Abstract
The Upf proteins are essential for nonsense-mediated mRNA decay (NMD). They have also been implicated in the modulation of translational fidelity at viral frameshift signals and premature termination codons. How these factors function in both mRNA turnover and translational control remains unclear. In this study, mono- and bicistronic reporter systems were used in the yeast Saccharomyces cerevisae to differentiate between effects at the levels of mRNA turnover and those at the level of translation. We confirm that upfΔ mutants do not affect programmed frameshifting, and show that this is also true for mutant forms of eIF1/Sui1p. Further, bicistronic reporters did not detect defects in translational readthrough due to deletion of the UPF genes, suggesting that their function in termination is not as general a phenomenon as was previously believed. The demonstration that upf sui1 double mutants are synthetically lethal demonstrates an important functional interaction between the NMD and translation initiation pathway.
Keywords: mRNA surveillance, frameshifting, termination, initiation, synthetic lethality
INTRODUCTION
Eukaryotic cells utilize numerous posttranscriptional mechanisms to regulate the fidelity of gene expression. Such quality control pathways are initiated during transcription, continue through pre-mRNA processing and export, and also affect the translation of the targeted transcripts (Maquat and Carmichael 2001). The nonsense-mediated decay (NMD) pathway functions to recognize and rapidly degrade transcripts containing premature termination codons (PTC) in their open reading frames (ORF) (Hilleren and Parker 1999; Gonzalez et al. 2001; Wagner and Lykke-Andersen 2002). Because faithful recognition of a PTC is essential for triggering the rapid removal of such mRNAs, this “mRNA surveillance” pathway represents a nexus between the cell’s machinery for mRNA turnover and translational fidelity (Ruiz-Echevarria et al. 1998a).
Molecular genetic analyses utilizing the yeast Saccharomyces cerevisiae have implicated a number of trans-acting factors in this mRNA surveillance pathway. These include the polypeptide release factors (eRF1 and eRF3), the Upf proteins (Upf1p, Upf2p, Upf3p), the RNA binding protein Hrp1p, the DEAD box helicase rRNA processing factor Dbp2p, eIF1 (Sui1p/Mof2p), the Prt1p subunit of eIF3, the decapping complex Dcp1p/2p, the 5′ → 3′exoribonuclease Xrn1p, and to a lesser degree, the cytoplasmic exosome (Leeds et al. 1991; Muhlrad and Parker 1994; Cui et al. 1995, 1998b; He and Jacobson 1995; Lee and Culbertson 1995; Dunckley and Parker 1999; Welch and Jacobson 1999; Gonzalez et al. 2000; Bond et al. 2001; Mitchell and Tollervey 2003; Takahashi et al. 2003). Whereas the protein products of the UPF1, UPF2/NMD2, and UPF3 genes are essential for the activity of NMD, these loci were initially identified in yeast by a genetic screen for allosuppressors of the his4–38 frameshift allele (Culbertson et al. 1980; Leeds et al. 1992). In addition to frameshift suppression, certain upf mutants also confer nonsense codon suppression phenotypes (nonsense suppression) (Wilusz et al. 2001). Suppression by the upf mutants has been hypothesized to be a consequence of the combined effects of stabilization of the PTC+ mRNAs and defects in termination fidelity within mutant cells. The finding that the Upf proteins coimmunoprecipitate with the eukaryotic release factors in yeast further supported a functional role in termination fidelity (Czaplinski et al. 1998; Wang et al. 2001). In addition, it has been reported that certain upf and sui1 mutants confer defects in programmed ribosomal frameshifting (PRF) (Cui et al. 1996, 1998a; Ruiz-Echevarria et al. 1998b).
A subsequent report suggested that the Upf proteins were not involved in programmed frameshifting in yeast (Bidou et al. 2000), provoking a controversy regarding their possible roles in translational fidelity (Dinman et al. 2000; Stahl et al. 2000). In the current study, mono- and bicistronic reporter systems were used to reexamine the roles of the surveillance complex proteins Upf1p, Upf2p, Upf3p, and eIF1/Mof2p proteins in PRF: Our findings here confirm those of Bidou et al. (2000), that is, that the upf mutants do not affect frameshifting. A bicistronic dual-luciferase reporter system was also used to reexamine the issue of stop codon readthrough efficiency in the upf mutants: Data are presented suggesting that the Upf proteins do not affect this process within the sequence context tested. Interestingly, upfΔ sui1 double mutants were found to be synthetically lethal, suggesting that the combined functions of the Upf1–3 proteins and eIF1 are required for cell viability.
RESULTS AND DISCUSSION
The Upf proteins do not regulate viral PRF
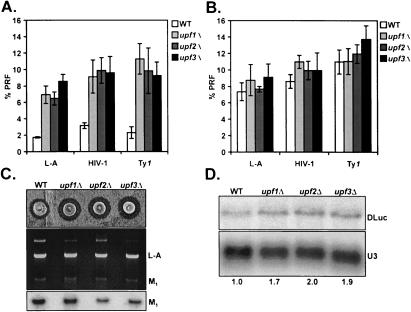
To reconcile the differences in opinion about Upf function in PRF, we monitored viral frameshifting efficiencies using several different methods in isogenic UPF gene deletion strain backgrounds. First, plasmid-borne monocistronic lacZ and bicistronic dual-luciferase reporter systems were used for quantitative, pairwise comparison of PRF efficiencies at several different viral signals in vivo (Fig. 1 ▶). As measured with the monocistronic system, PRF efficiencies (directed by L-A, HIV-1, and Ty1 frameshift signals) appeared to increase approximately threefold in each of the upfΔ mutants relative to wild type (Fig. 2A ▶). Introduction of plasmid-borne copies of the respective UPF genes complimented the L-A PRF phenotypes in each mutant, demonstrating that expression of the wild-type UPF alleles is required for normal PRF as determined by the lacZ system (data not shown). By contrast, no changes in PRF efficiency at any of these signals were observed using the bicistronic assay system (Fig. 2B ▶). The two reporter systems differ in that the bicistronic reporter is internally controlled to normalize for changes in mRNA translational efficiency and abundance (Harger and Dinman 2003) (see Materials and Methods), whereas both of these parameters may potentially affect the interpretation of results obtained using a monocistronic system. In addition, the ability to propagate the M1 satellite of the L-A virus was used as an independent means to monitor for changes in −1 PRF efficiencies in the mutants. The M1 genome encodes a secreted polypeptide toxin that kills cells not containing the satellite, resulting in a zone of growth inhibition around colonies growing over a lawn of uninfected yeast (Fig. 2C ▶). M1 propagation (killer activity) is exquisitely sensitive to changes in stoichiometry of L-A Gag and Gag-Pol proteins caused by changes in PRF (Dinman 1995; Harger et al. 2002). Therefore, propagation of the satellite, and thus killer activity, should diminish if −1 PRF is either increased or decreased in the upfΔ strains. However, no decreases in M1 maintenance or activity were observed in any of the mutants (Fig. 2C ▶). In fact, deletion of any of the UPF genes resulted in increased killer halo diameters (Fig. 2C ▶, top panel) without any apparent changes in L-A or M1 copy numbers (Fig. 2C ▶, middle and bottom panels). This apparent Ski−ype is similar to that observed upon deletion of XRN1/SKI1, the major cytoplasmic 5′→3′ exoribonuclease that is involved in degrading uncapped mRNAs (such as M1) (Masison et al. 1995), suggesting that the Upf gene products are required for optimal function of the exonuclease. The ability of the upf mutants to propagate the killer virus are consistent with the results from the dual-luciferase reporters, suggesting that the apparent increase in PRF observed with monocistronic lacZ reporters reflects something other than changes in recoding efficiency in the upfΔ strains. In sum, these data confirm the conclusions of Bidou et al. (2000), that is, that the Upf proteins are not involved in programmed ribosomal frameshifting.
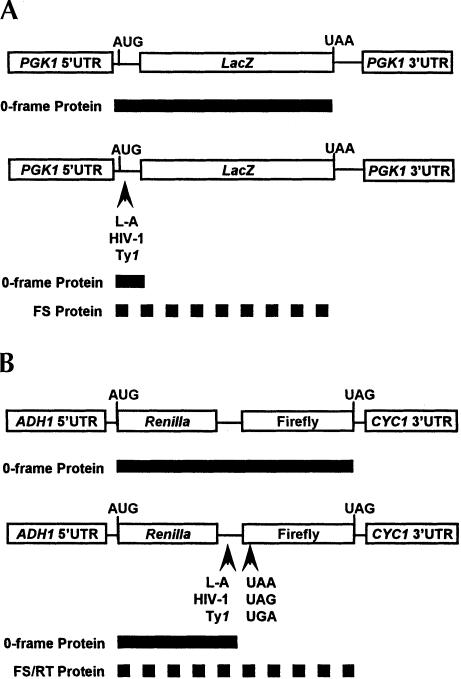
FIGURE 1.
A schematic of the lacZ monocistronic (A) and dual-luciferase bicistronic (B) in vivo reporter cassettes are shown. The 0-frame control constructs (A,B, top) contain either the lacZ or Renilla-Firefly coding regions in frame with regard to the AUG start codon. The solid bars indicate the predicted protein products of the control reporter mRNAs. The frameshift test reporters (A,B, bottom) are identical to each control construct except that the indicated viral frameshift signal sequences have been inserted between the translational start codon and the beginning of the lacZ or Firefly luciferase coding regions such that full-length reporter proteins are only produced consequent to programmed frameshift events (dashed bars). The termination readthrough reporters (B, bottom) are also identical to the control cassette except that in-frame UAA, UAG, or UGA termination codons were introduced by site-directed mutagenesis such that full-length Renilla-Firefly protein is only produced as a result of translational readthrough of the premature terminators in the 5′ end of the Firefly coding region (dashed bar).
FIGURE 2.
Programmed frameshifting efficiency and viral propagation phenotypes of the upfΔ mutants. Isogenic wild-type, upf1Δ , upf2Δ, and upf3Δmutants were transformed with the control, L-A, HIV-1, and Ty1 frameshift reporter plasmids. (A) Results from experiments using the monocistronic lacZ-based reporter system. (B) Results from experiments using the bicistronic dual-luciferase-based reporters. β-Galactosidase or dual-luciferase assays were performed and frameshifting efficiencies were calculated as described in the Materials and Methods. Mean efficiencies determined from at least three independent experiments are plotted and error bars represent the corresponding standard deviations. (C, top panel) The L-A and M1 viruses were introduced into the wild-type and mutants and viral activity assays were conducted. (Middle panel) dsRNAs were preferentially extracted from yeast, separated through a 1.2% nondenaturing agarose gel, and stained with ethidium bromide. The positions of the viral dsRNA species are indicated. Nucleic acids were transferred to a nylon membrane and hybridized to a probe specific to the M1 dsRNA (bottom panel). (D) Total RNA was isolated from cells harboring the L-A dual-luciferase frameshift reporter plasmid and subjected to Northern analysis to determine the steady-state abundance of the corresponding dual-luciferase (DLuc) mRNA in wild-type cells relative to the mutants. U3 snoRNA was used a loading control. The fold changes in DLuc mRNA steady-state abundances in mutants relative to wild-type are indicated at the bottom.
Apparent effects on frameshifting reflect reporter mRNA abundance
In addition to stabilizing PTC+ mRNAs, translational yields of certain nonsense-containing transcripts have been shown to increase upon deletion of UPF1, suggesting that one function of the Upf complex may be to inhibit translation of this class of mRNAs (Muhlrad and Parker 1999). Because the lacZ frameshift test reporters contain numerous in-frame PTCs that are only occasionally bypassed as a result of an upstream frameshift event (Fig. 1A ▶), the observed effects on PRF using the monocistronic reporters could have resulted from increased mRNA abundance and/or translational efficiency in the mutant cells. To determine whether the monocistronic frameshift reporter mRNA was regulated via the NMD pathway, its half-life was measured in wild-type and upf1Δ strains. In wild-type cells, the half-life of the L-A based-1 PRF reporter mRNA was less than 5 min, whereas deletion of UPF1 increased its half-life to 25 min (data not shown). This finding demonstrates that the monocistronic reporter is a substrate for the NMD pathway.
If the observed effect on -1 PRF in the upf1Δ mutant was only due to differences in the abundance of the two reporter mRNAs at steady state, then the apparent increase in frameshifting efficiencies should correspond to changes in the abundance of each reporter transcript between wild-type and upf1Δ cells. To test this hypothesis, β-galacto-sidase activities from wild-type and upf1Δ strains harboring the monocistronic control and L-A reporter plasmids were normalized to reporter mRNA levels as determined by nuclease protection analysis, and frameshifting efficiencies were recalculated with the adjusted values. In agreement with the half-life measurements, normalized L-A − PRF reporter mRNA abundance increased approximately threefold in the upf1Δ mutant relative to wild type (Table 1 ▶). After correction for LacZ mRNA abundance and for error in measurements, the efficiencies of L-A-directed -1 PRF in wild-type and upf1Δ were within 1.2-fold of each other (Table 1 ▶). Therefore, it is likely that the observed increases in PRF using our monocistronic system were the result of changes in reporter mRNA stability. Consistent with this, the abundance of the L-A dual-luciferase reporter mRNA harboring the L-A PRF signal was also increased approximately twofold in each of the upfΔ mutants (Fig. 2D ▶).
TABLE 1.
The effect of reporter mRNA abundance on PRF efficiency
| Strain | Reportera | β-Gal activityb | LacZ mRNA abundancec | Corrected β-Gald | % PRF correctede |
| Wild type | Control | 14.24 (± 1.14) | 1.88 | 7.57 (± 0.60) | |
| Wild type | L-A | 0.28 (± 0.04) | 0.88 | 0.31 (± 0.05) | 4.14 (± 0.74) |
| upf1Δ | Control | 15.88 (± 0.94) | 2.54 | 6.25 (± 0.37) | |
| upf1Δ | L-A | 1.14 (± 0.08) | 2.80 | 0.41 (± 0.05) | 6.52 (± 0.90) |
alacZ monocistronic reporter plasmids. Control: lacZ in-frame with AUG. L-A: contains L-A derived -1 PRF signal, lacZ in -1 frame with AUG.
bβ-Galacosidase assays were performed in triplicate on at least three separate occasions.
cAverage LacZ mRNA abundance calculated as the ratio of LacZ to 5S rRNA signal from two independent experiments.
dβ-Galacosidase activities were divided by the average LacZ mRNA abundance in each strain.
e% PRF = (corrected β-Gal L-A/corrected β-Gal control) ·100%.
Upf proteins and termination fidelity
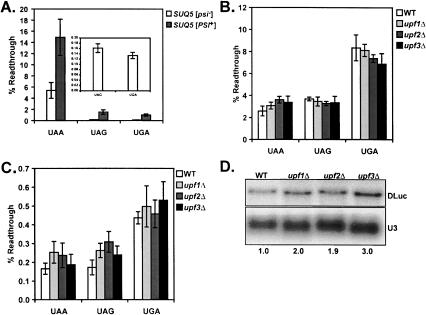
The ability of upf mutants to confer nonsense suppression phenotypes (Weng et al. 1996a, b, 1998; Maderazo et al. 2000), and their physical interactions with the termination release factors (Czaplinski et al. 1998; Wang et al. 2001), have implicated the Upf proteins in the process of termination in yeast. Use of mono- and bicistronic reporters to measure readthrough efficiency have yielded mixed results: Upf involvement in termination appears to vary with the upf mutant tested and termination codon contexts examined (Bidou et al. 2000; Keeling et al. 2004). To monitor termination readthrough efficiency in the upfΔ mutants, bicistronic dual-luciferase reporters were constructed similar to those used for assaying PRF. In-frame nonsense codons were introduced into the beginning of the firefly luciferase-coding region such that full-length dual-luciferase protein would only be produced by readthrough of the in-frame premature terminator (Fig. 1B ▶). The sensitivity of the assay system was validated using isogenic SUQ5 [psi−] and SUQ5 [PSI+] strains in a different genetic background (Fig. 3A ▶). Propagation of the cytoplasmic element [PSI+], the prion form of yeast polypeptide Release Factor 3 (eRF3), confers context-independent, global increases in stop codon readthrough (Wickner 1996). By contrast, the SUQ5 allele, which encodes a dominant ochre suppressor tRNA, promotes increased readthrough only at UAA codons (Waldron et al. 1981). The results demonstrated that the dual-luciferase system was sensitive to both the omnipotent increases in termination codon readthrough conferred by [PSI+], and the specific readthrough of the UAA codon conferred by SUQ5 (Fig. 3A ▶).
FIGURE 3.
Termination readthrough efficiency in the upfΔ mutants. (A) Isogenic [PSI+] and [psi−] yeast harboring the SUQ5 allele (a chromosomally encoded UAA stop codon suppressor tRNA) were transformed with the control, UAA, UAG, and UGA reporter plasmids (Fig. 1B ▶) and readthrough efficiencies at each terminator were determined as described in the Materials and Methods. (Inset) Readthrough efficiency in the SUQ5 [psi−] strains measured with the indicated reporters. Mean efficiencies determined from at least three independent experiments are plotted and error bars represent the corresponding standard deviations. Termination readthrough efficiency was determined in isogenic wild-type, upf1Δ, upf2Δ, and upf3Δcells as above, before and after passaging on media containing 3 mM GuCl (B and C, respectively). (D) Total RNA was isolated from wild-type, upf1Δ, upf2Δ, and upf3Δ cells containing the UAA dual-luciferase readthrough reporter plasmid and subjected to Northern analysis to determine the steady-state abundance of the DLuc mRNA in wild-type relative to the mutants as described in Figure 2D ▶.
Termination efficiency was next tested in the upfΔ mutants using the same reporters. Surprisingly, no differences in readthrough efficiency at the UAA, UAG, and UGA codons were observed between wild-type and upfΔ cells (Fig. 3B ▶). Interestingly the efficiency of termination readthrough in the SUQ5 [PSI+] strain was very similar to those observed in the wild-type and upfΔ cells (Fig. 3A ▶), suggesting that the latter strains were also propagating the prion form of eRF3. To address this possibility, the strains used in Figure 3B ▶ were cultured in medium containing guanidine hydrochloride (GuCl), which inhibits cytoplasmic inheritance of [PSI+], and readthrough efficiency was subsequently retested. Although overall efficiency of termination efficiency dropped ~10-fold with each nonsense reporter, no changes in efficiency were observed between the wild-type and mutant cells (Fig. 3C ▶). The dramatic decrease in termination readthrough observed after growth in GuCl suggests that the original strains were all [PSI+]. Further, these data demonstrate that the Upf proteins do not regulate termination efficiency at the stop codon contexts tested.
One difference between the reporter mRNAs used in the current study and other systems is the sequence context of the termination codons examined. In our system, the premature termination codons within the bicistronic reporter cassettes are preceded by an alanine codon (GCC) and followed by a histidine codon (CAC). It has been reported that sequences immediately upstream and downstream of stop codons can have a significant impact on termination efficiency (Bonetti et al. 1995), and translation termination in upf1Δ mutants was also recently shown to be context dependent (Keeling et al. 2004). Therefore, an attractive hypothesis is that these sequences may regulate termination efficiency by controlling the extent of involvement of accessory factors, such as the Upf proteins, in the termination process.
Another explanation for this result could be that the Upf protein complex did not interact with the readthrough reporter mRNAs in vivo. To test this possibility, the mRNA abundance of the UAA reporter was determined in the wild-type and upfΔ cells. Similar to the L-A frameshift reporter mRNA, the steady-state abundance of the UAA dual-luciferase mRNA was increased two- to threefold in the upfΔ mutants, suggesting that its stability is regulated in a Upf-dependent manner (Figs. 2D ▶, 3D ▶). These data indicate that the Upf proteins can regulate the mRNA stability of the UAA dual-luciferase reporter mRNA without modulating termination efficiency at the PTC during its translation.
eIF1 affects translation of the frameshift reporter mRNA but not PRF efficiency
The mof2-1 and sui1-1 mutants of eIF1/Sui1p/Mof2p were previously reported to confer apparent increases in L-A directed −1 PRF using a monocistronic lacZ reporter system (Cui et al. 1998a), and it was later reported that the steady-state abundance of certain naturally occurring and artificial PTC+ mRNAs are stabilized in the mof2-1 mutant (Cui et al. 1998b). At the time, it was postulated that eIF1 could function as part of the Upf surveillance complex in mRNA decay and translational fidelity. The SUI1 locus was originally identified in a genetic screen in yeast for mutants affecting the fidelity of translation initiation (Donahue and Cigan 1988), and it was subsequently found to encode an essential protein that functions during AUG recognition by tRNAiMet (Yoon and Donahue 1992). Moreover, recent biochemical analyses have demonstrated that the mammalian homolog of yeast eIF1/Sui1p is required for proper preinitiation complex assembly and confirmed a role for this factor in stringent start site selection (Pestova et al. 1998; Phan et al. 2001; Pestova and Kolupaeva 2002).
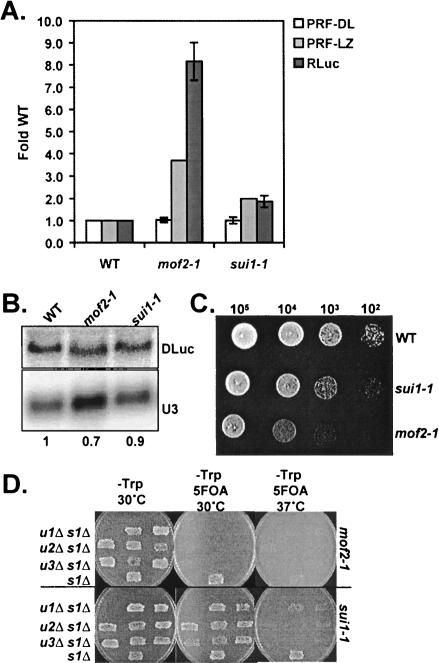
Given that the mof2-1 mutant was implicated in NMD (Cui et al. 1998b), we hypothesized that the apparent effect on frameshifting could also be explained by stabilization of the monocistronic −1 PRF reporter mRNA in the sui1-1 and mof2-1 strains. We therefore retested −1 PRF efficiency in these mutants using the dual-luciferase reporters. Similar to the upfΔ mutants, no significant increases in L-A directed -1 frameshifting were observed using the bicistronic system (Fig. 4A ▶). In contrast to upfΔ cells, however, the abundance of the dual-luciferase L-A reporter mRNA was actually slightly decreased in the mof2-1 and sui1-1 mutants relative to wild type (Fig. 4B ▶), demonstrating that the apparent defect in PRF efficiency was not attributable to increases in reporter mRNA stabilities.
FIGURE 4.
eIF1 mutants do not affect viral frameshifting, but do affect translation of reporter mRNA. (A) Isogenic wild-type, mof2-1, and sui1-1 mutants were transformed with the dual-luciferase control or L-A frameshift reporter plasmids and frameshifting efficiencies (PRF-DL) were determined. The translational efficiency of the L-A frameshift reporter mRNA, independent of PRF, was determined by normalizing Renilla luciferase (RLuc) activities to the total protein concentrations in the crude lysates used for the dual-luciferase assays and to relative mRNA abundances (determined below). Average fold changes relative to wild type are plotted for PRF-DL and Rluc from at least three independent experiments and the error bars represent the corresponding standard deviations. In addition, the average fold increases in PRF efficiency for the mof2-1 and sui1-1 mutants relative to wild type previously measured using a lacZ-based monocistronic system (PRF-LZ) were plotted (Cui et al. 1998a). (B) Total RNAs isolated from wild type and the eIF1 mutants containing the L-A dual-luciferase reporter were subjected to Northern analysis to determine the steady-state abundance of the DLuc mRNA as described in Figures 2D ▶ and 3D ▶. (C) wild-type, sui1-1, and mof2-1 mutants were grown to mid-log phase in liquid medium, and serial dilutions of cells were spotted onto plates and grown for an additional 2 d at 30°C. (D) A series of upfΔ sui1Δ double mutants were constructed by tetrad dissection of sporulated diploid yeast prepared by mating individual haploid upfΔ mutants with a sui1Δ strain. Two-upf1Δ sui1Δ , three-upf2Δ sui1Δ, and three-upf3Δ sui1Δ spore clones were isolated. One sui1Δ single mutant spore clone was used as a control. All spore clones initially contained the wild-type SUI1 gene on a low copy number plasmid with a URA3 selectable marker. The mof2-1 (top), or sui1-1 (bottom) alleles contained on low copy number plasmids with a TRP1 selectable marker were subsequently introduced into the mutants by transformation, after which growth was monitored in the presence the mutant alleles (by selecting against cells containing the wild-type SUI1-URA3 plasmid) on medium containing 5-fluoroorotic acid (5-FOA) and lacking tryptophan (-Trp) at the indicated temperatures.
The “closed-loop” model of translation posits that only mRNAs in which the 5′ and 3′ ends are bridged by a complex of trans-acting factors are translationally competent (i.e., can support multiple rounds of initiation on a single mRNA) (Tarun and Sachs 1996; Imataka et al. 1998; Otero et al. 1999). Recently, it has been demonstrated that the Upf2 protein contains conserved putative eIF4G homology (4GH) domains that are required for its activity in NMD in Schizosaccharomyces pombe (Mendell et al. 2000). In addition, the 4GH domains in hUpf2/rent2 have been shown by yeast two-hybrid analysis to mediate interactions with the human homologs of eIF4AI and Sui1p/eIF1, thus linking NMD function to factors involved in translation initiation (Mendell et al. 2000). Based on these findings, a model was proposed suggesting that activation of the Upf complex triggers remodeling of the 5′ and 3′ ends of PTC+ transcripts through competing interactions between the Upf2 4GH domains and eIF4G interacting proteins for binding to eIF4G. Thus, remodeling of the closed-loop structure of a PTC+ mRNA by the components of the surveillance complex could decrease its translational yield by down-regulating translation initiation on the transcript.
Because the mof2-1 and sui1-1 mutants have been reported to confer defects in translation initiation, we hypothesized that the apparent PRF defects in these strains could have been due to differential translation of the 0-frame and − 1 PRF reporter mRNAs in the wild-type and mutant cells. Because the Renilla coding region within the L-A dual-luciferase reporter plasmid lies upstream of the PTC-containing frameshift signal and is in frame with regard to the translational start codon (Fig. 1B ▶), we monitored Renilla luciferase specific activity (RLuc) as a measure of the translational efficiency for the entire reporter mRNA. Correction for reporter mRNA abundances revealed that RLuc specific activity from the nonsense-containing L-A reporter increased approximately nine- and twofold in the mof2-1 and sui1-1 mutants, respectively, as compared to wild-type cells (Fig. 4A ▶), whereas translation of the 0-frame control reporter decreased by twofold (data not shown). Notably, the increases in RLuc activity from the PTC+ L-A reporter agree well with the apparent increases in PRF observed using the lacZ monocistronic reporters in previous studies (Fig. 4A ▶; Dinman and Wickner 1994; Cui et al. 1998a). Moreover, the increase in translation of the PTC+ reporter mRNA correlated with the severity of the growth defect observed in the eIF1 mutants (Fig. 4C ▶). These data suggest that the eIF1/ Sui1/Mof2 protein may play a role in regulating translation of mRNAs targeted to the NMD pathway.
Mechanisms of NMD and translational fidelity are functionally redundant
A series of upfΔ sui1 double mutants were constructed to gain more insight into potential functional interactions between the Upf complex and eIF1 in the regulation of translation and stability of PTC+ mRNA in yeast. After introducing plasmid-borne copies of the mof2-1 and sui1-1 alleles into each of the double mutants, growth was monitored in the absence of the wild-type SUI1 gene at 25°C, 30°C, and 37°C. Surprisingly, deletion of the UPF genes was synthetically lethal with the eIF1 mutants, suggesting functional redundancy between these factors. The mof2-1 allele was synthetically lethal with upfΔ alleles at all of the temperatures tested (Fig. 4D ▶, top panels; data not shown), whereas the upfΔ sui1-1 double mutants were synthetically lethal only at 37°C (Fig. 4D ▶, bottom panels). The allele-specific extent of the synthetic lethality correlated with the growth phenotypes of mof2-1 and sui1-1, as well as with the observed increased translational efficiency of the frameshift reporter mRNAs in the single eIF1 mutants (Fig. 4A,C,D ▶). These data show that although the single sui1 and upf mutants significantly impair cellular function, the coordinated functions of eIF1 and the Upf complex are required for yeast cell viability.
The NMD pathway likely evolved to eliminate functionally aberrant mRNAs so that truncated polypeptides with potentially deleterious effects would not accumulate in the cell (Hentze and Kulozik 1999). Accordingly, a number of naturally occurring substrates of the NMD pathway have been reported in yeast, including transcripts prone to leaky scanning into an internal ORF or those containing short upstream ORFs (Welch and Jacobson 1999; Ruiz-Echevarria and Peltz 2000). Moreover, the efficiency of the NMD pathway has been proposed to underlie the recessive nature of many human genetic disorders resulting from nonsense mutations. However, in yeast, contrary to intuition, single or multiple deletions of the UPF genes do not confer measurable growth phenotypes under normal conditions (Weng et al. 1996a,b; Maderazo et al. 2000; Wang et al. 2001). Because both the sui1-1 and mof2-1 mutants affect the fidelity of initiation start site selection on reporter mRNAs (Yoon and Donahue 1992; Cui et al. 1998a), it is reasonable to assume that the frequency of initiation events at noncanonical sites on numerous other transcripts may be increased in these mutants as well. We hypothesize that one factor contributing to the synthetic lethality between the eIF1 and upf mutants was the combination of increased rates of “promiscuous initiation” coupled with the inability to clear the cell of the resulting PTC+ mRNAs, which would lead to accumulation of potentially toxic end products (Fig. 4D ▶). Moreover, there is a considerable amount of evidence that a large number of otherwise normal transcripts are regulated by the Upf proteins through indirect mechanisms (Lelivelt and Culbertson 1999; He et al. 2003), so it is also plausible that further deregulation of these mRNAs occurs in the upfΔ sui1 double mutants potentiates the growth defect of the single mutants.
Conclusions
Here, we have presented evidence demonstrating that neither the Upf proteins or eIF1 regulate viral PRF in yeast. These data represent an important clarification of early work by our laboratory and others’ that suggested a role for these proteins in PRF. As importantly, our analyses have identified eIF1 as a factor involved in the translational control of nonsense-containing mRNAs, and the demonstration of synthetic lethality in the upfΔ sui1 double mutants highlights a previously underappreciated functional interaction between the NMD pathway and the cellular translation initiation apparatus in yeast. Data were also presented bringing into question the generality of function of the Upf proteins in translation termination. In agreement with another recent report (Keeling et al. 2004), it appears that codon context dictates Upf involvement in this process. These data highlight the need for further study of the exact cis-acting determinants in an mRNA termination signal that are required to elicit Upf-mediated amplification of termination fidelity.
MATERIALS AND METHODS
Genetic methods and plasmids construction
Escherichia coli strains DH5α or SCS110 were used to amplify plasmids, and bacterial transformations were performed using the standard calcium chloride method. Isogenic yeast cells (Cui et al. 1998a; Wang et al. 2001) were transformed using the alkali cation method (Ito et al. 1983). Yeast cells were grown on rich (YPAD) and synthetic complete medium lacking the necessary nutrient (H-) (Wickner and Leibowitz 1976) at 30°C unless otherwise indicated. Sporulation and tetrad dissection were carried out by general methods (Rose et al. 1990). To cure yeast cells of [PSI+], cells were passaged twice on media containing 3 mM GuCl, selecting for single colonies each time. Single colonies were then used immediately for subsequent analyses. Radionucleotides were purchased from Perkin Elmer and Amersham. Restriction enzymes and in vitro transcription kits and reagents were purchased from MBI Fermentas. The plasmids containing wild-type copies of UPF1, UPF2, and UPF3 have been described previously (Wang et al. 2001).
To construct the yeast based dual-luciferase termination readthrough reporter plasmids (pYDL-UAA, pYDL-UAG, pYDL-UGA), site-directed mutagenesis was used to create in-frame UAAC, UAGC, or UGAC tetranucleotide sequences at the sixth codon in the firefly luciferase ORF using pYDL-control (pJD375) (Harger and Dinman 2003) as a template. The primers 5′-GGAGCT CATGGAAGACGCCTAACACATAAAGAAAGGC-3′ and 5′-GGCC TTTCTTTATGTGTTAGGCGTCTTCCATGAGCTCC-3′ were used to create pYDL-UAA. The primers 5′-GGAGCTCATGGAAGAC GCCTAGCACATAAAGAAAGGCC-3′ and 5′-GGCCTTTCTTTA TGTGCTAGGCGTCTTCCATGAGCTCC-3′ were used to create pYDL-UAG. The primers 5′-GGAGCTCATGGAAGACGCCTGA CACATAAAGAAAGGCC-3′ and 5′-GGCCTTTCTTTATGTGTC AGGCGTCTTCCATGAGCTCC-3′ were used to create pYDL-UGA. Reaction products were amplified in DH5α and mutant plasmids were isolated. Automated dye terminator sequencing at the UMBI sequencing facility confirmed the sequence of all plasmids.
Viral activity assays and dsRNA analysis
The L-A virus and its satellite, M1, were transferred to cells by cytoplasmic mixing (cytoduction) using a kar1-1 donor strain, and killer assays were performed as described previously (Dinman and Wickner 1994). Cytoductants were grown in YPAD to mid-log phase, harvested by centrifugation, and equal quantities were spotted onto 4.7-MB plates preseeded with a lawn of 5 × 47 diploid indicator cells. Plates were incubated at 20°C for 3–4 d and killer phenotypes were scored. Double-stranded RNA was extracted from cytoductants as described below and 5 μg/sample were separated through 1.2% TAE agarose gels. Nucleic acids were transferred to a nylon membrane by capillary action and cross-linked to the solid support by UV treatment. Immobilized nucleic acids were hybridized to a (+) sense α [32P]-CTP-labeled probe specific to M1 as previously described (Dinman and Wickner 1994).
RNA analysis
Yeast cells were grown in the appropriate media to an A595 nm of 0.5–0.8. Cells were harvested by centrifugation, washed once in dH2O, and resuspended in 0.6 ml of Smash & Grab Buffer (1% SDS, 2% Triton X-100, 100 mM NaCl, 10 mM Tris at pH 8.0, 1 mM EDTA) along with an equal volume of acid-phenol/chloroform 5:1 (Ambion). Glass beads (0.3 g of 0.5 mm; BioSpec) were added and cells were agitated continuously in a vortex minimixer for 1 min. The aqueous phase was reextracted with acid phenol/ chloroform 5:1 and extracted once more with phenol/chloroform/ IAA 25:24:1. RNA was ethanol precipitated and 15 μg/lane were separated through 1% MOPS/agarose-formaldehyde. Nucleic acids were transferred by capillary action to nylon membranes in 5× SSC and cross-linked to the membrane by UV treatment. Immobilized nucleic acids were hybridized to antisense riboprobes (α[32P]-CTP-labeled). Reactive species were visualized by phosphorimaging and quantitated using ImageQuant version 5.2 (Molecular Dynamics).
For ribonuclease protection analysis, total yeast RNA was extracted as above and 5 μg/sample were dried down and resus-pended in 19 μL of hybridization buffer (400 mM NaCl, 40 mM PIPES at pH 6.4, 1 mM EDTA, 80% Formamide). Test and control probes (1 μL each) were added to a final volume of 21 μL. Reactions were incubated at 80°C for 25 min, and then slowly brought to 50°C for overnight incubation. RNase Assay Buffer (300 mM NaCl, 10 mM Tris 7.5, 1 mM EDTA) was added to each reaction to a volume of 350 μL containing 0.3 μg of RNase A and 30 units of RNase T1 (MBI Fermentas). Reactions were incubated for 30 min at room temperature and then treated with 25 μL of a 1:4 solution of 10 mg/mL Proteinase K:10% SDS for 20 min at 39°C. Protected dsRNA fragments were extracted with an equal volume of phenol/chloroform/IAA 25:24:1. The aqueous phase was precipitated along with 20 μg of glycogen (Roche) and 2.5 volumes of 100% ethanol for 30 min at −80°C. Pellets were resuspended in 20 μL of RNA loading buffer (95% formamide, 10 mM EDTA, 0.25% xylene cyanol, 0.25% bromophenol blue), heated to 95°C for 5 min and separated through 6% polyacrylamide-7 M urea gels. Protected species were visualized by phosphorimaging and quantitated using ImageQuant version 5.2 (Molecular Dynamics).
For mRNA half-life measurements wild-type or upf1Δ yeast strains harboring the rpb1-1 allele and the necessary plasmids were grown to an A595 nm of 0.4–0.7 at room temperature (24°C) in selective media. To induce transcriptional arrest, 100-mL cultures were concentrated to 18 mL and combined with an equal volume of the same media preheated to 52°C. Cultures were incubated in a 37°C water bath with shaking. Aliquots of cells were collected at different times after the temperature shift and yeast cell pellets were frozen on liquid nitrogen. Time points were collected in duplicate and all experiments were repeated at least twice. RNAse protection was used to determine the relative abundance of each species at the indicated time. mRNA half-lives were determined by fitting [RNAt]/[RNAt0] values to double and single exponential rate equations for wild-type and upf1Δ-derived samples, respectively.
Frameshifting and nonsense suppression assays
Yeast cells harboring the dual-luciferase or lacZ reporter plasmids were grown to mid-log phase in selective media. β-Galactosidase assays were performed as described previously, and recoding efficiency was estimated by dividing the activities from cells harboring the test reporters by those from cells containing the control and multiplying by 100% [(αGal test/α Gal control) · (100%)] (Dinman and Wickner 1992). Luciferase assays were performed as described previously (Grentzmann et al. 1998; Harger and Dinman 2003). Cells (from 1–5-mL overnight cultures) were harvested by centrifugation, washed once with cold lysis buffer (PBS at pH 7.4, 1 mM PMSF) and resuspended in ~250 μL of the same buffer with 0.1–0.3 g of 0.5-mm glass beads (BioSpec). After chilling on ice, cells were disrupted by continuous agitation on a vortex mini-mixer for 4 min at 4°C. Cell lysates were clarified by centrifugation and typically 5 μL were used for measurement of the firefly/Renilla luciferase activity ratio using the Dual-Luciferase Assay system (Promega). Luminescence was measured using a TD20/20 luminometer (Turner Designs). The normalized activity ratios for each experiment were averaged and recoding efficiencies were calculated by dividing the mean activity ratios from the test reporters by those of the controls for each strain and multiplying by 100% [(Ratiotest/Ratiocontrol) · (100%)]. To determine Rluc specific activities, Renilla luciferase activities were normalized to total protein content in lysates as determined by the Bradford Assay. The specific activity of luciferase in the assay was calculated by the following equation: [(luminescence · assay volume)/(lysate concentration · lysate volume)]. All assays were performed in triplicate at least three times for each strain.
Acknowledgments
We thank Drs. Alan Jacobson, Stuart Peltz, and Daniel Masison for yeast strains and plasmids and Drs. Terri Goss Kinzy, Gary Brewer, and Samuel Gunderson for valuable feedback on this work. We also thank members of the Dinman laboratory for expert technical assistance and critical reading of the manuscript. This work was supported by grants to J.D.D. from the National Institutes of Health (GM58859 and GM62143).
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7120504.
REFERENCES
- Bidou, L., Stahl, G., Hatin, I., Namy, O., Rousset, J.P., and Farabaugh, P.J. 2000. Nonsense-mediated decay mutants do not affect programmed −1 frameshifting. RNA 6: 952–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond, A.T., Mangus, D.A., He, F., and Jacobson, A. 2001. Absence of Dbp2p alters both nonsense-mediated mRNA decay and rRNA processing. Mol. Cell. Biol. 21: 7366–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti, B., Fu, L., Moon, J., and Bedwell, D.M. 1995. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol. 251: 334–345. [DOI] [PubMed] [Google Scholar]
- Cui, Y., Hagan, K.W., Zhang, S., and Peltz, S.W. 1995. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes & Dev. 9: 423–436. [DOI] [PubMed] [Google Scholar]
- Cui, Y., Dinman, J.D., and Peltz, S.W. 1996. mof4-1 is an allele of the UPF1/IFS2 gene which affects both mRNA turnover and −1 ribosomal frameshifting efficiency. EMBO J. 15: 5726–5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, Y., Dinman, J.D., Kinzy, T.G., and Peltz, S.W. 1998a. The Mof2/ Sui1 protein is a general monitor of translational accuracy. Mol. Cell. Biol. 18: 1506–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, Y., Kinzy, T.G., Dinman, J.D., and Peltz, S.W. 1998b. Mutations in the MOF2/SUI1 gene affect both translation and nonsense-mediated mRNA decay. RNA 5: 794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson, M.R., Underbrink, K.M., and Fink, G.R. 1980. Frameshift suppression in Saccharomyces cerevisiae. II. Genetic properties of group II suppressors. Genetics 95: 833–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski, K., Ruiz-Echevarria, M.J., Paushkin, S.V., Weng, Y., Perlick, H.A., Dietz, H.C., Ter-Avanesyan, M.D., and Peltz, S.W. 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes & Dev. 12: 1665–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman, J.D. 1995. Ribosomal frameshifting in yeast viruses. Yeast 11: 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman, J.D. and Wickner, R.B. 1992. Ribosomal frameshifting efficiency and Gag/Gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 66: 3669–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1994. Translational maintenance of frame: Mutants of Saccharomyces cerevisiae with altered −1 ribosomal frameshifting efficiencies. Genetics 136: 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman, J.D., Ruiz-Echevarria, M.J., Wang, W., and Peltz, S.W. 2000. The case for the involvement of the Upf3p in programmed −1 ribosomal frameshifting. RNA 6: 1685–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue, T.F. and Cigan, A.M. 1988. Genetic selection for mutations that reduce or abolish ribosomal recognition of the HIS4 translational initiator region. Mol. Cell. Biol. 8: 2955–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley, T. and Parker, R. 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18: 5411–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, C.I., Ruiz-Echevarria, M.J., Vasudevan, S., Henry, M.F., and Peltz, S.W. 2000. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell 5: 489–499. [DOI] [PubMed] [Google Scholar]
- Gonzalez, C.I., Bhattacharya, A., Wang, W., and Peltz, S.W. 2001. Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene 274: 15–25. [DOI] [PubMed] [Google Scholar]
- Grentzmann, G., Ingram, J.A., Kelly, P.J., Gesteland, R.F., and Atkins, J.F. 1998. A dual-luciferase reporter system for studying recoding signals. RNA 4: 479–486. [PMC free article] [PubMed] [Google Scholar]
- Harger, J.W. and Dinman, J.D. 2003. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA 9: 1019–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harger, J.W., Meskauskas, A., and Dinman, J.D. 2002. An ‘integrated model’ of programmed ribosomal frameshifting and post-transcriptional surveillance. Trends Biol. Sci. 27: 448–454. [DOI] [PubMed] [Google Scholar]
- He, F. and Jacobson, A.J. 1995. Identification of a novel component of the nonsense-mediated mRNA decay pathway using an interacting protein screen. Genes & Dev. 9: 437–454. [DOI] [PubMed] [Google Scholar]
- He, F., Li, X., Spatrick, P., Casillo, R., Dong, S., and Jacobson, A. 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 12: 1439–1452. [DOI] [PubMed] [Google Scholar]
- Hentze, M.W. and Kulozik, A.E. 1999. A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96: 307–310. [DOI] [PubMed] [Google Scholar]
- Hilleren, P. and Parker, R. 1999. mRNA surveillance in eukaryotes: Kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA 5: 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka, H., Gradi, A., and Sonenberg, N. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17: 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, H., Fukuda, Y., Murata, K., and Kimura, A. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling, K.M., Lanier, J., Du, M., Salas-Marco, J., Gao, L., Kaenjak-Angeletti, A., and Bedwell, D.M. 2004. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10: 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B.S. and Culbertson, M.R. 1995. Identification of an additional gene required for eukaryotic nonsense mRNA turnover. Proc. Natl. Acad. Sci. 92: 10354–10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds, P., Peltz, S.W., Jacobson, A.J., and Culbertson, M.R. 1991. The product of the yeast UPF1 gene is required for rapid turnover on mRNAs containing a premature translational termination codon. Genes & Dev. 5: 2303–2314. [DOI] [PubMed] [Google Scholar]
- Leeds, P., Wood, J.M., Lee, B.-S., and Culbertson, M.R. 1992. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 2165–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelivelt, M.J. and Culbertson, M.R. 1999. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol. Cell. Biol. 19: 6710–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maderazo, A.B., He, F., Mangus, D.A., and Jacobson, A. 2000. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol. Cell. Biol. 20: 4591–4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison, D.C., Blanc, A., Ribas, J.C., Carroll, K., Sonenberg, N., and Wickner, R.B. 1995. Decoying the cap- mRNA degradation system by a double-stranded RNA virus and poly(A)- mRNA surveillance by a yeast antiviral system. Mol. Cell. Biol. 15: 2763–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat, L.E. and Carmichael, G.G. 2001. Quality control of mRNA function. Cell 104: 173–176. [DOI] [PubMed] [Google Scholar]
- Mendell, J.T., Medghalchi, S.M., Lake, R.G., Noensie, E.N., and Dietz, H.C. 2000. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell. Biol. 20: 8944–8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, P. and Tollervey, D. 2003. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′ → 5′ degradation. Mol. Cell 11: 1405–1413. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D. and Parker, R. 1994. Premature translational termination triggers mRNA decapping. Nature 370: 578–581. [DOI] [PubMed] [Google Scholar]
- ———. 1999. Recognition of yeast mRNAs as “nonsense containing” leads to both inhibition of mRNA translation and mRNA degradation: Implications for the control of mRNA decapping. Mol. Biol. Cell 10: 3971–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero, L.J., Ashe, M.P., and Sachs, A.B. 1999. The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J. 18: 3153–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V. and Kolupaeva, V.G. 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes & Dev. 16: 2906–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Borukhov, S.I., and Hellen, C.U.T. 1998. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394: 854–859. [DOI] [PubMed] [Google Scholar]
- Phan, L., Schoenfeld, L.W., Valasek, L., Nielsen, K.H., and Hinnebusch, A.G. 2001. A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNA(i)Met. EMBO J. 20: 2954–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M.D., Winston, F., and Hieter, P. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Ruiz-Echevarria, M.J. and Peltz, S.W. 2000. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell 101: 741–751. [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarria, M.J., Gonzalez, C.I., and Peltz, S.W. 1998a. Identifying the right stop: Determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 17: 575–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Echevarria, M.J., Yasenchak, J.M., Han, X., Dinman, J.D., and Peltz, S.W. 1998b. The Upf3p is a component of the surveillance complex that monitors both translation and mRNA turnover and affects viral maintenance. Proc. Natl. Acad. Sci. 95: 8721–8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, G., Bidou, L., Hatin, I., Namy, O., Rousset, J.P., and Farabaugh, P. 2000. The case against the involvement of the NMD proteins in programmed frameshifting. RNA 6: 1687–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, S., Araki, Y., Sakuno, T., and Katada, T. 2003. Interaction between Ski7p and Upf1p is required for nonsense-mediated 3′-to-5′ mRNA decay in yeast. EMBO J. 22: 3951–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun Jr., S.Z. and Sachs, A.B. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15: 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Wagner, E. and Lykke-Andersen, J. 2002. mRNA surveillance: The perfect persist. J. Cell Sci. 115: 3033–3038. [DOI] [PubMed] [Google Scholar]
- Waldron, C., Cox, B.S., Wills, N., Gesteland, R.F., Piper, P.W., Colby, D., and Guthrie, C. 1981. Yeast ochre suppressor SUQ5-ol is an altered tRNA Ser UCA. Nucleic Acids Res. 9: 3077–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Czaplinski, K., Rao, Y., and Peltz, S.W. 2001. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 20: 880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, E.M. and Jacobson, A. 1999. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 18: 6134–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, Y., Czaplinski, K., and Peltz, S.W. 1996a. Genetic and biochemical characterization of mutations in the ATPase and Helicase regions of the Upf1 protein. Mol. Cell. Biol. 16: 5477–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1996b. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol. Cell. Biol. 16: 5491–5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1998. ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA 4: 205–214. [PMC free article] [PubMed] [Google Scholar]
- Wickner, R.B. 1996. Prions and RNA viruses of Saccharomyces cerevisiae. Annu. Rev. Genet. 30: 109–139. [DOI] [PubMed] [Google Scholar]
- Wickner, R.B. and Leibowitz, M.J. 1976. Two chromosomal genes required for killing expression in killer strains of Saccharomyces cerevisiae. Genetics 82: 429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz, C.J., Wang, W., and Peltz, S.W. 2001. Curbing the nonsense: The activation and regulation of mRNA surveillance. Genes & Dev. 15: 2781–2785. [DOI] [PubMed] [Google Scholar]
- Yoon, H. and Donahue, T.F. 1992. The sui1 suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNAiMet recognition of the start codon. Mol. Cell. Biol. 12: 248–260. [DOI] [PMC free article] [PubMed] [Google Scholar]