Abstract
PURPOSE
To identify gene alterations in circulating tumor DNA (ctDNA) from palbociclib-treated patients with advanced or metastatic breast cancer (ABC) in POLARIS to identify potential mutagenic drivers of resistance.
METHODS
POLARIS was a prospective, real-world study of palbociclib in patients with hormone receptor–positive (HR+)/human epidermal growth factor receptor 2–negative (HER2–) ABC in the United States and Canada. Patients who received ≥1 palbociclib dose and had ≥1 ctDNA measurement were included in the biomarker analysis. ctDNA samples were analyzed using the Guardant360 platform (73 genes) at baseline, cycle 2 day 1 (C2D1), and end of treatment (EOT). Cox proportional hazard models were used to estimate hazard ratios (HRs) and 95% CIs.
RESULTS
A total of 344 patients were included in the biomarker analysis. Gene alterations were detected in 85% (286 of 336) of baseline samples, 72% (201 of 278) of C2D1 samples, and 85% (88 of 104) of EOT samples. The most frequently mutated genes were ESR1, PIK3CA, and TP53. CCND1, FGFR1, and EGFR were most frequently amplified. Real-world progression-free survival (rwPFS) was better in patients without baseline mutations in ESR1 (HR, 0.42) or PIK3CA (HR, 0.60) and amplifications in CCND1 (HR, 0.52) or FGFR1 (HR, 0.62) versus altered genes. Patients with undetectable versus detectable mutations at C2D1 also had better rwPFS (HR, 0.57).
CONCLUSION
Patients without altered ESR1, PIK3CA, CCND1, or FGFR1 at baseline had better rwPFS than patients with altered genes. Genotyping analysis of ctDNA over time highlights the emergence of mutations in estrogen receptor and cell cycle pathways under selective therapeutic pressure and could guide monitoring and therapeutic sequencing for patients with HR+/HER2– ABC.
INTRODUCTION
POLARIS was a real-world study of 1,250 patients with hormone receptor–positive (HR+)/human epidermal growth factor receptor 2–negative (HER2–) advanced or metastatic breast cancer (ABC) receiving treatment with palbociclib in routine clinical practice in the United States and Canada. After a median follow-up of 35.4 months, the median real-world progression-free survival (rwPFS) was 25.8 months (95% CI, 20.7 to 29.5) for palbociclib plus aromatase inhibitor (AI) and 17.1 months (95% CI, 12.9 to 19.4) for palbociclib plus fulvestrant in first-line treatment; in second or more lines (≥2L) of treatment, the median rwPFS was 13.2 months (95% CI, 8.5 to 17.3) and 13.5 months (95% CI, 9.5 to 17.5), respectively.1
CONTEXT
Key Objective
What gene alterations in circulating tumor DNA (ctDNA) from palbociclib-treated patients with advanced or metastatic breast cancer (ABC) in POLARIS might identify potential mutagenic drivers of resistance?
Knowledge Generated
This biomarker analysis of ctDNA samples from patients receiving palbociclib treatment for their hormone receptor–positive (HR+)/human epidermal growth factor receptor 2–negative (HER2–) ABC showed altered/amplified genes in cell cycle pathways (PIK3CA, TP53, CCND1, FGFR1, EGFR) and the ESR1 gene at baseline, cycle 2 day 1, and end of treatment. Real-world progression-free survival was worse in patients with baseline mutations or amplifications than those with wild-type genes, regardless of treatment or line of therapy.
Relevance
These results support the potential use of genotyping analysis of ctDNA over time to guide monitoring and therapeutic sequencing in patients with HR+/HER2– ABC.
A subset of patients with HR+/HER2– ABC show poor response to cyclin-dependent kinase 4/6 inhibitors (CDK4/6i) or experience rapid disease progression, which has spurred efforts to identify mechanisms of resistance to CDK4/6i.2-4 Data from preclinical and clinical studies have suggested several potential primary and acquired mutations in genes encoding components of cell cycle and estrogen and growth factor receptor signaling pathways that could be predictive of CDK4/6i response.4-6 In breast cancer (BC), circulating tumor DNA (ctDNA) profiling can temporally track the mutational landscape of primary tumors and metastases with the potential to predict responses to targeted therapies and adapt treatment.7 Herein, we present results from ctDNA tumor profiling in patients with ABC treated with palbociclib plus endocrine therapy (ET) in the first line or ≥2L of therapy in POLARIS. The aims of this study were to identify gene alterations that could potentially predict a patient's response to palbociclib, to determine if specific mutation losses or gains over time may elucidate mechanisms of resistance, and to investigate whether early changes in ctDNA after treatment with palbociclib were associated with different tumor responses and progression-free survival (PFS) in real-world settings.
METHODS
Study Design
POLARIS was a prospective, noninterventional, multicenter study (ClinicalTrials.gov identifier: NCT03280303). Details of the study design have been previously published.8 Briefly, adult patients with a diagnosis of HR+/HER2– ABC receiving palbociclib in routine clinical practice were enrolled at 123 sites across the United States and Canada from January 4, 2017, to October 3, 2019; the cutoff date for this analysis was January 9, 2023. All study participants signed an informed consent form. Separate consent was provided for serial blood sample acquisition for biomarker analyses. The study was conducted in accordance with local legal and regulatory requirements and reviewed and approved by applicable local institutional review boards.
Outcomes
rwPFS was defined as the duration from study treatment initiation until physician-documented disease progression (based on imaging, biopsies, biomarkers, and/or clinical judgment) or death because of any cause, whichever occurred first.1 Patients who did not have physician-documented disease progression or documented death were censored at the last date of response assessment. Analysis of biomarkers by ctDNA genotyping was an exploratory objective. Mutational variant allele fraction (VAF)9 was calculated from ctDNA genotyping at baseline and on cycle 2 day 1 (C2D1) of treatment.
ctDNA Profiling
Serial blood samples were collected from patients as part of routine clinical assessments at baseline, day 1 of every cycle for the first 6 months, day 1 of every third cycle thereafter, and at the end of treatment (EOT). Profiling of ctDNA was performed using the Guardant360 platform (Guardant Health, Redwood City, CA).10 Tumor gene alteration and DNA copy number amplification (more than two-fold increase) gene profiles were evaluated at baseline, C2D1, and EOT (including progression). Patient samples at EOT were compared with those at baseline to identify acquired and lost mutations. ctDNA clearance was defined as a lack of detectable ctDNA at C2D1 or EOT.
Statistical Analyses
The biomarker analysis set included patients who had consented to blood sample collection, received ≥1 dose of palbociclib combination treatment, and had ≥1 ctDNA measurement available. Median rwPFS estimates by biomarker subgroups were calculated using the Kaplan-Meier method and further analyzed by treatment and known prognostic clinical factors (ie, number of lines of therapy, visceral metastatic disease, bone-only metastatic disease, de novo disease and recurrent disease). Univariate Cox proportional hazards models were used to estimate hazard ratios (HRs) and 95% CIs. To further examine the effects of tumor genetic alterations on rwPFS, multivariable Cox regression analysis was conducted adjusting for the known prognostic clinical factors. In addition, rwPFS was examined by co-occurring genetic alterations.
To quantify changes in ctDNA levels, the maximum value of VAF (mVAF) of all detected nonsynonymous mutations was calculated for each patient at each visit and expressed as a percentage. For those with no alterations detected, the mVAF was set to 0%. The ratio of change in mVAF between C2D1 of treatment and baseline was calculated for each patient. To avoid division by zero, a value of 0.1 was added to the ratio's numerator and denominator. The association between early changes (defined as any increase or decrease from baseline to C2D1) in ctDNA mVAF and real-world best overall response groups (real-world complete response [rwCR]/real-world partial response [rwPR] v real-world stable disease [rwSD] v real-world progressive disease [rwPD]) was evaluated using the one-sided Jonckheere-Terpstra trend test. Tumor response was determined by treating physicians’ clinical judgment based on imaging, biopsies, and biomarkers.1 R version 4.1.0 was used for data analysis and figure generation.
RESULTS
Patient Baseline Demographics and Clinical Characteristics
Of the 1,250 patients in the POLARIS full analysis set (FAS), 476 provided informed consent for serial blood collections. Among these patients, 344 had ctDNA samples that passed quality control and were HER2–, and were included in the biomarker analysis set (Data Supplement, Fig S1). Similar patient characteristics were observed between the FAS and biomarker analysis set (Data Supplement, Table S1). The median (range) age of patients in the biomarker analysis set was 65 years (24-95), and the majority (84.3%) were White; most (95.6%) had metastatic BC (mBC), 4.4% had ABC without mBC, and 28.8% had de novo mBC.
Most Frequently Altered Genes and Gene Amplifications
ctDNA samples were available from 336 patients at baseline, 278 patients at C2D1, and 104 patients at EOT. Gene alterations were detected in 85% (n = 286) of baseline ctDNA samples. Of these patients, 28.0% (n = 80) was diagnosed with de novo mBC and 67.8% (n = 194) had recurrent BC, similar to the FAS and biomarker analysis set. Gene alterations were also detected in 72% (n = 201) of C2D1 samples and 85% (n = 88) of EOT samples. Most frequently mutated genes were PIK3CA (38.4%), TP53 (28.6%), and ESR1 (15.5%) at baseline; TP53 (27.7%), PIK3CA (24.5%), and NF1 (10.4%) at C2D1; and TP53 (40.4%), PIK3CA (39.4%), and ESR1 (31.7%) at EOT (Fig 1A). Most frequent gene amplifications were in CCND1 (8.3%), FGFR1 (7.7%), and EGFR (6.0%) at baseline; FGFR1 (5.0%), CCND1 (4.3%), and PIK3CA (3.6%) at C2D1; and CCND1 (13.5%), FGFR1 (9.6%), and EGFR (9.6%) at EOT (Fig 1B). OncoPrint heat maps of ctDNA alterations are shown in the Data Supplement (Fig S2). Similar results were observed in patients with visceral metastatic disease, bone-only disease, de novo disease, or recurrent disease (Data Supplement, Fig S3), except for patients with de novo disease, who appeared to have more PIK3CA mutations (50.0% v 39.4%) and PIK3CA (13.6% v 5.8%) and EGFR (22.7% v 9.6%) amplifications at EOT versus the overall population (Fig 1).
FIG 1.
Percentage of patients with (A) gene alterations and (B) gene amplifications by treatment stage. C2D1, cycle 2 day 1; EOT, end of treatment.
Identification of Acquired Gene Alterations
The most commonly acquired mutations by ≥5 patients at EOT versus baseline were in ESR1, TP53, ATM, RB1, and CDK12 genes. By contrast, most commonly lost (extinguished) mutations were in PIK3CA and NF1 genes (Fig 2A). ESR1 mutations were gained in 13 ctDNA samples (12.5%) and lost in five (4.8%) of 104 collected at disease progression. Most frequently acquired ESR1 mutations at EOT were D538G, Y537N, and Y537S (Fig 2B). OncoPrint heat maps of change in status at C2D1 and EOT are shown in the Data Supplement (Fig S4).
FIG 2.
(A) Genes most frequently identified with acquired and lost mutations at EOT and (B) most frequently acquired gene mutations in ESR1 at EOT. EOT, end of treatment.
Acquired Gene Alterations by Treatment
Acquired mutations at EOT were most frequently observed for ESR1 and TP53 in the palbociclib plus AI (n = 181; 54%) and palbociclib plus fulvestrant (n = 151; 45%) groups, with more acquired mutations observed in the palbociclib plus AI group (Fig 3A). Acquired mutations at EOT were also observed for ATM, RB1, and CDK12 in both treatment groups. In either treatment group, most frequently lost mutations at EOT were in PIK3CA and NF1 genes. The most frequently acquired mutations in ESR1 at EOT were D538G and Y537N with palbociclib plus AI treatment, whereas D538G and Y537S were the most frequently acquired mutations with palbociclib plus fulvestrant (Fig 3B).
FIG 3.
(A) Genes most frequently identified with acquired and lost mutations at EOT and (B) most frequently acquired gene mutations in ESR1 at EOT by treatment type. AI, aromatase inhibitor; EOT, end of treatment; FUL, fulvestrant; PAL, palbociclib.
Evaluation of rwPFS by Genotype
Mutations in ESR1 and PIK3CA at baseline were associated with shorter median rwPFS than wild-type (WT) genes (Fig 4A). Patients with WT ESR1 at baseline had longer median rwPFS than those with ESR1 mutations (22.3 v 12.0 months) and a 58% reduction in the risk of disease progression or death (HR, 0.42 [95% CI, 0.28 to 0.61]). Patients with WT PIK3CA also had longer median rwPFS than those with PIK3CA mutations (23.3 v 16.2 months), associated with a 40% reduction in the risk of disease progression or death (HR, 0.60 [95% CI, 0.44 to 0.81]). A similar trend toward longer median rwPFS was observed in patients with WT CCND1 (20.2 v 13.1 months) and FGFR1 (20.7 v 11.5 months) versus those with amplifications of those genes at baseline (Fig 4B), which were associated with a 48% (HR, 0.52 [95% CI, 0.32 to 0.84]) and 38% (HR, 0.62 [95% CI, 0.38 to 1.00]) reduction in risk of disease progression or death, respectively.
FIG 4.

Kaplan-Meier curves for rwPFS with baseline (A) mutations in ESR1 and PIK3CA and (B) amplifications of CCND1 and FGFR1. C2D1, cycle 2 day 1; HR, hazard ratio; rwPFS, real-world progression-free survival; WT, wild-type.
Baseline gene mutations and amplifications had similar effects on rwPFS when analyzed by treatment type and lines of therapy (Data Supplement, Fig S5; Fig 5) and when analyzed by other prognostic clinical factors (Data Supplement, Fig S6). Patients with WT ESR1 at baseline (Fig 5A; Data Supplement, Fig S6A) were more likely to have better rwPFS than those with mutated ESR1 when treated with palbociclib plus fulvestrant (HR, 0.59 [95% CI, 0.36 to 0.95]) or plus AI (HR, 0.25 [95% CI, 0.13 to 0.49]) or when they received palbociclib plus ET as a first-line treatment (HR, 0.45 [95% CI, 0.24 to 0.84]) or as ≥2L treatment (HR, 0.51 [95% CI, 0.31 to 0.84]), or when they had visceral metastatic disease (HR, 0.37 [95% CI, 0.21 to 0.65]), de novo disease (HR, 0.44 [95% CI, 0.22 to 0.89]), or recurrent disease (HR, 0.42 [95% CI, 0.28 to 0.61]). Similar findings in better rwPFS were observed with WT PIK3CA, CCND1, and FGFR1 (v mutant alleles) overall and regardless of the ET partner or line of treatment (Figs 5B-5D) or patients with visceral or bone-only metastases or with de novo or recurrent disease (Data Supplement, Figs S6B-S6D). Across patient subpopulations defined by different clinical factors, the associations observed between rwPFS and the genetic alterations are consistent with those in the overall population (Fig 5; Data Supplement, Fig S6). Although not all WT genes were statistically significantly associated with better rwPFS across all subgroups because of smaller sample sizes, their direction of effect remained the same as in the overall population. Multivariable Cox regression analysis adjusted for known prognostic clinical factors showed that the genetic alterations identified in the baseline ctDNA samples remained as independent prognostic factors for rwPFS (data not shown).
FIG 5.
Forest plot depicting effects of baseline mutations in (A) ESR1 and (B) PIK3CA and amplifications in (C) CCND1 and (D) FGFR1 on rwPFS by treatment type and by line of therapy. AI, aromatase inhibitor; FUL, fulvestrant; HR, hazard ratio; PAL, palbociclib; rwPFS, real-world progression-free survival; WT, wild-type.
In our examination of rwPFS by co-occurring genetic alterations (Data Supplement, Fig S7), patients with co-occurring baseline mutations in ESR1 and PIK3CA (two mutations) appeared to have worse rwPFS than those with only one mutation or no mutation (no mutation v two mutations: HR, 0.31 [95% CI, 0.18 to 0.51]; Data Supplement, Fig S7A) and those from either single-mutation analyses (ESR1 WT v mutation: HR, 0.42 [95% CI, 0.28 to 0.61]; Fig 4A; PIK3CA WT v mutation: HR, 0.60 [95% CI, 0.44 to 0.81]; Fig 4B). Co-occurring baseline mutations in TP53, ESR1, and PIK3CA also showed that patients with more baseline mutations consistently had worse rwPFS than those with fewer baseline mutations (Data Supplement, Fig S7B). Conversely, patients with coamplification of both CCND1 and FGFR1 had similar rwPFS to those with only one amplification (Data Supplement, Fig S7C). When analyzed by baseline ESR1 mutation and CCND1 and FGFR1 amplifications, patients with one alteration had similar rwPFS than those with two alterations (Data Supplement, Fig S7D); however, those with three alterations appeared to have worse rwPFS although the sample size was very small (n = 2).
rwPFS Association With the Change in mVAF at C2D1
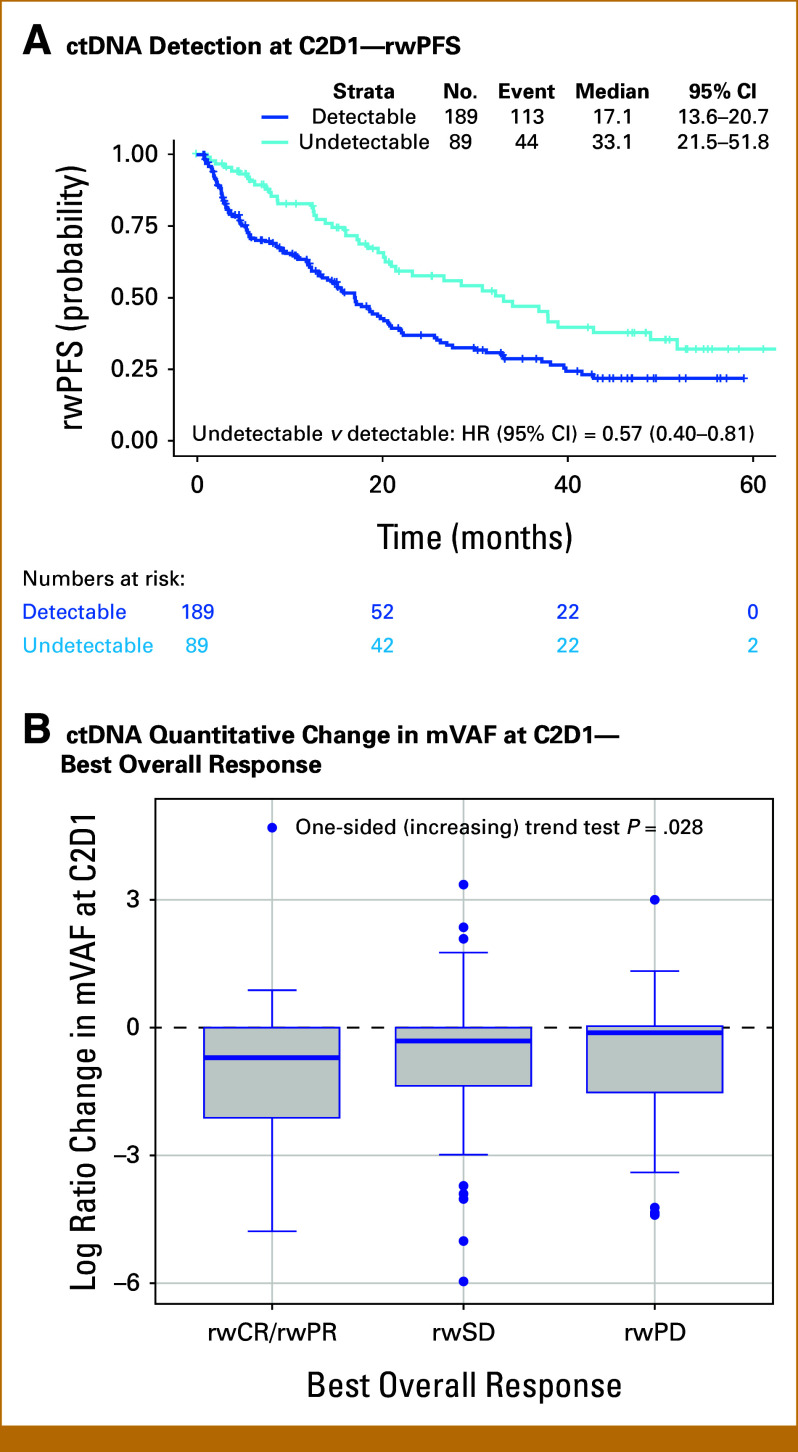
Patients with undetectable mutations at C2D1 had longer median rwPFS than those with detectable mutations (33.1 v 17.1 months), associated with a 43% decrease in the risk of disease progression or death (HR, 0.57 [95% CI, 0.40 to 0.81]; Fig 6A). Similarly, a longer median rwPFS was observed in patients with ctDNA clearance at C2D1 than those without ctDNA clearance (30.8 v 17.1 months; HR, 0.63 [95% CI, 0.41 to 0.97]) and in patients with no increase in mVAF at C2D1 from baseline compared with those with an increase in mVAF (21.5 v 12.4 months; HR, 0.57 [95% CI, 0.40 to 0.82]). A significant association was observed between an increasing ctDNA mutation level at C2D1 (log ratio change >0 in mVAF) and real-world best overall response (one-sided trend test; P = .028; Fig 6B). The median mVAF reduction was 50.4% in those achieving rwCR/rwPR, whereas patients with rwSD and rwPD had median mVAF reductions of 27.1% and 12.3%, respectively.
FIG 6.
Kaplan-Meier curve for rwPFS in patients with (A) detectable ctDNA at C2D1 versus those without and (B) tumor response associated with changes in ctDNA at C2D1. C2D1, cycle 2 day 1; ctDNA, circulating tumor DNA; HR, hazard ratio; mVAF, maximum value of variant allele fraction; rwCR, real-world complete response; rwPD, real-world progressive disease; rwPFS, real-world progression-free survival; rwPR, real-world partial response; rwSD, real-world stable disease.
DISCUSSION
To our knowledge, this analysis is the first to use longitudinal genotyping data to identify genetic alterations associated with resistance and response that are specific to palbociclib treatment in combination with ET in patients with HR+/HER2– ABC in a real-world setting using commercially available assays. Here, patients with HR+/HER2– ABC most frequently had mutations in the PIK3CA, TP53, and ESR1 genes and amplifications in the CCND1, EGFR, and FGFR1 genes before starting palbociclib treatment. Patients with mutations in ESR1 and PIK3CA or amplification of CCND1 and FGFR1 at baseline were also shown to have shorter rwPFS than those with WT genes. Furthermore, patients with co-occurring mutations at baseline had shorter rwPFS than those with only one mutation. These results are similar to previously reported prognostic markers of response to CDK4/6i in patients with hormone receptor–positive/HER2– ABC in clinical trials.3,11-13
Consistent with the genotyping results at baseline, mutations in ESR1 and TP53 were most frequently acquired by EOT in patients treated with palbociclib plus ET. Mutations in ESR1 and cell cycle pathway genes such as PIK3CA and CCND1 have been observed to confer resistance against ET and CDK4/6i in vitro and in tumor biopsies.14,15 In addition, this analysis showed that ATM and RB1 mutations were also frequently acquired; loss of RB1 function is a known mechanism of acquired resistance to CDK4/6i,5 and defects in DNA damage repair have been proposed to account for ET resistance in HR+ BC.16,17 PIK3CA mutations, on the other hand, were lost most frequently by EOT. The ESR1 mutations detected at EOT (Y537N, Y537S, and D538G) have been validated in vitro to confer resistance to ET in BC cell lines by promoting estrogen-independent cellular growth and division.18 Overall, the type of ET received (AI v fulvestrant) did not seem to affect the genomic landscape of acquired and lost mutations by EOT. However, the number of mutations acquired and lost was greater with AI versus fulvestrant. Updated ASCO guidelines recommend ESR1 mutation testing at recurrence or progression on ET in HR+/HER2– ABC although it is not yet clear which mutations drive fulvestrant versus AI treatment resistance.19
Changes in ctDNA can be indicative of tumor burden and can correlate with treatment effectiveness, which has led to the increasing use of ctDNA as a marker of treatment response,20,21 including in BC.9,20,22 This study evaluated early changes in ctDNA analysis and demonstrated that undetectable ctDNA at C2D1 is associated with longer rwPFS compared with the presence of detectable ctDNA during treatment with palbociclib. Tumor burden in those patients with undetectable ctDNA is likely to be low, which would account for the prolonged rwPFS compared with those with detectable ctDNA. In support of this, large reductions in ctDNA levels from baseline to C2D1 were associated with better tumor response than small reductions in ctDNA levels. These findings suggest that a decline in ctDNA levels after treatment could be indicative of better survival and tumor response.
Interpretations of real-world data and the findings presented here are subject to limitations. The observational study design, patient tumor assessments, and monitoring procedures were performed by treating physicians in routine clinical practice at study sites and were not dictated by protocol or disease criteria (ie, RECIST)8; therefore, some patient data could potentially be missing or incomplete. No formal hypothesis testing was conducted, and analyses were descriptive. Because all patients were receiving palbociclib plus ET, it is difficult to delineate and ascertain palbociclib-specific effects versus ET-specific effects. Study findings may not be applicable to ET in combination with other CDK4/6i. Inherent biases in prognostic factors between the mutation and genotyping groups could confound the observed differences in outcomes. We also note that the ctDNA analysis was limited by the detection sensitivity of the assay. For example, a lower proportion of patients with detectable mutations/amplifications at C2D1 compared with baseline might have been due to ctDNA levels being lower in responding patients and below the level of mutation detection as opposed to true loss of the mutation; thus, further research is warranted to validate any of these study findings.
Current ASCO guidelines recommend treatment according to pathologic and biomarker features when quality testing results following established guidelines are available.23 In this analysis, the comparison of rwPFS outcome by genotyping highlights the role of mutations in estrogen receptor and amplifications in cell cycle pathways in conferring resistance to palbociclib plus ET under selective therapeutic pressure. In the PADA-1 trial, patients on AI plus palbociclib therapy who acquired or showed increased ESR1 mutations were randomly assigned to continue on the same therapy or switch ET from AI to fulvestrant, with the switch group showing a significantly improved PFS (HR, 0.61).24 These genetic alterations could potentially serve as companion prognostic biomarkers for patient stratification based on treatment response with palbociclib plus ET and guide future monitoring and treatment sequencing for patients with HR+/HER2– ABC.
ACKNOWLEDGMENT
Medical writing support, conducted in accordance with Good Publication Practice (GPP 2022) and the International Committee of Medical Journal Editors (ICMJE) guidelines, was provided by Mahesh Chemudupati, PhD, and Dominique J. Verlaan, PhD, of Oxford PharmaGenesis Inc (Newtown, PA) and was funded by Pfizer Inc, USA. Monica Z. Montelongo is an employee of ICON, which was a paid consultant to Pfizer in connection with the development of this manuscript.
Debu Tripathy
Consulting or Advisory Role: Novartis, Pfizer, GlaxoSmithKline, Genomic Health, AstraZeneca, OncoPep, Sermonix Pharmaceuticals, Personalis, Ambrx, Roche, Gilead Sciences, Menarini, Jazz Pharmaceuticals, Zetagen, BeiGene
Research Funding: Novartis (Inst), Polyphor (Inst), Pfizer (Inst), Ambrx (Inst)
Travel, Accommodations, Expenses: Novartis, AstraZeneca
Joanne L. Blum
Honoraria: Tempus
Consulting or Advisory Role: Pfizer, Tempus
Speakers' Bureau: Tempus
Hong Zhang
Employment: Pfizer
Stock and Other Ownership Interests: Pfizer
Shibing Deng
Employment: Pfizer
Stock and Other Ownership Interests: Pfizer
Travel, Accommodations, Expenses: Pfizer
Steven L. McCune
Consulting or Advisory Role: Bristol Myers Squibb/Medarex
Speakers' Bureau: Bristol Myers Squibb
Research Funding: Roche/Genentech, Bristol Myers Squibb
Other Relationship: Roche (Inst)
Yao Wang
Employment: Pfizer
Stock and Other Ownership Interests: Pfizer
Travel, Accommodations, Expenses: Pfizer
Zhe Zhang
Employment: Pfizer
Stock and Other Ownership Interests: Pfizer
Chetan Deshpande
Stock and Other Ownership Interests: Pfizer
Consulting or Advisory Role: Pfizer, Danaher
Eric Gauthier
Employment: Pfizer
Stock and Other Ownership Interests: Pfizer
Travel, Accommodations, Expenses: Pfizer
Yuan Liu
Employment: Pfizer
Stock and Other Ownership Interests: Pfizer
Gabrielle B. Rocque
Employment: Atlas Oncology Partners
Consulting or Advisory Role: Pfizer, Gilead Sciences, Armada/Rubicon (Inst)
Research Funding: Genentech, Pfizer, Daiichi Sankyo/AstraZeneca, Gilead Sciences
Aditya Bardia
Consulting or Advisory Role: Novartis, Genentech, Pfizer, Merck, Novartis (Inst), Genentech/Roche (Inst), Pfizer (Inst), Radius Health (Inst), Innocrin Pharma (Inst), Sanofi, Daiichi Sankyo/AstraZeneca, Lilly, Lilly (Inst), Gilead Sciences, Gilead Sciences (Inst), Menarini, Menarini (Inst), Mersana, Alyssum
Research Funding: Genentech (Inst), Novartis (Inst), Pfizer (Inst), Merck (Inst), Sanofi (Inst), Radius Health (Inst), Immunomedics (Inst), AstraZeneca/Daiichi Sankyo (Inst)
Open Payments Link: https://openpaymentsdata.cms.gov/physician/523675
No other potential conflicts of interest were reported.
See accompanying Editorial, 10.1200/PO-25-00539
PRIOR PRESENTATION
Presented at the San Antonio Breast Cancer Symposium, San Antonio, TX, December 6-10, 2022.
SUPPORT
Supported by Pfizer (Pfizer Inc).
CLINICAL TRIAL INFORMATION
DATA SHARING STATEMENT
A data sharing statement provided by the authors is available with this article at DOI https://doi.org/10.1200/PO-24-00810. Upon request, and subject to review, Pfizer will provide the data that support the findings of this study. Subject to certain criteria, conditions, and exceptions, Pfizer may also provide access to the related individual deidentified participant data. See https://www.pfizer.com/science/clinical-trials/trial-data-and-results for more information.
AUTHOR CONTRIBUTIONS
Conception and design: Debu Tripathy, Joanne L. Blum, Steven L. McCune, Kamal Patel, Yao Wang, Shailendra Lakhanpal, Meghan S. Karuturi, Zhe Zhang, Chetan Deshpande, Monica Z. Montelongo, Eric Gauthier, Yuan Liu, Gabrielle B. Rocque, Aditya Bardia
Provision of study materials or patients: Joanne L. Blum, Steven L. McCune, Kamal Patel
Collection and assembly of data: Debu Tripathy, Joanne L. Blum, Kamal Patel, Yao Wang, Meghan S. Karuturi, Chetan Deshpande, Eric Gauthier, Yuan Liu, Aditya Bardia
Data analysis and interpretation: Debu Tripathy, Joanne L. Blum, Hong Zhang, Shibing Deng, Steven L. McCune, Yao Wang, Shailendra Lakhanpal, Zhe Zhang, Chetan Deshpande, Monica Z. Montelongo, Eric Gauthier, Yuan Liu, Gabrielle B. Rocque, Aditya Bardia
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Debu Tripathy
Consulting or Advisory Role: Novartis, Pfizer, GlaxoSmithKline, Genomic Health, AstraZeneca, OncoPep, Sermonix Pharmaceuticals, Personalis, Ambrx, Roche, Gilead Sciences, Menarini, Jazz Pharmaceuticals, Zetagen, BeiGene
Research Funding: Novartis (Inst), Polyphor (Inst), Pfizer (Inst), Ambrx (Inst)
Travel, Accommodations, Expenses: Novartis, AstraZeneca
Joanne L. Blum
Honoraria: Tempus
Consulting or Advisory Role: Pfizer, Tempus
Speakers' Bureau: Tempus
Hong Zhang
Employment: Pfizer
Stock and Other Ownership Interests: Pfizer
Shibing Deng
Employment: Pfizer
Stock and Other Ownership Interests: Pfizer
Travel, Accommodations, Expenses: Pfizer
Steven L. McCune
Consulting or Advisory Role: Bristol Myers Squibb/Medarex
Speakers' Bureau: Bristol Myers Squibb
Research Funding: Roche/Genentech, Bristol Myers Squibb
Other Relationship: Roche (Inst)
Yao Wang
Employment: Pfizer
Stock and Other Ownership Interests: Pfizer
Travel, Accommodations, Expenses: Pfizer
Zhe Zhang
Employment: Pfizer
Stock and Other Ownership Interests: Pfizer
Chetan Deshpande
Stock and Other Ownership Interests: Pfizer
Consulting or Advisory Role: Pfizer, Danaher
Eric Gauthier
Employment: Pfizer
Stock and Other Ownership Interests: Pfizer
Travel, Accommodations, Expenses: Pfizer
Yuan Liu
Employment: Pfizer
Stock and Other Ownership Interests: Pfizer
Gabrielle B. Rocque
Employment: Atlas Oncology Partners
Consulting or Advisory Role: Pfizer, Gilead Sciences, Armada/Rubicon (Inst)
Research Funding: Genentech, Pfizer, Daiichi Sankyo/AstraZeneca, Gilead Sciences
Aditya Bardia
Consulting or Advisory Role: Novartis, Genentech, Pfizer, Merck, Novartis (Inst), Genentech/Roche (Inst), Pfizer (Inst), Radius Health (Inst), Innocrin Pharma (Inst), Sanofi, Daiichi Sankyo/AstraZeneca, Lilly, Lilly (Inst), Gilead Sciences, Gilead Sciences (Inst), Menarini, Menarini (Inst), Mersana, Alyssum
Research Funding: Genentech (Inst), Novartis (Inst), Pfizer (Inst), Merck (Inst), Sanofi (Inst), Radius Health (Inst), Immunomedics (Inst), AstraZeneca/Daiichi Sankyo (Inst)
Open Payments Link: https://openpaymentsdata.cms.gov/physician/523675
No other potential conflicts of interest were reported.
REFERENCES
- 1. Tripathy D, Blum JL, Karuturi MS, et al. Real-world effectiveness of palbociclib plus endocrine therapy in HR+/HER2- advanced breast cancer: Final results from the POLARIS trial. Oncologist. 2024:oyae291. doi: 10.1093/oncolo/oyae291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Condorelli R, Spring L, O'Shaughnessy J, et al. Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann Oncol. 2018;29:640–645. doi: 10.1093/annonc/mdx784. [DOI] [PubMed] [Google Scholar]
- 3. O'Leary B, Cutts RJ, Liu Y, et al. The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 trial. Cancer Discov. 2018;8:1390–1403. doi: 10.1158/2159-8290.CD-18-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lloyd MR, Spring LM, Bardia A, et al. Mechanisms of resistance to CDK4/6 blockade in advanced hormone receptor-positive, HER2-negative breast cancer and emerging therapeutic opportunities. Clin Cancer Res. 2022;28:821–830. doi: 10.1158/1078-0432.CCR-21-2947. [DOI] [PubMed] [Google Scholar]
- 5. Asghar US, Kanani R, Roylance R, et al. Systematic review of molecular biomarkers predictive of resistance to CDK4/6 inhibition in metastatic breast cancer. JCO Precis Oncol. doi: 10.1200/PO.21.00002. 10.1200/PO.21.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watt AC, Goel S. Cellular mechanisms underlying response and resistance to CDK4/6 inhibitors in the treatment of hormone receptor-positive breast cancer. Breast Cancer Res. 2022;24:17. doi: 10.1186/s13058-022-01510-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fiste O, Liontos M, Koutsoukos K, et al. Circulating tumor DNA-based predictive biomarkers in breast cancer clinical trials: A narrative review. Ann Transl Med. 2020;8:1603. doi: 10.21037/atm-20-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tripathy D, Blum JL, Rocque GB, et al. POLARIS: A prospective, multicenter, noninterventional study assessing palbociclib in hormone receptor-positive advanced breast cancer. Future Oncol. 2020;16:2475–2485. doi: 10.2217/fon-2020-0573. [DOI] [PubMed] [Google Scholar]
- 9. Pairawan S, Hess KR, Janku F, et al. Cell-free circulating tumor DNA variant allele frequency associates with survival in metastatic cancer. Clin Cancer Res. 2020;26:1924–1931. doi: 10.1158/1078-0432.CCR-19-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.www.guardanthealth.com Guardant: Conquering cancer with data. 2025.
- 11. Bardia A, Su F, Solovieff N, et al. Genomic profiling of premenopausal HR+ and HER2- metastatic breast cancer by circulating tumor DNA and association of genetic alterations with therapeutic response to endocrine therapy and ribociclib. JCO Precis Oncol. doi: 10.1200/PO.20.00445. 10.1200/PO.20.00445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cristofanilli M, Turner NC, Bondarenko I, et al. Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone-receptor-positive, HER2-negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA-3): Final analysis of the multicentre, double-blind, phase 3 randomised controlled trial. Lancet Oncol. 2016;17:425–439. doi: 10.1016/S1470-2045(15)00613-0. [DOI] [PubMed] [Google Scholar]
- 13. Pascual J, Gil-Gil M, Proszek P, et al. Baseline mutations and ctDNA dynamics as prognostic and predictive factors in ER-positive/HER2-negative metastatic breast cancer patients. Clin Cancer Res. 2023;29:4166–4177. doi: 10.1158/1078-0432.CCR-23-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schoninger SF, Blain SW. The ongoing search for biomarkers of CDK4/6 inhibitor responsiveness in breast cancer. Mol Cancer Ther. 2020;19:3–12. doi: 10.1158/1535-7163.MCT-19-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brett JO, Spring LM, Bardia A, et al. ESR1 mutation as an emerging clinical biomarker in metastatic hormone receptor-positive breast cancer. Breast Cancer Res. 2021;23:85. doi: 10.1186/s13058-021-01462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haricharan S, Punturi N, Singh P, et al. Loss of MutL disrupts CHK2-dependent cell-cycle control through CDK4/6 to promote intrinsic endocrine therapy resistance in primary breast cancer. Cancer Discov. 2017;7:1168–1183. doi: 10.1158/2159-8290.CD-16-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Anurag M, Haricharan S, Ellis MJ. CDK4/6 inhibitor biomarker research: Are we barking up the wrong tree? Clin Cancer Res. 2020;26:3–5. doi: 10.1158/1078-0432.CCR-19-3119. [DOI] [PubMed] [Google Scholar]
- 18. Harrod A, Lai CF, Goldsbrough I, et al. Genome engineering for estrogen receptor mutations reveals differential responses to anti-estrogens and new prognostic gene signatures for breast cancer. Oncogene. 2022;41:4905–4915. doi: 10.1038/s41388-022-02483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Burstein HJ, DeMichele A, Somerfield MR, et al. Testing for ESR1 mutations to guide therapy for hormone receptor-positive, human epidermal growth factor receptor 2-negative metastatic breast cancer: ASCO guideline rapid recommendation update. J Clin Oncol. 2023;41:3423–3425. doi: 10.1200/JCO.23.00638. [DOI] [PubMed] [Google Scholar]
- 20. Sanz-Garcia E, Zhao E, Bratman SV, et al. Monitoring and adapting cancer treatment using circulating tumor DNA kinetics: Current research, opportunities, and challenges. Sci Adv. 2022;8:eabi8618. doi: 10.1126/sciadv.abi8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 22. Martinez-Saez O, Pascual T, Braso-Maristany F, et al. Circulating tumor DNA dynamics in advanced breast cancer treated with CDK4/6 inhibition and endocrine therapy. NPJ Breast Cancer. 2021;7:8. doi: 10.1038/s41523-021-00218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Al Sukhun S, Temin S, Barrios CH, et al. Systemic treatment of patients with metastatic breast cancer: ASCO resource-stratified guideline. JCO Glob Oncol. doi: 10.1200/GO.23.00411. 10.1200/GO.23.00285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bidard FC, Hardy-Bessard AC, Dalenc F, et al. Switch to fulvestrant and palbociclib versus no switch in advanced breast cancer with rising ESR1 mutation during aromatase inhibitor and palbociclib therapy (PADA-1): A randomised, open-label, multicentre, phase 3 trial. Lancet Oncol. 2022;23:1367–1377. doi: 10.1016/S1470-2045(22)00555-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A data sharing statement provided by the authors is available with this article at DOI https://doi.org/10.1200/PO-24-00810. Upon request, and subject to review, Pfizer will provide the data that support the findings of this study. Subject to certain criteria, conditions, and exceptions, Pfizer may also provide access to the related individual deidentified participant data. See https://www.pfizer.com/science/clinical-trials/trial-data-and-results for more information.