Abstract
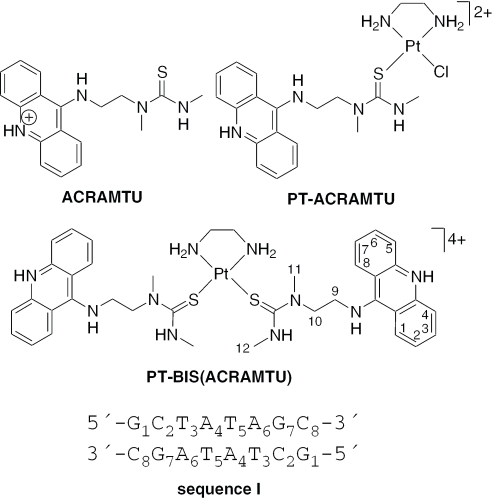
The DNA interactions of PT-BIS(ACRAMTU) ([Pt(en)(ACRAMTU)2](NO3)4; ACRAMTU = 1-[2-(acridin-9-ylamino)ethyl]-1,3-dimethylthiourea, en = ethylenediamine), a bifunctional platinum–acridine conjugate, have been studied in native and synthetic double-stranded DNAs and model duplexes using various biophysical techniques. These include ethidium-DNA fluorescence quenching and thermal melting experiments, circular dichroism (CD) spectroscopy and plasmid unwinding assays. In addition, the binding mode was studied in a short octamer by NMR spectroscopy in conjunction with molecular modeling. In alternating copolymers, PT-BIS(ACRAMTU) shows a distinct preference for poly(dA-dT)2, which is ∼3-fold higher than that of ACRAMTU. In the ligand-oligomer complex, d(GCTATAGC)2·PT-BIS(ACRAMTU) (complex I*), PT-BIS(ACRAMTU) increases the thermal stability of the B-form host duplex by ΔTm > 30 K (CD and UV melting experiments). The agent unwinds pSP73 plasmid DNA by 44(±2)° per bound molecule, indicating bisintercalative binding. A 2-D NMR study unequivocally demonstrates that PT-BIS(ACRAMTU)'s chromophores deeply bisintercalate into the 5′-TA/TA base pair steps in I*, while the platinum linker lies in the minor groove. An AMBER model reflecting the NMR results shows that bracketing of the central AT base pairs in a classical nearest neighbor excluded fashion is feasible. PT-BIS(ACRAMTU) inhibits DNA hydrolysis by BstZ17 I at the enzyme's restriction site, GTA↓TAC. Possible consequences for other relevant DNA–protein interactions, such as those involved in TATA-box-mediated transcription initiation and the utility of the platinum-intercalator technology for the design of sequence-specific agents are discussed.
INTRODUCTION
Among the DNA binders, bisintercalating molecules have attracted considerable attention in the bioorganic and drug development communities (1,2). Bisintercalators consist of two planar chromophores tethered by flexible linker groups, which are capable of simultaneously inserting into the DNA base stack (3). Since the discovery of echinomycin, a bioactive natural product consisting of two quinoxaline chromophores linked by a bicyclic peptide, bifunctional intercalating agents, both natural and synthetic, have demonstrated promising antimicrobial and antitumor properties (4). Several natural products structurally related to echinomycin and synthetic analogues have been studied for their biological activity and DNA interactions, including agents such as luzopeptin (5), TANDEM (6) and quinomycin (7). Motivated by the clinical utility of these agents and the desire to understand the fundamental biophysical implications of bisintercalation, ‘simple’ bifunctional molecules, in which the peptidic linkages have been replaced with polymethylene or polyamine chains, were also designed. 9-Aminoacridine (9-AA) derivatives (8,9), ethidium (10,11), thiazole orange (TO) (12) and naphthalimide (13) are some of the chromophores that have been studied in great detail.
In a search for novel hybrid inorganic–organic molecules for applications in cancer chemotherapy and transcription regulation, we developed a new class of platinum-intercalator conjugates based on the 9-aminoacridine derivative, 1-[2-(acridin-9-ylamino)ethyl]-1,3-dimethylthiourea (ACRAMTU, acridinium salt, Figure 1) (14). PT-ACRAMTU ([PtCl(en)(ACRAMTU)](NO3)2, en = ethylenediamine; Figure 1) and its derivatives show promising activity in vitro in several solid tumors, including ovarian and lung cancer cell lines (15–18). While the cellular biology of the parent conjugate is still under investigation, we proposed the unprecedented targeting of adenine residues at 5′-TA sites in the minor groove of DNA by this dual metalating-intercalating agent (19,20) may play an important role in its mechanism of action (21–23). Following our interest in small-molecule modulators of transcription, we have now begun to extend our studies to reversible bifunctional intercalators derived from PT-ACRAMTU. Here we have used various biochemical methods and spectroscopies to probe the DNA interactions of the prototypical conjugate, PT-BIS(ACRAMTU) ([Pt(en)(ACRAMTU)2](NO3)4, Figure 1). PT-BIS(ACRAMTU) is a cytotoxic agent shown previously to strongly bind to native DNA with a binding constant on the order of 107 M−1 and an excluded site size of n ≈ 4 bp, indicating the possibility of bisintercalative binding (24). Our results presented here demonstrate that this binding mode is, indeed, observed in the 5′-TATA sequence, which is known to play a crucial role in eukaryotic transcription initiation (25). PT-BIS(ACRAMTU) emerges as a structural motif that may offer a new approach to the design of sequence-specific inhibitors of transcription.
Figure 1.
Structures of PT-BIS(ACRAMTU) and related complexes and sequence I giving atom and residue numberings.
MATERIALS AND METHODS
Drugs and biochemicals
PT-BIS(ACRAMTU) and ACRAMTU (nitrate salts) were synthesized as described previously (15,24). Biochemical grade chemicals (Fisher/Acros and DNase-free) available were used for the preparation of biological buffers. All buffers were made from doubly distilled 0.22 µm-filtered DNase/RNase-free water obtained from a Milli-Q A10 synthesis water purification system. All other chemicals were purchased from common vendors and used without further purification. Stock solutions of PT-BIS(ACRAMTU) and ACRAMTU were stored at 277 K in the dark. Buffered solutions of oligodeoxyribonucleotide-ligand complex were lyophilized and stored at 253 K.
DNA
The oligodeoxyribonucleotides (ODNs) were purchased from Integrated DNA Technologies Inc. (Coralville, IA). All ODN sequences were synthesized using phosphoramidite chemistry and desalted by the vendor. The oligomer used in the NMR study and in circular dichroism (CD) and thermal melting experiments was further purified by size-exclusion chromatography (BioRad Biospin columns, Sephadex G-10 resin) and dialyzed at 277 K for 72 h against 10 mM phosphate buffer (pH 7.0) (Spectra/Por Float A Lyzer, 500 Da molecular weight cut-off). Oligomer purity was confirmed by 1H NMR spectroscopy (500 MHz). The synthetic 40mers used in the restriction enzyme cleavage assay were purified by high-performance liquid chromatography (HPLC) by the vendor. Quantification of ODNs was done spectrophotometrically in the appropriate buffers at 260 nm using the molar absorptivities provided by the vendor. Calf thymus DNA (lyophilized powder, Sigma) and the synthetic alternating copolymers poly(dG-dC)2 and poly(dA-dT)2 (Pharmacia Amersham Biotech) were used as supplied. DNA concentrations (base pairs) were determined from absorbances at 260 nm using Beer's law with ɛ260 (calf thymus) = 12 824 M−1cm−1 (bp), ɛ260 (GC) = 16 800 M−1cm−1 (bp) and ɛ260 (AT) = 13 100 M−1cm−1 (bp) (26). DNA stock solutions were stored at 277 K. Plasmid DNA (pSP73, 2464 bp) was isolated from transformed Escherichia coli Nova-blue cells using a Qiagen plasmid purification kit.
Fluorescence quenching assay
All measurements were performed in 1 cm quartz cuvettes at ambient temperature on a Perkin Elmer LS50B luminescence spectrometer. Calf thymus DNA, poly(dG-dC)2 and poly(dA-dT)2 were dissolved in a buffer containing 10 mM NaH2PO4, 0.1 mM Na2EDTA and 500 mM NaCl (pH 7.0) and annealed by slow cooling from 348 K to room temperature. Stock solutions of PT-BIS(ACRAMTU) (500 µM) and ACRAMTU (80 µM) were prepared in the same buffer. Quenching of ethidium-DNA fluorescence was studied according to a published assay (26). Samples (2000 µl) of ethidium-modified native and synthetic DNAs (30.0 µM of DNA (bp) and 5.0 µM of ethidium bromide) were titrated with suitable aliquots of the drug solutions. Samples were mixed thoroughly and equilibrated for 2 min before data acquisition. DNA-bound ethidium was excited at 505 nm (slit width 10 nm), and the fluorescence intensity was monitored at 603 nm (slit width 15 nm). PT-BIS(ACRAMTU) and ACRAMTU did not fluoresce at the emission of ethidium. The reported C50 values (concentration of drug required to reduce the initial ethidium fluorescence by 50%) were calculated from plots of dilution-corrected, normalized fluorescence intensities versus drug concentration using exponential curve fits (Origin 7, OriginLab, Northampton, MA). Reported values are averages of two titrations.
Titrations and sample preparations
The oligonucleotide 5′-GCTATAGC-3′ (sequence I) was dissolved in 1 ml of sodium phosphate buffer [10 mM NaH2PO4 and 10 mM NaCl (pH 7.0)]. The sequence was annealed by slow cooling from 348 K to room temperature and lyophilized. The DNA−ligand complex, d(GCTATAGC)2·PT-BIS(ACRAMTU) (I*) was generated by titrating suitable aliquots of a saturated solution of PT-BIS(ACRAMTU) in D2O to sequence I. Titrations were performed at 308 K in a 5 mm NMR tube, and a 1H NMR spectrum was recorded after each addition. Integration of the H2/H7 signal of acridine and thymine methyl protons was used to establish a 1:1 ligand-to-duplex ratio. The complex (I*) was lyophilized twice from 99.96% D2O and finally dissolved in 500 µl of 99.96% D2O to afford a concentration of ∼4 mM. The pH* (uncorrected pH meter reading) of the solution was 7.0−7.1. Solutions of I and I* for thermal melting and CD studies were prepared from the NMR sample by appropriate dilution. For NMR experiments at elevated temperatures, the samples were supplemented with additional NaCl to produce a total concentration of 0.1 M in Na+.
Optical studies of thermal denaturation
Melting experiments for the free oligomer 5′-GCTATAGC-3′ (I) and complex I* were carried out in phosphate buffer [10 mM NaH2PO4 and 10 mM NaCl (pH 7.0)]. Samples were annealed by slow cooling from 348 K to room temperature before data collection. The melting profiles were recorded on an HP 8453 diode array spectrophotometer equipped with a Peltier temperature-controlled cell-holder unit and an external temperature probe. Quartz cells with 1 cm path length were used in all experiments. Oligomer concentrations were maintained at 7 µM and 3.5 µM for I and I*, respectively. Absorbance as a function of temperature was measured at 260 nm. The temperature was raised in 0.5 K increments, and the stirred samples were allowed to equilibrate for 1 min at each temperature setting. Absorbance data for both heating and cooling traces were collected in the temperature range 293–343 K. Melting temperatures were determined from first derivatives of the melting curves and confirmed by fitting the data to the baseline-corrected two-state helix-coil transition model using Microcal Origin 7 (OriginLab Corp., Northampton, MA). Reported values are averages of three determinations.
CD spectropolarimetry
CD experiments were performed on an AVIV Model 215 spectrophotometer equipped with a thermoelectricallycontrolled cell-holder. Solutions of 5′-GCTATAGC-3′ (I) and PT-BIS(ACRAMTU) were prepared in the same buffer used for the melting experiments. Quartz cells with 1 cm path length were used for all experiments. Oligomer concentrations were 6.3 µM and 100 µM for CD spectra recorded in the UV (220–325 nm) and UV-visible (300–500 nm) regions, respectively. CD spectra were recorded at 298 K in 1 nm increments with an averaging time of 2 s.
Plasmid unwinding
Plasmid DNA, pSP73 [176 µM (bp)], was incubated at room temperature with varying concentrations of PT-BIS(ACRAMTU) (0−24 µM) in TE buffer [10 mM Tris and 1 mM EDTA (pH 7.5)] for 1 h in the dark. Unwinding of DNA was monitored by an electrophoretic mobility shift assay (EMSA) described previously (27). Gels were run at room temperature for 2−3 h at 70 V, stained twice with ethidium bromide (1 µg/ml), destained with 1% MgCl2 solution, and photo-documented using the ChemiDoc XRS imaging system (BioRad Laboratories, Hercules, CA). Unwinding angles were calculated from the following equation, ϕ = 18σ/rb(c), using the experimentally determined rb(c) values (drug-to-nucleotide ratios at which the drug completely unwinds supercoiled plasmid). The superhelical density (σ) of the plasmid under the present experimental conditions was −0.063, calculated on the basis of the known unwinding angle for the site-specific 1,2 intrastrand cross-link formed by cisplatin (28). The average rb(c) value was extracted from plots of band intensities versus ligand concentration using a parabolic curve fit (Origin 7, Originlab, Northhampton, MA). Band intensities were integrated with the Quantity One software (version 4.4.1; BioRad, Hercules, CA). Incubations and unwinding assays were performed in triplicates.
Acquisition and processing of NMR data
NMR spectra were acquired at 500 MHz on a Bruker DRX-500 spectrometer equipped with an inverse 3-channel Z-gradient TBI probe and a variable-temperature unit. The spectral width was 5000 Hz, and the carrier frequency was set to the frequency of the HDO signal for one-dimensional (1-D) spectra and two-dimensional (2-D) data sets collected in D2O. A spectral width of 12 530 Hz was used for spectra recorded in 90% H2O/10% D2O. The WATERGATE (water suppression by gradient-tailored excitation) and jump-and-return techniques (29) were used to suppress the (residual) water peak. Chemical shifts (δ, p.p.m.) were referenced to internal 3-(trimethyl-silyl)-1-propanesulfonic acid sodium salt (DSS) standard. 1-D and 2-D NMR data sets were acquired at various temperatures in the range 278–328 K. 1-D 1H NMR spectra were recorded with a total of 64 K data points, 16 transients, and a recycle delay of 1 s. The time domain data were apodized with an exponential window function using a line broadening of 0.3 Hz and then Fourier transformed. The g-COSY (gradient correlation spectroscopy) experiments were performed with 2048 complex points in t2, 512 points in t1 and 32 transients per t1 increment with a recycle delay of 1.5 s. 2-D NOESY (nuclear Overhauser enhancement spectroscopy) spectra were acquired with 2048 complex points in t2, 256 points in t1, 64 transients per t1 increment and a recycle delay of 2 s. The mixing time (τm) was 350 ms. All data were multiplied with optimized phase-shifted squared sinebell apodization functions and zero-filled to a final matrix of 2048 × 2048 data points before Fourier transformation. 1-D spectra were processed using the XWIN-NMR software (version 2.6, Bruker, Ettlingen, Germany), and 2-D data sets were processed with Felix 2000 (Accelrys, San Diego, CA) installed on a Silicon Graphics O2 workstation.
Modeling
Calculations were performed using the AMBER force field in the InsightII/Discover software (version 2000, Accelrys, San Diego, CA). Starting structures of the B-form octamer d(GCTATAGC)2 (I) and PT-BIS(ACRAMTU) were built using the Biopolymer module in InsightII. The duplex and the drug were energy-minimized separately, followed by ‘manual’ docking of the bisintercalator into the minor groove. To guide the drug's chromophores into the T3A6/T5A4 base pair steps, the following intermolecular drug-DNA distance restraints using a simple quadratic harmonic were initially included in the minimizations: ACRH3⋯T5CH3, ACRH4⋯T5CH3, ACRH5⋯T3CH3, and ACRH6⋯T3CH3, with d = 3.0 Å, based on the medium cross-peak intensities observed in the NOESY spectra, and force constants in the range 10–100 kcal mol−1 Å−2. Sodium (+1) counter ions were used to balance the negative charge of the DNA phosphate and solvent was simulated with a distance-dependent dielectric (ɛ = 4rij). Coulombic and 1−4 parameters were scaled by a factor of 0.5. Atom types, force constants, equilibrium bond lengths and angles, and charge distribution schemes were adopted from previous modeling studies of ACRAMTU and PT-ACRAMTU (19,30). Ad hoc parameters for the drug molecule were optimized to most accurately reproduce the structure of PT-BIS(ACRAMTU) in the solid state. (Details of the crystal structure of PT-BIS(ACRAMTU) ([Pt(N2C2H8)(C18H21N4S)2]Cl4·9H2O) used to validate the force field have been deposited with the Cambridge Crystallographic Data Centre as Supplementary publication no. CCDC 268383. Copies of the data can be obtained free of charge from the CCDC; 12 Union Road, Cambridge CB2 1EZ, UK; http://ccdc.cam.ac.uk.) The drug-DNA complex (I*) was subjected to steepest descent and conjugate gradient minimization until the convergence criteria of Δrms 0.001 kcal mol−1 Å−1 was satisfied. Molecular views of the model were generated from car files using DS ViewerPro (version 6.0, Accelrys, San Diego, CA).
Restriction enzyme cleavage assay
Stock solutions of the drugs and the single-stranded ODNs, 5′-GGCACGTGGAACTCTGGGTATACTCAGCGAGGCCGGCCTC-3′ (‘top strand’) and 5′-GAGGCCGGCCTCGCTGAGTATACCCAGAGTTCCACGTGCC-3′ (‘bottom strand’), were prepared in 10 mM Tris–HCl buffer (pH 7.5) and stored at 253 K in the dark until used. Concentrations of drugs and ODNs were determined spectrophotometrically. The top strand of the probe was labeled at the 5′ end using T4 polynucleotide kinase (EPICENTRE Biotechnologies, Madison, WI) and [γ-32P]ATP (Amersham Biosciences, Piscataway, NJ). Top and bottom strands were annealed in 50 mM NaCl by incubating both strands at 368 K for 5 min, followed by slow cooling to room temperature in a dry bath. Radiolabeled probe samples were purified using Sephadex G-25 Quick Spin Columns (Roche, Indianapolis, IN). The labeled probe (0.1 µM) was incubated at room temperature with PT-BIS(ACRAMTU) and ACRAMTU (0–100 µM), and the mixtures were allowed to equilibrate for 30 min. Drug-modified and unmodified probe samples were then allowed to react with 7.5 U of the restriction enzyme BstZ17 I (New England Biolabs, Beverly, MA) at 310 K for 45 min in enzyme buffer provided by the vendor. Reaction products were analyzed by PAGE under denaturing conditions (12% acrylamide, 8 M urea; SequaGel Sequencing System, National Diagnostics, Inc.). Electrophoretic separations were carried out at 190 V for 1 h, and the dried gels were analyzed on a BioRad FX-Pro Plus phosphorimager (Hercules, CA).
RESULTS
Biophysical characterization of the binding mode
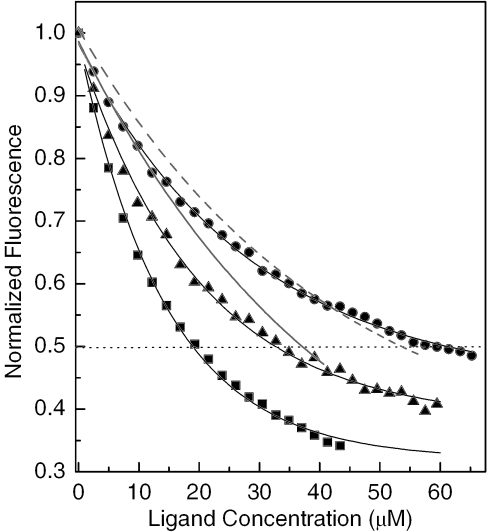
DNA-ethidium fluorescence assays are a useful tool for studying the binding affinity and global sequence selectivity of DNA binders. In these assays, the quenching is monitored by fluorescence originating from DNA-bound ethidium, which results from displacement of the chromophore by a competing ligand and/or direct contact between the reporter and probe ligands co-occupying the biopolymer (31). In the present study, the binding affinity of PT-BIS(ACRAMTU) to native DNA and synthetic alternating AT and GC copolymers was determined. The titrations were performed at neutral pH to ensure that acridine was present in its protonated form, and high-salt conditions (500 mM NaCl) were applied to reduce non-specific binding of the highly charged (4+) agent. ACRAMTU was included in this study to allow for a direct comparison between the effects of mono- and bisintercalation under the specific experimental conditions. The fluorescence data are plotted in Figure 2. The C50 values (concentration of acridine effecting 50% quenching of the ethidium fluorescence) are given in Figure 2 caption. Based on fluorescence quenching efficiencies, the order of binding affinities of PT-BIS(ACRAMTU) was found to be poly(dA-dT)2 > calf thymus DNA > poly(dG-dC)2. While an ∼4-fold higher affinity is observed in the alternating AT sequence compared to the GC sequence, the agent shows intermediate affinity to random-sequence DNA. In poly(dA-dT)2, PT-BIS(ACRAMTU) reduces the ethidium fluorescence to a greater extent than ACRAMTU (Figure 2), as can be expected for the increased number of chromophores and ‘bite size’ of a bifunctional molecule compared to a simple monointercalator. Surprisingly, the reverse situation is observed in poly(dG-dC)2, where PT-BIS(ACRAMTU) proves to be an even less efficient quencher than ACRAMTU (Figure 2). These findings may indicate that a bisintercalative binding mode is disfavored in alternating GC sequences. In essence, the linking of two ACRAMTU units in PT-BIS(ACRAMTU) leads to an ∼3-fold increase in AT selectivity, based on C50(GC)/C50(TA) ratios of 3.7 and 1.3 for PT-BIS(ACRAMTU) and ACRAMTU, respectively.
Figure 2.
DNA-ethidium fluorescence monitored at 603 nm in the presence of varying concentrations of intercalating agents (numbers in parentheses are C50 [µM] values). Squares: PT-BIS(ACRAMTU)/poly(dA-dT)2 (17.72); triangles: PT-BIS(ACRAMTU)/calf thymus DNA (32.65); circles: PT-BIS(ACRAMTU)/poly(dG-dC)2 (65.14); solid line: ACRAMTU/poly(dA-dT)2 (40.54); dashed line: ACRAMTU/poly(dG-dC)2 (53.57). For clarity, only the fitted curves are shown for ACRAMTU.
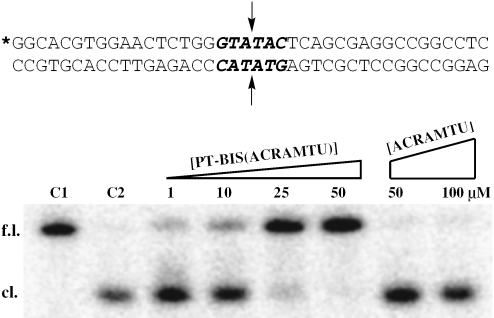
While the results of the fluorescence quenching experiments indicate that PT-BIS(ACRAMTU) binds to alternating AT most likely through bisintercalation, the exact site and mode of binding remained to be elucidated. Simple molecular models suggested that a conformation of PT-BIS(ACRAMTU) exists in which the two chromophores are coplanar to one another and separated by ∼10 Å, suitable for bracketing 2 bp in a bisintercalative binding mode. Previous studies of ACRAMTU and PT-ACRAMTU have shown that both agents intercalate into the 5′-TA step preferentially from the minor groove (21,30). To gain further insight into the binding preference within AT sequences we performed equilibrium dialysis experiments (see Supplementary Data) using short model duplexes as described previously for ACRAMTU (30).The results indicate that PT-BIS(ACRAMTU) binds to the 5′-TATA and 5′-ATAT sequences with similar affinity, whereas binding to 5′-AATT is clearly disfavored. This supports the notion that the two former sequences may be target sites for the proposed mode of bisintercalation of PT-BIS(ACRAMTU), in accordance with the classical sequence requirements of intercalation (32). Given the biological relevance of the 5′-TATA site, further biophysical and high-resolution structural work was performed using the sequence, 5′-GCTATAGC-3′ (I) (Figure 1). Similarly, a 40 bp probe containing the TATA site was used in the restriction enzyme cleavage assay (Figure 9).
Figure 9.
Restriction enzyme-mediated cleavage by BstZ17 I of a 40 bp duplex modified with PT-BIS(ACRAMTU) or ACRAMTU monitored by denaturing PAGE. Control lanes labeled C1 and C2 contain unmodified probe with no enzyme added and unmodified probe cleaved with enzyme, respectively. Bands are labeled ‘f.l.’ for full-length and ‘cl.’ for cleaved probe. The sequence of the probe is shown with the blunt-end restriction site highlighted in bold. The asterisk denotes the 32P-labeled 5′ end, and the arrows indicate the sites of hydrolytic cleavage.
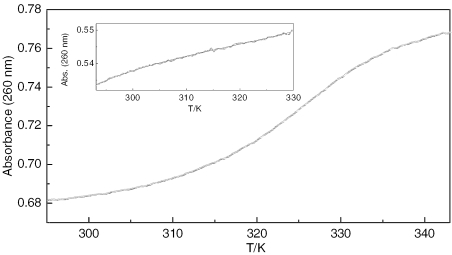
UV melting profiles were recorded of sequence I alone and in the presence of one equivalent of PT-BIS(ACRAMTU) (Figure 3). In our experiments, which used an oligomer concentration of 7 µM and a buffer containing 20 mM Na+, the unmodified sequence I was found to melt below 293 K. The theoretical melting temperature estimated for this oligomer is 294(±2) K (33). Addition of one equivalent of PT-BIS(ACRAMTU) (monitored by 1H NMR spectroscopy, see Materials and Methods section) to form the 1:1 complex, d(GCTATAGC)2·PT-BIS(ACRAMTU) (I*), causes a dramatic increase in the thermal stability of the duplex. Complex I* was found to melt at 326.6 K, indicating an unusual ΔTm of greater than 30 K. Under similar experimental conditions ACRAMTU increases the melting temperature of similar oligomer duplexes only insignificantly (ΔTm ≈ 2 K) (34). It is noteworthy to mention that PT-BIS(ACRAMTU) affects the thermal melting behavior of extended duplexes that are prone to hairpin formation. In d(GGTGGTATACCACC)2, for instance, PT-BIS(ACRAMTU) appears to completely eliminate the biphasic melting of the self-complementary duplex, which in its unmodified form shows distinct duplex → hairpin and hairpin → random coil transitions (35). These findings suggest that PT-BIS(ACRAMTU) stabilizes the duplex form through bisintercalation into the central (TATA)2 sequence, which is supported by the equilibrium dialysis experiments (see Suplementary Data).
Figure 3.
UV melting profile monitored at 260 nm for complex I*. The inset shows the melting trace recorded for the unmodified sequence, I. Experiments were performed at octamer concentrations of 3.5 and 7.0 µM, respectively [10 mM phosphate and 10 mM NaCl (pH 7.0)].
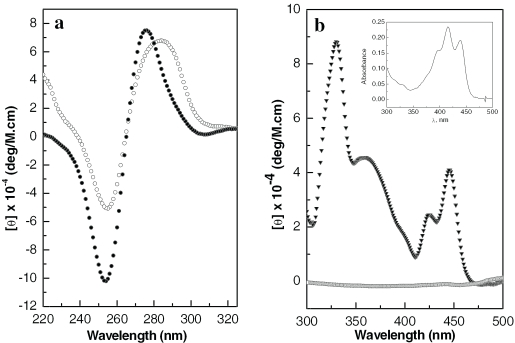
The CD spectrum recorded of the modified duplex (I*) in the 200–320 nm region, which is dominated by the CD of the DNA base pairs, shows a signal characteristic of a B-form helix with a negative band at 254 nm and a positive band at 275 nm (Figure 4a) (36). A comparison with the same feature in unmodified I suggests that PT-BIS(ACRAMTU) causes a conformational change in the oligomer structure upon binding, as evidenced by both the increase in band amplitudes and a pronounced blue-shift of the positive band by 7 nm. An explanation for the distinct shift of this CD band cannot be given at this point. In the 300–500 nm region of the CD spectrum of I*, a feature resulting from induced circular dichroism (ICD) is observed (Figure 4b). The ICD is the result of dipole–dipole interactions between the base pairs of the chiral duplex and the intercalated chromophores of intrinsically CD-inactive PT-BIS(ACRAMTU). The positive sign, amplitude and shape of the ICD are indicative of a bisintercalative binding mode, in which the long axes of the acridine chromophores are collinear with the long dimensions of the base pair pockets (37). However, the ICD of duplex-bound PT-BIS(ACRAMTU) does not resemble the absorption spectrum of acridine, implying that the alignment of the chromophores with adjacent base pairs deviates somewhat from that observed in DNA complexes of monomeric 9-aminoacridines (9). (The ICD of DNA-bound ACRAMTU, for instance, has been shown to mimic the UV-visible features of the acridine chromophore (34).) This deviation is most likely caused by the rigidity produced by the [Pt(en)] linkage connecting the two ACRAMTU molecules in the conjugate.
Figure 4.
CD spectra recorded at 298 K of the modified and unmodified octamer. (a) CD in the DNA region for I (open circles) and I* (filled circles). (b) ICD in the ligand region of I* (filled triangles). The trace close to baseline was recorded for a buffered solution of drug in the absence of oligomer. The inset shows the UV-visible feature of the acridine chromophores in PT-BIS(ACRAMTU). Concentrations of oligomers were 6.3 and 100 µM in (a) and (b), respectively [10 mM phosphate and 10 mM NaCl (pH 7.0)].
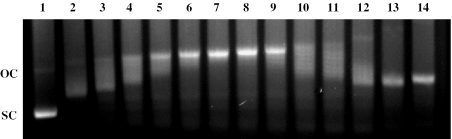
To discriminate between monofunctional and bifunctional intercalation, the ability of PT-BIS(ACRAMTU) to remove and reverse the supercoils in closed-circular pSP73 plasmid DNA was assessed by an EMSA (38). The negatively supercoiled plasmid was modified with varying concentrations of PT-BIS(ACRAMTU), and the samples were run on agarose gels. Figure 5 shows a representative gel resulting from a DNA-ligand titration. With increasing concentration of PT-BIS(ACRAMTU), supercoils in the plasmid are gradually removed until the DNA exists in its relaxed open-circular form. From the ligand-to-nucleotide ratio at which the relaxed form comigrates with the (naturally present) nicked open-circular form, rb(c) = 0.026(2), and the superhelical density of the plasmid (see Materials and Methods) an unwinding angle of 44(±2)° per bound molecule was determined. For comparison, under similar conditions, monointercalating PT-ACRAMTU has been shown to unwind DNA by 21° (19,27). Thus, it can be rationalized that the efficient unwinding of DNA by PT-BIS(ACRAMTU) is the result of simultaneous intercalation of both of its acridine chromophores. Unwinding angles of similar magnitude have been reported recently for a series of bis(acridinecarboxamides) (39).
Figure 5.
Unwinding of supercoiled pSP73 plasmid by PT-BIS(ACRAMTU). With increasing concentration of PT-BIS(ACRAMTU), relaxation of negatively supercoiled plasmid (sc) occurs resulting in the open-circular form (oc), as indicated by the change in electrophoretic mobility of the plasmid. At higher concentrations, PT-BIS(ACRAMTU) induces positive supercoils, thereby increasing the mobility of the modified plasmid. Drug-to-nucleotide ratios (rb) for lanes 1–14 are 0 (control), 0.011, 0.014, 0.017, 0.019, 0.022, 0.025, 0.028, 0.031, 0.034, 0.037, 0.045, 0.056 and 0.068 respectively.
High-resolution structural study
To gain insight into the DNA binding mode of PT-BIS(ACRAMTU) in complex I* at the molecular level, a 2-D NMR study was performed, and an AMBER model was generated based on the combined NMR data. Several chemical shift changes observed in 1-D 1H NMR spectra recorded during the titration of sequence I with one equivalent of drug suggested that the central 5′-TATA site is indeed involved in complex formation. Upon addition of PT-BIS(ACRAMTU), the A4H8 and A6H8 signals disappear and a new set of signals appears shifted upfield by ∼0.2 p.p.m.. Likewise, the signals assigned to T3CH3 and T5CH3 experience a distinct downfield shift of similar magnitude. In addition, a new set of broad signals is observed for PT-BIS(ACRAMTU). The change in line shapes during the NMR titrations indicated that complex I* is in intermediate chemical exchange (40), and unambiguous proton assignments from data sets acquired at 298 K proved to be unfeasible due to severe signal broadening. A similar dynamic behavior was observed previously for complexes of ACRAMTU (30). The observation that only one new set of signals is observed during the titration until a 1:1 duplex-to-bisintercalator stoichiometry is reached indicates that the drug binds site-specifically, which was confirmed by 2-D NMR experiments (vide infra). No further attempts were therefore made to investigate ligand-to-duplex ratios >1 for potential secondary binding sites. Variable-temperature 1H NMR spectra (Supporting Information) of I* recorded in the range 298–328 K indicate that with increasing temperature the line widths could be reduced and the signals resolved without compromising the stability of the duplex. In spectra recorded at 328 K in 90% H2O/10% D2O (not shown) the signals of the imino protons involved in interstrand hydrogen bonding were clearly observable, corroborating that the duplex was still intact. Thus, we took advantage of the unusual thermal stability of complex I* and acquired the 2-D NMR data sets at elevated temperatures.
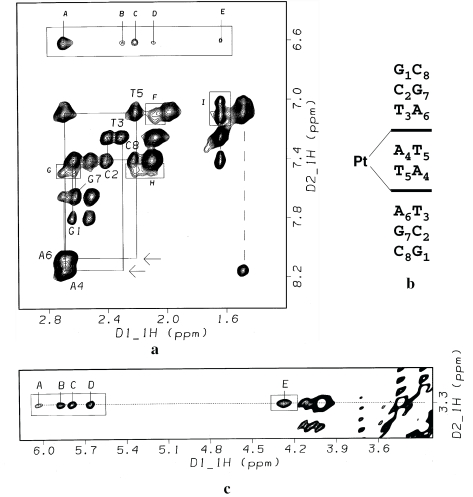
Routine strategies using 2-D NOESY and (magnitude) COSY spectra were applied to assign the non-exchangeable protons of the oligomer (Supplementary Table S1) and the protons of the drug (Supplementary Table S2). The assignment of the base and deoxyribose protons was accomplished using both H1′(i-1) → H8/H6(i) and H2′/H2″(i-1) → H8/H6(i) sequential NOE contacts (41). The section of the 2-D NOESY spectrum of I* depicted in Figure 6a shows that the ‘NOE walk’ is interrupted between T3H2′ and A4H8 and between T5H2′ and A6H8, as indicated by the complete absence of the corresponding cross-peaks. This important feature and the fact that a strong internucleotide cross-peak is observed for A4H8⋯T5CH3 (Figure 5a) is consistent with sequence-specific bisintercalation of the acridine chromophores into the 5′-T3A4/T5A6 base pair steps of the C2-symmetric duplex (Figure 6b). This contrasts the binding of ACRAMTU, which did not cause disruption of sequential NOE connectivities due to rapid association/dissociation (‘on/off’) rates and exchange between multiple sites (30). The aromatic protons of the acridine chromophores, H1–H8, appear significantly upfield shifted by as much as 0.8 p.p.m. relative to those in the unbound drug due to the ring current effect of the stacked DNA bases. On the other hand, H10–H12 of the linker in bound PT-BIS(ACRAMTU) are downfield shifted by the opposite ring current effect of the base pair edges (Supplementary Table S2). These findings are consistent with an intercalative binding mode of the planar acridine rings (42), while indicating extrahelical (groove) binding of the platinum−thiourea moiety (30).
Figure 6.
2-D NOESY study of the drug–octamer complex I* [500 MHz, 328 K, τm = 350 ms, buffer: D2O, 10 mM phosphate (pH* 7.0) and 90 mM NaCl]. (a) Section of the 2-D NOESY spectrum showing sequential connectivities in the aromatic-H2′/H2″ region (solid lines). Absent cross-peaks due to disruption of the T3H2′-A4H8 and T5H2′-A6H8 interresidue NOEs are indicated by arrows. The dashed vertical line indicates the connectivity between A4H8 and T5CH3 at the central base pair step. Assignment of intermolecular cross-peaks: (A) A4,A6H2′/H2″-ACRH2/H7; (B) T3H2″-ACRH7; (C) T5H2″-ACRH2; (D) T3H2′-ACRH7; (E) T3CH3-ACRH7; (F) T3H2′-ACRH6; (G) A6H2′/H2″-ACRH1; (H) T5H2″-ACRH1; (I) T3CH3-ACRH5 and T3CH3-ACRH6. The cross-peak for T5CH3- ACRH5 is partially overlapped with the intranucleobase cross-peak, T5CH3-T5H6 (top of the two cross-peaks connected by dashed line). (b) Illustration of the intercalation mode leading to interruption of the NOE connectivities at the 5′-T3A4/T5A6 base pair steps. (c) Section of the 2-D NOESY spectrum showing cross-peaks due to NOE contacts between the thiourea methyl groups (C11) and H1′ and H4′ deoxyribose protons in the minor groove of the octamer. Cross-peak assignments: (A) A4H1′-ACRH11; (B) A6H1′-ACRH11; (C) T3H1′-ACRH11; (D) T5H1′-ACRH11; (E) A6H4′- ACRH11.
At the site of intercalation, the aromatic acridine protons H1–H8 make NOE contacts with the H2′/H2″ deoxyribose protons of both strands. This observation is consistent with an intercalation geometry in which the drug's chromophores are aligned with the long dimensions of the base pairs defining the intercalation pocket. The NOESY spectrum also contains a wealth of information about the regioselectivity of the bisintercalation mode. Strong NOE cross-peaks are observed between H3–H6 of the acridine moieties and the thymine methyl groups (T3CH3 and T5CH3), which indicates PT-BIS(ACRAMTU)'s chromophores efficiently penetrate the host duplex from the minor groove. This notion is further supported by NOE contacts observed between the thiourea methyl groups (C11) and the H1′/H4′ deoxyribose protons adjacent to the intercalation sites (Figure 6c), confirming that the platinum linker makes close contact with the walls of the minor groove. Crucial intermolecular and interresidue NOE contacts at the drug binding site are summarized in Figure 7.
Figure 7.
Summary of crucial intermolecular and interresidue NOEs observed at the drug binding site in complex I*.
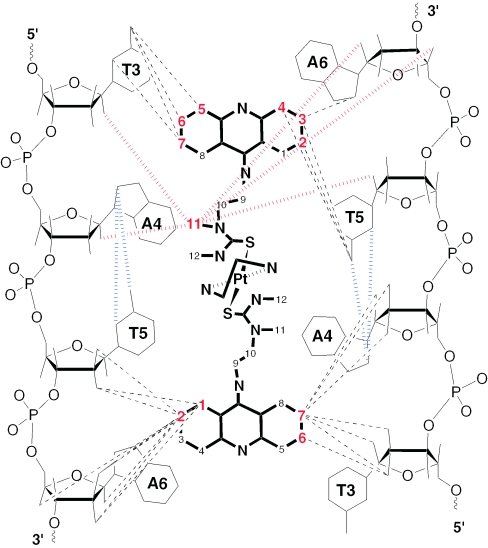
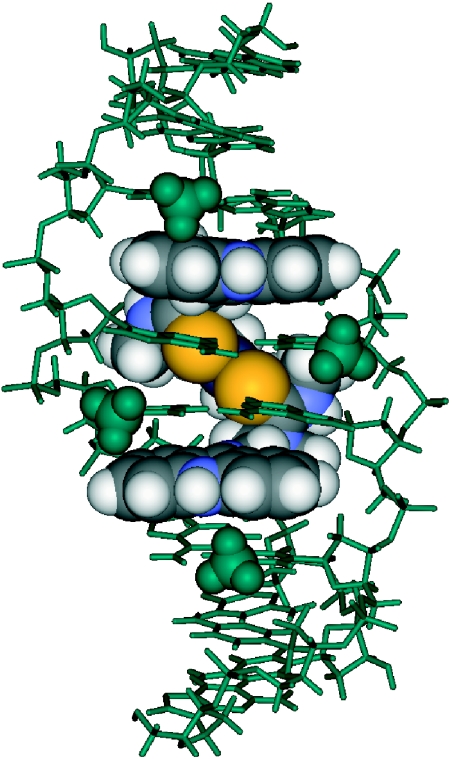
A strain-minimized AMBER model of complex I* was generated that reflects the results of the NMR study (Figure 8). In this model, the duplex accommodates bisintercalating PT-BIS(ACRAMTU) without major helical distortions and disruption of Watson–Crick hydrogen bonding. The rise at the T3A4/T5A6 base pair steps, however, has doubled to allow insertion of the two drug chromophores. The semirigid platinum–thiourea linker in PT-BIS(ACRAMTU) is positioned in the minor groove and produces the appropriate ‘bite’ to allow strainless bracketing of the two central AT base pairs. The binding mode produces short hydrophobic contacts between the linker and the walls of the minor groove. While the platinum-ethylenediamine moiety is directed away from the helical axis, the thiourea sulfur atoms make van der Waals contact with the edges of the central adenine residues at the bottom of the minor groove. This tight association allows deep penetration of PT-BIS(ACRAMTU)'s planar moieties into the base stack, concomitantly producing close contacts between the chromophores and the thymine methyl groups in the major groove.
Figure 8.
View into the major groove of an energy-minimized AMBER model of duplex I* based on NMR data. The oligomer is shown in green, and the drug molecule is represented by CPK rendering. The thymine methyl groups are highlighted as space-filling spheres.
Restriction enzyme inhibition
To investigate the effect of DNA-bound PT-BIS(ACRAMTU) on DNA-processing enzymes, a restriction enzyme cleavage assay was performed. BstZ17 I is a restriction endonuclease that produces a blunt-end cut in the recognition sequence, GTA↓TAC, where the arrow marks the site of hydrolytic cleavage. This restriction site was chosen because it contains the sequence 5′-TATA, a proposed target site for PT-BIS(ACRAMTU). Bisintercalation of PT-BIS(ACRAMTU) into this sequence can be expected to reduce the DNA-cleaving ability of the enzyme by conformational changes within the recognition site or by steric blockage. The 40 bp probe designed for this study containing a central GTATAC palindromic sequence is shown in Figure 9. PAGE analysis under denaturing conditions was used to monitor the cleavage of the probe by BstZ17 I in the presence of mono- and bisintercalator. Titration of the probe with PT-BIS(ACRAMTU) leads to virtually complete inhibition of endonucleolytic cleavage at a ligand concentration of 25 µM, as evidenced by the disappearance of the high-mobility band resulting from the 20 bp labeled fragment of the cleaved top strand. In contrast, ACRAMTU showed no effect, even at concentrations as high as 100 µM (Figure 9). These results suggest that bisintercalation, but not monointercalation, into the 5′-TATA sequence protects the probe duplex from cleavage at the central 5′-AT/AT base pair step of the restriction site.
DISCUSSION
The current study provides insight into the mechanism of action at the DNA level of PT-BIS(ACRAMTU). Our results suggest that bisintercalative binding, a potentially cytotoxic lesion in genomic DNA of this agent, is feasible and preferentially occurs in alternating TA sequences. Unlike the parent compound PT-ACRAMTU (20–22), PT-BIS(ACRAMTU) does not induce adducts with nucleobase nitrogen but binds to DNA in a reversible manner. (The inability of sulfur-bound ACRAMTU to act as a leaving group in reactions with double-stranded DNA has been amply demonstrated (21).) PT-BIS(ACRAMTU) shows a unique mode of bisintercalation that is more efficient in discriminating between AT and GC sequences than monointercalation by ACRAMTU (27). In the model complex d(GCTATAGC)2·PT-BIS(ACRAMTU) (I*), the drug associates tightly with the TATA sequence, a potential target site, through combined minor groove binding and bisintercalation, which leads to a pronounced increase in the thermal stability of the duplex. The platinum–thiourea linker aligns perfectly with the minor groove's curvature, providing the correct spacing between the two acridine chromophores for classical nearest neighbor excluded bisintercalative binding. The combined NMR and modeling results suggest that tight ‘clamping’ of the central AT/AT step and deep penetration of the base stack by the acridine chromophores in complex I* produces hydrophobic interactions between the drug and the minor groove. We believe the steric clash observed in computer models (not shown) at an analogous GC/GC step between the exocyclic NH2 group of guanine in the minor groove and thiourea sulfur may hinder insertion of the chromophores. This would explain the reduced binding affinity observed in poly(dG-dC)2.
The concept of linking two (or multiple) intercalators in a single molecule was inspired by the possibility of generating agents with greater DNA affinity and DNA structural impact compared to the corresponding monointercalators (2). Bisintercalation typically occurs with a minimum of one base pair step remaining vacant between intercalation sites, as implied by the nearest neighbor exclusion principle (43), and leads to local unwinding and lengthening of the host duplex (32). These structural distortions presumably interfere with DNA-processing enzymes to disrupt processes vital to cell function and proliferation, such as transcription and replication. The mechanism by which PT-BIS(ACRAMTU)'s DNA damage is recognized and the enzymes and DNA binding proteins involved in its processing remain to be determined. Recently, several new classes of anticancer active bisintercalators based on acridine- and phenazine-4-carboxamides have been described that target the cancer cell's polymerase and topoisomerase enzymes (39,44,45). These studies demonstrate that the specific signaling pathways through which these agents induce cancer cell death are not easily resolvable. Structure-activity relationship (SAR) studies of DNA-threading bisacridines show that tuning of the lifetimes of the intercalated state and the degree of duplex unwinding may alter the cell kill mechanism (39). These issues remain to be addressed in the case of PT-BIS(ACRAMTU) and its derivatives.
A recent study (46) shows that ‘covalent’ adducts formed by PT-ACRAMTU in the minor groove of a TATA box sequence inhibit the binding of human TATA-binding protein (hTBP). hTBP, which associates with its cognate sequence, TATA(A/T)A(A/T)N (N stands for any residue), through the DNA minor groove (47), is a key component of the transcription initiation complex (48). Thus, it was argued that platinum adducts, if formed at a high frequency in the promoter regions of genomic DNA, can be expected to down-modulate transcription, potentially contributing to the cytotoxicity of PT-ACRAMTU. In this study we have demonstrated the ability of PT-BIS(ACRAMTU) to inhibit endonucleolytic DNA cleavage, most likely by binding of the bisintercalator to the enzyme's TATA-containing restriction site. We suggest that strong reversible association of PT-BIS(ACRAMTU) (and suitably modified derivatives with enhanced DNA affinity) with TATA box sequences may produce similar effects. An important aspect of the DNA binding mode established in this study is related to our endeavors to design sequence-specific modulators of transcription using the platinum−intercalator technology. Unfortunately, PT-ACRAMTU lacks long-range sequence specificity and has been shown to promiscuously (and irreversibly) bind to nucleophilic sites of adenine and guanine in both grooves of double-stranded DNA, suggesting that this metalating agent may not be a good candidate for targeting the minor groove of the TATA sequence. In this regard, the PT-BIS(ACRAMTU) approach appears to be more promising. Earlier, it was rationalized that the increase in binding site sizes of bisintercalators compared to their monomeric precursors might be a viable strategy for producing agents with improved DNA binding selectivity through enhanced sequence and groove read-out (2). While the simple polymethylene- and polyamine-linked bisintercalators show no significant sequence specificity, recent work by Dervan et al. (49) suggests that specific minor groove recognition by designed polyamide linkers has the potential of turning bisintercalators into gene-specific agents. Because PT-BIS(ACRAMTU)'s high-affinity sequence, 5′-TATA, is found in the promoter region of most genes transcribed by eukaryotic RNA polymerase II (vide supra), our bisintercalator approach may offer a unique way of targeting the transcription machinery of specific genes. Naturally, this would require structural modification of the prototype to enhance its DNA binding affinity and to produce sequence specificity for, and beyond, the 5′-TATA tetranucleotide site. These studies are currently underway.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Dr M. W. Wright for assistance with the acquisition of the 2-D NMR data sets and Dr R. W. Alexander and M. E. Budiman for technical advice and the use of equipment. This work was supported by a grant from the National Institutes of Health (CA101880) and a Dean's Research Fellowship to J.R.C. Funding to pay the Open Access publication charges for this article was provided by the Publication and Research Fund of Wake Forest University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chaires J.B. Drug–DNA interactions. Curr. Opin. Struct. Biol. 1998;8:314–320. doi: 10.1016/s0959-440x(98)80064-x. [DOI] [PubMed] [Google Scholar]
- 2.Gago F. Stacking interactions and intercalative DNA binding. Methods. 1998;14:277–292. doi: 10.1006/meth.1998.0584. [DOI] [PubMed] [Google Scholar]
- 3.Le Pecq J.B., Le Bret M., Barbet J., Roques B. DNA polyintercalating drugs: DNA binding of diacridine derivatives. Proc. Natl Acad. Sci. USA. 1975;72:2915–2919. doi: 10.1073/pnas.72.8.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster B.J., Clagett-Carr K., Shoemaker D.D., Suffness M., Plowman J., Trissel L.A., Grieshaber C.K., Leyland-Jones B. Echinomycin: the first bifunctional intercalating agent in clinical trials. Invest. New Drugs. 1985;3:403–410. doi: 10.1007/BF00170766. [DOI] [PubMed] [Google Scholar]
- 5.Leroy J.L., Gao X.L., Misra V., Gueron M., Patel D.J. Proton exchange in DNA-luzopeptin and DNA-echinomycin bisintercalation complexes: rates and processes of base-pair opening. Biochemistry. 1992;31:1407–1415. doi: 10.1021/bi00120a017. [DOI] [PubMed] [Google Scholar]
- 6.Addess K.J., Sinsheimer J.S., Feigon J. Solution structure of a complex between [N-MeCys3,N-MeCys7]TANDEM and [d(GATATC)]2. Biochemistry. 1993;32:2498–2508. doi: 10.1021/bi00061a006. [DOI] [PubMed] [Google Scholar]
- 7.Chen H., Patel D.J. Solution structure of a quinomycin bisintercalator-DNA complex. J. Mol. Biol. 1995;246:164–179. doi: 10.1006/jmbi.1994.0074. [DOI] [PubMed] [Google Scholar]
- 8.Wakelin L.P., Creasy T.S., Waring M.J. Equilibrium constants for the binding of an homologous series of monofunctional and bifunctional intercalating diacridines to calf thymus DNA. FEBS Lett. 1979;104:261–265. doi: 10.1016/0014-5793(79)80828-5. [DOI] [PubMed] [Google Scholar]
- 9.Wirth M., Buchardt O., Koch T., Nielsen P.E., Norden B. Interactions between DNA and mono(aminoacridines), bis(aminoacridines), tris(aminoacridines), tetrakis(aminoacridines), and hexakis(aminoacridines) - a linear and circular-dichroism, electric orientation relaxation, viscometry, and equilibrium study. J. Am. Chem. Soc. 1988;110:932–939. [Google Scholar]
- 10.Gaugain B., Barbet J., Capelle N., Roques B.P., Le Pecq J.B. DNA Bifunctional intercalators. 2. Fluorescence properties and DNA binding interaction of an ethidium homodimer and an acridine ethidium heterodimer. Biochemistry. 1978;17:5078–5088. doi: 10.1021/bi00617a002. [DOI] [PubMed] [Google Scholar]
- 11.Gaugain B., Barbet J., Oberlin R., Roques B.P., Le Pecq J.B. DNA bifunctional intercalators. I. Synthesis and conformational properties of an ethidium homodimer and of an acridine ethidium heterodimer. Biochemistry. 1978;17:5071–5078. doi: 10.1021/bi00617a001. [DOI] [PubMed] [Google Scholar]
- 12.Jacobsen J.P., Pedersen J.B., Hansen L.F., Wemmer D.E. Site selective bis-intercalation of a homodimeric thiazole orange dye in DNA oligonucleotides. Nucleic Acids Res. 1995;23:753–760. doi: 10.1093/nar/23.5.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailly C., Brana M., Waring M.J. Sequence-selective intercalation of antitumour bis-naphthalimides into DNA - Evidence for an approach via the major groove. Eur. J. Biochem. 1996;240:195–208. doi: 10.1111/j.1432-1033.1996.0195h.x. [DOI] [PubMed] [Google Scholar]
- 14.Baruah H., Barry C.G., Bierbach U. Platinum-intercalator conjugates: from DNA-targeted cisplatin derivatives to adenine binding complexes as potential modulators of gene regulation. Curr. Topics Med. Chem. 2004;4:1537–1549. doi: 10.2174/1568026043387313. [DOI] [PubMed] [Google Scholar]
- 15.Martins E.T., Baruah H., Kramarczyk J., Saluta G., Day C.S., Kucera G.L., Bierbach U. Design, synthesis, and biological activity of a novel non-cisplatin-type platinum-acridine pharmacophore. J. Med. Chem. 2001;44:4492–4496. doi: 10.1021/jm010293m. [DOI] [PubMed] [Google Scholar]
- 16.Ackley M.C., Barry C.G., Mounce A.M., Farmer M.C., Springer B.E., Day C.S., Wright M.W., Berners-Price S.J., Hess S.M., Bierbach U. Structure-activity relationships in platinum-acridinylthiourea conjugates: effect of the thiourea nonleaving group on drug stability, nucleobase affinity, and in vitro cytotoxicity. J. Biol. Inorg. Chem. 2004;9:453–461. doi: 10.1007/s00775-004-0541-4. [DOI] [PubMed] [Google Scholar]
- 17.Hess S.M., Anderson J.G., Bierbach U. A non-crosslinking platinum-acridine hybrid agent shows enhanced cytotoxicity compared to clinical BCNU and cisplatin in glioblastoma cells. Bioorg. Med. Chem. Lett. 2005;15:443–446. doi: 10.1016/j.bmcl.2004.10.049. [DOI] [PubMed] [Google Scholar]
- 18.Hess S.M., Mounce A.M., Sequeira R.C., Augustus T.M., Ackley M.C., Bierbach U. Platinum-acridinylthiourea conjugates show cell line-specific cytotoxic enhancement in H460 lung carcinoma cells compared to cisplatin. Cancer Chemother. Pharmacol. 2005;56:337–343. doi: 10.1007/s00280-004-0987-7. [DOI] [PubMed] [Google Scholar]
- 19.Baruah H., Wright M.W., Bierbach U. Solution structural study of a DNA duplex containing the guanine-N7 adduct formed by a cytotoxic platinum-acridine hybrid agent. Biochemistry. 2005;44:6059–6070. doi: 10.1021/bi050021b. [DOI] [PubMed] [Google Scholar]
- 20.Baruah H., Day C.S., Wright M.W., Bierbach U. Metal-intercalator-mediated self-association and one-dimensional aggregation in the structure of the excised major DNA adduct of a platinum-acridine agent. J. Am. Chem. Soc. 2004;126:4492–4493. doi: 10.1021/ja038592j. [DOI] [PubMed] [Google Scholar]
- 21.Barry C.G., Baruah H., Bierbach U. Unprecedented monofunctional metalation of adenine nucleobase in guanine- and thymine-containing dinucleotide sequences by a cytotoxic platinum-acridine hybrid agent. J. Am. Chem. Soc. 2003;125:9629–9637. doi: 10.1021/ja0351443. [DOI] [PubMed] [Google Scholar]
- 22.Barry C.G., Day C.S., Bierbach U. Duplex-promoted platination of adenine-N3 in the minor groove of DNA: challenging a longstanding bioinorganic paradigm. J. Am. Chem. Soc. 2005;127:1160–1169. doi: 10.1021/ja0451620. [DOI] [PubMed] [Google Scholar]
- 23.Budiman M.E., Alexander R.W., Bierbach U. Unique base-step recognition by a platinum-acridinylthiourea conjugate leads to a DNA damage profile complementary to that of the anticancer drug cisplatin. Biochemistry. 2004;43:8560–8567. doi: 10.1021/bi049415d. [DOI] [PubMed] [Google Scholar]
- 24.Augustus T.M., Anderson J., Hess S.M., Bierbach U. Bis(acridinylthiourea)platinum(II) complexes: synthesis, DNA affinity, and biological activity in glioblastoma cells. Bioorg. Med. Chem. Lett. 2003;13:855–858. doi: 10.1016/s0960-894x(02)01078-8. [DOI] [PubMed] [Google Scholar]
- 25.Roeder R.G. The complexities of eukaryotic transcription initiation: regulation of preinitiation complex assembly. Trends Biochem. Sci. 1991;16:402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- 26.Jenkins T.C. Optical absorbance and fluorescence techniques for measuring DNA drug interactions. In: Fox K.R., editor. Drug DNA Interaction Protocols. vol. 90. Totowa: Humana Press; 1997. pp. 195–218. [DOI] [PubMed] [Google Scholar]
- 27.Baruah H., Rector C.L., Monnier S.M., Bierbach U. Mechanism of action of non-cisplatin type DNA-targeted platinum anticancer agents: DNA interactions of novel acridinylthioureas and their platinum conjugates. Biochem. Pharmacol. 2002;64:191–200. doi: 10.1016/s0006-2952(02)01107-3. [DOI] [PubMed] [Google Scholar]
- 28.Bellon S.F., Coleman J.H., Lippard S.J. DNA unwinding produced by site-specific intrastrand cross-links of the antitumor drug cis-diamminedichloroplatinum(II) Biochemistry. 1991;30:8026–8035. doi: 10.1021/bi00246a021. [DOI] [PubMed] [Google Scholar]
- 29.Berger S., Braun S. 200 and More NMR Experiments. Weinheim: Wiley-VCH; 2004. [Google Scholar]
- 30.Baruah H., Bierbach U. Unusual intercalation of acridin-9-ylthiourea into the 5′-GA/TC DNA base step from the minor groove: implications for the covalent DNA adduct profile of a novel platinum-intercalator conjugate. Nucleic Acids Res. 2003;31:4138–4146. doi: 10.1093/nar/gkg465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baguley B.C., Denny W.A., Atwell G.J., Cain B.F. Potential antitumor agents. 34. Quantitative relationships between DNA binding and molecular structure for 9-anilinoacridines substituted in the anilino ring. J. Med. Chem. 1981;24:170–177. doi: 10.1021/jm00134a009. [DOI] [PubMed] [Google Scholar]
- 32.Neidle S. Nucleic Acid Structure and Recognition. NY: Oxford University Press; 2002. pp. 89–138. [Google Scholar]
- 33.SantaLucia J., Allawi H.T., Seneviratne A. Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry. 1996;35:3555–3562. doi: 10.1021/bi951907q. [DOI] [PubMed] [Google Scholar]
- 34.Baruah H., Bierbach U. Biophysical characterization and molecular modeling of the coordinative-intercalative DNA monoadduct of a platinum-acridinylthiourea agent in a site-specifically modified dodecamer. J. Biol. Inorg. Chem. 2004;9:335–344. doi: 10.1007/s00775-004-0534-3. [DOI] [PubMed] [Google Scholar]
- 35.Kaushik M., Kukreti R., Grover D., Brahmachari S.K., Kukreti S. Hairpin-duplex equilibrium reflected in the A B transition in an undecamer quasi-palindrome present in the locus control region of the human beta-globin gene cluster. Nucleic Acids Res. 2003;31:6904–6915. doi: 10.1093/nar/gkg887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray D.M., Ratliff R.L., Vaughan M.R. Circular dichroism spectroscopy of DNA. Methods Enzymol. 1992;211:389–406. doi: 10.1016/0076-6879(92)11021-a. [DOI] [PubMed] [Google Scholar]
- 37.Schipper P.E., Norden B., Tjerneld F. Determination of binding geometry of DNA-adduct systems through induced circular-dichroism. Chem. Phys. Lett. 1980;70:17–21. [Google Scholar]
- 38.Keck M.V., Lippard S.J. Unwinding of supercoiled DNA by platinum ethidium and related complexes. J. Am. Chem. Soc. 1992;114:3386–3390. [Google Scholar]
- 39.Wakelin L.P., Bu X., Eleftheriou A., Parmar A., Hayek C., Stewart B.W. Bisintercalating threading diacridines: relationships between DNA binding, cytotoxicity, and cell cycle arrest. J. Med. Chem. 2003;46:5790–5802. doi: 10.1021/jm030253d. [DOI] [PubMed] [Google Scholar]
- 40.Lian L.-Y., Roberts G.C.K. Effects of chemical exchange on NMR spectra. In: Roberts G.C.K., editor. NMR of Macromolecules: A Practical Approach. NY: Oxford University Press; 1995. pp. 153–182. [Google Scholar]
- 41.Evans J.N.S. Biomolecular NMR Spectroscopy. NY: Oxford University Press; 1995. pp. 343–391. [Google Scholar]
- 42.Wilson W.D., Li Y., Veal J.M. NMR analysis of reversible nucleic acid small molecule complexes. In: Hurley L.H., editor. Advances in DNA Sequence Specific Agents. Vol. 1. NY: JAI Press; 1992. pp. 89–165. [Google Scholar]
- 43.McGhee J.D., von Hippel P.H. Theoretical aspects of DNA–protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 44.Antonini I., Polucci P., Magnano A., Gatto B., Palumbo M., Menta E., Pescalli N., Martelli S. Design, synthesis, and biological properties of new bis(acridine-4-carboxamides) as anticancer agents. J. Med. Chem. 2003;46:3109–3115. doi: 10.1021/jm030820x. [DOI] [PubMed] [Google Scholar]
- 45.Dai J., Punchihewa C., Mistry P., Ooi A.T., Yang D. Novel DNA bis-intercalation by MLN944, a potent clinical bisphenazine anticancer drug. J. Biol. Chem. 2004;279:46096–46103. doi: 10.1074/jbc.M404053200. [DOI] [PubMed] [Google Scholar]
- 46.Budiman M.E., Bierbach U., Alexander R.W. DNA minor groove adducts formed by a platinum−acridine conjugate inhibit association of TATA-binding protein with its cognate sequence. Biochemistry. 2005;44:11262–11268. doi: 10.1021/bi050745n. [DOI] [PubMed] [Google Scholar]
- 47.Juo Z.S., Chiu T.K., Leiberman P.M., Baikalov I., Berk A.J., Dickerson R.E. How proteins recognize the TATA box. J. Mol. Biol. 1996;261:239–254. doi: 10.1006/jmbi.1996.0456. [DOI] [PubMed] [Google Scholar]
- 48.Patikoglou G.A., Kim J.L., Sun L., Yang S.H., Kodadek T., Burley S.K. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999;13:3217–3230. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fechter E.J., Olenyuk B., Dervan P.B. Design of a sequence-specific DNA bisintercalator. Angew. Chem. Int. Ed. Engl. 2004;43:3591–3594. doi: 10.1002/anie.200454231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.