Abstract
Studies of the genetic network that controls the Caulobacter cell cycle have identified a response regulator, CtrA, that controls, directly or indirectly, one-quarter of the 553 cell cycle-regulated genes. We have performed in vivo genomic binding site analysis of the CtrA protein to identify which of these genes have regulatory regions bound directly by CtrA. By combining these data with previous global analysis of cell cycle transcription patterns and gene expression profiles of mutant ctrA strains, we have determined that CtrA directly regulates at least 95 genes. The total group of CtrA-regulated genes includes those involved in polar morphogenesis, DNA replication initiation, DNA methylation, cell division, and cell wall metabolism. Also among the genes in this notably large regulon are 14 that encode regulatory proteins, including 10 two-component signal transduction regulatory proteins. Identification of additional regulatory genes activated by CtrA will serve to directly connect new regulatory modules to the network controlling cell cycle progression.
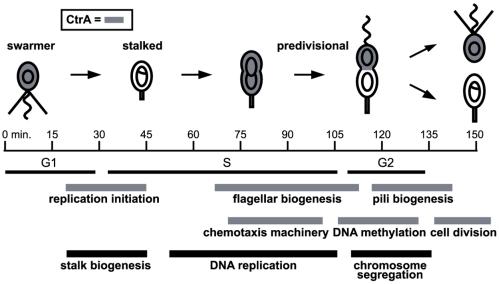
Regulated transcription plays a critical role in the cell cycle progression of nearly all organisms. Several recent studies have used DNA microarrays to systematically identify cell cycle-regulated mRNAs in yeast and human cells (1–3). Similar global analysis of cell cycle gene expression has also been done for bacterial cells, using the Gram-negative bacterium Caulobacter crescentus as a model system (4). Cell cycle progression in Caulobacter is marked by a series of morphological transitions leading to an asymmetric division that produces progeny cells with distinct cell fates (Fig. 1). G1-phase swarmer cells can be isolated easily and allowed to proceed synchronously through the cell cycle, facilitating temporal dissection of cell cycle events. Global analysis of expression profiles during the Caulobacter cell cycle showed that the mRNA levels of approximately 19% of the genes (553 of 2,966 assayed) are cell cycle-regulated (4). These temporal expression patterns revealed that cell cycle-regulated genes are maximally induced immediately before or coincident with the time of execution of the cell cycle event in which they participate, and genes that encode subunits of molecular complexes or that function in the same cellular process are coexpressed.
Figure 1.
Temporal coordination of Caulobacter cell cycle events. The swarmer cell has a nonreplicating chromosome and a polar flagellum and pili. At the swarmer-to-stalked cell transition, the pili and the flagellum are lost, and a stalk is formed at that same pole, coincident with the initiation of DNA replication. Construction of a new flagellum at the pole opposite the stalk occurs during S phase. The cell then divides asymmetrically, yielding two distinct daughter cells: a stalked cell that initiates a new round of DNA replication and a smaller swarmer cell that cannot replicate its chromosome until after it differentiates into a stalked cell. Timing of several key cell cycle-regulated events is indicated by the black and gray bars below. CtrA is present in cell types shaded gray and controls genes or events involved in the cell cycle processes with gray timing bars.
Identification of the transcription factors directly involved in controlling the temporal patterns of expression is critical to the mapping of global regulatory networks. A single regulatory factor, the CtrA response regulator (5), controls, directly or indirectly, at least 25% of the cell cycle-regulated genes (4). Response regulators and histidine kinases, the elements of two-component signal transduction systems, mediate a wide range of adaptive responses in bacteria. CtrA, like many response regulators, has a DNA-binding domain. The phosphorylated form of CtrA has been shown to act as a transcription factor that directly regulates genes involved in cell division, DNA methylation, flagellar biogenesis, and pili biogenesis (5–8); it has also been shown to repress initiation of DNA replication by binding to five sites in the origin of replication (9). The cell cycle-regulated expression of these CtrA target genes is effected by changes in active CtrA levels during the cell cycle (Fig. 1). CtrA∼P is present at relatively high levels in G1 swarmer cells before being dephosphorylated and rapidly proteolyzed at the swarmer-to-stalked cell transition (10). CtrA then accumulates in stalked and predivisional cells, a process accelerated by positive feedback on one of its two promoters (11). CtrA is selectively proteolyzed from the stalked half of the predivisional cell just before cell division, relieving CtrA's inhibition of DNA replication initiation in the stalked cell and maintaining replication inhibition in the progeny swarmer cell. Phosphorylation of CtrA is regulated by a phosphorylation cascade that is only partially characterized (12–14).
Global expression profiling of strains bearing temperature-sensitive alleles of ctrA showed that at least 144 of the 553 cell cycle-regulated genes in Caulobacter depend, either directly or indirectly, on CtrA for normal expression (4). Searching the upstream regulatory regions of these genes for consensus CtrA-binding sites (TTAA-N7-TTAAC) can help to distinguish between direct and indirect targets but is not definitive for two reasons. First, the mere presence of a CtrA consensus-binding site does not necessarily mean the site is bound by CtrA in vivo. Second, CtrA may bind in vivo to sites that do not conform exactly to the defined consensus site. To discriminate direct from indirect CtrA targets, we have used location analysis (15, 16) to map the in vivo binding sites of CtrA on a genome-wide level. We combined these data with expression profiling of wild-type (4) and ctrA mutant strains to identify 55 genes, of which 21 are the first genes in a potential operon, that are part of the CtrA regulon. This work lays the foundation for identification of the direct interactions in the transcriptional network governing cell cycle progression in a bacterium.
Materials and Methods
Formaldehyde Crosslinking and Immunoprecipitation (IP).
CB15N was grown in M2G minimal medium at 30°C to an optical density of 0.3–0.4 at 660 nm. Formaldehyde crosslinking and subsequent IP were done essentially as described in refs. 16 and 17, with minor modifications; protocols are published as supporting information on the PNAS web site, www.pnas.org.
DNA Microarrays.
Microarrays used in this study contained PCR amplicons for 3,767 predicted Caulobacter ORFs (18) and 1,615 intergenic regions. To detect candidate regulatory regions, we selected intergenic regions that satisfied three criteria: (i) no overlap with any of the predicted ORFs in the C. crescentus genome; (ii) greater than 70 bp in length; and (iii) upstream of one or both of the flanking ORFs. In the case of two divergent but overlapping genes, we amplified a 200- to 250-bp fragment centered around their overlapping regions and included these regions as well. Primer lists and protocols used for microarray construction are available at http://caulobacter.stanford.edu/CtrAIP.
Microarray Analysis of Genomic DNA.
Fluorescent labeling of genomic DNA, hybridization, and array scanning was done according to the protocol at http://microarrays.org/protocols.html. Array scanning and image processing were done as in ref. 4.
Microarray Analysis of the ctrA401ts Mutant.
Cultures of wild-type (CB15N) and ctrA401ts were grown in PYE at 28°C to an optical density of 0.1 at 660 nm and then were shifted to the restrictive temperature of 37°C. RNA was harvested from each culture at 0, 2, and 4 h after temperature shift and prepared for hybridization as described (4). Each sample was compared with a common reference derived from RNA of a logarithmic-phase culture of wild-type cells. The entire procedure was repeated twice and results averaged. We then calculated the ratios: ctrAts 0 h/wild type (wt) 0 h; ctrAts 2 h/wt 2 h; and ctrAts 4 h/wt 4 h. Genes that showed at least a 1.75-fold change in any of these three comparisons were selected as CtrA-dependent. The complete data set is published as supporting information on the PNAS web site.
Results
Mapping CtrA-Binding Sites.
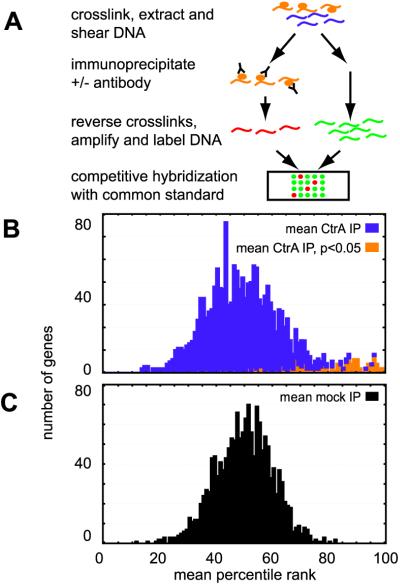
To identify the in vivo binding sites for CtrA, we used a recently developed genome-wide location analysis procedure (Fig. 2A). Briefly, DNA-binding proteins were crosslinked to their DNA targets in vivo with formaldehyde. DNA was extracted, fragmented, and then subjected to IP with (CtrA-IP) or without (mock-IP) anti-CtrA antibody. Crosslinks were then reversed, and immunoprecipitated DNA was PCR amplified, fluorescently labeled with Cy5, and compared on microarrays to a genomic DNA reference labeled with Cy3. A pre-IP sample (total) was also amplified, labeled with Cy5, and compared on a microarray to Cy3-labeled genomic DNA. Two ratios were then calculated for each intergenic region on the arrays: CtrA-IP/total and mock-IP/total. This entire procedure was repeated seven times for the CtrA-IP and eight times for the mock-IP. Genes that did not have reliable data in at least four of the seven CtrA-IP experiments and five of the eight mock-IP experiments were excluded from further analysis.
Figure 2.
Genome-wide analysis of in vivo CtrA-binding locations using IP and microarrays. (A) Schematic of the procedure for identifying intergenic regions to which CtrA is bound in vivo, as described in the text. Histograms are shown for the mean percentile ranks of the 1,549 intergenic regions in the CtrA-IP experiments (B) and in the mock-IP experiments (C). Intergenic regions enriched in the CtrA-IP experiments relative to the mock-IP experiments with a confidence level >95% are shown in orange in the CtrA-IP histogram (B).
DNA targets bound in vivo by CtrA should be enriched in the CtrA-IP sample but not the mock-IP or total DNA samples and thus should have higher CtrA-IP/total ratios than mock-IP/total ratios. Our data analysis method differs from that in previous location analysis reports (15, 16). We analyzed the data from the location analysis procedure after converting raw ratios to percentile ranks. For each repetition, the CtrA-IP/total or mock-IP/total ratios were assigned a percentile rank, with the 100th percentile being the largest. Using ranks provides two advantages over raw ratios: (i) ranks are more robust to variabilities and nonlinearities in the efficiency of crosslinking, sonication, PCR amplification, or labeling; and (ii) theoretical modeling of rank distributions is far easier. In theory, without any antibody in the mock-IP experiment, no DNA fragment should be consistently enriched. Indeed, a theoretical distribution assuming that each spot on a given array has equal probability of getting any rank fits well with the observed distribution of average percentile ranks for the mock-IP ratios (Fig. 2B), suggesting that there is virtually no bias in the mock-IP results (see supporting information on the PNAS web site). In addition, the large subpopulation of the CtrA-IP experiment (purple in Fig. 2B) also fits well with a random rank model. These observations show that addition of CtrA antibody has no effect on most DNA fragments but enriches a specific subpopulation (orange in Fig. 2B).
To systematically identify individual intergenic regions enriched by CtrA IP, we compared the percentile ranks of each intergenic fragment in the CtrA-IP and mock-IP experiments by using the Mann–Whitney test, a nonparametric statistical test that makes no assumptions about the underlying distribution of the data. By analyzing each intergenic region separately, we directly tested whether addition of the CtrA antibody led to a significant change in IP efficiency of the region. This test procedure eliminated a small number of regions that are inexplicably enriched by both the mock- and CtrA-IP procedures. See supporting information on the PNAS web site for a description of the statistical analysis.
Of 1,527 intergenic regions with valid data, 138 (shaded orange in Fig. 2B) were identified as enriched by the CtrA-IP procedure at a 95% confidence level (P < 0.05) by using the Mann–Whitney test. The raw ratios (available in supporting information) of these 138 regions showed an average of a 2.4-fold enrichment in the CtrA IP relative to the mock IP. We refer to these 138 significantly enriched intergenic regions as CtrA-bound regions. Fifty-nine of these regions are between divergent genes; in these cases, CtrA could participate in the regulation of both genes. Thus the number of genes flanking bound intergenic regions could be up to 197. The 138 CtrA-bound intergenic regions include promoters for six of the seven genes previously identified as directly controlled by CtrA (5–8). The gene not detected, fliX, has a CtrA-binding site that deviates significantly from the consensus-binding site sequence (19).
The CtrA Cell Cycle Regulon.
The temporal and spatial cell cycle regulation of active CtrA levels (Fig. 1) ensures that this protein activates or represses its target genes only at specific times during the cell cycle. Seven genes have previously been identified as directly controlled by CtrA by using in vitro footprinting analyses and expression assays of lacZ transcription fusions in ctrAts mutant strains (5–8). Further, all of these genes were shown to have cell cycle-regulated mRNAs, although maximal expression occurs at widely disparate times during the cell cycle (4). To identify the genes shown here to have flanking CtrA-bound intergenic regions that are also cell cycle-regulated and that depend on CtrA for normal expression level, we incorporated two additional genomic data sets in the analysis: (i) the 553 genes (of 2,966 assayed) that were identified as cell cycle-regulated during expression profiling of wild-type Caulobacter cells (4); and (ii) the 250 genes whose mRNA levels were determined to depend on functional CtrA for normal expression by expression profiling of a ctrA loss-of-function mutant strain, ctrA401ts (see Materials and Methods).
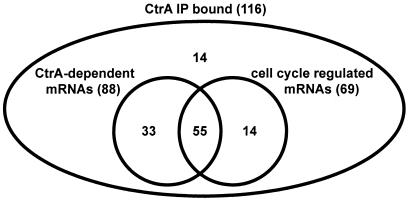
Valid microarray expression data are available from both the cell cycle profiling (4) and the ctrA mutant profiling for 116 of the 197 genes flanking CtrA-bound intergenic regions. The number of cell cycle-regulated genes and of CtrA-dependent mRNAs among these 116 genes is summarized in Fig. 3. For these 116 genes, 69 (59%) were previously identified as having cell cycle-regulated mRNAs, and 88 (76%) had mRNAs that changed significantly in response to loss of CtrA function. As some of the CtrA-bound regulatory regions fall between divergent genes and CtrA may control only one of the two genes, the percentages of cell cycle-regulated and CtrA-dependent genes, 59 and 76%, respectively, may be a conservative estimate.
Figure 3.
Intersections of genomic data sets. The diagram summarizes data for the 116 (of 197) genes whose upstream regulatory regions were enriched by the IP-based binding analysis and which had valid data from expression profiling of wild-type and ctrA mutant strains. Of 116 gene, 88 were identified as CtrA-dependent for normal expression levels, and 69 were identified as cell cycle-regulated. The 55 genes within the overlap of all three data sets are identified here as members of the CtrA cell cycle regulon, as described in the text.
Combining these three genomic data sets, we define members of the directly controlled CtrA cell cycle regulon as those genes satisfying three criteria: (i) cell cycle-regulated mRNA; (ii) mRNA level changes significantly in the ctrA loss-of-function strain; and (iii) upstream regulatory region significantly enriched in the CtrA-IP-based binding analysis. We found 55 genes that passed all three criteria (Fig. 3). These include 47 intergenic regions, with 8 regions between divergently transcribed genes. Although the 55 genes identified here are considered, with high confidence, to be directly controlled by CtrA, we note that ultimately, arbitrary thresholds were set for each of the three genomic data sets. To facilitate investigation of these data sets with more or less stringent thresholds, all data are available on the PNAS web site.
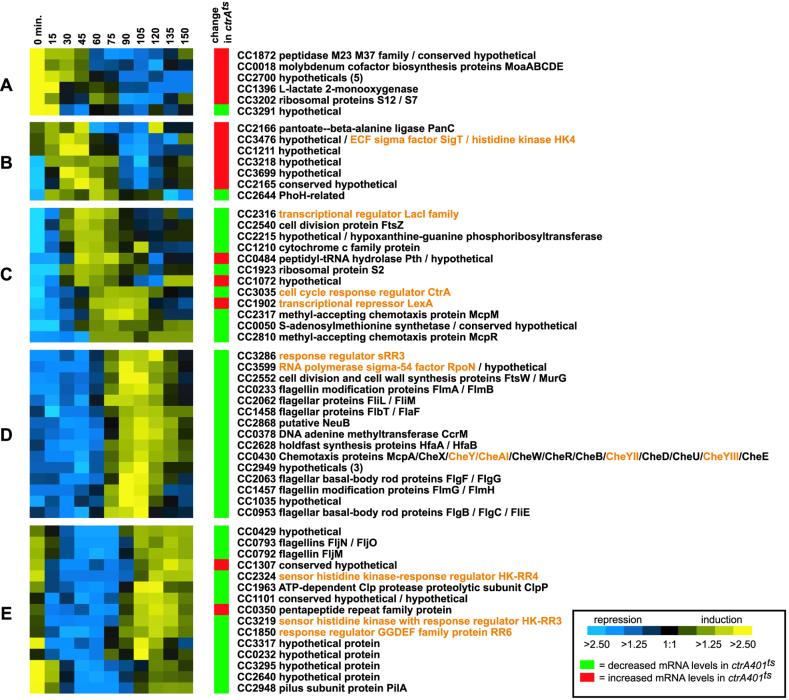
Of the 55 genes that are adjacent to an intergenic region bound by CtrA, 34 are single genes, and 21 are the first genes in potential operons (Fig. 4). Although only a fraction of the operons in the Caulobacter genome have been well characterized, for purposes of this analysis, we defined genes to be in an operon if they are read in the same direction, are temporally cotranscribed (4), and are separated by fewer than 55 bp. The majority of the genes in the operons are separated by less than 19 bp (18), and several are overlapping genes. These operons include the well characterized 12-gene chemotaxis operon (20). Thus, taking into account the genes in predicted operons, there are at least 95 genes in the CtrA cell cycle regulon. Wild-type cell cycle expression profiles and ctrA mutant expression data are shown in Fig. 4 for the 55 genes flanking the IP-enriched intergenic regions.
Figure 4.
Expression profiles for genes in the CtrA regulon. Wild-type and ctrAts expression data, along with CC number, are shown for the 55 genes satisfying all three criteria for members of the CtrA regulon (Fig. 3). Other genes potentially in the same operon are listed to the right, separated by slashes. For wild-type profiles, expression ratios of RNA from each time point have been compared with a common reference and are encoded using the scale at the bottom (4). The column labeled “change in ctrAts ” indicates, with either a green or red box, genes that were significantly decreased or increased, respectively, in response to loss of CtrA function in the ctrA401ts strain. Regulatory genes are listed in orange (see also Fig. 5).
Three sets of genes passed only two of the three criteria used for selecting the CtrA cell cycle regulon (Fig. 3). The first is a set of 33 genes that had upstream regulatory regions enriched in the CtrA-IP procedure and had CtrA-dependent mRNAs but did not meet all criteria used previously (4) to identify cell cycle-regulated genes. Some of these genes may also be regulated by an additional transcription factor or by some posttranscriptional mechanism that leads to constant mRNA levels during cell cycle progression.
The second set consists of 14 genes that were CtrA-IP-enriched and cell cycle-regulated but had mRNA levels that did not change significantly in the ctrA mutant strain. Three possible explanations for this set are: (i) the expression levels of these genes may respond to loss of CtrA but only if observed at time points other than the 2- and 4-hr time points we assayed; (ii) these genes may be regulated by additional transcription factors that masked the effects of CtrA; (iii) these genes may not be controlled by CtrA but are divergently transcribed from an actual CtrA-regulated gene and so were enriched in the IP-binding assay.
Finally, many genes were previously found to be cell cycle-regulated and CtrA-dependent but were not enriched by the CtrA-IP procedure reported here. These are likely to be genes indirectly regulated by CtrA.
Characterization of the CtrA Cell Cycle Regulon.
The wild-type expression profiles for the 55 CtrA cell cycle regulon members were subjected to hierarchical clustering, yielding five clusters, labeled Fig. 4 A–E. Of the genes in clusters D and E, 93% (28 of 30) were significantly decreased in expression in the ctrA loss-of-function strain. Expression of genes in these clusters is lowest in stalked cells when CtrA has been proteolytically cleared from the cell and then is induced in predivisional cells immediately after the rapid accumulation of active CtrA. These data suggest that the induction of genes in these clusters is driven predominantly by CtrA. For three of the genes in these two clusters (the ccrM gene encoding the CcrM DNA methyltransferase, the pilA gene encoding the major pilin subunit, and mcpA, encoding the McpA chemoreceptor), direct activation by CtrA has been verified (5, 6, 8, 21).
The majority of genes in cluster C (9 of 12) also show decreased RNA levels in response to loss of CtrA function, suggesting they are directly activated by CtrA. However, the genes in this cluster peak in expression either immediately before or coincident with the induction of ctrA itself, rather than after CtrA has accumulated, as might be expected for directly activated targets. Again, additional regulatory factors must play a role in the regulation of these genes. Alternatively, the promoters of some of these genes may be subject to more complicated regulation by CtrA. For example, this cluster includes two genes, ctrA and ftsZ, each of which is both activated and repressed by CtrA at different times during the cell cycle (7, 11).
The genes in cluster B peak in expression between 30 and 60 min of the cell cycle, approximately the time when CtrA is proteolytically cleared from the stalked cell. Six of the seven genes in this cluster also show increased mRNA levels in the ctrA loss-of-function experiment, suggesting they are directly repressed by CtrA.
The genes in cluster A show strong expression in swarmer cells when CtrA is present. However, five of six genes in this cluster are up-regulated in the ctrAts strain, suggesting that they are normally repressed by CtrA. The role of CtrA in regulating these genes is unclear.
Functional Distribution of the Genes in the CtrA Cell Cycle Regulon.
The 95 CtrA cell cycle regulon genes were classified by using the COG (Clusters of Orthologous Genes) classification scheme (18). Primary among these are five genes relating to cell division and cell wall metabolism, 14 regulatory genes, 29 genes relating to polar morphogenesis including genes for flagellar biogenesis, pili biogenesis, holdfast synthesis, and chemotaxis, and 25 genes of unknown function. Thus CtrA controls both essential cell cycle processes and nonessential polar morphogenesis processes.
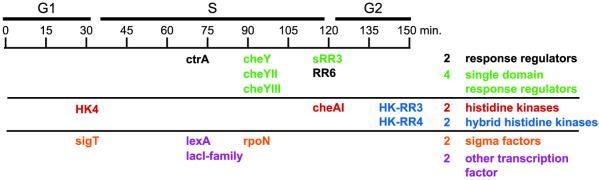
The 14 regulatory genes in the CtrA cell cycle regulon include four histidine kinases, six response regulators, two σ factors, the repressor lexA, and a LacI-family transcriptional regulator (Fig. 5). Two of these genes, encoding the histidine kinase HK4 and the σ factor SigT, appear to be in the same operon, are both induced to maximal expression during the G1–S transition, and show significant increases in mRNA expression levels in response to loss of CtrA function. These data suggest that CtrA directly represses these genes. The clearing of CtrA from stalked cells by proteolysis presumably relieves this inhibition, leads to rapid induction of these genes, and, at least in the case of the σ factor sigT, enables the expression of other genes. The 10 regulatory genes that peak in expression coincident with or after ctrA itself has peaked were all found to have significantly lower mRNA levels in the ctrAts mutant strain and so are likely to be regulatory genes directly activated by CtrA. Some of these genes, such as rpoN, cheY, cheYII, cheYIII, and cheA, play roles in coordinating polar morphogenesis events during the latter part of the cell cycle. The functions of the others, HK-RR3, HK-RR4, sRR3, and RR6, are currently unknown.
Figure 5.
Regulatory genes directly controlled by CtrA. The 14 regulatory genes in the CtrA cell cycle regulon are listed below the cell cycle timeline according to their approximate time of peak expression in wild-type cells. Numbering of the histidine kinases (HK), response regulators (RR), and hybrid histidine kinase (HK-RR) follows that in ref. 4. Hybrid histidine kinases are those kinases with a fused response regulator domain at their C-terminal ends.
CtrA-Binding Site Sequence Motifs.
Previous efforts to define a consensus-binding site for CtrA were based on in vitro protection footprinting and in vivo promoter mutagenesis for individual genes (5–8, 19). We used two programs, meme (http://meme.sdsc.edu) and bioprospector (http://bioprospector.stanford.edu), to search the intergenic sequences identified as upstream of the CtrA cell cycle regulon members for consensus-binding site motifs (as eight of the regions are between two divergent operons that are both in the CtrA cell cycle regulon, we searched 47 regions). Two consensus motifs were found: one gapped (TTAA-N7-TTAAC) and one ungapped (TTAACCAT). The most significant gapped motif corresponds to the previously published consensus of TTAA-N7-TTAAC, whereas the TTAACCAT motif suggests an extension of the previous consensus half-site for CtrA binding (see supporting information on the PNAS web site). Notably, not all intergenic regions bound by CtrA contained close matches to one of our two consensus motifs (29 of 47, or 62%, matched one or both of the two motifs), and not all intergenic regions with one of those binding motifs are bound by CtrA in our in vivo assay (of 1,440 intergenic regions that did not bind CtrA, 5% matched one of the two motifs). Further, there are intergenic regions such as the promoter for fliX whose best match to these motifs is TTAA but that still bind CtrA in footprinting assays in vitro (19). In sum, these results demonstrate that the DNA sequence-binding determinants of CtrA are still poorly understood.
Discussion
We have used location analysis to map the binding sites for the master regulator CtrA throughout the Caulobacter genome. We had shown previously that nearly 25% of all cell cycle-regulated genes show significant changes in mRNA levels in a ctrA loss-of-function strain (4). However, these data, like all expression profiling data, cannot distinguish direct from indirect targets, a critical distinction when mapping regulatory networks. In this report, we use a modified chromatin-IP procedure, often called location analysis, to experimentally identify sites in the genome bound by CtrA in vivo. Combining this binding location analysis with expression analysis of ctrA mutant strains, along with wild-type cell cycle expression profiling, we identified at least 55 genes (with an additional 40 genes in potential operons) that are members of the CtrA cell cycle regulon (Fig. 4). The function of nearly half of these genes is unknown. The remainder have known or predicted functions in a wide range of cellular processes but generally are either genes required for polar morphogenesis or genes involved in essential cell cycle processes.
Polar Morphogenesis.
Flagellar synthesis is tightly coordinated with cell cycle progression in Caulobacter and involves approximately 40 genes organized in a four-tier transcriptional cascade. ctrA is a class I flagellar gene that sits at the top of the cascade. CtrA is known to activate class II genes, which encode early structural components of the flagellum as well as regulatory genes required for transcription of later flagellar genes. We found that CtrA directly regulates several of the class II operons for which we have reliable data: flgBC, fliE, fliLM, and rpoN. Also included in the CtrA cell cycle regulon are several class III genes: flmAB, flmGH, flbT, flaF, and flgFG, as well as the class IV flagellin cluster fljMNO. According to the previous model of flagellar transcriptional control, CtrA should not be needed for activation of genes beyond the class II stage. Some overlap is to be expected, because at least three class II promoters are transcribed divergently from the same regulatory region as a class III promoter. However, recent reports have shown that the conventional model of the flagellar cascade may be oversimplified. For example, on the basis of the order of assembly, flgBC and fliE are expected to be Class III genes but are transcribed with Class II genes (22). Also, mutations in the flgBC and fliE locus do not prevent class III gene transcription, as would be expected for class II genes. Our finding that the pattern of CtrA binding is inconsistent with the conventional flagellar hierarchy is further evidence that flagellar gene regulation is more complex than previously thought.
The chemotaxis apparatus and pili are both spatially and temporally associated with the flagellum. Genes associated with both processes are expressed in the predivisional cell and, like the flagellar genes, were also found to be directly activated by CtrA. By activating flagellar, chemotactic, and pilus assembly and repressing chromosome replication, CtrA plays a central role in establishing the functionality of the daughter swarmer cell.
Essential Cell Cycle Processes.
CtrA activates critical genes for three core cell cycle processes that occur late in the cell cycle: DNA methylation, cell wall remodeling, and cell division. Consistent with previous results (5, 8), we found that CtrA directly controls expression of the essential gene ccrM, encoding an adenine DNA methyltransferase, late in the cell cycle. We also found that CtrA directly activates expression of the gene encoding S-adenosylmethionine (SAM) synthetase (CC0050), the primary enzyme responsible for production of SAM, the substrate used by CcrM for methylation of DNA. The expression of the SAM synthetase gene peaks immediately before expression of ccrM (4). Thus, CtrA may play a critical role in the timing and ordering of expression of multiple genes involved in DNA methylation.
Our results also confirm previous reports that CtrA directly controls the ftsZ gene, a tubulin-like GTPase critical for cell division in bacteria (7). In addition to ftsZ, CtrA also directly controls several other cell cycle-regulated genes, ftsQ, ftsA, and ftsW (divB), that are among the core set of genes required for initiation and progression of cell division. The regulatory region for ftsQ and ftsA, which are expressed predominantly as a single dicistronic message (23), is contained entirely within an upstream gene, ddlA. Thus, direct binding of CtrA with this regulatory region led to IP enrichment of the ddlA gene rather than an intergenic region. Cell wall remodeling is an inherent part of the cell division process. The cell division gene ftsW, activated by CtrA in Caulobacter, localizes in Escherichia coli to the cell division site and is thought to couple cell division to septal-specific peptidoglycan synthesis (24). CtrA also activates murG, which encodes a peptidoglycan biosynthesis enzyme.
CtrA regulates expression of two genes encoding, or predicted to encode, proteases: a peptidase (CC1872) and the catalytic domain of the ATP-dependent Clp protease, ClpP. The clpP gene has previously been shown to be essential for growth in Caulobacter and to be required for cell cycle-regulated proteolysis of CtrA itself (25). Our data suggest that CtrA, after accumulating to sufficiently high levels in late predivisional cells or in swarmer cells, may trigger its own destruction by activating expression and production of the ClpP protease.
CtrA has been shown previously to bind to five distinct sites in the origin of replication to repress replication initiation (9). Four partially overlapping regions of the minimal 680-bp origin of replication were represented on the microarrays used here. These four regions had the four highest mean percentile ranks in the CtrA-IP experiments, likely representing the strong and extensive binding of CtrA to this region of the chromosome.
Regulatory Genes.
Among the members of the CtrA cell cycle regulon are 14 genes encoding regulatory proteins, including members of the two component signal transduction family that likely regulate additional genetic modules. A recent report, using a IP-based binding analysis similar to that used here, analyzed 12 cell cycle regulators in the yeast Saccharomyces cerevisiae and found that each of these regulators activates several of the other regulators being investigated (26). That report further showed that these genes form a serially connected loop of activators that may control the oscillatory behavior of the cell cycle. The CtrA-controlled regulatory genes identified here are probably also part of a serially connected transcriptional network.
The genes identified here as directly controlled by CtrA extend the CtrA cell cycle regulon to a surprisingly large number of members. When we originally discovered that 144 of the 553 cell cycle-regulated genes were directly or indirectly controlled by CtrA (4), only seven of these were known from earlier work to be directly controlled by CtrA. Further, the complexity of the CtrA regulon is increased by the discovery that operons encoding at least 10 two-component signal transduction proteins are directly controlled by CtrA at specific times in the cell cycle. This discovery will now allow a dissection of the genetic subroutines controlled by each of these additional regulators, providing a genetic linkage among the regulatory modules that participate in cell cycle progression.
Supplementary Material
Acknowledgments
We thank Maliwan Meewan for help in constructing microarrays and Peter Roy for valuable discussions about statistical analysis. This work was supported by National Institutes of Health (NIH) Grant GM32506/5120 M2 (to L.S.), Office of Naval ResearchGrant N00014-99-1-0563-00002 and Defense Advanced Research Planning Agency Grant MDA972-00-1-0032 (to H.H.M.), a Howard Hughes Medical Institute Predoctoral Fellowship (to M.T.L.), and NIH Grant 2T32GM07365 to the Medical Scientist Training Program (S.L.C.).
Abbreviation
- IP
immunoprecipitation
References
- 1.Cho R J, Huang M, Campbell M J, Dong H, Steinmetz L, Sapinoso L, Hampton G, Elledge S J, Davis R W, Lockhart D J. Nat Genet. 2001;27:48–54. doi: 10.1038/83751. [DOI] [PubMed] [Google Scholar]
- 2.Cho R J, Campbell M J, Winzeler E A, Steinmetz L, Conway A, Wodicka L, Wolfsberg T G, Gabrielian A E, Landsman D, Lockhart D J, Davis R W. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 3.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laub M T, McAdams H H, Feldblyum T, Fraser C M, Shapiro L. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- 5.Quon K C, Marczynski G T, Shapiro L. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 6.Skerker J M, Shapiro L. EMBO J. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly A J, Sackett M J, Din N, Quardokus E, Brun Y V. Genes Dev. 1998;12:880–893. doi: 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reisenauer A, Quon K, Shapiro L. J Bacteriol. 1999;181:2430–2439. doi: 10.1128/jb.181.8.2430-2439.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quon K C, Yang B, Domian I J, Shapiro L, Marczynski G T. Proc Natl Acad Sci USA. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domian I J, Quon K C, Shapiro L. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 11.Domian I J, Reisenauer A, Shapiro L. Proc Natl Acad Sci USA. 1999;96:6648–6653. doi: 10.1073/pnas.96.12.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hung D, McAdams H, Shapiro L. In: Prokaryotic Development. Brun Y V, Shimkets L J, editors. Washington, DC: Am. Soc. Microbiol. Press; 2000. pp. 361–378. [Google Scholar]
- 13.Ohta N, Grebe T W, Newton A. In: Prokaryotic Development. Brun Y V, Shimkets L J, editors. Washington, DC: Am. Soc. Microbiol. Press; 2000. pp. 341–359. [Google Scholar]
- 14.Jacobs C, Domian I J, Maddock J R, Shapiro L. Cell. 1999;97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- 15.Iyer V R, Horak C E, Scafe C S, Botstein D, Snyder M, Brown P O. Nature (London) 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 16.Ren B, Robert F, Wyrick J J, Aparicio O, Jennings E G, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 17.Quisel J D, Lin D C, Grossman A D. Mol Cell. 1999;4:665–672. doi: 10.1016/s1097-2765(00)80377-9. [DOI] [PubMed] [Google Scholar]
- 18.Nierman W C, Feldblyum T V, Laub M T, Paulsen I T, Nelson K E, Eisen J, Heidelberg J F, Alley M R K, Ohta N, Maddock J R, et al. Proc Natl Acad Sci USA. 2001;98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohr C D, MacKichan J K, Shapiro L. J Bacteriol. 1998;180:2175–2185. doi: 10.1128/jb.180.8.2175-2185.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai J W, Alley M R. J Bacteriol. 2000;182:504–507. doi: 10.1128/jb.182.2.504-507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones S E, Ferguson N L, Alley M R. Microbiology. 2001;147:949–958. doi: 10.1099/00221287-147-4-949. [DOI] [PubMed] [Google Scholar]
- 22.Boyd C H, Gober J W. J Bacteriol. 2001;183:725–735. doi: 10.1128/JB.183.2.725-735.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sackett M J, Kelly A J, Brun Y V. Mol Microbiol. 1998;28:421–434. doi: 10.1046/j.1365-2958.1998.00753.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Khattar M K, Donachie W D, Lutkenhaus J. J Bacteriol. 1998;180:2810–2816. doi: 10.1128/jb.180.11.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenal U, Fuchs T. EMBO J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon I, Barnett J, Hannett N, Harbison C T, Rinaldi N J, Volkert T L, Wyrick J J, Zeitlinger J, Gifford D K, Jaakkola T S, Young R A. Cell. 2001;106:697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.