methylation defines a singular group of colon cancers, we used restriction landmark genomic scanning (RLGS) analysis (24) to compare the patterns of global genome methylation in a group of 12 colon cancer cell lines, six of which demonstrated HLTF methylation and silencing and in six of which HLTF was unmethylated and expressed. A total of 497 loci demonstrated methylation present in at least one of the 12 colon cancer cell lines (D.S., unpublished data). However, none of these loci demonstrated a pattern of methylation matching that of HLTF. That is, no other locus demonstrated the presence of methylation across the six colon cancers in which HLTF was methylated and silenced, plus the absence of methylation across the six colon cancers in which HLTF was unmethylated and expressed. Although RLGS analysis samples only a portion of the genome, these data independently suggest the uniqueness of HLTF methylation, and the potential that if hypermethylated tumors derive from a general class they may also potentially evolve along multiple distinct pathways.
HLTF Reconstitution Induces Growth Suppression.
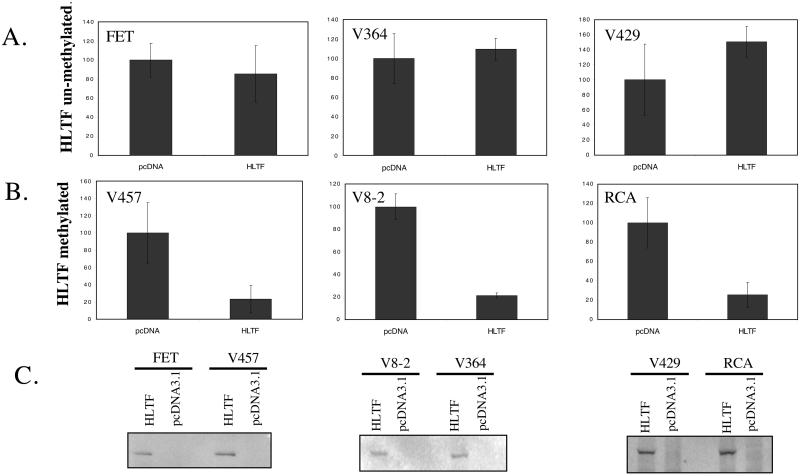
The high frequency of HLTF methylation observed in colon cancer suggested that inactivation of this gene might confer a selective advantage. To assay for such an advantage we examined the effect of HLTF transfection on colony formation in three HLTF-methylated and nonexpressing colon cancer cell lines (V457, V8–2, and RCA) as compared with three HLTF unmethylated and HLTF-expressing colon cancer cell lines (FET, V364, and V429). Reconstitution of HLTF expression in HLTF-methylated cells suppressed colony-forming ability by 75% in each of the three lines tested (P < 0.0001 for each) (Fig. 5B). In contrast, transfection of HLTF did not show significant colony suppression in the any of the three cell lines that already expressed endogenous HLTF (Fig. 5A). Growth suppression by exogenous HLTF was thus specific to colon cancers that had silenced the endogenous alleles (P < 0.01 for the difference in effect of HLTF transfection in HLTF-methylated versus unmethylated cell lines). Transient transfections showed that both HLTF-methylated and unmethylated cells were well able to express exogenous HLTF, as determined by Western analysis for a V5 epitope tag attached to HLTF in the expression vector (Fig. 5C). These findings suggest that HLTF methylation and silencing indeed confers a growth advantage in a distinct subclass of colon cancers.
Figure 5.
HLTF colony suppressor activity. Shown are the number of G418-resistant colonies arising from transfection with an HLTF expression vector (HLTF) or a control empty expression vector (pcDNA) in HLTF unmethylated and expressing FET, V364, and V429 cells (A) as compared with HLTF-methylated and -deficient V457, V8–2, and RCA cells (B). (C) Anti-V5 Western blot assay of V5 epitope-tagged HLTF introduced by transient transfection into HLTF-methylated versus unmethylated cells. Control cells were transfected with an empty expression vector (pcDNA3.1).
Discussion
In this study, we have identified HLTF, a SWI/SNF family gene, as a target gene for methylation and silencing in human colon cancer. HLTF locus methylation was detected in 43% of primary tumors, suggesting that the inactivation of HLTF gene is a frequent event in the process of tumorogenesis in the colon. The methylation of the HLTF promoter region was associated with loss of HLTF gene expression in colon cancer cell lines. Moreover, transfection of HLTF suppresses growth of HLTF-deficient colon cancers, but does not alter growth of HLFT-proficient cancers. These observations suggest that HLTF silencing may confer a growth advantage on some colon cancers, and hence that HLTF may be a colon cancer tumor suppressor gene. No loss of HLTF expression was seen in lung cancer or breast cancer samples that were also examined, consistent with HLTF silencing being a specific event selected for in colon cancer.
Alterations of DNA methylation patterns are widely documented in human neoplasia. Cancerous cells, as compared with normal tissues, frequently show global genome hypomethylation accompanied by regional increases in DNA methylation that are often within CpG-dense promoter regions of specific genes (25). In those colon cancers that show hMLH1 gene promoter methylation and silencing, the somatic hMLH1 methylation plays a causal event in carcinogenesis, as it induces microsatellite instability and perfectly recapitulates the effects of hMLH1 germ-line mutations in kindreds with the syndrome of hereditary nonpolyposis colon cancer (19, 26). In other instances, such as the methylation in some colon cancers of p16 (27, 28), the established tumor suppressor properties of p16 in noncolonic cancers are strongly suggestive of a direct contribution to colon cancer pathogenesis. In yet other instances, for example the estrogen receptor, a high rate of age-associated methylation and the absence of clear colon cancer tumor suppressor activity suggest that promoter methylation may be an accompaniment rather than a cause of colon carcinogenesis (29). We did not detect mutational inactivation of HLTF in colon cancer; however, the high frequency of HLTF methylation, the finding of growth suppression by HLTF, the emerging role of other SWI/SNF family genes in human cancer, and the general absence of HLTF methylation in normal colon when detected in colon cancer all are supportive of HLTF inactivation contributing directly to colon carcinogenesis. The SWI/SNF gene family of chromatin remodeling engines has been associated with both gene transcription and gene silencing, and elucidating downstream targets for HLTF function may provide further evidence for its playing such a direct role in colon cancer.
HLTF promoter methylation correlated strongly with CIMP+. However, unlike colon cancers that demonstrate methylation of the p16 and hMLH1 promoters, which are associated almost exclusively with the CIMP+ tumor class, a significant number of HLTF-methylated tumors fall into the CIMP− group. It is unclear at this point whether this represents two different subsets of tumors with potentially different underlying predispositions for aberrant methylation of the HLTF locus. We noted that a 300-bp fragment of an Alu repeat is present 2 kb upstream of the putative HLTF promoter, and that in normal colon tissue this repeat is heavily methylated (W.C., unpublished data). It is possible that this structure may act as a methylation center from which in some cells methylation spreads outward to the adjacent HLTF promoter.
Age-related methylation in various normal tissues, including colon, has been reported for a number of genes, including ER, MYOD1, IGF1, and N33 (30). Weak methylation of the HLTF locus also was detected in a small subset of normal tissues, mostly in older individuals (median age of 81 years), indicating that the HLTF locus can be silenced during aging in some colon cells of some individuals. As aging is one of the predisposing factors for many cancers, it has been suggested that, at least for some genes, age-related methylation in a precancerous cell could be an early event leading to tumorigenesis, for example by silencing a tumor suppressor gene and thus conferring a selective growth advantage (30). Although we cannot exclude a similar mechanism for HLTF gene, our data indicate that in general HLTF methylation happens subsequent to cancer initiation, most likely in advanced adenomas or frank carcinomas, suggesting that perhaps age-related methylation and HLTF gene silencing in cancers are independent events.
HLTF methylation was observed with a higher frequency in proximal colon and was very rarely observed in rectal cancers. This observation is similar to the previously reported association of CIMP+ and hMLH1 methylator phenotypes with proximal tumors and may reflect increased exposure of the proximal colon to certain classes of carcinogens (29).
In addition to HLTF-methylated tumors appearing to be more often proximal in location, these tumors were less likely to be metastatic to distant sites. Although the mechanism underlying this difference between HLTF-methylated and unmethylated cancers is unclear, it is possible that tumors that have silenced the HLTF gene grow more slowly than tumors arising by means of alternative pathways.
Certainly future studies can be expected to further elucidate the presumptive pathogenetic role that we suggest for HLTF inactivation in colon cancer. Moreover, the high frequency of HLTF methylation in colon cancer may also be useful in potential translational applications. We and others have shown that methylated promoter DNA can be detected in the blood of some cancer patients (31). Thus, it will also be attractive to explore the possibility that assays for methylation of HLTF in body fluids may be of future value for early detection of colon cancer incidence, relapse, or prognosis.
Acknowledgments
We thank Dr. Katherine Jones for helpful discussions. This work is supported by Public Health Service Grants RO1-CA67409, RO1-CA72160, U01-CA88130, P30-CA43703, and P50-CA70907 and gifts from the National Colon Cancer Research Alliance and the G. Harold and Leila Y. Mathers Charitable Foundation. S.D.M. is an Associate Investigator of the Howard Hughes Medical Institute.
Abbreviations
- HLTF
helicase-like transcription factor
- RT-PCR
reverse transcription–PCR
- MS-PCR
methylation-specific PCR
- MLH1
MutL homolog1
- CIMP
CpG island methylator phenotype
- 5-azaC
azacytidine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hiramoto T, Nakanishi T, Sumiyoshi T, Fukuda T, Matsuura S, Tauchi H, Komatsu K, Shibasaki Y, Inui H, Watatani M, et al. Oncogene. 1999;18:3422–3426. doi: 10.1038/sj.onc.1202691. [DOI] [PubMed] [Google Scholar]
- 2.Schmitz U, Mueller W, Weber M, Sevenet N, Delattre O, von Deimling A. Br J Cancer. 2001;84:199–201. doi: 10.1054/bjoc.2000.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Versteege I, Sevenet N, Lange J, Rousseau-Merck M F, Ambros P, Handgretinger R, Aurias A, Delattre O. Nature (London) 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 4.Muchardt C, Yaniv M. J Mol Biol. 1999;293:187–198. doi: 10.1006/jmbi.1999.2999. [DOI] [PubMed] [Google Scholar]
- 5.Sudarsanam P, Winston F. Trends Genet. 2000;16:345–351. doi: 10.1016/s0168-9525(00)02060-6. [DOI] [PubMed] [Google Scholar]
- 6.Girdham C H, Glover D M. Genes Dev. 1991;5:1786–1799. doi: 10.1101/gad.5.10.1786. [DOI] [PubMed] [Google Scholar]
- 7.Eisen J A, Sweder K S, Hanawalt P C. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adamkewicz J I, Hansen K E, Prud'homme W A, Davis J L, Thorner J. J Biol Chem. 2001;276:11883–11894. doi: 10.1074/jbc.M010665200. [DOI] [PubMed] [Google Scholar]
- 9.Ding H, Descheemaeker K, Marynen P, Nelles L, Carvalho T, Carmo-Fonseca M, Collen D, Belayew A. DNA Cell Biol. 1996;15:429–442. doi: 10.1089/dna.1996.15.429. [DOI] [PubMed] [Google Scholar]
- 10.Sheridan P L, Schorpp M, Voz M L, Jones K A. J Biol Chem. 1995;270:4575–4587. doi: 10.1074/jbc.270.9.4575. [DOI] [PubMed] [Google Scholar]
- 11.Gong X, Kaushal S, Ceccarelli E, Bogdanova N, Neville C, Nguyen T, Clark H, Khatib Z A, Valentine M, Look A T, Rosenthal N. Dev Biol. 1997;183:166–182. doi: 10.1006/dbio.1996.8486. [DOI] [PubMed] [Google Scholar]
- 12.Stokes D G, Perry R P. Mol Cell Biol. 1995;15:2745–2753. doi: 10.1128/mcb.15.5.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding H, Benotmane A M, Suske G, Collen D, Belayew A. J Biol Chem. 1999;274:19573–19580. doi: 10.1074/jbc.274.28.19573. [DOI] [PubMed] [Google Scholar]
- 14.Yuge M, Nagai H, Uchida T, Murate T, Hayashi Y, Hotta T, Saito H, Kinoshita T. Cancer Genet Cytogenet. 2000;122:37–42. doi: 10.1016/s0165-4608(00)00274-0. [DOI] [PubMed] [Google Scholar]
- 15.Wong A K, Shanahan F, Chen Y, Lian L, Ha P, Hendricks K, Ghaffari S, Iliev D, Penn B, Woodland A M, Smith R, et al. Cancer Res. 2000;60:6171–6177. [PubMed] [Google Scholar]
- 16.Herman J G, Graff J R, Myohanen S, Nelkin B D, Baylin S B. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, et al. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 18.Wu S P, Theodorescu D, Kerbel R S, Willson J K, Mulder K M, Humphrey L E, Brattain M G. J Cell Biol. 1992;116:187–196. doi: 10.1083/jcb.116.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veigl M L, Kasturi L, Olechnowicz J, Ma A H, Lutterbaugh J D, Periyasamy S, Li G M, Drummond J, Modrich P L, Sedwick W D, Markowitz S D. Proc Natl Acad Sci USA. 1998;95:8698–8702. doi: 10.1073/pnas.95.15.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Y, Sheridan P L, Jones K A, Evans G A. Genomics. 1995;27:381–382. doi: 10.1006/geno.1995.1064. [DOI] [PubMed] [Google Scholar]
- 21.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 22.Toyota M, Ahuja N, Ohe-Toyota M, Herman J G, Baylin S B, Issa J P. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toyota M, Ohe-Toyota M, Ahuja N, Issa J P. Proc Natl Acad Sci USA. 2000;97:710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costello J F, Fruhwald M C, Smiraglia D J, Rush L J, Robertson G P, Gao X, Wright F A, Feramisco J D, Peltomaki P, Lang J C, et al. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 25.Issa J P. Ann NY Acad Sci. 2000;910:140–153. doi: 10.1111/j.1749-6632.2000.tb06706.x. ; discussion 153–155. [DOI] [PubMed] [Google Scholar]
- 26.Herman J G, Umar A, Polyak K, Graff J R, Ahuja N, Issa J P, Markowitz S, Willson J K, Hamilton S R, Kinzler K W, et al. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burri N, Shaw P, Bouzourene H, Sordat I, Sordat B, Gillet M, Schorderet D, Bosman F T, Chaubert P. Lab Invest. 2001;81:217–229. doi: 10.1038/labinvest.3780230. [DOI] [PubMed] [Google Scholar]
- 28.Herman J G, Jen J, Merlo A, Baylin S B. Cancer Res. 1996;56:722–727. [PubMed] [Google Scholar]
- 29.Issa J P. Curr Top Microbiol Immunol. 2000;249:101–118. doi: 10.1007/978-3-642-59696-4_7. [DOI] [PubMed] [Google Scholar]
- 30.Issa J P. Crit Rev Oncol Hematol. 1999;32:31–43. doi: 10.1016/s1040-8428(99)00019-0. [DOI] [PubMed] [Google Scholar]
- 31.Grady W M, Rajput A, Lutterbaugh J D, Markowitz S D. Cancer Res. 2001;61:900–902. [PubMed] [Google Scholar]