Abstract
A signal turnover system is an essential component of many genetic regulatory mechanisms. The best-known example is the ubiquitin-dependent protein degradation system that exists in many organisms. We found that Agrobacterium tumefaciens adopts a unique signal turnover system to control exiting from a quorum-sensing mode. A. tumefaciens regulates Ti plasmid conjugal transfer by a quorum-sensing signal, N-3-oxo-octanoyl homoserine lactone (3OC8HSL), also known as Agrobacterium autoinducer. By using Tn5 mutagenesis and a functional cloning approach, we identified two genes that are involved in switching from a conjugal quorum-sensing mode to a nonconjugal mode at the onset of stationary phase. First, we located attJ, which codes for an IclR-type suppressor that regulates the second gene attM. The latter encodes a homologue of N-acylhomoserine lactone (AHL)-lactonase. Mass spectrometry analysis shows that the enzyme encoded by attM is an AHL-lactonase that hydrolyzes the lactone ring of 3OC8HSL. In wild-type A. tumefaciens, attM expression is initially suppressed by AttJ but significantly elevated at the stationary phase accompanied a sharp decline in 3OC8HSL. DNA gel retardation analysis shows that AttJ specifically binds to the promoter that controls AHL-lactonase expression. Mutation of attJ resulted in constitutive production of AHL-lactonase that abolishes 3OC8HSL accumulation and Ti plasmid transfer. These data suggest that A. tumefaciens has a sophisticated multicomponent quorum-sensing signal turnover system, allowing the cell to sense a change in growth and adjust cellular activities accordingly.
Bacterial cells are able to sense and respond to a change of their population by communication with small signal molecules. This cell-to-cell communication mechanism is now generally known as quorum sensing (1). Most of the quorum-sensing signals identified so far in Gram-negative bacteria are N-acylhomoserine lactones (AHLs), which contain the same homoserine lactone ring but differ in the length and structure of the acyl chain (2–5). The AHL-mediated quorum-sensing systems regulate diverse biological functions ranging from bioluminescence (2), Ti plasmid conjugal transfer (4, 6), virulence (7–9), biofilm formation (10), antibiotics production (11), and swarming mobility (12). Studies during the past few years have outlined how bacterial cells enter the quorum-sensing phase. Initially, each cell in the bacterial population produces a basal level of the diffusible AHL by way of the activity of AHL synthase (13–15). At a sufficiently high population density, the accumulated AHL interacts with a transcription regulator, usually a member of the LuxR proteins, to form a complex that acts to trigger the expression of target genes (16–18). In consequence, bacterial cells switch to a new biological process. However, little is known of how bacterial cells exit the quorum-sensing phase.
Plasmid transfer in a bacterial population is usually a transient process, which ends within hours (19). The quorum-sensing-dependent Ti plasmid conjugal transfer system of the plant pathogen Agrobacterium tumefaciens seems to be an excellent model to study how bacterial cells enter and exit from quorum-sensing mode for a particular biological process. In the presence of opine, the conjugation inducers, A. tumefaciens produces a diffusible conjugation signal that promotes conjugation (20). The signal was later identified as N-(3-oxooctanoyl)-L-homoserine lactone (3OC8HSL), also known as Agrobacterium autoinducer (4). In conjunction with the transcription factor TraR, 3OC8HSL positively regulates the expression of genes required for Ti plasmid transfer (Tra genes) (4, 6, 13, 21). Growth phase of donor cells is very important in determining the efficiency of Ti plasmid transfer. A. tumefaciens donor cells initiate Ti plasmid transfer at the midlag phase, maximize at the midexponential phase, and stop at the stationary phase (22). Our preliminary experiment found that production of 3OC8HSL in donor cells displays the same growth phase-dependent pattern; the concentration of 3OC8HSL in culture decreases dramatically when bacterial cells enter stationary phase. These data suggest that bacterial cells might exit from the quorum-sensing phase by terminating biosynthesis of 3OC8HSL or eliminating the 3OC8HSL quorum-sensing signal or both.
In this study, we have identified a gene, designated previously as attM (23), which encodes a homologue of the AHL-lactonase encoded by aiiA (24, 25). The enzyme encoded by attM inactivates 3OC8HSL quorum-sensing signals by hydrolysis of its homoserine lactone ring. Furthermore, we present evidence that attM is negatively controlled by a transcription factor, AttJ, a homologue of the IclR family. Expression of the AHL-lactonase is growth phase-dependent. Its expression is enhanced substantially when A. tumefaciens bacterial cells enter the stationary phase; the enzyme degrades 3OC8HSL and terminates the conjugation-related quorum sensing.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
A. tumefaciens strains A6, K588, NT1 (traR, tra∷lacZ749), and K749 were used in this study. A6 is a wild-type strain, in which 3OC8HSL production and Ti plasmid conjugal transfer are induced by octopine (20). K588, with pTiB6S3Trac in the genetic background of C58C1 (pTi−, EryR, ChlR), is constitutive in 3OC8HSL production and Ti plasmid conjugal transfer. NT1 (traR, tra∷lacZ749) is an AHL biosensor strain (6). The conjugation recipient strain K749 (pTi−, pAt−, RifR) is a derivative of strain C58 (20). Unless otherwise stated, Escherichia coli DH5α was used as a host for DNA cloning. E. coli M15 (pREP4) from Qiagen (Chatsworth, CA) was used as the host strain for protein expression. The plasmid pREP4 constitutively expresses the Lac repressor protein encoded by lacI and thus provides tight control of the tac promoter-mediated transcription (Qiagen). A. tumefaciens strains were grown at 28°C in YEB (5 g/liter yeast extract/10 g/liter tryptone/5 g/liter NaCl/5 g/liter sucrose/0.5 g/liter MgSO4⋅7H2O, pH 7.0), or in BM medium [basic minimal (BM) nutrient with mannitol (0.2%) added as sole carbon source] (20). When necessary, glucose or sucrose was added to replace mannitol as sole carbon source. Octopine at a final concentration of 0.2 mg/ml was added to the BM medium when required. E. coli strains were grown at 37°C in LB medium (per liter contains 10 g Bacto-tryptone/5 g yeast extract/10 g NaCl, pH 7.0). Antibiotics were added at the following concentrations when required: kanamycin, 50 μg/ml; tetracycline, 5 μg/ml (A. tumefaciens), or 10 μg/ml (E. coli); rifampin, 50 μg/ml; streptomycin, 100 μg/ml; ampicillin, 100 μg/ml. X-Gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) (Promega) was included in media at 50 μg/ml for detection of β-galactosidase activity.
Tn5 Insertion Mutagenesis of A. tumefaciens.
Transposon Tn5 carried by the suicide plasmid pSUP10 (26) in E. coli SM10 was introduced into the genome of A. tumefaciens strain A6 as described (27), except that after mating the bacterial suspensions were spread onto BM plates containing kanamycin for selection of tranconjugants.
3OC8HSL Bioassay and Ti Plasmid Conjugal Transfer Assay.
Soluble protein samples were prepared from exponential cultures (12 h after inoculation) with 0.067 M phosphate buffer (pH 7.4). Equal volume (100 μl) of soluble proteins (1.46 mg/ml) was mixed with 3OC8HSL (final concentration, 5 μM) and incubated at 28°C. Control protein samples were denatured by boiling for 3 min before reaction. An aliquot of reaction mixture was collected at different times and the reaction was stopped by boiling. The remaining 3OC8HSL in the reaction mixture was determined as described (24). The Ti plasmid conjugal transfer assay was performed with the drop-mating method as described (20).
Cosmid Library Construction of Mutant Strain M103.
A cosmid library of mutant M103 was constructed with the cosmid vector pLAFR3. Total DNA of M103 was partially digested with EcoRI, then the 20- to 30-kb fragments were purified from low-melting-point agarose gel as described (24). After ligation of the purified fragments with the EcoRI-linearized vector pLAFR3, the ligation mixture was packaged with Gigapack III XL Packaging Extract (Stratagene) and then transfected to E. coli DH5α.
Reverse Transcription (RT)–PCR Analysis.
Strains A6 and M103 were grown in BM medium to exponential growth phase (12 h after inoculation). Total RNA was isolated from A. tumefaciens as described (28) and then the RNA sample was treated with RNase-free DNase I to remove any contaminating DNA. The cDNA samples of the attK-attL-attM operon were prepared from strains A6 and M103, respectively, by using a primer complementary to the 3′ end of attM RNA and a reverse transcriptase. One microgram of RNA sample was added in the RT reactions at a final reaction volume of 20 μl. A 2-μl aliquot of the synthesized cDNA was used as PCR template with relevant PCR primers in a 50-μl reaction volume. The RNA samples were used as control in PCR to verify the absence of DNA contamination. PCR was conducted by using the PCR Core Kit as recommended (Qiagen).
Protein Expression and Purification.
The attJ coding sequence was amplified by PCR with primers: 5′-CTCGG ATCCA TGGGC CAAAG GGGCC AA-3′ and 5′-AACTG CAGCT AGTCT TTCTG CGATC-3′, then fused into 6 × His tagging expression vector pQE-9 (Qiagen) by BamHI and PstI digestion. We then transformed the resultant construct pQE-AttJ into expression host strain E. coli M15 (pREP4) after sequencing confirmation. The isopropyl β-D-thiogalactoside-induced E. coli M15 (pREP4, pQE-AttJ) cells were collected by centrifugation, and the cell-free extracts were prepared by ultrasonification (29). The cell-free extracts were applied to the Ni-NTA affinity column for purification of (His)6-AttJ as recommended (Qiagen).
The AHL-lactonase encoded by attM was expressed and purified by using GST Gene Fusion System (Amersham Pharmacia). The AHL-lactonase coding sequence was amplified with primers 5′-TCCCC CGGGA GTGAC CGATA TCAGA CTT-3′and 5′-TCGGA ATTCT TACGC GTAAA ATTCG GG-3′, then fused into expression vector pGEX-2T digested by SmaI and EcoRI. The construct pGST-attM was verified by DNA sequencing and transferred to the host E. coli BL21 (DE3). The AHL-lactonase was purified as described (29), and the AttJ polyclonal antibody was raised in rabbit according to standard procedures.
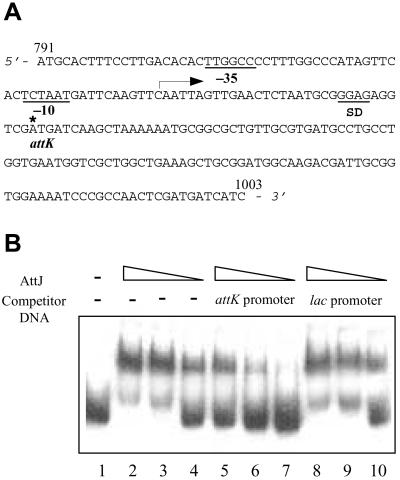
Gel Retardation Assay.
The 214-bp putative AttJ-binding promoter region, corresponding to 791-1003 of the attJ-attM operon (see Fig. 5A), was amplified by PCR with primers 5′-CGGGA TCCAT GCACT TTCCT TGACA C-3′ (forward) and 5′-CGGAA TTCGA TGATC ATCGA GTTGG C-3′ (reverse). After sequencing confirmation, the purified PCR product was labeled at the 3′ end with [α-32P]dCTP by Klenow enzyme. The DNA-binding reaction was conducted as described (30). The reaction samples were separated by electrophoresis at 4°C on a 4.5% native polyacrylamide gel in 2× TAE buffer (80 mM Tris⋅acetate/2 mM EDTA, pH 8.0). After electrophoresis, the gel was dried before detection of AttJ-promoter DNA complex and free DNA fragments by autoradiograph.
Figure 5.
AttJ specifically binds to the promoter of the attK-attL-attM operon. (A) The promoter sequence of attK-attL-attM operon. The start (791) and end (1003) sites of this fragment correspond to the designations in Fig. 4A. The arrow indicates the direction of transcription, and the star indicates the translation start site of attK. (B) DNA gel retardation analysis. Lane 1 is the negative control of labeled probe without AttJ protein. Purified AttJ was incubated at decreasing concentrations (100, 50, and 10 nM) with the labeled attK-attL-attM promoter probe (lanes 2–4); labeled probe plus a 100-fold excess of unlabeled probe (lanes 5–7); labeled probe plus 100-fold excess of unlabeled 213-bp lac promoter fragment (lanes 8–10).
HPLC and Electrospray Ionization (ESI)-MS Analysis.
3OC8HSL digestion by AHL-lactonase and the subsequent HPLC analysis and ESI-MS characterization have been described (25).
Results
Mutation of attJ Gene Abolishes 3OC8HSL Production.
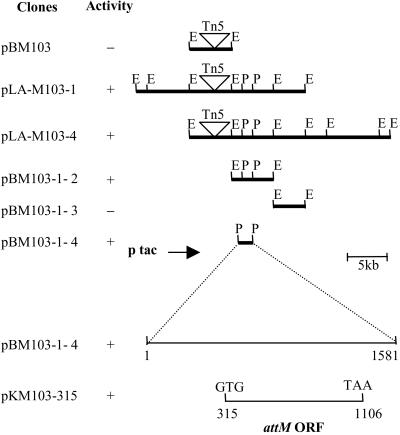
To search for the genes involved in regulation of quorum-sensing signal biosynthesis and signal turnover in A. tumefaciens, we screened for 3OC8HSL-deficient mutants of A. tumefaciens strain A6 generated by Tn5 random insertion, with the AHL biosensor strain A. tumefaciens NT1 (traR, tra∷lacZ749) (6). Of 1,600 Tn5 mutants tested, three showed no, or a reduction, in 3OC8HSL accumulation. One of the mutants, M103, which was completely deficient in 3OC8HSL accumulation (3OC8HSL−) and Ti plasmid transfer (Tra−), was selected for further study. On the basis of the kanamycin resistance conferred by Tn5, we identified the clone pBM103 that contains a 11-kb EcoRI DNA fragment including Tn5 (Fig. 1). Sequence analysis indicates that the Tn5 insertion is located at the att gene region, which is involved in attachment of A. tumefaciens cells to plant cells (23, 31). The exact Tn5 insertion site is at a position corresponding to the 43rd base pair of the 828-bp encoding sequence of attJ gene of strain C58 (GenBank accession no. U59485). Sequence comparison suggests that attJ is homologous to members of the iclR negative transcription regulator gene family (30, 32). AttJ shares 25–30% identity with other members of the IclR family (32, 33).
Figure 1.
Cloning attM from A. tumefaciens strain M103. Symbols: +, with 3OC8HSL inactivation activity; −, without 3OC8HSL inactivation activity; E, EcoRI; P, PstI. pLA-M103–1 and pLA-M103–4 are two cosmid clones in vector pLAFR3. pKM103–315 was constructed by cloning the attM-coding sequence in the expression vector pKK223–3, the numbers indicating the positions corresponding to the 1.5-kb region of clone pBM103–1-4. The other subclones are in the plasmid vector pBluescript II SK(+). The arrow indicates the direction of tac promoter in these clones.
attJ− Mutant Expresses 3OC8HSL Inactivation Enzyme.
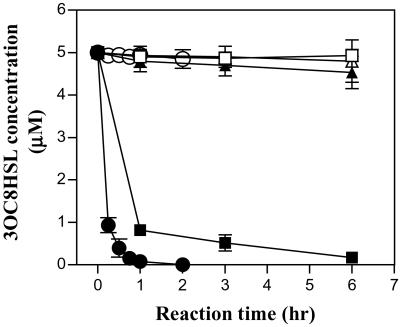
How does a Tn5 insertional mutation in the negative transcription factor AttJ cause 3OC8HSL signal deficiency? One possibility is that a target gene(s) involved in 3OC8HSL degradation is negatively regulated by the transcription factor AttJ. To test this hypothesis, we prepared total protein samples from early-exponential-phase cultures of strains M103 and A6 and incubated them with 3OC8HSL. As shown in Fig. 2, the total protein extracted from M103 inactivated 3OC8HSL progressively, whereas that from A6 and the denatured total proteins of M103 showed no or very weak enzyme activity, respectively, suggesting that the attJ− mutant M103 expresses an enzyme responsible for 3OC8HSL degradation.
Figure 2.
Time course of 3OC8HSL inactivation by protein extracts and the AHL-lactonase encoded by attM. Symbols: ■, M103 total protein; □, denatured M103 total protein; ▴, A6 total protein; ▵, denatured A6 total protein. ●, purified AHL-lactonase encoded by attM (1 μM final concentration in 1 × PBS buffer, pH7.4); ○, denatured AHL-lactonase.
Cloning of attM Coding for the 3OC8HSL Inactivation Enzyme.
It is plausible that the gene coding for 3OC8HSL inactivation is located at the vicinity of the attJ gene. Because the E. coli strain harboring clone pBM103 did not show any 3OC8HSL inactivation activity (Fig. 1), it seems that the gene for 3OC8HSL inactivation is not in the immediate vicinity of attJ. We then constructed a cosmid library of the mutant M103 with insert size ranging from 20 to 35 kb, and screened for cosmid clones containing Tn5. We identified two cosmid clones, pLA-M103–1 and pLA-M103–4, which contain Tn5 and confer 3OC8HSL inactivation function (Fig. 1). Further subcloning of the cosmid clone pLA-M103–1 narrowed down the region to a 1.5-kb PstI fragment contained in clone pBM103–1-4 (Fig. 1). Sequence analysis and deletion analysis identified one ORF that encodes 3OC8HSL inactivation in clone pKM103–1-5 (Fig. 1). The ORF starts with GTG, which has 21 more base pairs in front of the predicted attM-coding region of A. tumefaciens strain C58 (GenBank accession no. U59485) (23, 31). The predicted protein contains seven more amino acids at the N-terminal region than the reported AttM protein, which was apparently an annotation error and has been corrected at the recently released genome sequence of A. tumefaciens strain C58 (http://www.ncbi.nlm. nih.gov/PMGifs/Genomes/micr.html).
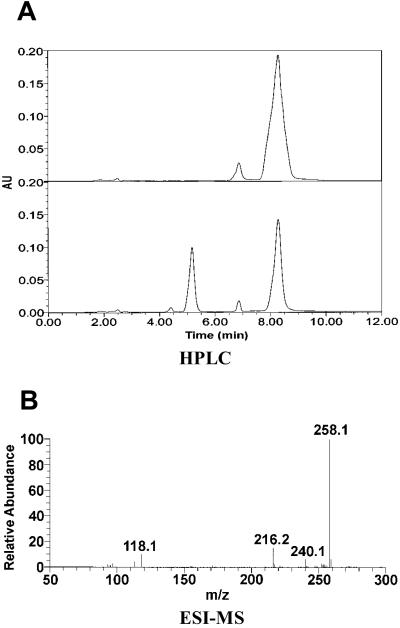
The product of attM is predicted to be a 264-aa protein with a calculated Mr of 29,302. Sequence comparison indicates that attM and aiiA (24) share 24.8% identity at the DNA level and 31.3% at the peptide level. In addition, the two enzymes share a conserved motif, HXDH≈60 aa≈H (data not shown), which was proven essential for enzyme activity (24). To analyze how the enzyme encoded by attM inactivates 3OC8HSL, we digested 3OC8HSL with the purified enzyme encoded by attM (Fig. 2) and analyzed the reaction products with HPLC and MS. The enzyme digestion of 3OC8HSL resulted in only one product with a retention time at 5.3 min (Fig. 3A). The control experiment with the AHL-lactonase encoded by aiiA showed the same result (data not shown), suggesting that the enzyme encoded by attM is likely to be an AHL-lactonase. We collected the enzyme-digested product fraction of 5.3 min from HPLC for ESI-MS analysis. Fig. 3B shows that molecular mass of 3OC8HSL (M–H ion m/z 240) was increased by 18 after AHL-lactonase digestion, which is identical with that of the same substrate after digestion by the AHL-lactonase encoded by aiiA (25). These data confirm that the attM encodes an AHL-lactonase that hydrolyzes the homoserine lactone ring of 3OC8HSL to produce N-(3-oxooctanoyl)-L-homoserine (M–H ion m/z 258).
Figure 3.
Demonstration that attM encodes an AHL-lactonase. (A) HPLC analysis of the AHL-lactonase digestion product of 3OC8HSL. (Upper) HPLC profile of 3OC8HSL (retention time = 8.3 min), the minor peak at 7.7 min was the sodium salt of 3OC8HSL based on MS analysis (data not shown). (Lower) Profile shows that 3OC8HSL digestion by AHL-lactonase results in one product with a retention time of 5.3 min. (B) ESI-MS analysis of the hydrolysis product of 3OC8HSL. The fraction at 5.3 min from HPLC was collected and analyzed by ESI-MS.
Sequence Analysis of the attJ Upstream Region.
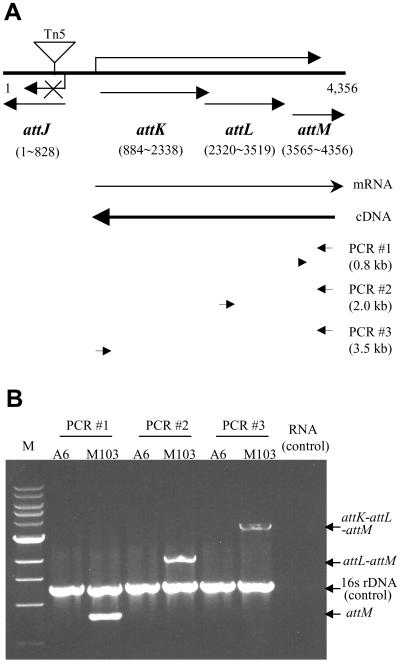
We determined the DNA sequence of the region containing attJ and attM of A. tumefaciens strain A6, which has been submitted to the GenBank database (accession no. AY052389). The sequence has 98% identity at the nucleotide level with the same region of strain C58 (31). As shown in Fig. 4A, between attJ and attM, two other ORFs, known as attK and attL, exist (31). As reported, attK and attL share 65.7% and 35.8% identities at the peptide level, respectively, with the genes for succinic semialdehyde dehydrogenase and alcohol dehydrogenase of E. coli (31). Sequence analysis of the attK-attL-attM fragment indicated that the sequences TCTAAT and TTGGCC at positions −10 and −35 upstream of the attK-coding sequence, match to the TATAAT and TTGACA promoter regions of E. coli, respectively. The ribosomal binding sequence SD (GGAG) is located 6 bp upstream of the start codon ATG (Fig. 5A). However, these promoter elements, except the SD sequence, have not been found either upstream of attL or attM. Besides, the distances between the ORFs of attK and attL, and that of attL and attM are either very small or overlapping (Fig. 4A). It is likely that attK, attL, and attM are the components of one operon and are subjected to the control of the common promoter located upstream of attK.
Figure 4.
RT-PCR analysis of the attK-attL-attM operon. (A) Map positions of attJ, attK, attL, and attM genes and the primers for RT-PCR analysis. (B) RT-PCR analysis of expression profiles of the attK-attL-attM operon in strain A6 and M103. The RT-PCRs were performed by using three primer-pair combinations as illustrated by short arrows in A. To each RT-PCR, a pair of PCR primers specific for amplification of a 1.2-kb 16S rDNA fragment were added as an internal control to indicate quality and relative quantity of cDNA samples. Symbols: M, 1-kb DNA marker; RNA, RT-PCR profile when RNA sample from either A6 or M103 was used as a template.
AttJ Negatively Regulates Its Upstream Operon.
We used RT-PCR to test whether attK, attL, and attM belong to the same operon and to analyze the mechanism of gene regulation. We prepared total RNA samples from exponential cultures of strains M103 and A6, and then prepared cDNA fragments by using a primer specific to the terminal region of the attM gene. As shown in Fig. 4A, we designed three sets of PCR primers for amplification of attM, attL-attM, and attK-attL-attM, respectively. Data in Fig. 4B show that in strain M103 these three genes and their combination gene blocks can be amplified from the same cDNA sample, indicating that the three genes comprise one operon. In contrast, RT-PCR products were hardly detectable when the cDNA sample from the early exponential culture of strain A6 is used (Fig. 4B), demonstrating that AttJ suppresses the expression of its upstream operon in the wild-type strain of A. tumefaciens.
To determine whether transcription factor AttJ could specifically bind to the putative promoter DNA fragment, we conducted a gel retardation assay with purified AttJ protein. We amplified the attK promoter DNA fragment as shown in Fig. 5A, and labeled it with isotope (see Materials and Methods). The result shows formation of the labeled AttJ-[attK promoter] complex when AttJ was added in concentrations ranging from 10 to 100 nM in the reaction mixture (Fig. 5B). To test the binding specificity of AttJ, we used the unlabeled attK promoter and the lacZ gene promoter from the cloning vector pGEM-7zf(+) as competitors in gel retardation analysis. Addition of the unlabeled attK promoter DNA to the reaction mixture significantly reduced the formation of the labeled AttJ-[attK promoter] complex, whereas the unrelated lacZ gene promoter failed to compete against the labeled attK promoter probe (Fig. 5B).
AHL-Lactonase Expression Is Growth Phase-Dependent.
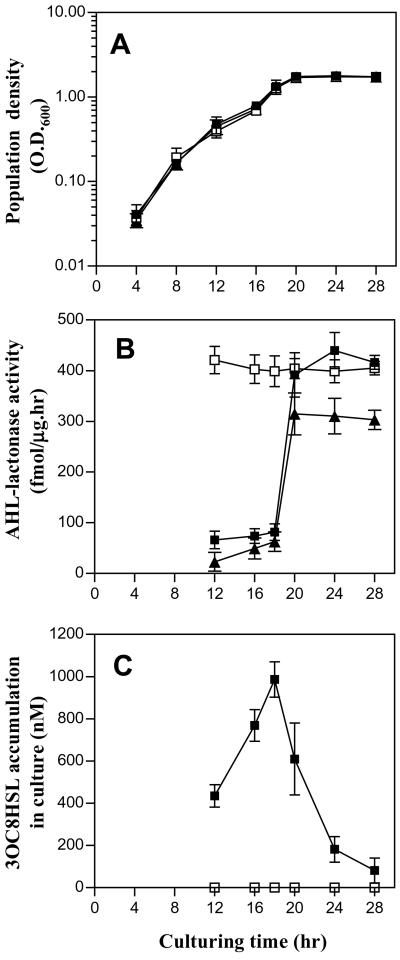
We determined AHL-lactonase activities at different growth phases of bacterial culture. Fig. 6A shows that strains A6, K588, and M103 have a similar growth pattern; bacterial cells grew exponentially until about 18 h after inoculation when they moved into stationary phase. AHL-lactonase activities of strains A6 and K588 dramatically increased when growth entered stationary phase (Fig. 6B). Correspondingly, in strain K588, 3OC8HSL concentration was decreased significantly after entering the stationary phase (Fig. 6C). Besides mannitol, we also tested glucose and sucrose as the sole carbon source in BM medium and observed a similar AHL-lactonase expression pattern (data not shown). Therefore, it is unlikely that catabolite repression is involved in control of the AHL-lactonase expression. We were unable to quantify 3OC8HSL production in strain A6 because it failed to produce 3OC8HSL in liquid culture (20). In contrast, the attJ− mutant strain M103 shows a high level of 3OC8HSL inactivation throughout all growth phases, suggesting that AHL-lactonase is constitutively produced, inactivates 3OC8HSL, and abolishes quorum-dependent 3OC8HSL biosynthesis (Fig. 6C).
Figure 6.
AHL-lactonase expression patterns and 3OC8HSL accumulation profiles of A. tumefaciens strains A6, K588, and M103. Symbols: ▴, A6; ■, K588; □, M103. (A) The bacterial growth curves in minimal medium. (B) AHL-lactonase activities at different growth stages. (C) 3OC8HSL accumulation in liquid culture.
Discussion
The best-known signal-turnover system is perhaps selective protein degradation (i.e., ubiquitin-dependent protein degradation), which is almost always a component of genetic regulatory mechanisms that are involved in developmental control. Timely degradation of protein signals and other component proteins is of critical importance to many biological processes, such as cell cycle progression, circadian rhythms, and embryogenesis (34). In this study, we show that A. tumefaciens adopts a signal-turnover system to exit from quorum-sensing-dependent Ti plasmid conjugal transfer. The system, targeting a small AHL quorum-sensing molecule rather than proteins, comprises at least an AHL degradation enzyme encoded by attM, and a negative transcription factor AttJ. Expression of attM, which is controlled by the negative transcription factor AttJ at the early stages of growth, is enhanced significantly when bacterial growth enters stationary phase, likely because of the relieving AttJ control by an unidentified factor(s). Identification of this growth phase-dependent AHL quorum-sensing signal-turnover system provides a logical explanation for the early finding that Ti plasmid conjugal transfer in A. tumefaciens occurs from midlag phase of bacterial growth and stops when bacterial cells enter stationary phase (22). This is an example of a quorum-sensing signal turnover system that controls bacterial cells exiting from the quorum-sensing mode.
The attM gene shows similarity to aiiA, the gene encoding an AHL inactivation enzyme (24), at both nucleotide and peptide sequence level. Similar to AiiA, it contains a conserved motif “HXDH≈60aa≈H” which was proven essential for enzyme activity (24). More recently, it has been shown that the enzyme encoded by aiiA is an AHL-lactonase, which inactivates AHLs by hydrolyzing the ester bond of the homoserine lactone ring (25). By HPLC and MS analysis, we showed that attM encodes an AHL-lactonase, which hydrolyzes AHLs and produces N-acylhomoserines.
Our results show that AHL-lactonase expression is under control of the negative transcription factor AttJ. Genetic and biochemical data demonstrate that attM, attK, and attL genes comprise one transcriptional unit that is negatively regulated by AttJ. AttJ is a member of the IclR negative transcription factor family. In E. coli, adaptation to growth on acetate or fatty acids requires the induction of the aceBAK operon, which includes genes coding for isocitrate lyase (aceA), malate synthase (aceB), and isocitrate dehydrogenase kinase-phosphatase (aceK). Expression of aceBAK is repressed by IclR (35) and activated by integration host factor in the presence of acetate (36). The binding site of the IclR repressor protein, which is a palindromic repeat of 7 nt separated by 1 nt (TGGAAAT × A/gTTTCA/g), was identified at the −35 element region of the promoter of aceBAK (35). We showed that AttJ binds to the promoter of attK-attL-attM operon, but did not find a nucleotide sequence with obvious homology to the IclR-binding site sequence. Because AttJ shares only about 25% identity with IclR, it is probable that the two repressors target different nucleotide sequences.
We showed that AttJ suppresses expression of AHL-lactonase before bacterial cells of A. tumefaciens enter stationary phase. Apparently, the suppression is relieved when bacterial cells enter the stationary phase. Consequently, AHL-lactonase initiation leads to elimination of 3OC8HSL signal molecules. AttJ is constitutively expressed throughout all growth phases (H.B.Z., unpublished data), suggesting that another factor(s) is involved in control of attM expression at the stationary phase. We tested whether 3OC8HSL accumulation in culture triggers expression of AHL-lactonase. Supplementation of 3OC8HSL in medium at a final concentration of 50 μM and 100 μM, however, did not enhance expression level of AHL-lactonase or 3OC8HSL inactivation during the exponential growth phase of strain K588, indicating that another factor is involved in control of attM.
The quorum-sensing turnover system in A. tumefaciens may not be a unique case. The quorum-sensing signal of Erwinia carotovora, N-(oxohexanoyl)-L-homoserine lactone, was significantly decreased when bacteria moved into stationary phase (37). The diffusible signal factor of Xanthomonas campestris, which is required for pathogenicity, accumulates in early stationary phase and its level subsequently declines sharply (38). Although genetic components of quorum-sensing signal-turnover systems in different bacterial species may be varied, the effective control of quorum-sensing signal turnover may be a common mechanism in different bacterial quorum-sensing systems.
AttJ and AttM were reported to be involved in attachment of A. tumefaciens cells to plant cells (31). The att gene cluster contains 33 genes. Mutants of 24 att genes showed a nonattachment phenotype and decreased virulence, although the exact role of each att gene product in attachment has not been identified (31). Genetic analysis indicated that Ti plasmid conjugal transfer between A. tumefaciens bacterial cells does not require the vir system that is essential for T-DNA transfer from A. tumefaciens Ti plasmid to plant cells (39, 40). However, T-DNA transfer is a multistep process requiring expression of at least three genetic components each consisting of many genes (41). It is possible that a certain degree of coordination is required for the maximal efficiencies of Ti plasmid conjugal transfer and T-DNA transfer.
Acknowledgments
We thank Stephen K. Farrand for critical reviews of the manuscript. Funding was provided by the National Science and Technology Board of Singapore.
Abbreviations
- 3OC8HSL
N-3-oxo-octanoyl homoserine lactone
- AHL
N-acylhomoserine lactone
- RT
reverse transcription
- BM
basic minimal
- ESI
electrospray ionization
Footnotes
Data deposition: The sequence reported in this paper has been submitted to the GenBank database (accession no. AY052389).
References
- 1.Fuqua C, Winans S C, Greenberg E P. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 2.Eberhard A, Burlingame A L, Eberhard C, Kenyon G L, Nealson K H, Oppenheimer N J. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 3.Cao J G, Meighen E A. J Biol Chem. 1989;264:21670–21676. [PubMed] [Google Scholar]
- 4.Zhang L H, Murphy P J, Kerr A, Tate M E. Nature (London) 1993;362:446–448. doi: 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]
- 5.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piper K R, von Bodman S B, Farrand S K. Nature (London) 1993;362:448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- 7.Jones S, Yu B, Bainton N J, Birdsall M, Bycroft B W, Chhabra S R, Cox A J, Golby P, Reeves P J, Stephens S, et al. EMBO J. 1993;12:2477–2482. doi: 10.1002/j.1460-2075.1993.tb05902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Passador L, Cook L M, Gambello M J, Rust L, Iglewski B H. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 9.Pirhonen M, Flego D, Heikinheimo R, Palva E A. EMBO J. 1993;12:2467–2476. doi: 10.1002/j.1460-2075.1993.tb05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 11.Pierson L S, Keppenne V D, Wood D W. J Bacteriol. 1994;176:3966–3974. doi: 10.1128/jb.176.13.3966-3974.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eberl L, Winson M K, Sternberg C, Steward G S A B, Christiansen G, Chhabra S R, Bycroft B, Williams P, Molin S, Givskov M. Mol Microbiol. 1996;20:127–136. doi: 10.1111/j.1365-2958.1996.tb02495.x. [DOI] [PubMed] [Google Scholar]
- 13.Hwang I, Li P L, Zhang L H, Piper K R, Cook D M, Tate M E, Farrand S K. Proc Natl Acad Sci USA. 1994;91:4639–4643. doi: 10.1073/pnas.91.11.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.More M I, Finger L D, Stryker J L, Fuqua C, Eberhard A, Winans S C. Science. 1996;272:1655–1658. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 15.Schaefer A L, Val D L, Hanzelka B L, Cronan J E, Greenberg E P. Proc Natl Acad Sci USA. 1996;93:9505–9509. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu J, Winans S C. Proc Natl Acad Sci USA. 1999;96:4832–4837. doi: 10.1073/pnas.96.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welch M, Todd D E, Whitehead N A, McGowan S J, Bycroft B W, Salmond G P C. EMBO J. 2000;19:631–641. doi: 10.1093/emboj/19.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin Y, Luo Z Q, Smyth A J, Gao P, von Bodman S B, Farrand S K. EMBO J. 2000;19:5212–5221. doi: 10.1093/emboj/19.19.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willetts N. In: Methods in Microbiology. Bennett P M, Grinsted J, editors. New York: Academic; 1984. pp. 33–59. [Google Scholar]
- 20.Zhang L H, Kerr A. J Bacteriol. 1991;173:1867–1872. doi: 10.1128/jb.173.6.1867-1872.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piper K R, von Bodman S B, Hwang I, Farrand S K. Mol Microbiol. 1999;32:1077–1089. doi: 10.1046/j.1365-2958.1999.01422.x. [DOI] [PubMed] [Google Scholar]
- 22.Tempé J, Estrade C, Petit A. Proc Int Conf Plant Pathog Bact. 1978;4:153–160. [Google Scholar]
- 23.Matthysse A G, Yarnall H A, Young N. J Bacteriol. 1996;178:5302–5308. doi: 10.1128/jb.178.17.5302-5308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong Y H, Xu J L, Li X Z, Zhang L H. Proc Natl Acad Sci USA. 2000;97:3526–3531. doi: 10.1073/pnas.060023897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong Y H, Wang L H, Xu J L, Zhang H B, Zhang X F, Zhang L H. Nature (London) 2001;411:813–817. doi: 10.1038/35081101. [DOI] [PubMed] [Google Scholar]
- 26.Simon R, Priefer U, Pûhler A. Bio/Technology. 1983;11:784–791. [Google Scholar]
- 27.Garfinkel D J, Nester E W. J Bacteriol. 1980;144:732–743. doi: 10.1128/jb.144.2.732-743.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong X, Delacruz N, Reznikoff W S. J Bacteriol. 1991;173:4570–4577. doi: 10.1128/jb.173.15.4570-4577.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L H, Xu J L, Birch R G. Microbiology. 1998;144:555–559. doi: 10.1099/00221287-144-2-555. [DOI] [PubMed] [Google Scholar]
- 30.Gui L, Sunnarborg A, Pan B, LaPorte D C. J Bacteriol. 1996;178:321–324. doi: 10.1128/jb.178.1.321-324.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthysse A G, Yarnall H, Boles S B, McMahan S. Biochim Biophys Acta. 2000;1490:208–212. doi: 10.1016/s0167-4781(99)00250-x. [DOI] [PubMed] [Google Scholar]
- 32.Sunnarborg A, Klumpp D J, Chung T, LaPorte D C. J Bacteriol. 1990;172:2642–2649. doi: 10.1128/jb.172.5.2642-2649.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galinier A, Negre D, Cortay J C, Marcandier S, Maloy S R, Cozzone A J. Nucleic Acids Res. 1990;18:3656. doi: 10.1093/nar/18.12.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hochstrasser M. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 35.Pan B, Unnikrishnan I, LaPorte D C. J Bacteriol. 1996;178:3982–3984. doi: 10.1128/jb.178.13.3982-3984.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Resnik E, Pan B, Ramani N, Freundlich M, LaPorte D C. J Bacteriol. 1996;178:2715–2717. doi: 10.1128/jb.178.9.2715-2717.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holden M T G, McGowan S J, Bycroft B W, Gordon S A B, Williams P, Salmond G PC. Microbiology. 1998;144:1495–1508. doi: 10.1099/00221287-144-6-1495. [DOI] [PubMed] [Google Scholar]
- 38.Barber C E, Tang J L, Feng J X, Pan M Q, Wilson T J G, Slater H, Dow J W, Williams P, Daniels M J. Mol Microbiol. 1997;24:555–566. doi: 10.1046/j.1365-2958.1997.3721736.x. [DOI] [PubMed] [Google Scholar]
- 39.Alt-Morbe J, Stryker J L, Fuqua C, Li P, Farrand S K, Winans S C. J Bacteriol. 1996;178:4248–4257. doi: 10.1128/jb.178.14.4248-4257.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cook D M, Li P-L, Ruchaud F, Padden S, Farrand S K. J Bacteriol. 1997;179:1291–1297. doi: 10.1128/jb.179.4.1291-1297.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheng J, Citovsky V. Plant Cell. 1996;8:1699–1710. doi: 10.1105/tpc.8.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]