Introduction
The fibroblast growth factor receptor (FGFR) family, consisting of FGFR1 to FGFR4, belongs to the receptor tyrosine kinase superfamily. FGFR2 amplification is detected in approximately 5% of patients with advanced gastric cancer (AGC) and is recognized as a potential therapeutic target.1 Although FGFR inhibitors have shown clinical efficacy in these patients, the resistance mechanisms involved remain poorly understood.2-5
Understanding the molecular basis of FGFR2 amplification has become increasingly important for optimizing therapeutic strategies. Extrachromosomal DNA (ecDNA) has recently emerged as a critical mechanism driving oncogene amplification and therapeutic resistance across various cancers. ecDNA, characterized as circular DNA elements existing outside chromosomes, exhibits unique properties such as uncontrolled replication and non-Mendelian inheritance patterns; these features contribute to elevated oncogene copy number (CN) and intratumoral heterogeneity, ultimately leading to treatment resistance.6,7
The clinical relevance of ecDNA in AGC is substantial. ecDNA-based amplification in AGC occurs in approximately 37.9% of patients and correlates with poorer prognosis than focal amplification.8 Furthermore, a whole-genome sequencing of 170 gastric cancer genomes suggest that FGFR2 amplification is associated with ecDNA.9 However, to our knowledge, there have been no clinical reports to date investigating the therapeutic effects and resistance of FGFR-targeted therapy in patients with FGFR2 amplification on ecDNA.
Herein, we present a case of an AGC patient with FGFR2 amplification on ecDNA and discuss its potential role in therapeutic response and resistance. Additionally, to elucidate the prevalence and clinical significance of this phenomenon, we used fluorescence in situ hybridization (FISH) to determine the ecDNA status of patients with FGFR2-amplified AGC who were enrolled in the MONSTAR-SCREEN-2 study (UMIN000043899), a multicenter biomarker development initiative that used artificial intelligence–driven multiomics analysis for a variety of solid tumors.10
The clinical information presented in this report was collected throughout the patient's course of care. The patient provided informed consent for the use and publication of their health information and medical imaging.
Case
A 48-year-old man was diagnosed with AGC after staging laparoscopy that revealed positive peritoneal cytology. The patient received systemic chemotherapy with S-1 plus oxaliplatin and intraperitoneal chemotherapy with paclitaxel for 5 months at the previous hospital, followed by total gastrectomy. The patient developed para-aortic lymph node (PALN) metastases 2 months after surgery and was treated with systemic chemotherapy using S-1 plus paclitaxel. After transferring to our institution, computed tomography (CT) was performed, revealing obvious disease progression with further enlargement and increase of the PALN, multiple bone metastases, and lung lymphangitis. The patient received a combination of capecitabine, oxaliplatin, plus nivolumab while undergoing next-generation sequencing (NGS) using Caris MI Profile (Caris Life Sciences, Phoenix, AZ) for tumor tissue resected at surgery and Caris ASSURE (Caris Life Sciences) for plasma samples. Before the NGS results were available, his condition deteriorated with worsening of the lymphangitis, prompting the initiation of irinotecan. However, 2 days later, the patient was hospitalized because of respiratory distress and the onset of disseminated intravascular coagulation (DIC). NGS revealed FGFR2 amplification (CN, 264.9) along with pathogenic short variants in CDH1, SMAD4, and TP53 in the tumor tissue. Liquid genotyping identified FGFR2 amplification (plasma CN [pCN], 16.6) along with the pathogenic short variants of CDH1 and FGFR2. Histologic evaluation with immunohistochemical staining (IHC) for FGFR2 demonstrated cytoplasmic staining with 10% IHC 3+ and 5% IHC 2+ without membrane positivity (Figs 1A and 1B). On the basis of the NGS findings, off-label treatment with pemigatinib, a selective FGFR1-3 inhibitor, was initiated. Although the patient required platelet and fresh-frozen plasma transfusions for 3 days, the DIC resolved, and the respiratory condition improved. Resistance to irinotecan could not be fully assessed because of its short duration of use; therefore, pemigatinib was combined with irinotecan (initiated 2 days pre-pemigatinib) after approval from the institutional review board. The patient's condition gradually improved, and he was discharged 3 weeks after admission. Liquid NGS revealed a decrease in the detected FGFR2 pCN (Table 1). Positron emission tomography-CT (PET-CT) 7 weeks later from the initiation of pemigatinib showed a marked reduction of [18F] fluorodeoxyglucose uptake in multiple bone metastases, accompanied by decreased tumor marker levels (Fig 1C).
FIG 1.

Clinical and pathologic findings of the case. (A) Hematoxylin-and-eosin–stained biopsy specimen of the primary tumor. (B) IHC analysis of FGFR2 showing 10% IHC 3+ and 5% IHC 2+ in the cytoplasm without membrane positivity. (C) Clinical course depicting changes in tumor markers (CEA and CA 19-9) and PET-CT scans before and after pemigatinib initiation at our hospital. The arrows indicate the timing of liquid biopsy sampling at −6, 2, 8, and 13 weeks, with reference to the pemigatinib initiation. CA 19-9, carbohydrate antigen 19-9; CAPOX, capecitabine plus oxaliplatin; CEA, carcinoembryonic antigen; FGFR, fibroblast growth factor receptor; IHC, immunohistochemistry; IRI, irinotecan; NGS, next-generation sequencing; nivo, nivolumab; PET-CT, positron emission tomography-computed tomography.
TABLE 1.
Genomic Dynamics in Gene Alterations at Each Time Point
| Specimen | Pre-Pemigatinib (6 weeks before pemigatinib) | PR Post-Pemigatinib (2 weeks after peimigatinib) | PD Post-Pemigatinib (8 weeks after pemigatinib) | PD Post-Erdafitinib (13 weeks after pemigatinib) | ||||
|---|---|---|---|---|---|---|---|---|
| Liquid | Caris Assure | Caris Assure | Guardant360 | Guardant360 | ||||
| Amplification | pCN | Amplification | pCN | Amplification | pCN | Amplification | pCN | |
| FGFR2 | 16.6 | FGFR2 | 5.2 | FGFR2 | 110.1 | FGFR2 | 34.3 | |
| Mutation | VAF | Mutation | VAF | MYC | 45.4 | MYC | 11.2 | |
| CDH1, L214P | 46 | CDH1, L214P | 0.6 | KRAS | 3.0 | KRAS | ND | |
| FGFR2, C382R | 0.1 | CDH1, W409* | 0.6 | Mutation | VAF | Mutation | VAF | |
| FGFR2, L617V | 8.4 | FGFR2, L617V | 7.4 | |||||
| FGFR2, E565A | 7.1 | FGFR2, E565A | 11.6 | |||||
| FGFR2, V564L | 2.3 | FGFR2, V564L | 6.6 | |||||
| FGFR2, N549H | 0.8 | FGFR2, N549H | 1.1 | |||||
| FGFR2, N549K | 0.7 | FGFR2, N549K | 0.8 | |||||
| FGFR2, V564F | 0.2 | FGFR2, V564F | 0.2 | |||||
| Fusion | FGFR2, V564I | 0.3 | ||||||
| FGFR2-PRNCR1 | 0.08 | FGFR2, E565G | 1.3 | |||||
| FGFR2, N549D | 0.7 | |||||||
| FGFR2, E565K | 0.6 | |||||||
| TP53, E339fs | 12.5 | |||||||
| Fusion | ||||||||
| FGFR2-PRNCR1 | 0.05 | |||||||
| Tissuea | MI Profile | |||||||
| Amplification | CN | |||||||
| FGFR2 | 264 | |||||||
| Mutation | VAF | |||||||
| CDH1, L214P | 58 | |||||||
| SMAD4, E330K | 44 | |||||||
| TP53, E339fs | 46 | |||||||
Abbreviations: CN, copy number; FGFR, fibroblast growth factor receptor; ND, not detected; PD, progressive disease; pCN, plasma CN; PR, partial response; VAF, variant allele frequency.
Tumor tissue resected at surgery.
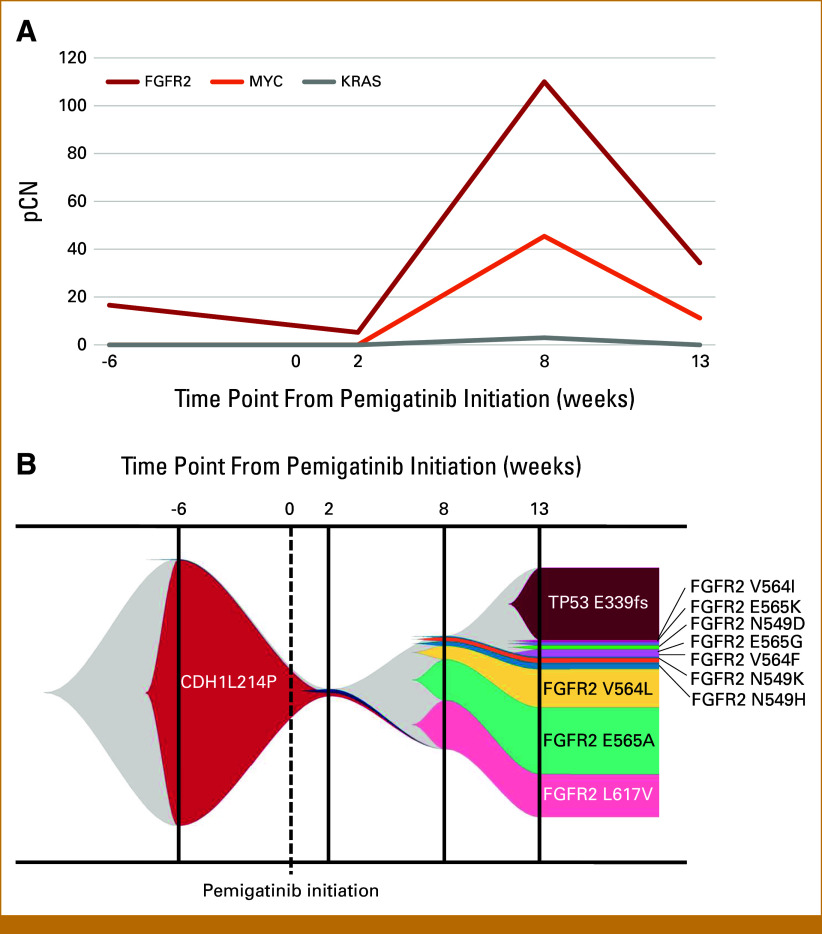
However, disease progression occurred 2 months after pemigatinib initiation, with worsening lower back pain attributed to progressive bone metastases, confirmed by PET-CT at 9 weeks after pemigatinib initiation (Fig 1C). Plasma NGS using Guardant360 (Guardant Health, Redwood City, CA) revealed a marked increase in FGFR2 pCN (110) and the emergence of MYC amplification (pCN 45.4), KRAS amplification (pCN 3.0), various FGFR2 mutations (L617V, N549H, E565A, V564L, N549K, and V564F), and an FGFR2-PRNCR1 fusion. Progressive DIC necessitated continuous platelet and plasma transfusions. Subsequent off-label use of other FGFR inhibitor therapy with erdafitinib followed by futibatinib was ineffective, resulting in disease progression and the patient's death. The final liquid NGS analysis showed an expanded array of gene alterations, including increased FGFR2 mutations (Figs 2A and 2B; Table 1).
FIG 2.
Genomic dynamics in liquid NGS analysis. (A) The pCN variations of FGFR2, MYC, and KRAS monitored by liquid NGS using Caris Assure (first two time points) and Guardant360 (subsequent time points) were correlated with the treatment response. (B) Fish plot visualization of clonal evolution depicting the emergence of multiple acquired FGFR2 mutations during disease progression. The time points are indicated relative to pemigatinib initiation. FGFR, fibroblast growth factor receptor; IRI, irinotecan; NGS, next-generation sequencing; pCN, plasma copy number; VAF, variant allele frequency.
ecDNA Evaluation in AGC Patients With FGFR2 Amplification
We assessed AGC patients with FGFR2 amplification enrolled in the MONSTAR-SCREEN-2 study in our institution. This study was conducted in accordance with the Declaration of Helsinki and Ethical Guidelines for Medical and Biological Research Involving Human Subjects, and approved by the Institutional Review Board of the National Cancer Center, Japan. Written informed consent was obtained from all participants. Of 207 patients with AGC enrolled at our institution from May 2021 to March 2024, 34 were excluded because of insufficient tissue samples or failed analysis. Tissue NGS detected FGFR2 amplification in 2.9% (5/173) of the patients, including Patient 1 presented in this report. FISH analysis was performed in all five patients, with repeat testing performed in Patients 2 and 3. The analysis revealed scattered extrachromosomal FGFR2 signals, consistent with ecDNA, in four patients (Patients 1-4). By contrast, Patient 5 exhibited patterns consistent with homogeneously staining region (Fig 3).
FIG 3.

FISH in AGC patients with FGFR2 amplification enrolled in the MONSTAR-SCREEN-2 study. The yellow arrowheads indicate scattered extrachromosomal FGFR2 signals suggesting ecDNA in patients 1-4. The white arrows in patient 5 indicate HSRs. AGC, advanced gastric cancer; ecDNA, extrachromosomal DNA; FGFR, fibroblast growth factor receptor; FISH, fluorescence in situ hybridization; HSR, homogeneously staining region.
Discussion
We present a patient with an extraordinarily high CN of FGFR2 amplifications driven by ecDNA, treated with FGFR-targeted therapies. The patient received concurrent pemigatinib and irinotecan, on the basis of the limited exposure to previous irinotecan and the preclinical evidence that combining cytotoxic chemotherapy with FGFR-targeted tyrosine kinase inhibitors enhances antitumor effects.11 However, the duration of response was short, and disease progression was marked by the emergence of numerous genetic alterations.
A high CN is a reported characteristic of ecDNA, with amplified oncogenes on ecDNA showing significantly higher CNs (mean CN ≥50) compared with those on intrachromosomal DNA.12 Previous studies demonstrated tumor responses to FGFR-targeted therapy in patients with FGFR2 amplification, especially in those with a high CN, although the duration of response was generally short.2,13 These findings may suggest ecDNA involvement. However, in our analysis of the MONSTAR-SCREEN2 cohort, ecDNA was also detected in patients with low CN, indicating that high CN is neither necessary nor sufficient for the definitive diagnosis of ecDNA. Another characteristic of ecDNA is early therapeutic resistance through unique mechanisms such as increased genetic heterogeneity, rapid genome changes driven by super-enhancers, and amplification of resistance genes, leading to a poor prognosis.7,14,15The rapid emergence and diversity of acquired FGFR2 mutations and a fusion observed in our patient would be highly unusual if confined to chromosomal DNA, given conventional mutation rates and selective pressures.16,17 These alterations are thought to occur on ecDNA, accumulating unrepaired genetic alterations during its rapid amplification and replication processes. We previously reported an AGC patient with markedly high EGFR amplification in ctDNA who acquired multiple EGFR mutations and MET amplification after treatment with cetuximab.18 The clinical course of this patient closely resembled that of our present patient. Therefore, FISH analysis was performed in this AGC patient with EGFR amplification, revealing the presence of ecDNA (Appendix Fig A1).
Additionally, we investigated the association between FGFR-amplified AGC and ecDNA. A previous study using whole-genome sequencing analysis of gastric cancer revealed association between FGFR2 amplification and self-joining amplicons, structural variant clusters characteristic of ecDNA.9 In fact, four of the five patients with FGFR2-amplified AGC in our cohort of MONSTAR-SCREEN-2 also exhibited ecDNA. Although the cancer types were different, this association may underlie the limited efficacy of FGFR inhibitors in FGFR2-amplified AGC compared with biliary tract cancer with FGFR2 fusions. In a phase II study of futibatinib in FGFR-amplified AGC, the overall response rate (ORR) and progression-free survival (PFS) were 18% and 2.9 months, whereas in another study on biliary tract cancer with FGFR fusion, ORR and PFS were 42% and 9.0 months, respectively.19,20
These findings highlight the need to overcome ecDNA-driven resistance. A study from Memorial Sloan Kettering Cancer Center showed improved survival in AGC patients with ERBB2 ecDNA treated with HER2-targeted therapy plus first-line fluoropyrimidine and oxaliplatin.21 Although high CN may confound results, DNA damage from multiple cytotoxic agents appears effective against ecDNA-positive tumors. Irinotecan, a topoisomerase I inhibitor, may have potential efficacy against ecDNA; however, we did not observe such an effect in our patient, suggesting combination chemotherapy may be required. Recent evidence shows that ecDNA causes transcription-replication conflict, replication stress, and DNA damage, activating the S-phase checkpoint. In preclinical AGC models with FGFR2 amplification on ecDNA, the checkpoint kinase 1 (CHK1) inhibitor BBI-2779 suppressed tumor growth and prevented resistance to the FGFR inhibitor infigratinib.22 Additionally, the phase I/II POTENTIATE trial evaluated BBI-355, a selective CHK1 inhibitor, as monotherapy and in combination with futibatinib or erlotinib in metastatic solid tumors with FGFR or EGFR amplifications driven by ecDNA (ClinicalTrials.gov identifier: NCT05827614). However, these trial arms were discontinued because of tolerability issues, including hematologic toxicity and a narrow therapeutic index with continuous dosing regimens. Given the expected synergistic effect and nonoverlapping toxicity profiles, a novel combination of BBI-355 with BBI-825, a selective ribonucleotide reductase inhibitor, is being developed, with this combination arm of the POTENTIATE trial planned to be initiated in the second half of 2025.23
Important caveats of our study include the absence of standardized ecDNA diagnostic criteria and the inability to reliably identify ecDNA using IHC. Although we explored IHC as a more accessible diagnostic approach, FGFR2 often exhibits nonspecific cytoplasmic staining as an artifact, even in cases without FGFR2 amplification, which complicates interpretation. Our analysis suggests that IHC alone cannot definitively identify ecDNA-based amplification. These findings emphasize that the development of more specific diagnostic approaches for ecDNA detection remains a critical area for future research.
In conclusion, this case report provides valuable insights into FGFR2-amplified AGC driven by ecDNA. The distinct patterns of response and resistance highlight the need for novel therapeutic strategies. Targeting the primary oncogenic driver and the underlying mechanisms of ecDNA-mediated heterogeneity is critical for achieving more durable treatment outcomes.
ACKNOWLEDGMENT
The authors thank all patients and families for participating in the study. The authors would like to thank Enago (www.enago.jp) for the English language review.
APPENDIX
FIG A1.
FISH analysis of an AGC patient with EGFR amplification. This patient exhibited a markedly high EGFR plasma copy number in ctDNA, as previously reported by our institution. After treatment with cetuximab, multiple EGFR mutations and MET amplification were acquired (Nakamura et al18). FISH analysis revealed a scattered EGFR staining pattern, indicating the presence of ecDNA. AGC, advanced gastric cancer; ecDNA, extrachromosomal DNA; FISH, fluorescence in situ hybridization.
Yu Aoki
Honoraria: Guardant Health, Daiichi Sankyo, Ono Pharmaceutical
Tadayoshi Hashimoto
Honoraria: Cytogen, Takata Pharmaceutical
Saori Mishima
Honoraria: Chugai Pharma, Taiho Pharmaceutical, Lilly Japan
Consulting or Advisory Role: Exact Sciences
Daisuke Kotani
Honoraria: Takeda, Chugai Pharma, Lilly Japan, Merck Serono, Sysmex, Taiho Pharmaceutical, Ono Pharmaceutical, Bristol Myers Squibb Japan, MSD, Daiichi Sankyo/UCB Japan, Pfizer, Eisai, Novartis, Guardant Health
Consulting or Advisory Role: Takeda, Seagen, MSD K.K
Research Funding: Ono Pharmaceutical (Inst), MSD (Inst), Novartis (Inst), Janssen (Inst), IQvia (Inst), Syneos Health (Inst), CMIC (Inst), SERVIER (Inst)
Akihito Kawazoe
Honoraria: Ono Pharmaceutical, Taiho Pharmaceutical, Bristol Myers Squibb, Daiichi Sankyo/UCB Japan, Lilly
Consulting or Advisory Role: Zymeworks, Revolution Medicines, MSD, AbbVie, GlaxoSmithKline
Speakers' Bureau: Taiho Pharmaceutical, Ono Pharmaceutical, Bristol Myers Squibb, Daiichi Sankyo/UCB Japan, Lilly
Research Funding: Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), AstraZeneca (Inst), MSD (Inst)
Yoshiaki Nakamura
Consulting or Advisory Role: Natera, Inc, Roche Ltd, Seagen, Inc, Premo Partners, Inc, Daiichi Sankyo Co, Ltd, Takeda, Exact Sciences, Gilead Sciences, Guardant Health Pte Ltd
Speakers' Bureau: MSD K.K, Eisai, Zeria Pharmaceutical, MIYARISAN Pharmaceutical, Merck, CareNet, Inc, Hisamitsu Pharmaceutical, Taiho Pharmaceutical, Daiichi Sankyo Co, Ltd, Chugai Pharma, Becton Dickinson, Guardant Health Japan Corp, Guardant Health Pte Ltd
Research Funding: Seagen, Inc (Inst), Genomedia (Inst), Guardant Health AMEA, Inc (Inst), Guardant Health (Inst), Tempus (Inst), Roche Diagnostics K.K (Inst), Daiichi Sankyo Co, Ltd (Inst), Chugai Pharma (Inst)
Izuma Nakayama
Honoraria: Astellas Pharma, MSD, Ono Pharmaceutical, Bristol Myers Squibb Japan, Taiho Pharmaceutical
Research Funding: Boehringer Ingelheim (Inst), Daiichi Sankyo/UCB Japan (Inst), MSD (Inst), Chugai/Roche (Inst), Ono Pharmaceutical (Inst)
Yasutoshi Kuboki
Honoraria: Taiho Pharmaceutical, Lilly Japan, Kyowa Kirin, Amgen
Consulting or Advisory Role: Takeda, Amgen, Incyte, AbbVie, Noile-Immune Biotech, Inc
Research Funding: Taiho Pharmaceutical (Inst), Takeda (Inst), Incyte (Inst), Ono Pharmaceutical (Inst), Boehringer Ingelheim (Inst), Amgen (Inst), Chugai Pharma (Inst), Astellas Pharma (Inst), Genmab (Inst), AbbVie (Inst), Lilly (Inst), AstraZeneca (Inst), Merck Serono (Inst), Jiangsu Hengrui Pharmaceuticals (Inst), Novartis (Inst), Carna Biosciences (Inst), Daiichi Sankyo/UCB Japan (Inst), Kyowa Kirin (Inst), Bristol Myers Squibb Japan (Inst)
Travel, Accommodations, Expenses: Amgen, Incyte, Chugai Pharma
Hideaki Bando
Honoraria: Taiho Pharmaceutical, Chugai Pharma, Ono Pharmaceutical
Takashi Kojima
Honoraria: Bristol Myers Squibb, MSD, Ono Pharmaceutical, Japanese Society of Pharmaceutical Health Care and Sciences, Japan Esophageal Society, Taiho Pharmaceutical, TFBS Bioscience, Inc, BeiGene Japan
Consulting or Advisory Role: Boehringer Ingelheim, Nippon Boehringer Ingelheim, BeiGene Japan
Research Funding: Ono Pharmaceutical (Inst), MSD (Inst), Chugai Pharma (Inst), Taiho Pharmaceutical (Inst), BeiGene (Inst), AstraZeneca (Inst), Parexel International (Inst)
Naoya Sakamoto
Speakers' Bureau: Astellas Pharma
Takao Fujisawa
Honoraria: Merck Serono, AmelieF, AstraZeneca
Takayuki Yoshino
Honoraria: Chugai Pharma, Takeda, Merck, Ono Pharmaceutical
Consulting or Advisory Role: Sumitomo Corp, Indivumed
Research Funding: MSD (Inst), Daiichi Sankyo Company, Limited (Inst), Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), Pfizer (Inst), Sysmex (Inst), Chugai Pharma (Inst), Eisai (Inst), FALCO Biosystems Ltd (Inst), Merus (Inst), Bristol Myers Squibb Japan (Inst), Medical & Biological Laboratories Co, Ltd (Inst), Takeda (Inst), Caris MPI (Inst), Exact Sciences (Inst), MIYARISAN Pharmaceutical (Inst), Natera (Inst), Nippon Boehringer Ingelheim (Inst)
Kohei Shitara
Honoraria: Bristol Myers Squibb, Janssen, AstraZeneca, Lilly, Ono Pharmaceutical, Astellas Pharma
Consulting or Advisory Role: Bristol Myers Squibb, Takeda, Ono Pharmaceutical, MSD, Novartis, Daiichi Sankyo, Amgen, Astellas Pharma, Guardant Health, Bayer, Zymeworks, AstraZeneca, ALX Oncology, GlaxoSmithKline K.K, Janssen, Healios, Moderna, Inc, Arcus Biosciences Inc
Research Funding: MSD (Inst), Daiichi Sankyo (Inst), Taiho Pharmaceutical (Inst), Chugai Pharma (Inst), Ono Pharmaceutical (Inst), Astellas Pharma (Inst), Eisai (Inst), Amgen (Inst), PRA Health Sciences (Inst), AstraZeneca (Inst), PPD-SNBL (Inst), Toray Industries (Inst)
No other potential conflicts of interest were reported.
SUPPORT
Supported by SCRUM-Japan Funds (http://www.scrum-japan.ncc.go.jp/index.html) and research funding from Department of Gastroenterology and Gastrointestinal Oncology, National Cancer Center Hospital East.
AUTHOR CONTRIBUTIONS
Conception and design: Yu Aoki, Tadayoshi Hashimoto, Nobuyuki Takahashi, Saori Mishima, Yoshiaki Nakamura, Izuma Nakayama, Nobuyuki Nakamura, Naoya Sakamoto, Genichiro Ishii, Yasuhito Arai, Tatsuhiro Shibata, Kohei Shitara
Financial support: Kohei Shitara
Administrative support: Tadayoshi Hashimoto, Nobuyuki Nakamura, Kohei Shitara
Provision of study materials or patients: All authors
Collection and assembly of data: Yu Aoki, Tadayoshi Hashimoto, Hideaki Bando, Nobuyuki Nakamura, Naoya Sakamoto, Takao Fujisawa, Takayuki Yoshino, Kohei Shitara
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Yu Aoki
Honoraria: Guardant Health, Daiichi Sankyo, Ono Pharmaceutical
Tadayoshi Hashimoto
Honoraria: Cytogen, Takata Pharmaceutical
Saori Mishima
Honoraria: Chugai Pharma, Taiho Pharmaceutical, Lilly Japan
Consulting or Advisory Role: Exact Sciences
Daisuke Kotani
Honoraria: Takeda, Chugai Pharma, Lilly Japan, Merck Serono, Sysmex, Taiho Pharmaceutical, Ono Pharmaceutical, Bristol Myers Squibb Japan, MSD, Daiichi Sankyo/UCB Japan, Pfizer, Eisai, Novartis, Guardant Health
Consulting or Advisory Role: Takeda, Seagen, MSD K.K
Research Funding: Ono Pharmaceutical (Inst), MSD (Inst), Novartis (Inst), Janssen (Inst), IQvia (Inst), Syneos Health (Inst), CMIC (Inst), SERVIER (Inst)
Akihito Kawazoe
Honoraria: Ono Pharmaceutical, Taiho Pharmaceutical, Bristol Myers Squibb, Daiichi Sankyo/UCB Japan, Lilly
Consulting or Advisory Role: Zymeworks, Revolution Medicines, MSD, AbbVie, GlaxoSmithKline
Speakers' Bureau: Taiho Pharmaceutical, Ono Pharmaceutical, Bristol Myers Squibb, Daiichi Sankyo/UCB Japan, Lilly
Research Funding: Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), AstraZeneca (Inst), MSD (Inst)
Yoshiaki Nakamura
Consulting or Advisory Role: Natera, Inc, Roche Ltd, Seagen, Inc, Premo Partners, Inc, Daiichi Sankyo Co, Ltd, Takeda, Exact Sciences, Gilead Sciences, Guardant Health Pte Ltd
Speakers' Bureau: MSD K.K, Eisai, Zeria Pharmaceutical, MIYARISAN Pharmaceutical, Merck, CareNet, Inc, Hisamitsu Pharmaceutical, Taiho Pharmaceutical, Daiichi Sankyo Co, Ltd, Chugai Pharma, Becton Dickinson, Guardant Health Japan Corp, Guardant Health Pte Ltd
Research Funding: Seagen, Inc (Inst), Genomedia (Inst), Guardant Health AMEA, Inc (Inst), Guardant Health (Inst), Tempus (Inst), Roche Diagnostics K.K (Inst), Daiichi Sankyo Co, Ltd (Inst), Chugai Pharma (Inst)
Izuma Nakayama
Honoraria: Astellas Pharma, MSD, Ono Pharmaceutical, Bristol Myers Squibb Japan, Taiho Pharmaceutical
Research Funding: Boehringer Ingelheim (Inst), Daiichi Sankyo/UCB Japan (Inst), MSD (Inst), Chugai/Roche (Inst), Ono Pharmaceutical (Inst)
Yasutoshi Kuboki
Honoraria: Taiho Pharmaceutical, Lilly Japan, Kyowa Kirin, Amgen
Consulting or Advisory Role: Takeda, Amgen, Incyte, AbbVie, Noile-Immune Biotech, Inc
Research Funding: Taiho Pharmaceutical (Inst), Takeda (Inst), Incyte (Inst), Ono Pharmaceutical (Inst), Boehringer Ingelheim (Inst), Amgen (Inst), Chugai Pharma (Inst), Astellas Pharma (Inst), Genmab (Inst), AbbVie (Inst), Lilly (Inst), AstraZeneca (Inst), Merck Serono (Inst), Jiangsu Hengrui Pharmaceuticals (Inst), Novartis (Inst), Carna Biosciences (Inst), Daiichi Sankyo/UCB Japan (Inst), Kyowa Kirin (Inst), Bristol Myers Squibb Japan (Inst)
Travel, Accommodations, Expenses: Amgen, Incyte, Chugai Pharma
Hideaki Bando
Honoraria: Taiho Pharmaceutical, Chugai Pharma, Ono Pharmaceutical
Takashi Kojima
Honoraria: Bristol Myers Squibb, MSD, Ono Pharmaceutical, Japanese Society of Pharmaceutical Health Care and Sciences, Japan Esophageal Society, Taiho Pharmaceutical, TFBS Bioscience, Inc, BeiGene Japan
Consulting or Advisory Role: Boehringer Ingelheim, Nippon Boehringer Ingelheim, BeiGene Japan
Research Funding: Ono Pharmaceutical (Inst), MSD (Inst), Chugai Pharma (Inst), Taiho Pharmaceutical (Inst), BeiGene (Inst), AstraZeneca (Inst), Parexel International (Inst)
Naoya Sakamoto
Speakers' Bureau: Astellas Pharma
Takao Fujisawa
Honoraria: Merck Serono, AmelieF, AstraZeneca
Takayuki Yoshino
Honoraria: Chugai Pharma, Takeda, Merck, Ono Pharmaceutical
Consulting or Advisory Role: Sumitomo Corp, Indivumed
Research Funding: MSD (Inst), Daiichi Sankyo Company, Limited (Inst), Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), Pfizer (Inst), Sysmex (Inst), Chugai Pharma (Inst), Eisai (Inst), FALCO Biosystems Ltd (Inst), Merus (Inst), Bristol Myers Squibb Japan (Inst), Medical & Biological Laboratories Co, Ltd (Inst), Takeda (Inst), Caris MPI (Inst), Exact Sciences (Inst), MIYARISAN Pharmaceutical (Inst), Natera (Inst), Nippon Boehringer Ingelheim (Inst)
Kohei Shitara
Honoraria: Bristol Myers Squibb, Janssen, AstraZeneca, Lilly, Ono Pharmaceutical, Astellas Pharma
Consulting or Advisory Role: Bristol Myers Squibb, Takeda, Ono Pharmaceutical, MSD, Novartis, Daiichi Sankyo, Amgen, Astellas Pharma, Guardant Health, Bayer, Zymeworks, AstraZeneca, ALX Oncology, GlaxoSmithKline K.K, Janssen, Healios, Moderna, Inc, Arcus Biosciences Inc
Research Funding: MSD (Inst), Daiichi Sankyo (Inst), Taiho Pharmaceutical (Inst), Chugai Pharma (Inst), Ono Pharmaceutical (Inst), Astellas Pharma (Inst), Eisai (Inst), Amgen (Inst), PRA Health Sciences (Inst), AstraZeneca (Inst), PPD-SNBL (Inst), Toray Industries (Inst)
No other potential conflicts of interest were reported.
REFERENCES
- 1. Lau DK, Collin JP, Mariadason JM. Clinical developments and challenges in treating FGFR2-Driven gastric cancer. Biomedicines. 2024;12:1117. doi: 10.3390/biomedicines12051117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jogo T, Nakamura Y, Shitara K, et al. Circulating tumor DNA analysis detects FGFR2 amplification and concurrent genomic alterations associated with FGFR inhibitor efficacy in advanced gastric cancer. Clin Cancer Res. 2021;27:5619–5627. doi: 10.1158/1078-0432.CCR-21-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Doi T, Shitara K, Kojima T, et al. Phase I study of the irreversible fibroblast growth factor receptor 1–4 inhibitor futibatinib in Japanese patients with advanced solid tumors. Cancer Sci. 2023;114:574–585. doi: 10.1111/cas.15486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Catenacci DVT, Kang Y-K, Saeed A, et al. FIGHT: A randomized, double-blind, placebo-controlled, phase II study of bemarituzumab (bema) combined with modified FOLFOX6 in 1L FGFR2b+ advanced gastric/gastroesophageal junction adenocarcinoma (GC) J Clin Oncol. 2021;39 (15_suppl; abstr 4010) [Google Scholar]
- 5. Cutsem EV, Bang Y-J, Mansoor W, et al. A randomized, open-label study of the efficacy and safety of AZD4547 monotherapy versus paclitaxel for the treatment of advanced gastric adenocarcinoma with FGFR2 polysomy or gene amplification. Ann Oncol. 2017;28:1316–1324. doi: 10.1093/annonc/mdx107. [DOI] [PubMed] [Google Scholar]
- 6. Wu S, Bafna V, Mischel PS. Extrachromosomal DNA (ecDNA) in cancer pathogenesis. Curr Opin Genet Dev. 2021;66:78–82. doi: 10.1016/j.gde.2021.01.001. [DOI] [PubMed] [Google Scholar]
- 7. Huang Q, Zhang S, Wang G, et al. Insight on ecDNA-mediated tumorigenesis and drug resistance. Heliyon. 2024;10:e27733. doi: 10.1016/j.heliyon.2024.e27733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bailey C, Pich O, Thol K, et al. Origins and impact of extrachromosomal DNA. Nature. 2024;635:193–200. doi: 10.1038/s41586-024-08107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saito-Adachi M, Hama N, Totoki Y, et al. Oncogenic structural aberration landscape in gastric cancer genomes. Nat Commun. 2023;14:3688. doi: 10.1038/s41467-023-39263-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hashimoto T, Nakamura Y, Fujisawa T, et al. The SCRUM-MONSTAR cancer-omics ecosystem: Striving for a quantum leap in precision medicine. Cancer Discov. 2024;14:2243–2261. doi: 10.1158/2159-8290.CD-24-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xie L, Su X, Zhang L, et al. FGFR2 gene amplification in gastric cancer predicts sensitivity to the selective FGFR inhibitor AZD4547. Clin Cancer Res. 2013;19:2572–2583. doi: 10.1158/1078-0432.CCR-12-3898. [DOI] [PubMed] [Google Scholar]
- 12. Turner KM, Deshpande V, Beyter D, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543:122–125. doi: 10.1038/nature21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pearson A, Smyth E, Babina IS, et al. High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. Cancer Discov. 2016;6:838–851. doi: 10.1158/2159-8290.CD-15-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim H, Nguyen N-P, Turner K, et al. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat Genet. 2020;52:891–897. doi: 10.1038/s41588-020-0678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nathanson DA, Gini B, Mottahedeh J, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343:72–76. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chell V, Balmanno K, Little AS, et al. Tumour cell responses to new fibroblast growth factor receptor tyrosine kinase inhibitors and identification of a gatekeeper mutation in FGFR3 as a mechanism of acquired resistance. Oncogene. 2013;32:3059–3070. doi: 10.1038/onc.2012.319. [DOI] [PubMed] [Google Scholar]
- 17. Goyal L, DiToro D, Facchinetti F, et al. A model for decoding resistance in precision oncology: Acquired resistance to FGFR inhibitors in cholangiocarcinoma. Ann Oncol. 2025;36:426–443. doi: 10.1016/j.annonc.2024.12.011. [DOI] [PubMed] [Google Scholar]
- 18. Nakamura Y, Sasaki A, Yukami H, et al. Emergence of concurrent multiple EGFR mutations and MET amplification in a patient with EGFR—Amplified advanced gastric cancer treated with cetuximab. JCO Precis Oncol. doi: 10.1200/PO.20.00263. 10.1200/PO.20.00263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goyal L, Meric-Bernstam F, Hollebecque A, et al. Futibatinib for FGFR2-rearranged intrahepatic cholangiocarcinoma. N Engl J Med. 2023;388:228–239. doi: 10.1056/NEJMoa2206834. [DOI] [PubMed] [Google Scholar]
- 20. Satoh T, Barthélémy P, Nogova L, et al. Phase 2 study of futibatinib in patients with gastric or gastroesophageal junction cancer harboring FGFR2 amplifications. Eur J Cancer. 2025;218:115262. doi: 10.1016/j.ejca.2025.115262. [DOI] [PubMed] [Google Scholar]
- 21. Tsai C, Suman S, Chu C, et al. Association of ERBB2 extrachromosomal DNA (ecDNA) with first-line HER2-targeted treatment outcomes in advanced esophagogastric adenocarcinoma (EGC) J Clin Oncol. 2024;42 (16_suppl; abstr 4041) [Google Scholar]
- 22. Tang J, Weiser NE, Wang G, et al. Enhancing transcription–replication conflict targets ecDNA-positive cancers. Nature. 2024;635:210–218. doi: 10.1038/s41586-024-07802-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boundless Bio Announces Portfolio Prioritization and Runway Extension. https://investors.boundlessbio.com/node/7421/pdf [Google Scholar]