Abstract
HutP is an RNA-binding protein that regulates the expression of the histidine utilization (hut) operon in Bacillus subtilis, by binding to cis-acting regulatory sequences on hut mRNA. It requires L-histidine and an Mg2+ ion for binding to the specific sequence within the hut mRNA. In the present study, we show that several divalent cations can mediate the HutP–RNA interactions. The best divalent cations were Mn2+, Zn2+ and Cd2+, followed by Mg2+, Co2+ and Ni2+, while Cu2+, Yb2+ and Hg2+ were ineffective. In the HutP–RNA interactions, divalent cations cannot be replaced by monovalent cations, suggesting that a divalent metal ion is required for mediating the protein–RNA interactions. To clarify their importance, we have crystallized HutP in the presence of three different metal ions (Mg2+, Mn2+ and Ba2+), which revealed the importance of the metal ion binding site. Furthermore, these analyses clearly demonstrated how the metal ions cause the structural rearrangements that are required for the hut mRNA recognition.
INTRODUCTION
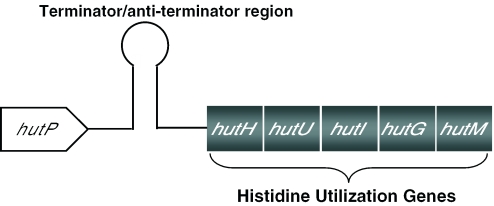
Genes responsible for the degradation and utilization of L-histidine, as a carbon and nitrogen source under nutrient-limiting conditions, are located within the hut operon in Bacillus subtilis. The operon consists of six open reading frames; the hutP gene is located near the promoter, and five other genes, hutH, hutU, hutI, hutG and hutM, are located far downstream from the promoter. Previous biochemical and genetic studies suggested that the hut mRNA forms a terminator structure (+459 to +571 nt, Figure 1) in between the hutP and hutH genes, and regulates the transcription of the hut operon by an anti-termination mechanism (1–3). Several bacterial proteins that regulate anti-terminator/terminator structures have been described, including TRAP, PyrR, LacT, BglG, SacT and GlpP (4–10). One such protein is the hutP gene product, HutP, of B.subtilis. HutP is a 16.2 kDa protein that shares ∼60% sequence identity with the HutP proteins found in other Bacillus species, such as Bacillus halodurans (11), Bacillus cereus (12) and Bacillus anthracis (13).
Figure 1.
Histidine utilizing (hut) operon of B.subtilis. Schematic representation of the hut operon, showing the arrangement of the hutP, terminator/anti-terminator (+459 to +571 nt), and structural genes.
HutP is a positive regulatory protein that interacts specifically with the terminator region (3) of hut mRNA. Similar to the other transcriptional regulatory proteins mentioned above, it requires a ligand for its activation; HutP requires L-histidine as a ligand for binding to the terminator RNA (3). For complete activation of HutP, a concentration of ∼10 mM L-histidine was sufficient for binding to the cognate RNA (14). Using various analogs of L-histidine, we determined that the imidazole ring and the backbone of L-histidine are important for supporting RNA binding in vitro (14,15). Obviously, it is intriguing how the ligand (L-histidine) modulates HutP for binding to the terminator. In order to obtain insights into these interactions, we recently solved the crystal structure of the HutP–HBN (L-histidine analog, L-histidine β-naphthylamide) complex (14). The complex structure revealed that HutP forms a hexamer, which consists of three dimers of HutP. The L-histidine was located in between the two monomers of HutP and participates in an extensive hydrogen bond network within the hydrophobic pocket (14). Furthermore, we mapped the HutP binding sites within the terminator region, as the UAGNNNNUAGNNNNUAG recognition motif, where N indicates any base, and also identified the important chemical groups of the bases within the core region (UAG) (16).
To analyze the ability of HutP to bind to mRNA, the reactions are carried out in the presence of L-histidine (10 mM) and Mg2+ ions (5 mM). However, we do not know what function the metal ions play in anti-termination complex formation. In order to understand the role of metal ions in the formation of this complex, we used a gel mobility shift assay, and these studies suggested that Mg2+ ions are important for the HutP–RNA interactions (17). Furthermore, our quaternary complex analyses revealed that the Mg2+ ions were simultaneously coordinated within the histidine cluster of HutP and the L-histidine ligand. Upon these interactions, the Mg2+ ion was found to facilitate appropriate structural rearrangements, especially at the L-histidine binding site and the loop regions of HutP, thus leading to the recognition of the cognate RNA sequence (17). All of the structural rearrangements caused by the divalent metal ions are essential for the RNA recognition. The L-histidine movement only occurred in the presence of the metal ion. The side chain of Arg88 moved in the opposite direction of the L-histidine and formed a salt bridge, leading to the disruption of the hydrophobic pocket formed by the salt bridge between Arg88 and Glu81 in the HutP–HBN complex, and formed a new salt bridge with Arg98. In order to establish this new salt bridge interaction with the L-histidine ligand, the conformation of the residue Arg98 changes drastically, and this is accompanied by a large shift in the Cα position of the next residue, Thr99. Owing to these structural rearrangements caused by the divalent metal ion, the Thr99 side chain forms two hydrogen bonds with the N3 and 2′-OH of the A base in the UAG motif. The involvement of the metal ions in the histidine cluster causes the His138 residue to reorient its imidazole ring to coordinate with the divalent metal ion. This critical contribution for the metal ion coordination causes a large conformational change in the local backbone chain, particularly in the loop regions L3 and L5 (17). Therefore, we suggest that the divalent metal ions play a major role in activating the HutP protein through the structural rearrangements specifically required for the RNA recognition. Divalent and monovalent cations play several important roles in mediating the catalysis and folding of bio-molecules (18–21). However, in RNA–protein interactions, the affinity of a protein for its RNA ligand generally decreases with increasing ionic strength, reflecting the contribution of ionic contacts to the RNA–protein interaction.
The effects of several cations on RNA–protein complexes, including the R17 coat protein and its translational operator complex (22), the Escherichia coli ribosomal protein S4–16S ribosomal RNA complex (23), the signal recognition particle (SRP)–Ffh complex (24), the C5 protein–M1 RNA complex (25) and the E.coli L11–23S rRNA ribosomal protein complex (26), have been evaluated. Interestingly, these RNA–protein complexes involve longer RNAs (S4–16S, SPR–Ffh, C5 protein–M1 RNA and L11–23S rRNA complexes) within the complex, and they specifically require divalent metal ions for the interactions. The metal ions in these complexes are primarily needed for folding/stabilizing alternative RNA conformations for recognition by the cognate protein, rather than directly mediating the interactions. On the other hand, RNA-binding proteins that bind to single-stranded RNA, such as Sex-lethal (27), Poly(A) binding protein (28), transcription termination factor Rho (29) and TRAP (30), recognize and bind their RNAs even in the absence of metal ions. Thus, based on these examples, it appears that the divalent metal ions are primarily required in the RNA–protein complexes when longer RNAs (>55 nt) are involved, to obtain a specific secondary structure for complex formation [e.g. the L11–RNA complex (31)]. Alternatively, it is possible in some cases that the metal ions may be required to mediate specific interactions between the RNA and the protein, without interacting with the RNA.
In the present study, we show that a single-stranded RNA-binding protein, HutP, is an L-histidine and divalent metal ion-dependent anti-termination protein that binds to the RNA sequences within the terminator region of hut mRNA. Among the 15 divalent metal ions tested, only Cu2+, Yb2+ and Hg2+ failed to support the HutP–RNA interactions, while Mn2+, Zn2+ and Cd2+ were the best metal ions. Monovalent metal ions, such as, Na+ and K+, could not substitute for the divalent metal ions in the HutP–RNA interaction. These analyses suggest that a metal ion binding pocket exists within the HutP protein that specifically coordinates with its counterparts. To determine whether specific divalent metal ion interactions occur with HutP, we solved the crystal structures of HutP in the presence of three metal ions, Mn2+, Ba2+ and Mg2+. Interestingly, all three divalent ions reside in a precise position and interact specifically with the HutP residues His73, His77 and His138, and the L-histidine ligand. A comparison of the complex structures of HutP in the presence and absence of divalent metal ions suggests that the metal ions cause structural rearrangements, especially at the loop regions, by interacting with the histidine cluster and L-histidine ligand. These structural rearrangements of the HutP protein might be required for recognizing the specific sequences in the terminator region of the hut mRNA. Thus, the divalent metal ion plays a major role in the regulation of HutP–RNA interactions.
MATERIALS AND METHODS
Expression and purification of the HutP protein
The nucleotide sequence encoding HutP, containing the Val 51 Ile mutation, was amplified and cloned into the pET5a vector (Promega). The resultant plasmid was transformed into E.coli strain BL21 (DE3). The HutP protein was overexpressed at mid-log phase by the addition of isopropyl-β-d-thiogalactopyranoside (1 mM) and was purified as described previously (32).
RNA synthesis and 5′ end labeling
A minimal RNA element, 5′-CAUAGAUCUUAGACGAUAGGG-3′, which binds to HutP, was chemically synthesized on an RNA/DNA synthesizer (Model 394; Applied Biosystems Inc.) using phosphoramidite chemistry. All amidites were purchased from Glen Corporation, USA. The chemical groups of the RNA were deprotected by established protocols (ABI Manual), and the resulting RNA was purified on a 20% acrylamide gel containing 7 M urea. The RNA was located, eluted from the gel pieces and recovered by ethanol precipitation, as described previously (14). The RNAs were labeled at the 5′ end with [γ-32P]ATP (Amersham Biosciences) in the presence of T4 polynucleotide kinase (Takara, Japan), and the labeled products were recovered after fractionation by 15% denaturing PAGE.
Gel-shift assay
To evaluate the role of metal ions in the protein–RNA interactions, we carried out a gel-shift assay. The 21mer RNA was labeled as described above. The calculated concentrations of HutP were based on the hexamer (96 000 Da). The purified HutP was extensively dialyzed in binding buffer (15 mM HEPES, pH 7.5, 30 mM NaCl and 10 µg of yeast tRNA) to remove the MgCl2. The HutP protein (1200 nM concentration) was mixed with the RNA (21mer) probe with different concentrations of metal ions, in the presence of 10 mM L-histidine. The resulting reaction mixture was incubated for 15 min at room temperature, and then was mixed with 2 µl of 90% glycerol before loading onto the gel. The free RNA and HutP–RNA complexes were resolved using a 10% polyacrylamide gel (running buffer, 0.5× TBE; constant voltage, 15 mA; temperature, 4°C). The resulting autoradiograms were analyzed with a bio-image analyzer, BAS 2500 (Fuji Film). The equilibrium dissociation (Kd) constants were determined for the metal ions mediating protein–RNA complex formation, which were fit to the following equation: Y = Bmax X/Kd + X
A nonlinear regression analysis was carried out using the GraphPad Prism software (GraphPad Software).
Crystallization and data collection
The HutP–L-histidine–Mn2+, HutP–L-histidine–Ba2+ and HutP–Mg2+ (crystallized in the absence of L-histidine) complex crystals were grown at 20°C by the hanging drop vapor diffusion method. Drops consisted of 2 µl protein, 1 µl 0.1 M L-histidine, 1 µl RNA and 2 µl well solution (100 mM HEPES, pH 7.4, 0.1 M MgCl2 or MnCl2 or BaCl2, 40% MPD). In order to crystallize the HutP in the presence of only Mg2+, we omitted the L-histidine from the above experiments. Diffraction data were collected using a CCD (ADSC) detector at the Photon Factory, Tsukuba, Japan. Datasets for the HutP–L-histidine–Mn2+ and HutP–L-histidine–Ba2+ and HutP–Mg2+ crystals structures were processed up to 2.20, 2.35 and 2.60 Å, respectively, with the HKL2000 suite of programs (33). The HutP–L-histidine–Mn2+, HutP–L-histidine–Ba2+ crystals belonged to the P21212, space group, and the HutP–Mg2+ crystals belonged to the P213 space group (Table 2).
Table 2.
Data collection and refinement statistics of the HutP–L-histidine–Mn2+, HutP–L-histidine–Ba2+ and HutP–Mg2+ complex structures
| HutP–L-histidine–Mn2+ complex | HutP–L-histidine–Ba2+ complex | HutP–Mg2+ | |
|---|---|---|---|
| Data collection | |||
| Space group | P21212 | P21212 | P213 |
| Unit cell | a = 77.76, b = 81.40, c = 76.04 | a = 78.22, b = 81.14, c = 75.97 | a = b = c = 95.41 |
| Solvent content (%) | 50.6 | 50.6 | 45.2 |
| Number of molecules in ASU | 3 | 3 | 2 |
| Resolution range | 50.0–2.20 Å | 50.0–2.35 Å | 50.0–2.60 Å |
| (Outer shell) | (2.28–2.20 Å) | (2.43–2.35 Å) | (2.69–2.60 Å) |
| No. of independent reflections | 24 576 | 20 744 | 8514 |
| Redundancy | 7.3 (7.5) | 7.1 (6.8) | 12.2 (12.8) |
| Completeness (%) | 98.0 (97.9) | 99.8 (100.0) | 93.1 (100.0) |
| Rmerge | 0.036 (0.258) | 0.061 (0.384) | 0.041 (0.350) |
| Refinement and model correlation | |||
| Resolution | 20.0–2.30 Å | 20.0–2.35 Å | 20.0–2.70 Å |
| No. of reflections used for refinement | 21 584 | 20 669 | 7492 |
| Rfactor | 0.254 | 0.247 | 0.228 |
| No. of reflections used for Rfree | 1079 | 1048 | 542 |
| Rfree | 0.305 | 0.300 | 0.286 |
| No. of protein atoms | 3211 | 3330 | 2144 |
| No. ligand atoms | 72 | 33 | – |
| No. of metal ions | 3 | 6 | – |
| No. water molecules | 367 | 240 | 67 |
| Average B-factor (Å2) | 56.6 | 38.9 | 63.0 |
| RMS deviation from ideal geometry | |||
| Bond lengths (Å) | 0.007 | 0.007 | 0.007 |
| Bond angles (degree) | 1.50 | 1.30 | 1.30 |
| Ramachandran statistics | |||
| Residues in most favored regions (%) | 89.3 | 88.9 | 85.0 |
| Residues in additional allowed regions (%) | 10.7 | 11.1 | 15.0 |
| PDB code | 1WRN | 1WRO | 1WPT |
Structure determination
The HutP–L-histidine–Mn2+, HutP–L-histidine–Ba2+ and HutP–Mg2+ (crystallized in the absence of L-histidine) structures were determined by molecular replacement, using the HutP–HBN complex as a search model (PDB ID 1VEA). Both molecular replacement and model refinement were carried out using CNS (34). The protein model was built using the program Quanta (35). The structures of the HutP–L-histidine–Mn2+, HutP–L-histidine–Ba2+ and HutP–Mg2+ complexes were refined to 2.30, 2.35 and 2.70 Å, respectively, and the overall refinements and model statistics are given in Table 2. The HutP–Mg2+ crystals contain two molecules in the asymmetric unit, whereas the HutP–L-histidine–Mn2+ and HutP–L-histidine–Ba2+ crystals contain three molecules. The final HutP–L-histidine–Mn2+ complex model contains three monomers (A, B and C) of HutP (except for residue 22 of molecule A, residue 22 of molecule B and residues 21–24 of molecule C), three Mn2+ ions, three L-histidines, 6 PEGs and 367 water molecules in the asymmetric unit. Residues 2–4 of molecules A and B in the final model of HutP–Mg2+ and the N-terminal residues of all of the molecules in these analyses were not modeled, because of the absence of electron density. Also, A5Lys, A23Glu, A24Ser, A26Gln, A28Glu, A29Glu, A32Arg, A67Glu, A113Glu, A134Lys, B4His, B5Lys, B20Glu, B23Glu, B24Ser, B26Gln, B28Glu, B29Glu, B32Arg, B36Lys, B41Lys, B67Glu, B71Glu, B113Glu, B115Glu, B134Lys, C5Lys, C26Gln, C28Glu, C29Glu, C60Lys, C115Glu, and C134Lys of the HutP–L-histidine–Mn2+ complex, 5Lys, 22Glu, 23Glu, 24Ser, 60Lys and 134Lys of all three molecules, A, B and C, of the HutP–L-histidine–Ba2+ complex, and A5Lys, A60Lys, A113Glu, A114Ser, A115Glu, A133Ile, A134Lys, B5Lys, B49Lys, B68Tyr, B88Arg, B96Leu, B97Leu, B113Glu, B114Ser, B115Glu, B117Glu and B134Lys of the HutP–Mg2+ complex were modeled with Ala, because the side chain densities were lacking for these residues. Figures were prepared using the Ribbons (36) and XtalView (37) programs.
RESULTS AND DISCUSSION
Our previous analyses suggested that the HutP protein binds to its cognate RNA only upon activation by L-histidine (14). Recently, we showed that the HutP protein also requires Mg2+ ions for RNA recognition (17). Taken together, HutP requires divalent cations and L-histidine to recognize its cognate RNA sequences in the hut mRNA. To assess the importance of metal ions in the HutP–RNA interactions, in the present study, we analyzed their interactions in the presence of various divalent and monovalent metal ions and their ability to cause the structural rearrangements required for the HutP–RNA interactions.
Requirement of Mg2+ ions for HutP–RNA interactions
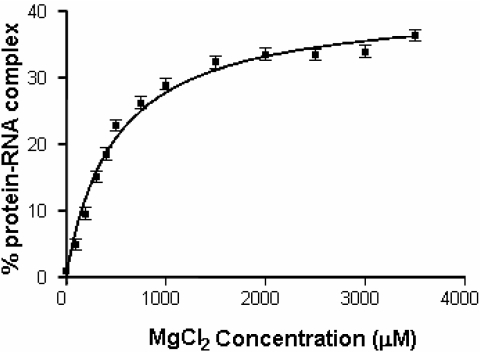
Although Mg2+ ions are essential for the HutP–RNA interactions, to recognize the specific sequence within the terminator region, their affinity for the ternary complex is unknown. Therefore, we carried out binding reactions with various Mg2+ concentrations (0–3.5 mM) in the presence of the HutP–L-histidine–RNA. The resulting complexes were separated, and the amount of complex formed was quantitated and fitted into a saturation binding equation, using Graphpad Prism. Two independent analyses suggested that MgCl2 has a 489 ± 51 µM Kd for binding the HutP–RNA complex (Figure 2). The Kd value for the HutP–RNA interactions appeared to be more efficient (>10-fold) as compared with those of the metal ions involved in other protein–RNA interactions, suggesting the existence of an efficient metal ion binding pocket. Therefore, HutP represents the first example of a single-stranded RNA-binding protein that requires metal ions for mediating the RNA–protein interactions.
Figure 2.
Determination of the equilibrium dissociation constant of Mg2+ ions for mediating the protein–RNA complex, using a gel mobility shift assay. The equilibrium dissociation (Kd) constants were determined for the metal ions mediating protein–RNA complex formation, using a non-linear curve-fitting algorithm (GraphPad Prism 2.0, GraphPad Software).
Although the importance of Mg2+ in the stabilization of RNA conformations is known, most of the RNA–protein interactions studied thus far are only influenced weakly by Mg2+. Aminoacyl-tRNA synthetase binding to tRNA and ribosomal protein S8 binding to rRNA are enhanced only several-fold by Mg2+ (38–41). The R17 coat protein recognition of hairpin RNA is unaffected by Mg2+ (22). However, the formation of the L11–RNA protein complex strongly required Mg2+ (23). In this complex, Mg2+ binds to RNA with higher affinity (Kd ∼ 3 mM). Recently, the importance of metal ions in this complex was revealed by X-ray analyses, which showed that the divalent cation occupies a crucial location at the center of a four-way junction, where the ion appears to stabilize the sharp turns (1056–1057 and 1086–1087) of the junction. Thus, the metal ion seems to play a crucial role in determining the overall structure of the four-way junction (31). Interestingly, in the case of HutP, the divalent metal ions interact with the histidine cluster and the L-histidine ligand, but not with the RNA, in contrast to other protein–RNA complexes.
Analysis of HutP–RNA interactions in the presence of various divalent cations
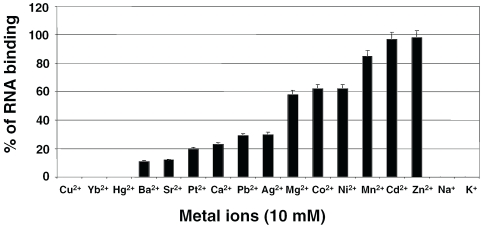
From the aforementioned studies, it is clear that divalent metal ions, Mg2+ ions, are essential for mediating the HutP–RNA interactions. To substantiate the requirement for divalent metal ions and also to identify the best divalent metal ions that support the interactions, we performed binding reactions in the presence of various divalent metal ions. The properties of the divalent metal ions used in the present study including their ionic radii, preferred coordination and concentrations present within the bacterial cells, are summarized in Table 1. Of the 15 different divalent metal ions tested, 12 were able to mediate the HutP–RNA interactions. The only metal ions that failed to support the interactions were Cu2+, Yb2+ and Hg2+ (Figure 3). Among the 12 divalent ions that participated in the interactions, Mn2+, Zn2+ and Cd2+ were more efficient, followed by Mg2+, Co2+ and Ni2+ ions (Figure 3). Interestingly, the divalent metal ions that are less abundant in the bacterial cell, such as Mn2+, Zn2+ and Cd2+, were the active divalent metal ions, whereas the more commonly found divalent metal ions, Mg2+, Ag2+ and Ca2+, were weakly efficient.
Table 1.
Selected monovalent and divalent metal ions and its properties (18)
| Metal ion (compound used) | Ionic radii | Preferred coordination | Concentration in bacterial cells (mg/kg) |
|---|---|---|---|
| Divalent | |||
| Mg (MgCl2) | 0.72 | 6 | 7 × 103 |
| Ca (CaCl2) | 0.99, 1.12 | 6, 8 | 5.1 × 103 |
| Mn (MnCl2) | 0.83 | 6 | 260 |
| Cu (CuCl2) | 0.57, 0.73 | 4, 6 | 150 |
| Zn (ZnCl2) | 1.02, 1.08 | 4, 6 | 83 |
| Co (CoCl2) | 0.74 | 6 | 7.9 |
| Cd (CdCl2) | 0.95 | 4–7 | 0.31 |
| Ba (BaCl2) | 1.35, 1.38 | 6, 7 | – |
| Sr (SrCl2) | 1.13 | 6 | – |
| Yb (YbCl2) | 1.02–01.14 | 6–8 | – |
| Ni (NiCl2) | 0.69 | 6 | – |
| Pb (PbCl2) | 1.19–11.49 | 4–12 | – |
| Ag (AgNO3) | 0.94 | 6 | – |
| Hg(Hg(CN)2) | 0.69–61.14 | 2,4,6,8 | – |
| Pt (K2PtCl4) | 0.80 | 6 | – |
| Monovalent | |||
| Na (NaCl) | 0.99–91.39 | 4–12 | 4.6 × 103 |
| K (KCl) | 1.37–31.64 | 4–12 | 115 × 103 |
Figure 3.
Analysis of the abilities of various metal ions to mediate the HutP–RNA interactions. Fifteen divalent and two monovalent cations were analyzed by a gel-shift assay, as described in Figure 2. The amounts of complexed and free RNA were used to calculate the percentage of metal ion interactions involved in making the ternary complex. The percentages from three independent experiments were averaged (standard errors, ±5), are plotted against the metal ions.
Interestingly, Ni2+ ions, which are reportedly not present within bacterial cells, also mediate the complex formation better than Mg2+, and other metals that are not found in bacteria, such as Ba2+, Pb2+, Pt2+ and Sr2+, also participated in the complex. We compared the atomic radii of the metal ions that support the HutP–RNA interactions, in order to understand the pocket that accommodates the metal ions. When we compared the ionic radii of the metal ions, based on their preferred hexameric coordination, we found that the radii for Cu2+ (0.73), Mg2+ (0.72) and Co2+ (0.74) were nearly the same size. However, Cu2+ failed to interact with the complex. In a similar way, although the radii between the Yb2+ and Zn2+ ions were essentially the same size, Yb2+ was non-interactive, whereas Zn2+ showed the highest interactions with the protein–RNA, complex. The Ba2+ ion, which has the longest ionic radius, showed the lowest support for the HutP–RNA interactions. This could be due to the increase of coordination distances between Ba2+ and N atoms of the histidine residues (∼0.3 Å larger) than those between Mg2+ (and also Mn2+) and N atoms of the histidine residues, which leads widening of the dimer interface. Although these studies suggested that the atomic radii of the divalent metal ions are important for their support in mediating the HutP–RNA interactions, it is possible that the mediating mechanism might be much more complicated.
We next analyzed whether the monovalent cations could substitute for divalent cations in the HutP–RNA interactions. For this, binding reactions were carried out in the binding buffer in the presence of 10 and 100 mM of NaCl or KCl and analyzed for the ternary complex formation. Both of the monovalent cations failed to mediate the interactions at the above concentrations and in the absence of divalent cations. These results suggested that divalent cations are required for the interactions.
Crystal structures of HutP–L-histidine–Mn2+, HutP–L-histidine–Ba2+ and HutP–Mg2+
The previous analyses revealed that divalent metal ions are important for the HutP–RNA interactions. In order to evaluate the metal ion coordinations within the HutP and L-histidine ligand complex, we chose two other metal ions (Ba2+ and Mn2+), in addition to Mg2+, which showed lower and higher levels of binding influence on HutP, representatively. All of the crystallization trials were carried out with HutP, L-histidine, and BaCl2, MnCl2 or MgCl2 ions. Well-ordered crystals were appeared within a week and reached to a size of 0.2 × 0.1 × 0.2 mm. Two of the crystals (HutP–L-histidine–Mn2+ and HutP–L-histidine–Ba2+) belong to the space group P21212 and contain three complexes per asymmetric unit, whereas the HutP–Mg2+ (crystallized in the absence of L-histidine) crystal belongs to the P213 space group, with two molecules in the asymmetric unit. All of the structures, HutP–L-histidine–Mn2+, HutP–L-histidine–Ba2+ and HutP–Mg2+, were solved by molecular replacement with HutP–HBN (PDB 1VEA) as a search model and were refined to 2.30, 2.35 and 2.70 Å resolutions with R factors of 25.4% (Rfree = 30.5%), 24.7% (Rfree = 30%) and 22.8 (Rfree = 28.6%), respectively. The refinement statistics are given in Table 2, together with further details of the X-ray analysis.
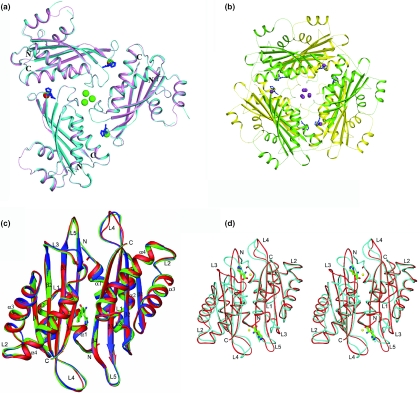
The overall structures of the HutP–L-histidine–Mn2+ and HutP–L-histidine–Ba2+ complexes are superimposed in Figure 4a. Each asymmetric unit contains three molecules of HutP and three L-histidines related by non-crystallographic 3-fold axis, forming a tight trimer, with each monomeric HutP molecule consisting of four α-helices and four β-strands, arranged in the order α–α–β–α–α–β–β–β in the primary structure, and the four antiparallel β-strands form a β-sheet in the order β1–β2–β3–β4, with two α-helices each on the front and the back (Figure 4a). Consistent with previous X-ray and biochemical analyses, these complexes form a hexameric structure along the 2-fold axis (Figure 4b). The root-mean-square deviation (RMSD) of superposition between these two complexes was 0.32 Å. However, these complex structures correlated well with our recently solved structures of the HutP–L-histidine–Mg2+ complex (RMSD, 0.25 Å) and the HutP–L-histidine–Mg2+–21mer complex (RMSD, 0.35 Å) (17). The overall structure of the HutP–Mg2+ complex (crystallized in the absence of L-histidine) was superimposed on our recently reported structures, HutP–HBN (RMSD, 0.18 Å) and uncomplexed HutP (RMSD, 0.31 Å), as shown in Figure 4c, and from this figure it is clear that the HutP–Mg2+ structure correlates well with those two structures (14,17).
Figure 4.
Crystal structures of the HutP-divalent metal ion complexes. (A) Structure of the HutP–L-histidine–Mn2+ (cyan) and HutP–L-histidine–Ba2+ (pink) complexes superimposed and shown in a ribbon diagram, with labels for the N and C termini only. The bound L-histidines are shown as a ball-and-stick model colored in blue and the divalent metal ions are represented by cpk models (Mn2+ and Ba2+ are red and green, respectively). (B) Hexameric formation of the divalent metal complexes. For clarity, we only show the HutP–L-histidine–Ba2+ complex. One trimer is shown in yellow, and the other is green. The bound L-histidines are shown as a ball-and-stick model colored in blue and the Ba2+ ions are represented by the cpk model (Ba2+ ions are colored magenta). (C) The crystal structure of the HutP–Mg2+ complex (crystallized in the absence of L-histidine, green) superimposed along with the uncomplexed HutP (red) and the HutP–HBN complex (blue) shown in a ribbon diagram with labels for α-helices, β-strands and coil regions. The bound HBN is shown as a ball-and-stick model colored by atom type (nitrogen, blue; carbon, green; oxygen, red). (D) Superimposition of two crystal structures, showing conformational changes at loop regions. In this ribbon model, the uncomplex HutP and ternary complex (HutP–L-his–Mg2+) are shown in red and blue, respectively. The L-histidines are represented by ball-and-stick models in green and the Mg2+ ions are represented by cpk models, colored in yellow.
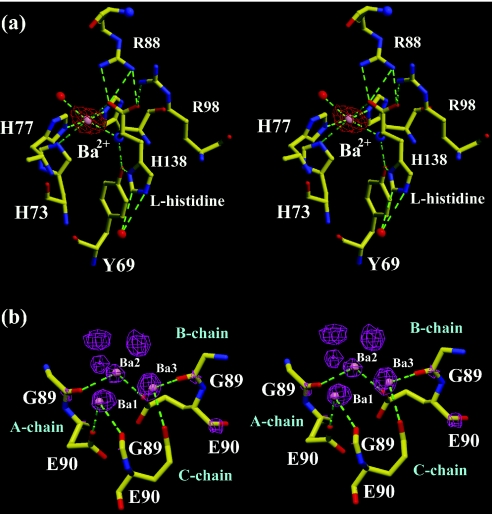
The 2|Fobs| − |Fcalc| maps of HutP–L-histidine–Mn2+ and HutP–L-histidine–Ba2+ showed clear electron densities corresponding to divalent metals in their binding sites (Figure 5a and b). These sites were further confirmed using anomalous difference Fourier maps (Supplementary Figures 1 and 2). These specific binding sites, identified, were consistent in the present HutP–L-histidine–Mn2+ and HutP–L-histidine–Ba2+, and our recently reported HutP–L-histidine–Mg2+, and HutP–L-histidine–Mg2+–21mer RNA complexes. The bound metal ions (Mn2+, Ba2+ and Mg2+) were located at the dimer or dimer–dimer interface with a similar recognition motif, critically forming the hexa-coordination with the L-histidine ligand and the histidine cluster of the HutP protein (Figure 5a). Of the six coordinations, two were with the amino and carboxyl groups of the L-histidine ligand, and the three others were with the imidazole nitrogens of His138, His73 and His77. The sixth coordination of the Mg2+ ion was that with a water molecule, which was anchored by a hydrogen bond to the side chain of Glu81. This typical hexa-coordination with the L-histidine and histidine cluster may not be possible with the monovalent metal ions (K+ and Na+), and hence these metal ions could not mediate the protein–RNA interactions (Figure 3). In the case of the HutP–L-histidine–Ba2+ complex, we found three additional non-specific Ba2+ ions located at the center of the HutP trimer (Figure 4a and b). However, these three Ba2+ are related by non-crystallographic 3-fold axis and may be occurred as alternative positions of a disordered Ba2+ ion. A view of these Ba2+ positions and their interactions (Figure 5b) shows that each Ba2+ bonded with two coordinations derived from the two molecules, i.e. the O atom of Gly89 of one molecule and the other from the OE2 of Glu90 from the adjacent molecule. However, this Ba2+-binding site is not required for the RNA binding or structural rearrangements, because without this metal ion interactions, Mn2+ and Mg2+ can mediate the interactions as well.
Figure 5.
Divalent metal ion coordinations in the complex structures. (A) A close up stereo view of the Ba2+ ion binding site in the HutP–L-histidine–Ba2+ complex. Hydrogen bonds are indicated by broken lines. The L-histidine ligand and the protein residues are represented by ball-and-stick models colored by atom type, as shown in Figure 4c. The Ba2+ and water molecules are represented by cpk models in magenta and red, respectively. The electron density around the metal ion was contoured at 3 σ level. (B) A close-up stereo view of the non-specific Ba2+ ion binding site and its interactions. The electron density around the metal ions was contoured at 3 σ level. Hydrogen bonds and the color scheme are described in Figure 5a.
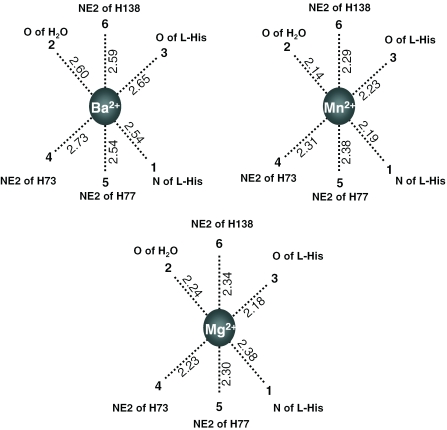
In order to substantiate the importance of the histidine cluster in the specific divalent metal ion coordination site, we mutated all three histidines (His73, His77 and His138) individually and analyzed the RNA binding. These binding analyses showed that all of the histidines involved in the metal ion interactions were required for the coordination as observed recently with Mg2+ ions (17). Since all three of the divalent metal ions (Mn2+, Ba2+ and Mg2+) were able to bind to HutP, are there any differences in the metal ion coordinations with their counterparts? Why did the Ba2+ ion weakly facilitate the activation? The metal ion coordinations were essentially the same in all of the cases, whereas the coordination distance changed slightly, depending on the metal ion radius. We averaged each coordination distance individually in all three of the observed sites, and these values are depicted in Figure 6. Although the bonding distances varied slightly among the metal ions, the average coordination distances for Mn2+, Ba2+ and Mg2+ were 2.26, 2.60 and 2.28 Å, respectively. The maximum difference in coordination distances observed for Ba2+ was 0.33 Å, in comparison with Mg2+, and this may be due to the difference in ionic radii (Table 1). Owing to the larger ionic radius of the Ba2+ ions, all six of the metal coordinating functional groups moved away (∼0.3–0.4 Å) from the center of the ion, as compared with the Mn2+ and Mg2+ ions. The Kd of Ba2+ is probably much higher than those of the Mn2+ and Mg2+ ions; however, we have not determined the Kd, yet. Even though the ionic radius of Mn2+ is slightly higher than that of Mg2+, the coordination distances were almost equal (0.02 Å). Since the metal ion resides in between the HutP monomers the interface may accept a wide range of divalent metal ions with different ionic radii (0.72–1.35 Å).
Figure 6.
Divalent metal ion coordination distance comparison for different metal ions observed in the complex structures. A schematic hexa-coordination of the metal ions, drawn and numbered as in Figure 5a. The metal ion binding sites observed in the asymmetric unit were averaged individually and are depicted in the figures.
Role of divalent metal ions in the HutP–RNA interactions
Interestingly, when we analyzed the recently reported structures of HutP (uncomplexed HutP, HutP–HBN complex, HutP–L-histidine–Mg2+ and HutP–L-histidine–Mg2+–21mer complex) and the current complex structures of HutP (HutP–L-histidine–Mn2+, HutP–L-histidine–Ba2+ and HutP–Mg2+), obviously, it was quite clear that the specific metal ion coordination with the histidine cluster of HutP and the L-histidine ligand are essential for the rearrangements. The uncomplexed HutP, HutP–Mg2+ (crystallized in the absence of L-histidine) and HutP–HBN complexes assumed similar conformations (inactive/preactive), suggesting that the Mg2+ ion or L-histidine ligand alone may not be sufficient for the activation of HutP. This is consistent with our biochemical analyses (17). In the absence of L-histidine, two interactions with the metal ions are lost in the HutP–Mg2+ complex, and that is why the metal ion itself may not reside in the complex to facilitate the required structural rearrangements. When we introduced the divalent metal ion into the HutP–L-histidine complex, the metal ion bound to the L-histidine moved away from the original location (12 Å) and changed the HutP conformation, by settling within the specific binding site and interacting with the histidine cluster and the L-histidine ligand. In order to extend the metal ion coordination, with the HutP, the HutP protein undergoes a significant conformational change, especially in the loop regions, L3, L4 and L5, in addition to the L-histidine binding site (Figure 4d). This conformational change becomes evident upon a comparison of the inactive/preactive conformations and the divalent metal ion complexes. The divalent metal ion complexes represent the activated conformation of HutP, which is ready to recognize the hut mRNA. Also, it is clear that there are no further conformational changes of the activated HutP protein upon RNA binding (17).
In summary, the present results suggest that many divalent metal ions can mediate the HutP–RNA interactions, but monovalent cations cannot. These new crystal structures also revealed the existence of a specific divalent metal ion binding site that can accommodate different divalent cations with ionic radii ranging from 0.72 to 1.35 Å. Furthermore, as previously observed with Mg2+ ions, other metal ions, such as Mn2+ and Ba2+, ions together with L-histidine (ligand) can modulate the HutP structure at the loop regions to facilitate binding to the hut mRNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
This work was supported by funds from the National Institute of Industrial Science and Technology (AIST) to P.K.R.K. and T.S.K. was supported by an AIST fellowship. We thank our colleague, Dr D. Balasundaresan, for help in preparing the figures. Funding to pay the Open Access publication charges for this article was provided by AIST.
Conflict of interest statement. None declared.
REFERENCES
- 1.Oda M., Katagai T., Tomura D., Shoun H., Hoshino T., Furukawa K. Analysis of the transcriptional activity of the hut promoter in Bacillus subtilis and identification of a cis-acting regulatory region associated with catabolite repression downstream from the site of transcription. Mol. Microbiol. 1992;6:2573–2582. doi: 10.1111/j.1365-2958.1992.tb01434.x. [DOI] [PubMed] [Google Scholar]
- 2.Wray L.V., Jr, Fisher S.H. Analysis of Bacillus subtilis hut operon expression indicates that histidine-dependent induction is mediated primarily by transcriptional antitermination and that amino acid repression is mediated by two mechanisms: regulation of transcription and inhibition of histidine transport. J. Bacteriol. 1994;176:5466–5473. doi: 10.1128/jb.176.17.5466-5473.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oda M., Kobayashi N., Ito A., Kurusu Y., Taira K. Cis-acting regulatory sequences for antitermination in the transcript of the Bacillus subtilis hut operon and histidine-dependent binding of HutP to the transcript containing the regulatory sequences. Mol. Microbiol. 2000;35:1244–1254. doi: 10.1046/j.1365-2958.2000.01795.x. [DOI] [PubMed] [Google Scholar]
- 4.Houman F., Diaz-Torres M.R., Wright A. Transcriptional antitermination in the bgl operon of E.coli is modulated by a specific RNA binding protein. Cell. 1990;62:1153–1163. doi: 10.1016/0092-8674(90)90392-r. [DOI] [PubMed] [Google Scholar]
- 5.Aymerich S., Steinmetz M. Specificity determinants and structural features in the RNA target of the bacterial anti-terminator proteins of the Bgl/SacY family. Proc. Natl Acad. Sci. USA. 1992;89:10410–10414. doi: 10.1073/pnas.89.21.10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babitzke P., Yanofsky C. Reconstruction of Bacillus subtilis trp attenuation in vitro with TRAP, the trp RNA-binding attenuation protein. Proc. Natl Acad. Sci. USA. 1993;90:133–137. doi: 10.1073/pnas.90.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnaud M.D., Debarbouille M., Rapport G., Saier M.H., Jr, Reizer J. In vitro reconstitution of transcriptional attenuation by the SacT and SacY proteins of Bacillus subtilis. J. Biol. Chem. 1996;271:18966–18972. doi: 10.1074/jbc.271.31.18966. [DOI] [PubMed] [Google Scholar]
- 8.Lu Y., Turner R.J., Switzer R.L. Function of RNA secondary structures in transcriptional attenuation of the Bacillus subtilis pyr operon. Proc. Natl Acad. Sci. USA. 1996;93:14462–14467. doi: 10.1073/pnas.93.25.14462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alpert C.A., Siebers U. The lac operon of Lactobacillus casei contains lacT, a gene coding for a protein of the BglG family of transcriptional anti-terminators. J. Bacteriol. 1997;179:1555–1562. doi: 10.1128/jb.179.5.1555-1562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glatz E., Nilsson R.P., Rutberg B. A dual role for the Bacillus subtilis glpD leader and the GlpP protein in the regulated expression of glpD: antitermination and control of mRNA stability. Mol. Microbiol. 1996;19:319–328. doi: 10.1046/j.1365-2958.1996.376903.x. [DOI] [PubMed] [Google Scholar]
- 11.Takami H., Nakasone K., Takaki Y., Maeno G., Sasaki R., Masui N., Fuji F., Hirama C., Nakamura Y., Ogasawara N., et al. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 2000;28:4317–4331. doi: 10.1093/nar/28.21.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivanova N., Sorokin A., Anderson I., Galleron N., Candelon B., Kapatral V., Bhattacharyya A., Reznik G., Mikhailova N., Lapidus A., et al. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature. 2003;423:87–91. doi: 10.1038/nature01582. [DOI] [PubMed] [Google Scholar]
- 13.Read T.D., Peterson S.N., Tourasse N., Baillie L.W., Paulsen I.T., Nelson K.E., Tettelin H., Fouts D.E., Eisen J.A., et al. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature. 2003;423:81–86. doi: 10.1038/nature01586. [DOI] [PubMed] [Google Scholar]
- 14.Kumarevel T.S., Fujimoto Z., Karthe P., Oda M., Mizuno H., Kumar P.K.R. Crystal structure of active HutP: an RNA binding protein that regulates the transcription of hut operon in Bacillus subtilis. Structure. 2004;12:1269–1280. doi: 10.1016/j.str.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Kumarevel T.S., Mizuno H., Kumar P.K.R. Allosteric activation of HutP protein that regulates transcription of hut operon in Bacillus subtilis, mediated by various analogs of L-histidine. Nucleic Acids Res. Suppl. 2003;3:199–200. doi: 10.1093/nass/3.1.199. [DOI] [PubMed] [Google Scholar]
- 16.Kumarevel T.S., Gopinath S.C.B., Mizuno H., Kumar P.K.R. Identification of important chemical groups of the hut mRNA for HutP interactions that regulate the hut operon in Bacillus subtilis. Nucleic Acids Res. 2004;32:3904–3912. doi: 10.1093/nar/gkh725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumarevel T.S., Mizuno H., Kumar P.K.R. Structural basis of the HutP-mediated anti-termination and roles of the Mg2+ ion and L-histidine ligand. Nature. 2005;434:183–191. doi: 10.1038/nature03355. [DOI] [PubMed] [Google Scholar]
- 18.Pan T., Long D.M., Uhlenbeck O.C. Divalent metal ions in RNA folding and catalysis. In: Gesteland R.F., Atkins J.F., editors. The RNA World. New York, USA: CSHL press; 1993. pp. 271–302. [Google Scholar]
- 19.Weber T.P., Widger W.R., Kohn H. Metal dependency for transcription factor Rho activation. Biochemistry. 2003;42:1652–1659. doi: 10.1021/bi020601y. [DOI] [PubMed] [Google Scholar]
- 20.Niefind K., Muller J., Riebel B., Hummel W., Schomburg D. The crystal structure of R-specific alcohol dehydrogenase from Lactobacillus brevis suggests the structural basis of its metal dependency. J. Mol. Biol. 2003;327:317–328. doi: 10.1016/s0022-2836(03)00081-0. [DOI] [PubMed] [Google Scholar]
- 21.Eswaramoorthy S., Kumaran D., Keller J., Swaminathan S. Role of metals in the biological activity of Clostridium botulinum neurotoxins. Biochemistry. 2004;43:2209–2216. doi: 10.1021/bi035844k. [DOI] [PubMed] [Google Scholar]
- 22.Carey J., Uhlenbeck O.C. Kinetic and thermodynamic characterization of the R17 coat protein–ribonucleic acid interactions. Biochemistry. 1983;22:2610–2615. doi: 10.1021/bi00280a003. [DOI] [PubMed] [Google Scholar]
- 23.Vartikar J.V., Draper D.E. S4-16S ribosomal RNA complex, binding constant measurements and specific recognition of a 460-nucleotide region. J. Mol. Biol. 1989;209:221–234. doi: 10.1016/0022-2836(89)90274-x. [DOI] [PubMed] [Google Scholar]
- 24.Batey R.T., Doudna J.A. Structural and energetic analysis of metal ions essential to SPR signal recognition domain assembly. Biochemistry. 2002;41:11703–11710. doi: 10.1021/bi026163c. [DOI] [PubMed] [Google Scholar]
- 25.Talbot S.J., Altman S. Kinetics and thermodynamic analysis of RNA–protein interactions in the RNase P Holoenzyme from Escherichia coli. Biochemistry. 1994;33:1406–1411. doi: 10.1021/bi00172a017. [DOI] [PubMed] [Google Scholar]
- 26.Ryan P.C., Draper D.E. Thermodynamics of protein–RNA recognition in a highly conserved region of the large-subunit ribosomal RNA. Biochemistry. 1989;28:9949–9956. doi: 10.1021/bi00452a012. [DOI] [PubMed] [Google Scholar]
- 27.Handa N., Nureki O., Kurimoto K., Kim I., Sakamoto H., Shimura Y., Muto Y., Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the sex-lethal protein. Nature. 1999;398:579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- 28.Deo R.C., Bonanno J.B., Sonenberg N., Burley S.K. Recognition of polyadenylate RNA by the poly (A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 29.Bogden C.E., Fass D., Bergman N., Nichols M.D., Berger J.M. The structural basis for terminator recognition by the Rho transcription termination factor. Mol. Cell. 1999;3:487–493. doi: 10.1016/s1097-2765(00)80476-1. [DOI] [PubMed] [Google Scholar]
- 30.Antson A.A., Dodson E.J., Dodson G., Greaves R.B., Chen X.P., Gollnick P. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 1999;401:235–242. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- 31.Wimberly B.T., Guymon R., McCutcheon J.P., White S.W., Ramakrishnan V. A detailed view of a ribosomal active site: the structure of the L11–RNA complex. Cell. 1999;97:491–502. doi: 10.1016/s0092-8674(00)80759-x. [DOI] [PubMed] [Google Scholar]
- 32.Kumarevel T.S., Fujimoto Z., Padmanabhan B., Oda M., Nishikawa S., Mizuno H., Kumar P.K.R. Crystallization and preliminary X-ray diffraction studies of HutP protein: an RNA-binding protein that regulates the transcription of hut operon in Bacillus subtilis. J. Struct. Biol. 2002;138:237–240. doi: 10.1016/s1047-8477(02)00024-2. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Brunger A., Adams P.D., Clore G.M., Gros P., Grosse-Kuntsleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., Read R.J. Crystallography and NMR system: a new software system for macromolecular structure determination. Acta. Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 35.Oldfield T.J. A number of real-space torsion-angle refinement techniques for proteins, nucleic acids, ligands and solvent. Acta. Crystallogr. D. 2001;57:82–94. doi: 10.1107/s0907444900014098. [DOI] [PubMed] [Google Scholar]
- 36.Carson M. Ribbons. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
- 37.McRee D.E. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 38.Helene C., Brun F., Yaniv M. Fluorescence studies of interactions between Escherichia coli valyl-tRNA synthetase and its substrates. J. Mol. Biol. 1971;58:349–365. doi: 10.1016/0022-2836(71)90251-8. [DOI] [PubMed] [Google Scholar]
- 39.Yarus M. Binding of isoleucyl transfer ribonucleic acid by isoleucyl transfer ribonucleic acid synthetase: solvent, the strength of interaction, and a proposed source of specificity. Biochemistry. 1972;11:2050–2060. doi: 10.1021/bi00761a009. [DOI] [PubMed] [Google Scholar]
- 40.Lam S.S.M., Schimmel P.R. Equilibrium measurements of cognate and noncognate interactions between aminoacyl tRNA synthetases and transfer RNA. Biochemistry. 1975;14:2775–2780. doi: 10.1021/bi00683a034. [DOI] [PubMed] [Google Scholar]
- 41.Mougel M., Ehresmann B., Ehresmann C. Binding of Escherichia coli ribosomal protein S8 to 16 S rRNA: kinetic and thermodynamic characterization. Biochemistry. 1986;25:2756–2765. doi: 10.1021/bi00358a003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.