Abstract
A small RNA, RyhB, was found as part of a genomewide search for novel small RNAs in Escherichia coli. The RyhB 90-nt RNA down-regulates a set of iron-storage and iron-using proteins when iron is limiting; it is itself negatively regulated by the ferric uptake repressor protein, Fur (Ferric uptake regulator). RyhB RNA levels are inversely correlated with mRNA levels for the sdhCDAB operon, encoding succinate dehydrogenase, as well as five other genes previously shown to be positively regulated by Fur by an unknown mechanism. These include two other genes encoding enzymes in the tricarboxylic acid cycle, acnA and fumA, two ferritin genes, ftnA and bfr, and a gene for superoxide dismutase, sodB. Fur positive regulation of all these genes is fully reversed in an ryhB mutant. Our results explain the previously observed inability of fur mutants to grow on succinate. RyhB requires the RNA-binding protein, Hfq, for activity. Sequences within RyhB are complementary to regions within each of the target genes, suggesting that RyhB acts as an antisense RNA. In sdhCDAB, the complementary region is at the end of the first gene of the sdhCDAB operon; full-length sdhCDAB message disappears and a truncated message, equivalent in size to the region upstream of the complementarity, is detected when RyhB is expressed. RyhB provides a mechanism for the cell to down-regulate iron-storage proteins and nonessential ironcontaining proteins when iron is limiting, thus modulating intracellular iron usage to supplement mechanisms for iron uptake directly regulated by Fur.
Keywords: Fur‖Hfq‖posttranscriptional regulation
Small RNAs (sRNAs; noncoding RNAs usually smaller than 300 nucleotides) in Escherichia coli are involved in a variety of cellular functions such as modulation of RNA polymerase activity (6S RNA) (1), protein tagging for degradation (SsrA or tmRNA) (2), and regulation of translation (3–5). Some of these sRNAs use sequence-specific RNA–RNA interactions to regulate mRNA utilization. For example, the sRNA DsrA is involved in the regulation of two global transcriptional regulators, RpoS and HNS. DsrA acts at the translation level to stimulate RpoS and inhibit HNS translation. In both cases, a region of DsrA is complementary to the target mRNAs (6, 7). DsrA and many other sRNAs bind to Hfq, an RNA-binding protein, which has been found to participate in many RNA transactions in the cell, including mRNA stability, mRNA polyadenylation, and translation (8). Hfq functions also include mRNA targeting for degradation either by increasing polyadenylation (9) or by interfering with ribosome binding (10).
A recent genomewide search for genes encoding short nontranslated RNAs yielded 17 new sRNAs, many of which bind Hfq (8). Independent approaches identified some of the same sRNAs and a number of additional ones (11, 12). In total, these studies identified about 28 new sRNAs. One of these newly discovered sRNAs, RyhB (encoded by the ryhB gene), was found to be complementary to a portion of the sdhCDAB operon encoding succinate dehydrogenase. When RyhB is overproduced, the cells show poor growth on media containing succinate as a sole carbon source (8).
In addition, it was noted that a study of regulation by Fur (Ferric uptake regulator) had identified Fur-binding sites and Fur regulation of a promoter that we now find is the promoter for ryhB (13). Fur is a repressor that binds to specific DNA sequences called Fur boxes, usually located in the promoter region of a target gene. This binding is Fe2+-dependent and blocks access of the RNA polymerase to the promoter (14). When iron becomes scarce in the cell, Fur is inactivated by the release of the iron cofactor, and genes under Fur control are transcribed. Essentially all genes involved in iron acquisition are Fur-regulated (14). Other genes involved in general metabolism, pathogenicity, and defense against oxidative and acid stresses are also regulated by Fur (14). Previously unexplained was how Fur positively regulates some genes (15, 16) and why fur mutants fail to grow on succinate or fumarate (17).
Here, we describe the regulation of genes at the posttranscriptional initiation level by the sRNA RyhB. We observed that RyhB down-regulates the mRNA levels for the genes that are known to be positively regulated by Fur (14, 18), including ferritins, a superoxide dismutase, and some genes of the tricarboxylic acid (TCA) cycle. All these genes encode iron-containing proteins, some of which are involved in iron storage. Our data suggest that the iron-sensor protein Fur positively regulates intracellular iron use and storage indirectly, by repression of RyhB in the presence of iron.
Materials and Methods
Bacterial Strains.
An isogenic set of strains carrying wild-type or mutant alleles of fur [fur∷kan, obtained from G. Storz (19)] and ryhB (ΔryhB1∷cat) were derived from DJ480, a ΔX74lac derivative of MG1655 from D. Jin (National Institutes of Health), were constructed by P1 transduction and used for the experiments described in this work. A deletion/insertion mutation in ryhB, ΔryhB1∷cat was constructed by using the method described by Yu et al. (20). A PCR fragment was obtained by amplifying the chloramphenicol resistance cassette of strain NC397, obtained from D. Court (National Institutes of Health), with EM2 (5′-acatcattgactttcaaatgcgagtcaaatgcatttttttgcaaaaagtgaaaatgagacgttgatcggcacg-3′) and EM3 (5′-tggataaattgagaacgaaagatcaaaaaaaaagccagcacccggctggctaccagcaatagacataagcggc-3′) oligonucleotides. The underlined sequences of EM2 and EM3 are identical to the 5′ and the 3′ ends, respectively, of the chloramphenicol cassette. The rest of EM2 is homologous to the sequence upstream of the −5 region of ryhB, whereas EM3 is complementary to the last 22 nucleotides of the ryhB gene (see Fig. 1). The resulting PCR product was transformed into DY330 after induction of λred, according to Yu et al., selecting for chloramphenicol resistance. Recombinant product was verified by sequencing.
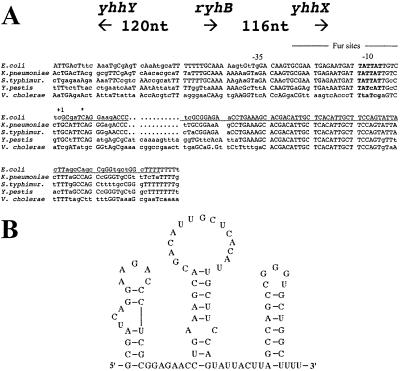
Figure 1.
(A) RyhB is oriented counterclockwise on the E. coli chromosome. Residues identical for 3/5 positions are shown with capital letters. The +1 site is as predicted by Argaman et al. (11) and agrees with our predictions from the alignment of the promoter. The asterisk is the start site of the lacZ fusion used in ref. 13. The predicted Fur sites are from ref. 13. The accession number for ryhB is AF480876. (B) Predicted secondary structure for ryhB from MFOLD (21).
Sequence Comparison.
The sequence of ryhB was originally used in a blast search against the Unfinished Microbial Genome Database (National Center for Biotechnology Information). Sequences with high matches were found for Salmonella species, Klebsiella pneumoniae, Yersinia pestis, and Vibrio cholerae. Contigs for these matched sequences were used to construct the full alignment to ryhB and its promoter, using gap and pileup (GCG; ref. 21). Further analysis was carried out with the finished Salmonella typhimurium (22), Y. pestis (23), and V. cholerae (24) genome sequences.
Growth Tests.
Strains were grown in LB or M63 media supplemented with 0.2% glycerol at 37°C. To test the ability of strains to use different carbon sources, cells were grown until saturation at 37°C in M63 liquid media with 0.2% glucose. Two microliters of the culture were streaked on solid M63 media supplemented at 0.2% with one carbon source (glucose, glycerol, succinate, or fumarate) and incubated for 36 h at 37°C.
RNA Extraction and Northern Blot Analysis.
Overnight cultures were diluted 1/1000 in fresh media and grown at 37°C until they reached an OD600 of 0.3. The culture was split in two, and one portion was treated with 250 μM of 2,2′-dipyridyl for 15 min before they were recovered by centrifugation at 2,600 × g for 10 min. RNA was then extracted by the hot phenol method (25).
For detection of RyhB RNA, the protocol used was as described in Majdalani et al. (4). For detection of acnA, fumA, sdhCDAB, bfr, ftnA, and sodB mRNAs, 10 μg of RNA of each sample, extracted as described above, was loaded on a 1.2% denaturing gel. After migration, the RNA was transferred to a charged nylon membrane (Nytran supercharged, Schleicher & Schuell) by using the passive transfer method described by Ambion (Austin, TX). Prehybridization and hybridization were carried out as described in Majdalani et al.. The 5′ biotinylated DNA probe used for RyhB detection was EM1. The other probes used for detection of acnA, fumA, sdhCDAB, bfr, ftnA, sodB, and icd were EM26, EM23, EM8, EM32, EM31, EM33, and EM28, respectively (Table 1).
Table 1.
Oligonucleotide probes
| Probes (genes) | Biotinylated oligonucleotide probes |
|---|---|
| EM8 (sdhC) | 5′-GGGGAACCGGATGGTCTGTAGGTCCAGATTAACAGGTC-3′ |
| EM9 (sdhD) | 5′-GCGTCAGGACGATAGCGGTAGCGCGAACGAGGATGAAAT-3′ |
| EM1 (RyhB) | 5′-AAGTAATACTGGAAGCAATGTGAGCAATGTCGTGCTTTCAGGTTCTC-3′ |
| EM26 (acnA) | 5′-GCGATCCGCCAACGGTAGTGAATCCAGACCATCACCATAA-3′ |
| EM23 (fumA) | 5′-CAGATCCCCTGACGGTTGATCTTCGCTTTGATATTACGGT-3′ |
| EM32 (bfr) | 5′-GCCAGATCAGAACGCAGCATTTCCTCAACATCTTCACCAA-3′ |
| EM31 (ftn) | 5′-TTCTTTGTCGATAAAATACAGACCTTCGCCGCTTTTGCCT-3′ |
| EM33 (sodB) | 5′-TGCGATAGTCGATGTAATAAGCGTGTTCCCAGACATCAAC-3′ |
| EM28 (icd) | 5′-TCTTCACCCCCATCTCTTCACGCAGGAATTTAATCACTTT-3′ |
Results
A New Conserved sRNA.
RyhB was initially identified as a well conserved sequence in the 332-nt intergenic region between yhhX and yhhY, two genes of unknown function at minute 77 on the E. coli map (Fig. 1A). It was expressed as a 90-nt RNA when cells were grown in minimal medium. Independently, the same sRNA was identified by Argaman et al. (11) and named sraI; they also observed highest expression in minimal media and in late stationary phase. Fig. 1A shows the arrangement of the genes flanking ryhB and the alignment of the sequence of ryhB and its upstream regions among a number of bacteria, as well as the start and stop points, deduced by us from size and conservation information, and experimentally determined by Argaman et al.; these are in full agreement. Also shown are predicted consensus-binding sites for Fur, the Fe-responsive repressor. Vassinoya and Kozyrev (13) identified these sites and demonstrated the regulation of a lac fusion downstream of these sites by the Fur repressor. Their fusion begins at a GATC site just 5 nucleotides beyond the +1 site for ryhB; thus, their fusion data are relevant for the ryhB promoter rather than the downstream yhhX gene, as originally described. The sequence of the ryhB RNA, as well as the immediate upstream region, including the −35, −10, and Fur sites, is very well conserved in E. coli, Salmonella, and Klebsiella, and is somewhat less conserved in Y. pestis and V. cholerae. Other sequences were also noted that share promoter elements with ryhB, including Fur sites, and also contain the highly conserved core of RyhB (nucleotides 35–61), 5′ stem-loops, and ρ-independent terminators, but are less closely related in overall sequence to ryhB and are flanked by genes other than yhhX and yhhY. One such related sRNA was found in the Salmonella and Yersinia genomes, but not in E. coli (data not shown); thus, Salmonella and Yersinia seem to encode two different RyhB-like molecules. RyhB is predicted to fold as shown in Fig. 1B; this structure has not thus far been experimentally confirmed.
Phenotype of RyhB Overproduction.
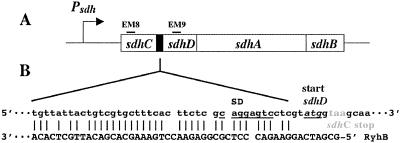
Two pieces of information from our initial analysis of RyhB suggested that it might regulate the sdhCDAB (succinate dehydrogenase) operon. The sequences of the new sRNAs were used to search the E. coli genome for possible complementary sequences that might be targets for antisense regulation; ryhB showed the most extensive match found, to a region within the sdhCDAB operon, near the end of sdhC and just before the start of the second gene in the operon, sdhD (Fig. 2). The expression of the four genes of the operon is required for synthesis of the functional enzyme (26), which is essential for growth on minimal media with succinate. We found that cells carrying a plasmid encoding RyhB (pRS-c25) were unable to grow on succinate minimal plates (8). Cells containing vector alone grew well, and there was no significant difference in the growth of these strains on glucose media and only a slight effect of the ryhB plasmid on growth on glycerol.
Figure 2.
Complementarity between the sdhCDAB operon and RyhB. Genes of the sdhCDAB operon are shown in A. Lines marked EM8 and EM9 show the position of the oligonucleotide probes used for Northern blots (Fig. 3). B shows the predicted interaction between RyhB and the sdhCDAB sense strand. The ribosome binding site for sdhD is underlined. The start codon for sdhD is shown underlined and in italics, and the stop codon for sdhC is shown in gray.
Regulation of ryhB Synthesis by the Fur Protein and Iron.
As noted above, fusion experiments suggested that the ryhB promoter should be efficiently repressed by Fur protein in the presence of iron (13). Thus, a fur− mutant is predicted to derepress RyhB synthesis. We show that this is the case below. In addition, it has been noted that fur− mutants fail to grow on succinate and fumarate (17), an observation we confirmed in our strains (Table 2). We constructed an insertion mutant in ryhB (ΔryhB1∷cat) and introduced the ryhB mutation into the fur∷kan host. As shown in Table 2, the fur− ryhB− double mutant was now able to grow on the TCA cycle carbon sources. Thus, the inhibition of growth in a fur− strain is entirely attributable to overproduction of RyhB. Overproduction of RyhB either from a plasmid (Fur repressor may be titrated in this case, leading to further overproduction) or from the chromosome in a fur− mutant leads to restriction of growth on these TCA cycle compounds.
Table 2.
Growth of fur and fur ryhB mutants
| Strains | M63 + Glucose | M63 + Succinate | M63 + Fumarate |
|---|---|---|---|
| Wild-type | ++++ | +++ | +++ |
| ΔryhB1∷cat | ++++ | +++ | +++ |
| hfq∷cat | +++ | +++ | +++ |
| Δfur∷kan | +++ | − | − |
| Δfur∷kan ΔryhB1∷cat | ++++ | +++ | +++ |
| Δfur∷kan hfq∷cat | +++ | +++ | ++ |
We confirmed the negative regulation of ryhB by Fur by directly measuring RyhB RNA in the fur mutant. As shown in Fig. 3 (compare lanes 1 and 5), the amount of RyhB is dramatically increased in a fur− mutant. As mentioned above, Fur represses only when iron is present. Thus, an iron chelator should act to induce RyhB expression. The expected increase is shown in Fig. 3 (lane 3); addition of the iron chelator 2,2′-dipyridyl to rich broth rapidly induces RyhB expression. Thus, ryhB repression requires both Fur protein and iron.
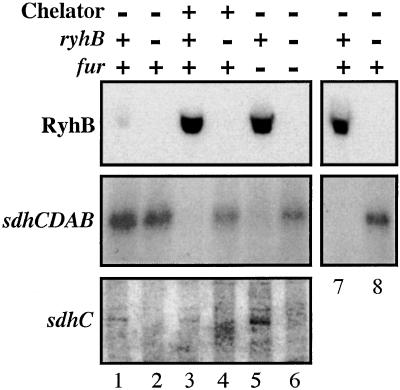
Figure 3.
Cells were grown in LB (RyhB and sdhCDAB, lanes 1–6) or M63 glycerol (RyhB and sdhCDAB , lanes 7 and 8; and sdhC, lanes 1–6) to an OD600 0.3 and 2,2′-dipyridyl was added to a final concentration of 250 μM where indicated. A sample was removed 15′ after chelator addition; RNA was isolated as described in Materials and Methods. For lanes 1, 3, and 7, a fur+ ryhB+ strain (EM1055) was used; for lanes 2, 4, and 8, a fur+ ΔryhB1∷cat strain, EM1238, was used. Cells for lanes 5 (fur− ryhB+) and 6 (fur− ΔryhB1∷cat) were strains EM1256 and EM1257, respectively. [Top (RyhB)] For determination of RyhB amount, 3 μg of total RNA samples extracted from cells grown in LB (lanes 1–6) or M63 glycerol media (lanes 7 and 8) were loaded on an 8% PAGE gel. After migration, a Northern blot analysis was performed with oligoprobe EM1 (Table 1). [Middle (sdhCDAB)] Ten micrograms of the same total RNA extracts as in Top were loaded on a denaturating 1.2% agarose gel. After migration, a Northern blot hybridization was performed with a specific oligoprobe for sdhC (probe EM8 shown in Fig. 2, Table 1). [Bottom (sdhC)] Three micrograms of RNA extracted from cells grown in M63 glycerol media were loaded on a 4% PAGE gel. A Northern blot was performed by using oligonucleotide probe EM8 within sdhC.
RyhB Level Inversely Correlates with the mRNA Level of sdhCDAB.
To test whether the predicted antisense pairing between RyhB and sdhCDAB had an effect on sdhCDAB mRNA levels, we carried out Northern blots on total RNA isolated from cells grown under a number of conditions. When cells are grown in LB medium, relatively little RyhB is made from the chromosome unless cells are either treated with an iron chelator or lack Fur repressor (Fig. 3). Under conditions where little RyhB is made (wild-type cells grown in LB; Fig. 3, lane 1), the full-length sdhCDAB mRNA can be detected; it increases somewhat in a ΔryhB mutant (Fig. 3, lane 2). However, the full-length sdhCDAB mRNA disappears within 15 min after addition of an iron chelator (Fig. 3, lane 3). This loss of the full-length message is not observed in a ΔryhB1∷cat mutant (Fig. 3, lane 4). In a fur− mutant, very little full-length sdhCDAB mRNA is detected (Fig. 3, lane 5); once again, synthesis is restored in a fur− ΔryhB− host (Fig. 3, lane 6).
In minimal medium with either glucose or glycerol as the carbon source, RyhB is easily detected (refs. 8 and 11; Fig. 3, lane 7). We assume this induction reflects relatively low iron levels in this medium. Studies done by others with an sdhCDAB operon fusion demonstrated that the sdh promoter is poorly expressed in glucose but well expressed in glycerol (27); we therefore used glycerol in our studies. The fusion point in these experiments was located upstream of the predicted site of the RyhB–sdhCDAB interaction and therefore would not have been expected to measure RyhB effects. In our Northern blot experiments, when cells were grown in minimal media with glycerol as the carbon source, little or no sdhCDAB mRNA was detected unless cells carried an ryhB mutation (Fig. 3, compare lanes 7 and 8). Thus, the presence of RyhB sRNA correlates with the absence of sdhCDAB target mRNA. We note that, although essentially no sdh mRNA was detected in minimal medium with glycerol as a carbon source, wild-type cells are able to grow on the same medium with succinate as a carbon source, suggesting that the negative regulation of sdh may not be absolute under these conditions.
RyhB Regulates the Target Gene at a Postinitiation Level.
During the course of these experiments, we observed that the presence of RyhB in the cell, while interfering with accumulation of full-length sdhCDAB mRNA, correlated with the presence of a smaller RNA product specific to the sdhC part of the operon (Fig. 3 Bottom). The truncated mRNA was only present in samples extracted from ryhB+ strains grown in minimal media supplemented with glycerol. The molecular weight (≈600 nucleotides) of this particular band corresponds to a transcript from the expected promoter up to the site of interaction with RyhB. Because there is no such signal in the ΔryhB1∷cat mutant we can conclude that transcription elongation is not intrinsically stopped at this site. This finding, as well as findings with the sdhC–lac fusion described above, suggest that the promoter of the sdhCDAB operon is not affected by the presence of RyhB. Thus, mRNA is interfered with by RyhB at the site of complementarity without affecting initiation. Possible modes of action are discussed below.
RyhB Requires Hfq for Activity.
Hfq is an RNA-binding protein with similarities in structure to eukaryotic Sm proteins (28, 29). It has been found to be necessary for the action of many sRNAs that use RNA pairing to act (refs. 30 and 31; N. Majdalani and S.G., unpublished data) and was found to bind to these sRNAs as well as to RyhB in immunoprecipitation experiments (8). To confirm a role for Hfq in RyhB action, we tested the ability of an hfq mutant to reverse the effect of fur− mutants for growth on succinate and fumarate. fur− cells cannot grow on succinate or fumarate. This growth defect is reversed by mutations in ryhB and by hfq mutants (Table 2). Thus, an hfq mutant acts phenotypically like an ryhB− strain, consistent with a requirement for Hfq for RyhB activity.
Positive Regulation of Genes by Fur Repressor Is RyhB-Dependent.
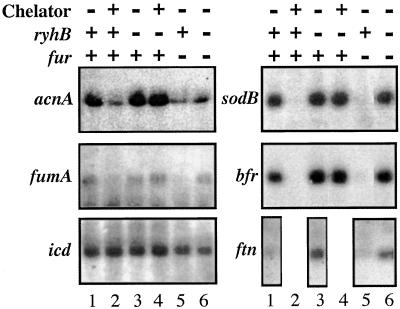
Formally speaking, the sdhCDAB operon is positively regulated by Fur repressor and iron. sdhCDAB mRNA is present only when both Fur and iron are available, and growth on succinate requires fur+ (Fig. 3, Table 2). This pattern of positive regulation by Fur was similar to that reported for a group of other genes [acnA, fumA, bfr, ftn, and sodB (18)]. acnA and fumA encode aconitase and fumarase, two other TCA cycle enzymes. bfr and ftn encode bacterial ferritins, or iron-storage proteins. sodB encodes an iron-containing superoxide dismutase. There is no evidence of a direct interaction between Fur (or iron) and the regulatory region of these genes. Studies on sodB regulation by Fur suggested it acted at a posttranscriptional initiation level (16). We asked whether RyhB could be responsible for the positive regulation of these genes by Fur, as it is for sdhCDAB. The level of acnA, fumA, bfr, ftn, and sodB mRNAs was determined for wild-type and ryhB mutant cells grown with or without chelator, and for fur− and fur− ryhB− mutants. The mRNAs for all of these genes that are positively regulated by Fur can be detected in the wild-type background in the absence of chelator (Fig. 4, lane 1). Consistent with positive regulation by Fur and iron, mRNA amounts decrease within 15′ of chelator addition (Fig. 4, compare lanes 1 and 2) and in the fur− mutant (lane 5). As seen for sdhCDAB mRNA, the effect of both the iron chelator and the fur mutant on the mRNA level depends on the presence of the functional ryhB allele (Fig. 4, lanes 4 and 6). Because there were already three TCA cycle genes regulated by RyhB, we tested for other genes of the cycle (sucAB, sucCD, gltA, mdh, and icd). No RyhB-dependent regulation was observed for these mRNAs (Fig. 4 and data not shown). We note that the TCA cycle proteins aconitase (encoded by acnA), fumarase (encoded by fumA), and succinate dehydrogenase all contain iron, whereas succinyl-CoA synthetase (encoded by suc), citrate synthase (encoded by gltA), malate dehydrogenase (encoded by mdh), and isocitrate dehydrogenase (encoded by icd) do not.
Figure 4.
The same RNA samples used in lanes 1–6 of Fig. 3 were loaded on 1.2% agarose gels and probed with oligoprobes for acnA (probe EM26), fumA (probe EM23), icd (probe EM28), sodB (probe EM33), and bfr (probe EM32). FtnA expression was found to be very low in cells grown in LB, even in the absence of ryhB. Therefore, cells were grown in M63 glycerol medium and their total RNA was extracted and migrated as for the other blots, and probed with probe EM31 (ftn). Because RyhB expression is already high in this medium, induction with chelator was not carried out.
Discussion
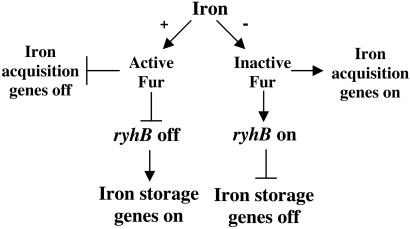
Fur protein has long been recognized as a central regulator of iron metabolism in the cell, repressing various iron acquisition systems when iron is abundant. Release of repression when iron is limiting leads to induction of these systems and the ability of these cells to scavenge iron from various extracellular environments. We have now found that Fur represses the synthesis of the sRNA ryhB, and RyhB in turn negatively regulates synthesis of at least six proteins that bind iron in the cell. Thus, Fur indirectly regulates intracellular iron storage and utilization as well as iron uptake (Fig. 5). This new circuit is fully consistent with the somewhat unexpected finding that fur mutants, with increased expression of iron acquisition systems, have low intracellular iron content (32), presumably because they have lost many iron-binding proteins. Although the regulation of some iron-storage and intracellular iron-usage genes by Fur was previously recognized, the basis for this regulation is now explained by Fur regulation of RyhB synthesis.
Figure 5.
Model of Fur and RyhB interaction to regulate iron utilization.
Of the six genes identified thus far as targets for RyhB regulation, two clearly encode iron-storage proteins, ferritin and bacterioferritin. They help prevent Fe-dependent damage by removing free iron from the cytoplasm and also serve as a source of iron when it becomes limiting. Cells devoid of ferritin grow poorly when transferred from iron-rich to iron-poor medium (32). In addition, the availability of this stored iron can help to repair damaged Fe-containing proteins after oxidative damage. Thus, the described positive regulation of these genes by Fur and iron was not surprising; however, the mechanism was unclear before the work presented here (33). bfr is the second gene in an operon and is preceded by bfd, a gene that encodes a 64-aa protein with a [2Fe-2S] site; it may act as a regulatory component for the storage of iron by helping the release and/or delivery of iron to bacterioferritin (34). We do not yet know whether bfd is also down-regulated by RyhB.
Another of the regulated genes is sodB, encoding one of three superoxide dismutases in E. coli. Superoxide dismutase functions to lessen the load of superoxide in the cell, a source of oxidative damage. Free iron, in particular in the presence of oxygen, causes such damage via the Fenton reaction, and superoxide dismutases play an important role in protecting from this damage (35, 36). The cell seems to adjust the relative amounts of the three superoxide dismutases to fit the state of its nutrition and growth. SodB is the only one of the three E. coli enzymes that uses Fe in its active site. When iron is limiting, it is more useful for the cell to make an enzyme that does not depend on using the limited iron. In fact, whereas sodB is positively regulated by Fur, sodA (MnSOD) is negatively regulated (37). The positive Fur regulation of sodB has been studied recently (16). The presence of a palindrome and an AT-rich region located in the 5′ untranslated region of the mRNA was necessary for the full positive regulation by Fur. This region is immediately upstream of a possible RyhB interaction site with sodB mRNA. Thus, the details of the regulation of sodB by RyhB may be complex and may provide the possibility of other levels of regulation. Consistent with the role of RyhB in regulating sodB, Hfq has been found to be necessary for the Fur regulation of sodB (D. Touati and S. Dubrac, personal communication).
The other three enzymes that were down-regulated by RyhB are in the TCA cycle: succinate dehydrogenase, encoded by the sdh operon; aconitase, encoded by acnA; and fumarase, encoded by fumA. They all contain [4Fe-4S]2+ clusters; other enzymes in the TCA cycle that do not contain these clusters were not regulated by RyhB. However, not all iron-containing proteins are regulated in this manner. Fur was found to positively regulate acnA and fumA, but not acnB or fumB, two homologs of acnA and fumA (15). Why this regulation differs remains to be explained. However, when acnA and fumA are down-regulated by RyhB, other enzymes capable of carrying out the same reaction may still be active. Our experiments provide an explanation for the long-mysterious observation that fur mutants fail to grow on succinate (17).
What is the advantage to the cell of this two-tiered system, in which Fur protein is involved in the regulation of genes involved in the uptake of iron while RyhB would be regulating nonessential iron-containing proteins and proteins involved in iron storage? Using an sRNA does not require protein synthesis and could be one of the most economical and efficient ways to globally repress genes. Because it may act on preexisting message, it also should act more quickly than regulation at the level of the promoter, which leaves preexisting message active. In the case of a sudden drop in iron availability, the cell can rapidly stop the synthesis of targeted protein(s) by using an sRNA. Switching off synthesis would immediately decrease the use of iron by these proteins, which would then help the cell to reorient free Fe toward more critical functions. Alternatively, the use of an sRNA may simply be a mechanistically simple way to convert the Fur-negative regulator into a positive regulator of a subset of genes, many already under complex transcriptional control.
The discovery of this switch suggests that, in addition, there might be a mechanism for releasing the iron from preexisting stores of these same proteins. At the same time, inactivation of Fur repression in low iron will allow the synthesis of iron uptake genes. It would make sense if the cell specifically decreases dispensable intracellular iron usage before seeking extracellular sources of iron. If so, we would expect that RyhB would be a particularly sensitive indicator of Fe limitation. We note that it gave the most dramatic induction ratio in the presence of iron in the experiments of Vassinova and Kozyrev (13).
Use of an sRNA also opens the possibility of affecting the relative levels of proteins within an operon. At least one sRNA, spot 42, has recently been shown to mediate cAMP-dependent changes in the ratios of the proteins of the gal operon by specifically down-regulating expression of the third gene in the operon (29). We do not yet know whether the fact that sdhC mRNA is not fully down-regulated by this mechanism leads to synthesis of functional SdhC protein; if so, this would suggest that SdhC has another role in the cell, independent of that in the succinate dehydrogenase enzyme complex. Similarly, the bfr gene is the second gene in an operon; we have not thus far examined the synthesis of the upstream gene (bfd).
There is no reason to believe that the six genes identified here as regulated by RyhB are its only targets. Two-dimensional gel experiments in Salmonella uncovered 15 protein spots positively regulated by Fur, 9 of them also depend on iron (38). It seems likely that at least these 9 proteins, and possibly all 15, are regulated by RyhB or the second RyhB-like sRNA present in Salmonella. Many proteins are negatively regulated by Fur. It is possible that in some cases, regulation is at both transcription initiation, by direct Fur repression, and postinitiation, by positive regulation by RyhB. Two other sRNAs, DsrA and RprA, act positively to stimulate RpoS mRNA translation (4, 6).
We thus far have relatively little information on the mechanism of RyhB action. When RyhB is made in high quantities, the full-length message for sdhCDAB disappears and is replaced by a short message encoding the 5′ untranslated region and first gene of the operon. The end of the short message corresponds to the region that is complementary between RyhB and sdh. Possible short complementarity between RyhB and the other five genes regulated by RyhB can be found; most are near the ribosome-binding site and ATG (data not shown). We assume that the general mechanism of RyhB action on all six of its demonstrated targets will be similar.
RyhB depends on the Hfq RNA-binding protein for function. Given the previous demonstration that RyhB is efficiently immunoprecipitated with Hfq, this is not surprising. Thus far, there is a complete correlation between sRNAs that act by means of RNA–RNA pairing and Hfq use, consistent with RNA–RNA pairing playing a role in RyhB action as well. Recent experiments suggest that Hfq directly stimulates such RNA–RNA pairing, apparently acting as a general RNA chaperone (28, 29). Among the general mechanisms that might explain the polarity observed in the sdhCDAB operon, most depend on RyhB RNA–mRNA pairing. Such pairing could stop translation by blocking the elongating ribosomes on the mRNA. This translation stop might, in turn, allow access of the Rho transcription terminator to the untranslated message beyond the block site; in this model, a Rho site should be present soon after the region of complementarity. It is also possible that RyhB and Hfq actively recruit Rho when they interact with a target mRNA or otherwise lead directly to rapid termination of transcription by interaction with the growing mRNA. Alternatively, the annealing of RyhB to the message could be recognized by a double-strand specific RNA endoribonuclease, initiating rapid degradation, particularly of the downstream mRNA. We were not able to detect any signal with an sdhD probe (data not shown). Although the upstream mRNA is detected (Fig. 3), we do not know how much of it is present compared with the full-length message seen in a ΔryhB mutant. It seems likely it is also subject to degradation, a possible explanation for our inability to detect it at all in cells grown in rich medium.
It is also possible that RyhB anneals with the DNA, and not the RNA, creating an RNA–DNA hybrid (R-loop) with the nontemplate strand. This structure would eventually block the transcription machinery from proceeding further (39). To test this possibility, we introduced a plasmid overproducing RNase H in a Δfur∷kan mutant. This endoribonuclease is specific for degradation of RNA in RNA–DNA hybrids. No improved growth was observed on succinate-minimal media (data not shown).
The discovery of RyhB and its central role in regulating intracellular iron use reinforces the importance of sRNAs for regulating key metabolic switches. Iron acquisition and storage have long been recognized as a complex problem for bacteria. They must avoid excess free iron that leads to various sorts of damage, and they must find adequate sources of iron to allow growth. Complex systems for bacterial pathogens to acquire iron from their hosts have developed and have been demonstrated to be important pathogenicity components in many cases (40). We can now add a complex and well conserved system for setting intracellular iron-usage priorities to the mechanisms that regulate iron flux. It seems likely that parallel mechanisms will exist in many, if not all, organisms likely to be iron-deficient at some point in their growth.
Acknowledgments
We thank J. A. Imlay and G. Storz for their comments on the manuscript and insights into the regulatory system, and D. Touati for sharing unpublished results. The members of our laboratory, particularly F. Repoila, N. Majdalani, and Y.-N. Zhou, are thanked for their advice throughout this work. We thank M. Drolet, M. Maurizi, F. Repoila, and G. Storz for chemicals, strains, and plasmids. E.M. was supported by a postdoctoral fellowship from the Canadian Institutes of Health Research.
Abbreviations
- sRNA
small RNA
- TCA
tricarboxylic acid
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF480876).
References
- 1.Wassarman K M, Storz G. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 2.Keiler K C, Waller P R H, Sauer R T. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 3.Sledjeski D D, Gupta A, Gottesman S. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 4.Majdalani N, Chen S, Murrow J, St. John K, Gottesman S. Mol Microbiol. 2001;39:1382–1394. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
- 5.Wassarman K M, Zhang A, Storz G. Trends Microbiol. 1999;7:37–45. doi: 10.1016/s0966-842x(98)01379-1. [DOI] [PubMed] [Google Scholar]
- 6.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. Proc Natl Acad Sci USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lease R A, Cusick M, Belfort M. Proc Natl Acad Sci USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wassarman K M, Repoila F, Rosenow C, Storz G, Gottesman S. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hajndsorf E, Regnier P. Proc Natl Acad Sci USA. 2000;97:1501–1505. doi: 10.1073/pnas.040549897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vytvytska O, Jakobsen J, Balcunaite G, Andersen J, Baccarini M, von Gabain A. Proc Natl Acad Sci USA. 1998;95:14118–14123. doi: 10.1073/pnas.95.24.14118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner E G H, Margalit H, Altuvia S. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 12.Rivas E, Klein R J, Jones T A, Eddy S R. Curr Biol. 2001;11:1369–1373. doi: 10.1016/s0960-9822(01)00401-8. [DOI] [PubMed] [Google Scholar]
- 13.Vassinova N, Kozyrev D. Microbiology. 2000;146:3171–3182. doi: 10.1099/00221287-146-12-3171. [DOI] [PubMed] [Google Scholar]
- 14.Escolar L, Pérez-Martin J, de Lorenzo V. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruer M J, Guest J R. Microbiology. 1994;140:2531–2541. doi: 10.1099/00221287-140-10-2531. [DOI] [PubMed] [Google Scholar]
- 16.Dubrac S, Touati D. J Bacteriol. 2000;182:3802–3808. doi: 10.1128/jb.182.13.3802-3808.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hantke K. Mol Gen Genet. 1987;210:135–139. doi: 10.1007/BF00337769. [DOI] [PubMed] [Google Scholar]
- 18.Hantke K. Curr Opin Microbiol. 2001;4:172–177. doi: 10.1016/s1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- 19.Zheng M, Doan B, Schneider T D, Storz G. J Bacteriol. 1999;181:4639–4643. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu D G, Ellis H M, Lee E C, Jenkins N A, Copeland N G, Court D L. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClelland M, Sanderson K E, Spieth J, Clifton S W, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, et al. Nature (London) 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 23.Parkhill J, Wren B W, Thomson N R, Titball R W, Holden M T, Prentice M B, Sebaihia M, James K D, Churcher C, Mungall K L, et al. Nature (London) 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- 24.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, et al. Nature (London) 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aiba H, Adhya S, de Crombrugghe B. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 26.Ackrell B A, Johnson M K, Gunsalus R P, Cecchini G. In: Chemistry and Biochemistry of Flavoenzymes. Muller F, editor. Vol. 3. Boca Raton, FL: CRC; 1992. pp. 229–297. [Google Scholar]
- 27.Park S-J, Tseng C-P, Gunsalus R P. Mol Microbiol. 1995;15:473–482. doi: 10.1111/j.1365-2958.1995.tb02261.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhang A, Wassarman K M, Ortega J, Steven A C, Storz G. Mol Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 29.Moller T, Franch T, Hojrup P, Keene D R, Bachinger H P, Brennan R, Valentin-Hansen P. Mol Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 30.Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sledjeski D D, Whitman C, Zhang A. J Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abdul-Tehrani H, Hudson A J, Chang Y-S, Timms A R, Hawkins C, Williams J M, Harrison P M, Guest J R, Andrews S C. J Bacteriol. 1999;181:1415–1428. doi: 10.1128/jb.181.5.1415-1428.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrews S C, Harrison P M, Guest J R. J Bacteriol. 1989;171:3940–3947. doi: 10.1128/jb.171.7.3940-3947.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garg R P, Vargo C J, Cui X, Kurtz D M J. Biochemistry. 1996;35:6297–6301. doi: 10.1021/bi9600862. [DOI] [PubMed] [Google Scholar]
- 35.Imlay J A, Linn S. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 36.Maringanti S, Imlay J A. J Bacteriol. 1999;181:3792–3802. doi: 10.1128/jb.181.12.3792-3802.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Compan I, Touati D. J Bacteriol. 1993;175:1687–1696. doi: 10.1128/jb.175.6.1687-1696.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foster J W, Hall H K. J Bacteriol. 1992;174:4317–4323. doi: 10.1128/jb.174.13.4317-4323.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hraiky C, Raymond M A, Drolet M. J Biol Chem. 2000;275:11257–11263. doi: 10.1074/jbc.275.15.11257. [DOI] [PubMed] [Google Scholar]
- 40.Ratledge C, Dover L G. Annu Rev Microbiol. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]