Abstract
By comparing sequences of human, mouse and rat orthologous genes, we show that in 5′-untranslated regions (5′-UTRs) of mammalian cDNAs but not in 3′-UTRs or coding sequences, AUG is conserved to a significantly greater extent than any of the other 63 nt triplets. This effect is likely to reflect, primarily, bona fide evolutionary conservation, rather than cDNA annotation artifacts, because the excess of conserved upstream AUGs (uAUGs) is seen in 5′-UTRs containing stop codons in-frame with the start AUG and many of the conserved AUGs are found in different frames, consistent with the location in authentic non-coding sequences. Altogether, conserved uAUGs are present in at least 20–30% of mammalian genes. Qualitatively similar results were obtained by comparison of orthologous genes from different species of the yeast genus Saccharomyces. Together with the observation that mammalian and yeast 5′-UTRs are significantly depleted in overall AUG content, these findings suggest that AUG triplets in 5′-UTRs are subject to the pressure of purifying selection in two opposite directions: the uAUGs that have no specific function tend to be deleterious and get eliminated during evolution, whereas those uAUGs that do serve a function are conserved. Most probably, the principal role of the conserved uAUGs is attenuation of translation at the initiation stage, which is often additionally regulated by alternative splicing in the mammalian 5′-UTRs. Consistent with this hypothesis, we found that open reading frames starting from conserved uAUGs are significantly shorter than those starting from non-conserved uAUGs, possibly, owing to selection for optimization of the level of attenuation.
INTRODUCTION
Translational efficiency of eukaryotic mRNAs depends on the structural features of the 5′-untranslated region (5′-UTR, or leader sequence) and the nucleotide sequence flanking the translation start codon (start codon context). Both experimental (1–5) and statistical (6–17) approaches have been used to reveal the characteristics of the 5′-UTRs that influence translation. In particular, 5′-UTR length (18), secondary structure (19) and the presence of AUG triplets upstream of the true translation start in mRNA, known as upstream AUGs (20) (hereinafter uAUGs; for the sake of uniformity, we designate start codons AUG disregarding the fact that it should be rendered as ATG when DNA sequences are considered), have been shown to affect the efficiency of translation. Although, in most cases, translation of eukaryotic mRNAs is initiated at the first AUG of the 5′-UTR, according to the scanning model of Kozak, many exceptions to this rule have been described (2,3,19–25). The context of the start codon is another important regulatory factor. A consensus sequence, (GCC)GCCRCCAUGG (where R = G or A), has been derived for the start codon context of mammalian mRNAs (6,8). The most critical sites near the start AUG codon (sAUG) are −3 (the A of the sAUG is position +1), usually occupied by a purine, and +4, typically occupied by a guanine (19,26). Other sites near the start codon seem to be less important, although the influence of nucleotides in different positions may vary from species to species (27–30).
Generally, uAUGs decrease mRNA translation efficiency and may be considered strong negative translational regulatory signals (3,31,32), since the major mechanism of translation initiation in eukaryotes appears to be the scanning model, according to which eukaryotic ribosomes initiate translation at the 5′-proximal AUG codon (1,6). This model is supported by the observation that mammalian 5′-UTRs contain significantly fewer uAUGs than expected by chance, suggesting that purifying selection caused elimination of uAUGs in many 5′-UTRs (13,17). Nevertheless, a substantial fraction of 5′-UTRs contain uAUGs (13,33–35). The presence of uAUGs correlates with a ‘weak’ start codon context (low information content) and conversely, high information content of the start codon context is typical of short 5′-UTRs containing no uAUGs (13). This finding seems to be compatible with the possibility that uAUGs function as attenuators of translation initiation by engaging ribosomes in futile initiation and precluding them from reaching the sAUG. However, the alternative possibility remains that cDNAs with AUG-containing 5′-UTRs might represent mainly non-functional sequences, such as incorrectly processed transcripts and transcripts of pseudogenes, artifacts of cloning and coding region annotation, such that the purported uAUGs actually belong to misannotated 5′-portions of coding regions, or mRNAs of genes with multiple transcription start sites (2,13,19,31,32). Indeed, follow-up studies have shown that some cDNA sequences containing many AUGs in the 5′-UTR do not correspond to functional mRNAs (31,32,36).
A natural approach to address the issue of the functional significance of uAUGs is to examine the evolutionary conservation of these triplets on genome scale. By comparing sequences of human, mouse and rat orthologous genes, we show here that in 5′-UTRs of mammalian cDNAs but not in 3′-UTRs or coding sequences, AUG is conserved to a significantly greater extent than any of the other 63 nt triplets. This observation can be hardly explained by annotation errors because pronounced excess of conserved uAUGs was detected in 5′-UTRs containing an in-frame stop codon and because many of the conserved AUGs are found in different frames, which is consistent with the location in non-coding sequences. A similar excess of conserved uAUGs was detected in yeast 5′-UTRs. These observations strongly suggest that at least some of the conserved uAUGs are functional elements of translation initiation regulation.
MATERIALS AND METHODS
Mammalian cDNA sequences were extracted from the Refseq database (16 773 human cDNAs, 19 777 mouse cDNAs and 21 178 rat cDNAs). All 5′-UTRs that showed statistically significant BLASTX (37) hits (E-value < 10−5) to the non-redundant protein database (National Center for Biotechnology Information, NIH, Bethesda) and, accordingly, were probable to represent that undetected coding sequences were removed from the datasets. In addition, the identity between the first 50 amino acids of the Refseq proteins encoded in the analyzed genes and the corresponding Swissprot entries were checked. All redundant 5′-UTR sequences were removed from the datasets. Two 5′-UTRs were considered redundant if their masked sequences (RepeatView, www.itb.cnr.it/webgene) (38) produced a bidirectional BLAST hit with an E-value < 10−6. Of all redundant 5′-UTRs, only the shortest versions were retained to exclude cloning artifacts and unspliced introns. Probable one-to-one orthologs were identified by comparing symmetrical best hits (39) between proteins from the respective genomes using the BLASTP program (40). The nucleotide sequence alignments for identified orthologous pairs of cDNAs were produced using the BLASTN program (37). Only long pairwise alignments (high-scoring segment pairs) with length ≥50 bases and with a low expectation value (E-value < 10−10) were retained for analysis. This ensures the reliability of nucleotide sequence alignments since BLASTN is known to produce conservative alignments, at least when applied with restrictive cut-off values as done here (41,42). The sequence lists and alignments from this dataset are available at ftp://ftp.ncbi.nih.gov/pub/koonin/uAUG/.
Aligned sequences of orthologous genes from four yeast species, Saccharomyces cerevisiae, Saccharomyces paradoxus, Saccharomyces mikatae and Saccharomyces bayanus were extracted from the SGD database (ftp://genome-ftp.stanford.edu/pub/yeast/data_download/sequence/fungal_genomes) (43). The alignment of start and stop codons was fixed in order to ensure the correct partitioning of 5′-UTR, the CDS (protein-coding sequence) and the 3′-UTR. Saccharomyces cDNA sequences were extracted from GenBank, and the reliability of annotation of the 5′-UTRs and CDS was assessed as described above. The evolutionary conservation was calculated as the fraction of triplets that are conserved in pairwise (for comparisons of mammalian sequences) or multiple (for comparisons of yeast sequences) alignments of 5′-UTRs.
RESULTS
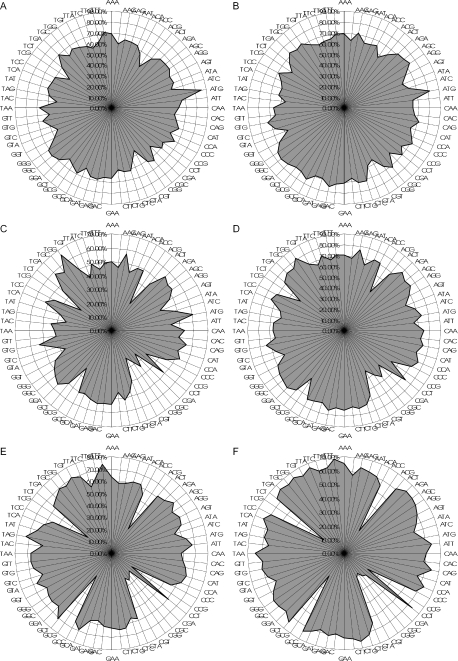
We analyzed evolutionary conservation of uAUGs in pairwise alignments of human–mouse and mouse–rat orthologs. Conservative criteria were adopted to select reliable alignments for further analysis (Materials and Methods). It was found that 33% of the 2241 selected human–mouse 5′-UTR alignments and 38% of the 2312 mouse–rat 5′-UTR alignments contained at least one conserved uAUG. These fractions of uAUG-containing 5′-UTRs are consistent with previous observations (13,33–35). Examination of the evolutionary conservation of each of the 64 nt triplets in the aligned 5′-UTRs showed that AUG was the most conserved triplet in both human–mouse and mouse–rat comparisons (Figure 1A and B). Tables 1 and 2 compare the frequencies of conserved uAUGs with those of the five triplets that are permutations of AUG, in order to exclude any possible effect of nucleotide composition; in both cases, the fraction of conserved AUGs was markedly greater than those of the other triplets (P < 10−42 by the Fisher's exact test). In contrast, no excessive conservation of AUG compared with the other 63 triplets was detected in 3′-UTRs and CDSs (excluding the start AUG) of mammalian mRNAs (Figure 1C–F), indicating that AUG conservation was specific for 5′-UTRs. The conservation of uAUGs seemed to be spread throughout the 5′-UTRs: a significant excess of conserved uAUGs was observed both in the proximal (region −100: −1 nt) and the distal (region −200: −101 nt) parts of 5′-UTR alignments, without a significant enrichment in either (Supplementary Tables S1–S4).
Figure 1.
Plots of nucleotide triplet conservation in mammalian cDNAs. (A) Human–mouse 5′-UTRs. (B) Mouse–rat 5′-UTRs. (C) Human–mouse CDS. (D) Mouse–rat CDS. (E) Human–mouse 3′-UTR. (F) Mouse–rat 3′-UTR.
Table 1.
Preferential conservation of uAUGs in orthologous human and mouse 5′-UTRs
| Trinucleotide | Triplet present in human but not in mouse (%) | Triplet conserved in human and mouse (%) | Triplet present in mouse but not in human (%) |
|---|---|---|---|
| AUG | 8 | 85 | 7 |
| AGU | 19 | 61 | 20 |
| GUA | 24 | 54 | 23 |
| GAU | 17 | 68 | 15 |
| UAG | 21 | 59 | 20 |
| UGA | 16 | 69 | 15 |
Fisher's exact test: 1266 conserved uAUGs, 218 non-conserved uAUGs, 5690 conserved AGU/GUA/GAU/UAG/UGAs, 3219 non-conserved AGU/GUA/GAU/UAG/UGAs and P = 1.4 × 10−66.
Table 2.
Preferential conservation of uAUGs in mouse and rat orthologous 5′-UTRs
| Trinucleotide | Triplet present in mouse but not in rat (%) | Triplet conserved in mouse and rat (%) | Triplet present in rat but not in mouse (%) |
|---|---|---|---|
| AUG | 9 | 81 | 10 |
| AGU | 17 | 65 | 18 |
| GUA | 20 | 57 | 23 |
| GAU | 15 | 69 | 16 |
| UAG | 18 | 62 | 20 |
| UGA | 13 | 72 | 15 |
Fishers's exact test: 1539 conserved uAUGs, 359 non-conserved uAUGs, 11 218 conserved AGU/GUA/GAU/UAG/UGAs, 5703 non-conserved AGU/GUA/GAU/UAG/UGAs and P = 2.9 × 10−42.
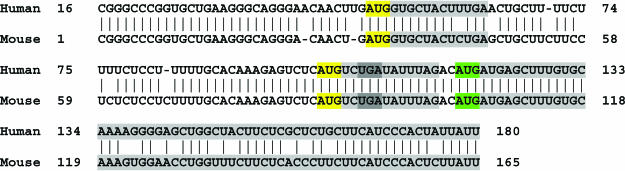
An issue of concern with this observation is that the excess of conserved uAUGs could be an artifact of erroneous annotation of coding regions, with the sequences annotated as 5′-UTR actually containing the upstream portion of the CDS, including the typically conserved authentic start AUG and, possibly, conserved methionine codons as well (36). This did not seem to be a major factor because the observed excess of conserved uAUG did not depend on whether we used Refseq annotations (Tables 1 and 2) or only those Refseq annotations that were consistent with the corresponding SwissProt annotations (Supplementary Tables S5 and S6). Nevertheless, to control such an artifact in a more direct fashion, we examined 5′-UTRs containing an upstream conserved stop codon(s) in the same frame with the sAUG (Supplementary Figure S1); 5′-UTRs containing in-phase stop codons are unlikely to represent undetected parts of the CDS. Figure 2 shows an example of a conserved in-frame stop codon in a human–mouse 5′-UTR alignment with two conserved uAUGs. The main open reading frame (ORF) encoding the α1 type III collagen proprotein cannot be extended in the 5′ direction because, in both human and mouse 5′-UTRs, there is an in-frame stop codon UGA. Thus, the start codon of this gene apparently is annotated correctly, and the conserved uAUGs are, indeed, located in the 5′-UTR (Figure 2). Analysis of the portions of 5′-UTRs located upstream of conserved in-frame stop codons (5′-UTR< stop) yielded results that were fully compatible with those for complete 5′-UTRs. Specifically, 19 and 22% of the human–mouse and mouse–rat alignments, respectively, contained conserved uAUGs, and there was a highly significant excess of conserved uAUGs compared with the five shuffled triplets (Tables 3 and 4).
Figure 2.
A mammalian 5′-UTR with conserved uAUGs and an in-frame stop codon. The alignment of the 5′UTRs of human and mouse α1 type III collagen proprotein (GI numbers: 15 149 480 and 33 859 525, respectively). uAUGs are colored yellow, the collagen starting codon is colored green, and the open reading frames are colored grey. The protein-coding region cannot be extended in the 5′ direction because of the presence of a conserved in-frame UGA stop codon (dark gray).
Table 3.
Preferential conservation of uAUGs in human–mouse stop-codon-bounded 5′-UTR alignments (5′-UTR<stop set)
| Trinucleotide | Triplet present in human, but not in mouse (%) | Triplet conserved in human and mouse (%) | Triplet present in mouse, but not in human (%) |
|---|---|---|---|
| AUG | 9 | 84 | 7 |
| AGU | 17 | 69 | 14 |
| GUA | 20 | 62 | 18 |
| GAU | 17 | 70 | 13 |
| UAG | 15 | 70 | 15 |
| UGA | 16 | 72 | 12 |
Fishers's exact test: 255 conserved uAUGs, 49 non-conserved uAUGs, 1124 conserved AGU/GUA/GAU/UAG/UGAs, 491 non-conserved AGU/GUA/GAU/UAG/UGAs and P = 1.52 × 10−7.
Table 4.
Preferential conservation of uAUGs in mouse–rat stop-codon-bounded 5′-UTR alignments (5′-UTR<stop set)
| Trinucleotide | Triplet present in mouse, but not in rat (%) | Triplet conserved in mouse and rat (%) | Triplet present in rat, but not in mouse (%) |
|---|---|---|---|
| AUG | 11 | 80 | 9 |
| AGU | 15 | 67 | 18 |
| GUA | 18 | 57 | 25 |
| GAU | 16 | 68 | 16 |
| UAG | 17 | 62 | 21 |
| UGA | 14 | 70 | 16 |
Fishers's exact test: 499 conserved uAUGs, 124 non-conserved uAUGs, 3262 conserved AGU/GUA/GAU/UAG/UGAs, 1675 non-conserved AGU/GUA/GAU/UAG/UGAs and P = 3.53 × 10−13.
Further evidence of bona fide evolutionary conservation of uAUGs was obtained by analysis of the phase distribution of the conserved uAUGs (phase 0 is the frame of the start codon, and phases 1 and 2 are frames shifted by 1 or 2 nt, respectively) (Supplementary Figure S2). We found that human–mouse and mouse–rat alignments have a significant excess of conserved uAUGs in the same phase (0–0, 1–1 or 2–2 in Table 5). However, the fraction of conserved uAUGs in different phases was also substantial (0–1, 1–2 and so on; Table 5) which is not the expectation for protein-coding regions where frameshift mutations are, generally, not tolerated. It should be noted that BLASTN alignments are extremely conservative with respect to deletions/insertions, so the actual fraction of conserved uAUGs in different phases might be even greater. The phase distribution of the conserved uAUGs showed a significant excess of phase 1–1 and phase 2–2 uAUGs compared with phase 0–0 uAUGs (Table 5). This may reflect the greater chance for uORFs in phase 0–0 to merge with the main ORFs, thereby extending the main ORFs in the 5′ direction, which could be a mechanism of evolution of protein-coding regions yielding alternative translation starts (44,45). However, the possibility cannot be ruled out that, at least in part, the deficit of phase 0–0 is caused by the erroneous annotation of the 5′-terminal AUG as the start codon in a subset of mRNAs, resulting in depletion of phase 0 uAUGs. Indeed, analysis of the 5′-UTR<stop regions which are, obviously, refractory to this artifact, showed a nearly uniform phase distribution (Table 6). The lower excess of same-phase uAUGs in the 5′-UTR<stop data set compared to the entire set of uAUGs (compare the data in Tables 5 and 6) is probably owing to the lower sequence conservation and greater prevalence of indels in the distal parts of the 5′-UTRs (16). Thus, the phase distribution of the conserved uAUGs is best compatible with their localization in non-coding regions. Taken together, these results show that the observed exceptional conservation of uAUGs in the 5′-UTRs of mammalian genes is not an artifact and strongly suggests that the uAUGs are subject to purifying selection and hence have a function(s), most probably, related to the regulation of translation initiation.
Table 5.
Phase distribution of conserved uAUGs in alignments of mammalian 5′-UTRs
| Phase 0 | Phase 1 | Phase 2 | |
|---|---|---|---|
| Human/mouse | |||
| Phase 0 | 221 | 60 | 72 |
| Phase 1 | 94 | 282 | 94 |
| Phase 2 | 54 | 89 | 350 |
| Mouse/rat | |||
| Phase 0 | 267 | 88 | 100 |
| Phase 1 | 66 | 350 | 93 |
| Phase 2 | 81 | 99 | 395 |
Table 6.
Phase distribution of conserved uAUGs in stop-codon-bounded alignments of mammalian 5′-UTRs (5′-UTR<stop set)
| Phase 0 | Phase 1 | Phase 2 | |
|---|---|---|---|
| Human/mouse | |||
| Phase 0 | 50 | 23 | 34 |
| Phase 1 | 36 | 42 | 22 |
| Phase 2 | 30 | 32 | 36 |
| Mouse/rat | |||
| Phase 0 | 64 | 60 | 56 |
| Phase 1 | 59 | 65 | 54 |
| Phase 2 | 58 | 37 | 72 |
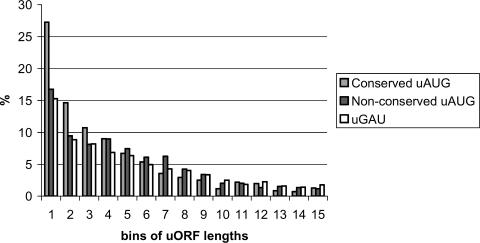
Obviously, each uAUG starts an ORF, typically, a short one. It has been reported recently that uORFs are, on average, slightly but significantly shorter than random ORFs (17). Figure 3 compares the length distributions of uORFs starting from conserved uAUGs, non-conserved uAUGs and uGAUs (GAU triplet, a permutation of AUG, located in the 5′-UTRs).
Figure 3.
Length distributions of human uORFs starting with conserved uAUGs, non-conserved uAUGs and pseudo-ORFs starting with uGAUs. The ORF length is represented by bins, each including 10 codons (i.e. bin 1 includes ORFs from 0 to 10 codons, bin 2 ORFs from 11 to 20 codons and so on).
A noticeable and statistically highly significant excess of very short uORFs was observed for conserved uAUGs: the length distribution of uORFs starting with conserved uAUGs differed from the other two distributions with P < 10−4 (χ2 test), and the excess of the shortest uORFs (0–20 codons) was significant at P < 10−6 (Fisher's exact test). The length distributions of uORF starting with conserved uAUG were very similar for all three phases (Supplementary Figure S3). In contrast, the distributions of uORF lengths starting from non-conserved uAUGs and GAUs were statistically indistinguishable (similar results were obtained with other permutations of AUG; data not shown). Conceivably, the uORFs that start from conserved uAUGs and may be engaged in the regulation of translation initiation were selected for short length optimization of the level of attenuation and prevent complete shutdown of initiation from the sAUG.
There was no significant difference between the nucleotide contexts of the conserved and non-conserved uAUGs (data not shown); in both cases, the information content of the uAUG context was lower than that of the sAUG context (13,17). Thus, uORFs do not appear to be optimized for translation efficiency.
Having shown that uAUGs are exceptionally conserved in 5′-UTRs of mammalian mRNAs, we sought to investigate how general this pattern might be. To this end, we examined the conservation of uAUGs in orthologous gene sets from four species of yeasts. Since precise mRNA mapping is unavailable for most yeast genes, we had to analyze, as a proxy for 5′-UTRs, genomic sequences (50 nt) located upstream of sAUGs codons of yeast genes. This analysis revealed a highly significant excess of conserved uAUGs compared with the permuted triplets (Table 7). This result was confirmed also with genomic alignments of 5′-UTR<stop sequences (see above) and with 5′-UTRs of the available yeast cDNA sequences (Table 7). Furthermore, 11% of the 253 genomic alignments of 5′-UTRs for known yeast cDNA sequences contained uAUGs conserved in all four yeast species for which genome sequences are available (data not shown).
Table 7.
Preferential conservation of uAUGs in yeast orthologous 5′-UTRs
| Trinucleotide | Conserved triplets (%) | Non-conserved triplets (%) |
|---|---|---|
| AUG | 30 | 70 |
| AUG (5′-UTR<stop) | 28 | 72 |
| AUG (5′-UTR<stop + cDNA) | 36 | 64 |
| AGU | 9 | 91 |
| GUA | 7 | 93 |
| GAU | 9 | 91 |
| UAG | 10 | 90 |
| UGA | 10 | 90 |
Triplets were considered when present in the aligned sequences from all four yeast species. The AUG data set consisted of alignments of genomic sequences located upstream (50 nt) of sAUGs. The 5′-UTR<stop dataset consisted of the respective sequences containing in-frame stop codons. The AUG(5′-UTR< stop + cDNA) dataset consisted of genomic alignments of 5′-UTR<stop sets that matched 5′-UTRs of the available of yeast cDNA sequences. Fisher's exact test for the fraction of conserved AUG triplets versus the fraction of conserved AGU/GUA/GAU/UAG/UGA triplets produced highly significant results (P < 10−13) for each of the three datasets. The shuffled triplet frequencies are shown for genomic sequences located upstream of sAUGs codons of yeast genes, this corresponds to the first row of the table (the AUG dataset).
DISCUSSION AND CONCLUSIONS
The comparative genomic analysis reported here shows that the 5′-UTRs of at least 20–30% of mammalian mRNAs contain conserved uAUGs; qualitatively similar results were obtained with yeast 5′-UTRs. Strikingly, in both mammals and yeasts, AUG is by far the most conserved nucleotide triplet in 5′-UTR but not in 3′-UTRs or coding regions. These findings suggest that at least a fraction of the conserved uAUGs perform a function related to the regulation of translation initiation. Compatible with this conclusion, we found that uORFs starting from conserved (and, potentially, functional) uAUGs are, on average, significantly shorter than those that start from non-conserved (probably, non-functional) AUGs. There was no single dominant functional trend among the genes containing multiple conserved uAUGs but many of these genes encode transcription factors, receptors and other proteins involved in various forms of signal transduction (Supplementary Table S7 and data not shown). These genes are subject to complex regulation which seems to be compatible with the proposed role of uAUGs in modulation of translation initiation.
The conclusion of the present work on the probably functional importance of conserved uAUGs has to be considered in conjunction with the more or less orthogonal previous observation that mammalian 5′-UTRs are significantly depleted of AUGs, a trend that was confirmed in this study and is even stronger in yeast 5′-UTRs (Table 8). Apparently, purifying selection acts on the uAUGs in two opposite directions: the uAUGs that have no specific function tend to be deleterious and are eliminated during evolution; in contrast, those uAUGs that do serve a function are maintained. More specifically, what could be the regulatory functions of uAUGs? Probably, the leading hypothesis is that the uAUGs attenuate translation by diverting scanning ribosomes from the authentic sAUG. Kozak (2,19) noticed that mRNAs coding for regulatory proteins often contain multiple uAUGs in the 5′-UTRs and proposed that this is an adaptation to prevent excessive and deleterious production of proto-oncogenes, transcription and growth factors, and other proteins that require tight regulation of expression. Double suppression of mRNA translational activity by uAUGs and weak start codon context also might be employed for this purpose (12,13,46). Apparently, selection also operates at the level of the regulatory uORFs by shortening their length, possibly, to optimize the level of translation attenuation. An important role for down-regulation signals affecting translation initiation (uAUGs in 5′-UTR and weak sAUG context) is supported by experimental data: deletion of AUG-containing fragments of 5′-UTR has been shown to greatly increase the mRNA translation rate and protein expression levels (47,48).
Table 8.
Expected and observed numbers of uAUG triplets and shuffled triplets per 1000 nt in mammalian and yeast 5′-UTRs
| Species, triplets | Expected | Observed |
|---|---|---|
| Human uAUGs Human | 12.6 | 7.4 |
| AGU/GUA/GAU/UAG/UGAs | 63.0 | 47.7 |
| Mouse uAUGs | 12.6 | 6.9 |
| Mouse AGU/GUA/GAU/UAG/UGAs | 63.0 | 48.2 |
| Rat uAUGs | 12.7 | 7.6 |
| Rat AGU/GUA/GAU/UAG/UGAs | 63.5 | 49.4 |
| Yeast uAUGs | 17.7 | 10.6 |
| Yeast AGU/GUA/GAU/UAG/UGAs | 88.5 | 73.6 |
The statistical significance of the differences between expected and observed frequencies of uAUG and shuffled triplets were estimated using the χ2 test (2 × 2 tables). In all cases, the difference for uAUGs was significantly greater than that for the combined shuffled triplets (P < 0.001).
An attractive hypothesis adding another level of complexity to the regulation of translation initiation is that, in eukaryotic cells, the AUG-containing portions of 5′-UTRs are similarly removed by alternative splicing, thus converting poorly translatable (in some cases, virtually inactive) mRNAs into active ones. Mironov et al. (49) found that the majority of alternative splicing events in human genes occurred in 5′-UTR regions. This observation suggests that AUG-containing regions are removed by alternative splicing in many human mRNAs and that the structures of cDNAs, which reflect the hypothetical major forms of mRNAs, and the corresponding functional mRNAs (hypothetical minor forms) for certain genes (especially those expressed at low levels) are substantially different. If, indeed, alternative splicing is a common mechanism for the activation of AUG-containing 5′-UTRs, then such 5′-UTRs are expected to be typical of weakly expressed proteins. In accord with this hypothesis, the density of uAUGs is notably lower in yeast genes (Table 8) where alternative splicing does not seem to be a possibility, given the scarcity of introns and partial degradation of the splicing machinery (50,51). Conceivably, yeast genes are subject to stronger purifying selection against uAUGs compared with mammalian genes, owing to the lack of the alternative splicing option in the former. However, the pronounced conservation of uAUGs in yeast genes (Table 7) suggests the existence of a different regulatory mechanism, which is common to a broad variety if not all eukaryotes, possibly, sequestration of ribosomes by the uAUGs.
The conventional scanning mechanism (6) is likely to be completely or at least partially suppressed by uAUGs. Apart from the possibility of alternative splicing, several molecular mechanisms might provide for efficient translation of mRNAs containing uAUGs including leaky scanning, reinitiation (1) or internal initiation of translation (52–55). The relative contributions of these mechanisms remain uncertain (31,56,57) but several recent studies suggest that the impact of at least some of them might be substantial (3,5,17,23,24,32,58–66).
Additional functional signals, such as specific secondary structure elements within the 5′-UTR, might compensate for the negative effects of uAUGs on translation initiation. Such hypothetical signals, which might interact with still unknown regulatory proteins, could be used for regulation of gene expression in response to various stimuli. Under this hypothesis, the uAUGs may be regarded as regulatory elements that maintain the low constitutive level of expression of proteins that need to be sharply up-regulated in response to specific cues. It has been shown that the efficiency of translation of certain mRNAs depends on unknown 5′-UTR elements other than the −3 or +4 positions of the start codon context (30) although there is no clear evidence of a wide distribution of such regulatory signals (2,4,19,31,56). A recent genome-wide analysis of uAUGs and uORFs in a curated set of human and rodent cDNAs has suggested that the ribosome-shunt mechanism, in which the small ribosome subunit binds to the mRNA in a cap-dependent manner but then jumps over a large region of the 5′-UTR containing secondary structures, uORFs and uAUGs to land near the sAUG, might be a widespread mode of translation regulation (17).
Generally, the results of our analysis are compatible with ribosome scanning being the major mechanism of translation initiation in eukaryotes but they also indicate that, in a substantial fraction of eukaryotic mRNAs, uAUGs and uORFs might confer an additional level of translation regulation. The comparative-genomic results presented here suggest that there are thousands of genes in mammalian genomes that might be subject to translation initiation regulation involving uAUGs. This finding is expected to stimulate experimental study of the repertoire and impact of such regulatory mechanisms.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
Acknowledgments
We thank Yuri Wolf, Svetlana Shabalina, Fyodor Kondrashov, King Jordan and Aleksey Kochetov for helpful discussions. This work was supported in part by the Intramural Research Program of the National Library of Medicine at National Institutes of Health/DHHS and NIH grant number P20 RR16469 from the INBRE program of the National Center for Research Resources. Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health, USA.
Conflict of interest statement. None declared
REFERENCES
- 1.Kozak M. The scanning model for translation: an update. J. Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozak M. Interpreting cDNA sequences: some insights from studies on translation. Mamm. Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 3.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pain V.M. Initiation of protein synthesis in eukaryotic cells. Eur. J. Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- 5.Wilkie G.S., Dickson K.S., Gray N.K. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 6.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joshi C.P. An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res. 1987;15:6643–6653. doi: 10.1093/nar/15.16.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavener D.R., Ray S.C. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991;19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pesole G., Fiormarino G., Saccone C. Sequence analysis and compositional properties of untranslated regions of human mRNAs. Gene. 1994;140:219–225. doi: 10.1016/0378-1119(94)90547-9. [DOI] [PubMed] [Google Scholar]
- 10.Joshi C.P., Zhou H., Huang X., Chiang V.L. Context sequences of translation initiation codon in plants. Plant Mol. Biol. 1997;35:993–1001. doi: 10.1023/a:1005816823636. [DOI] [PubMed] [Google Scholar]
- 11.Pesole G., Liuni S., Grillo G., Saccone C. Structural and compositional features of untranslated regions of eukaryotic mRNAs. Gene. 1997;205:95–102. doi: 10.1016/s0378-1119(97)00407-1. [DOI] [PubMed] [Google Scholar]
- 12.Kochetov A.V., Ischenko I.V., Vorobiev D.G., Kel A.E., Babenko V.N., Kisselev L.L., Kolchanov N.A. Eukaryotic mRNAs encoding abundant and scarce proteins are statistically dissimilar in many structural features. FEBS Lett. 1998;440:351–355. doi: 10.1016/s0014-5793(98)01482-3. [DOI] [PubMed] [Google Scholar]
- 13.Rogozin I.B., Kochetov A.V., Kondrashov F.A., Koonin E.V., Milanesi L. Presence of ATG triplets in 5′ untranslated regions of eukaryotic cDNAs correlates with a ‘weak’ context of the start codon. Bioinformatics. 2001;17:890–900. doi: 10.1093/bioinformatics/17.10.890. [DOI] [PubMed] [Google Scholar]
- 14.Kochetov A.V., Syrnik O.A., Rogozin I.B., Glazko G.V., Komarova M.L., Shumnyi V.K. Context organization of mRNA 5′-untranslated regions of higher plants. Mol. Biol. 2002;36:649–656. [PubMed] [Google Scholar]
- 15.Larizza A., Makalowski W., Pesole G., Saccone C. Evolutionary dynamics of mammalian mRNA untranslated regions by comparative analysis of orthologous human, artiodactyl and rodent gene pairs. Comput. Chem. 2002;26:479–490. doi: 10.1016/s0097-8485(02)00009-8. [DOI] [PubMed] [Google Scholar]
- 16.Shabalina S.A., Ogurtsov A.Y., Rogozin I.B., Koonin E.V., Lipman D.J. Comparative analysis of orthologous eukaryotic mRNAs: potential hidden functional signals. Nucleic Acids Res. 2004;32:1774–1782. doi: 10.1093/nar/gkh313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iacono M., Mignone F., Pesole G. uAUG and uORFs in human and rodent 5′untranslated mRNAs. Gene. 2005;349:97–105. doi: 10.1016/j.gene.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr. 1991;1:111–115. [PMC free article] [PubMed] [Google Scholar]
- 19.Kozak M. Determinants of translational fidelity and efficiency in vertebrate mRNAs. Biochimie. 1994;76:815–821. doi: 10.1016/0300-9084(94)90182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geballe A.P., Morris D.R. Initiation codons within 5′-leaders of mRNAs as regulators of translation. Trends Biochem. Sci. 1994;19:159–164. doi: 10.1016/0968-0004(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 21.McCarthy J.E. Posttranscriptional control of gene expression in yeast. Microbiol. Mol. Biol. Rev. 1998;62:1492–1553. doi: 10.1128/mmbr.62.4.1492-1553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willis A.E. Translational control of growth factor and proto-oncogene expression. Int. J. Biochem. Cell. Biol. 1999;31:73–86. doi: 10.1016/s1357-2725(98)00133-2. [DOI] [PubMed] [Google Scholar]
- 23.Pestova T.V., Kolupaeva V.G., Lomakin I.B., Pilipenko E.V., Shatsky I.N., Agol V.I., Hellen C.U. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl Acad. Sci. USA. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider R., Agol V.I., Andino R., Bayard F., Cavener D.R., Chappell S.A., Chen J.J., Darlix J.L., Dasgupta A., Donze O., et al. New ways of initiating translation in eukaryotes. Mol. Cell. Biol. 2001;21:8238–8246. doi: 10.1128/MCB.21.23.8238-8246.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hellen C.U., Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 26.Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–2492. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cigan A.M., Pabich E.K., Donahue T.F. Mutational analysis of the HIS4 translational initiator region in Saccharomyces cerevisiae. Mol. Cell. Biol. 1988;8:2964–2975. doi: 10.1128/mcb.8.7.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yun D.F., Laz T.M., Clements J.M., Sherman F. mRNA sequences influencing translation and the selection of AUG initiator codons in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 1996;19:1225–1239. doi: 10.1111/j.1365-2958.1996.tb02468.x. [DOI] [PubMed] [Google Scholar]
- 29.Hamilton R., Watanabe C.K., de Boer H.A. Compilation and comparison of the sequence context around the AUG startcodons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res. 1987;15:3581–3593. doi: 10.1093/nar/15.8.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Futterer J., Hohn T. Translation in plants—rules and exceptions. Plant Mol. Biol. 1996;32:159–189. doi: 10.1007/BF00039382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozak M. Do the 5′untranslated domains of human cDNAs challenge the rules for initiation of translation (or is it vice versa)? Genomics. 2000;70:396–406. doi: 10.1006/geno.2000.6412. [DOI] [PubMed] [Google Scholar]
- 32.Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001;29:5226–5232. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki Y., Ishihara D., Sasaki M., Nakagawa H., Hata H., Tsunoda T., Watanabe M., Komatsu T., Ota T., Isogai T., et al. Statistical analysis of the 5′ untranslated region of human mRNA using ‘Oligo-Capped’ cDNA libraries. Genomics. 2000;64:286–297. doi: 10.1006/geno.2000.6076. [DOI] [PubMed] [Google Scholar]
- 34.Pesole G., Gissi C., Grillo G., Licciulli F., Liuni S., Saccone C. Analysis of oligonucleotide AUG start codon context in eukariotic mRNAs. Gene. 2000;261:85–91. doi: 10.1016/s0378-1119(00)00471-6. [DOI] [PubMed] [Google Scholar]
- 35.Davuluri R.V., Suzuki Y., Sugano S., Zhang M.Q. CART classification of human 5′-UTR sequences. Genome Res. 2000;10:1807–1816. doi: 10.1101/gr.gr-1460r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casadei R., Strippoli P., D'Addabbo P., Canaider S., Lenzi L., Vitale L., Giannone S., Frabetti F., Facchin F., Carinci P., et al. mRNA 5′ region sequence incompleteness: a potential source of systematic errors in translation initiation codon assignment in human mRNAs. Gene. 2003;321:185–193. doi: 10.1016/s0378-1119(03)00835-7. [DOI] [PubMed] [Google Scholar]
- 37.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Rogozin I.B., Mayorov V.I., Lavrentieva M.V., Milanesi L., Adkison L.R. Prediction and phylogenetic analysis of mammalian short interspersed elements (SINEs) Brief. Bioinform. 2000;1:260–274. doi: 10.1093/bib/1.3.260. [DOI] [PubMed] [Google Scholar]
- 39.Koonin E.V. Orthologs, paralogs and evolutionary genomics. Annu. Rev. Genet. 2005 doi: 10.1146/annurev.genet.39.073003.114725. in press. [DOI] [PubMed] [Google Scholar]
- 40.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogozin I.B., D'Angelo D., Milanesi L. Protein-coding regions prediction combining similarity searches and conservative evolutionary properties of protein-coding sequences. Gene. 1999;226:129–137. doi: 10.1016/s0378-1119(98)00509-5. [DOI] [PubMed] [Google Scholar]
- 42.Rivas E., Eddy S.R. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001;2:8. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 44.Kochetov A.V. AUG codons at the beginning of protein coding sequences are frequent in eukaryotic mRNAs with a suboptimal start codon context. Bioinformatics. 2005;21:837–840. doi: 10.1093/bioinformatics/bti136. [DOI] [PubMed] [Google Scholar]
- 45.Kochetov A.V., Sarai A., Rogozin I.B., Shumny V.K., Kolchanov N.A. The role of alternative translation start sites in the generation of human protein diversity. Mol. Genet. Genomics. 2005;273:491–496. doi: 10.1007/s00438-005-1152-7. [DOI] [PubMed] [Google Scholar]
- 46.Kochetov A.V., Ponomarenko M.P., Frolov A.S., Kisselev L.L., Kolchanov N.A. Prediction of eukaryotic mRNA translational properties. Bioinformatics. 1999;15:704–712. doi: 10.1093/bioinformatics/15.7.704. [DOI] [PubMed] [Google Scholar]
- 47.Marth J.D., Overell R.W., Meier K.E., Krebs E.G., Perlmutter R.M. Translational activation of the lck proto-oncogene. Nature. 1988;332:171–173. doi: 10.1038/332171a0. [DOI] [PubMed] [Google Scholar]
- 48.van der Velden A.W., Thomas A.A. The role of the 5′ untranslated region of an mRNA in translation regulation during development. Int. J. Biochem. Cell. Biol. 1999;31:87–106. doi: 10.1016/s1357-2725(98)00134-4. [DOI] [PubMed] [Google Scholar]
- 49.Mironov A.A., Fickett J.W., Gelfand M.S. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goffeau A., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., et al. Life with 6000 genes. Science. 1996;274:546. doi: 10.1126/science.274.5287.546. 563–547. [DOI] [PubMed] [Google Scholar]
- 51.Aravind L., Watanabe H., Lipman D.J., Koonin E.V. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl Acad. Sci. USA. 2000;97:11319–11324. doi: 10.1073/pnas.200346997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macejak D.G., Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 53.Ye X., Fong P., Iizuka N., Choate D., Cavener D.R. Ultrabithorax and Antennapedia 5′ untranslated regions promote developmentally regulated internal translation initiation. Mol. Cell. Biol. 1997;17:1714–1721. doi: 10.1128/mcb.17.3.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le S.Y., Maizel J.V., Jr A common RNA structural motif involved in the internal initiation of translation of cellular mRNAs. Nucleic Acids Res. 1997;25:362–369. doi: 10.1093/nar/25.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sachs A.B. Cell cycle-dependent translation initiation: IRES elements prevail. Cell. 2000;101:243–245. doi: 10.1016/s0092-8674(00)80834-x. [DOI] [PubMed] [Google Scholar]
- 56.Kozak M. New ways of initiating translation in eukaryotes? Mol. Cell. Biol. 2001;21:1899–1907. doi: 10.1128/MCB.21.6.1899-1907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kozak M. Alternative ways to think about mRNA sequences and proteins that appear to promote internal initiation of translation. Gene. 2003;318:1–23. doi: 10.1016/s0378-1119(03)00774-1. [DOI] [PubMed] [Google Scholar]
- 58.Morris D.R., Geballe A.P. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meijer H.A., Thomas A.A. Control of eukaryotic protein synthesis by upstream open reading frames in the 5′-untranslated region of an mRNA. Biochem. J. 2002;367:1–11. doi: 10.1042/BJ20011706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shin D., Park S., Park C. A splice variant acquiring an extra transcript leader region decreases the translation of glutamine synthetase gene. Biochem. J. 2003;374:175–184. doi: 10.1042/BJ20030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X.Q., Rothnagel J.A. 5′-untranslated regions with multiple upstream AUG codons can support low-level translation via leaky scanning and reinitiation. Nucleic Acids Res. 2004;32:1382–1391. doi: 10.1093/nar/gkh305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dorokhov Y.L., Skulachev M.V., Ivanov P.A., Zvereva S.D., Tjulkina L.G., Merits A., Gleba Y.Y., Hohn T., Atabekov J.G. Polypurine (A)-rich sequences promote cross-kingdom conservation of internal ribosome entry. Proc. Natl Acad. Sci. USA. 2002;99:5301–5306. doi: 10.1073/pnas.082107599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hernandez-Sanchez C., Mansilla A., de la Rosa E.J., Pollerberg G.E., Martinez-Salas E., de Pablo F. Upstream AUGs in embryonic proinsulin mRNA control its low translation level. EMBO J. 2003;22:5582–5592. doi: 10.1093/emboj/cdg515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chappell S.A., Edelman G.M., Mauro V.P. Biochemical and functional analysis of a 9-nt RNA sequence that affects translation efficiency in eukaryotic cells. Proc. Natl Acad. Sci. USA. 2004;101:9590–9594. doi: 10.1073/pnas.0308759101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gebauer F., Hentze M.W. Molecular mechanisms of translational control. Nature Rev. Mol. Cell. Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stoneley M., Willis A.E. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]