Abstract
Here we describe the isolation and characterization of the mouse homolog of the human zinc-finger transcription factor INSM1 (IA-1) and identify an interacting protein. A 2.9-kb cDNA with an open reading frame of 1563 nucleotides, corresponding to a translated protein of 521 amino acids, was isolated from a mouse βTC-1 cDNA library. Mouse INSM1 was found to be 86% identical to human INSM1 and both proteins contain proline-rich regions and multiple zinc-finger DNA-binding motifs. Sequencing of mouse Insm1 genomic DNA revealed that it is an intronless gene. Chromosomal mapping localized Insm1 to chromosome 2. Northern blot analysis showed that mouse Insm1 expression begins at 10.5 days in the embryo, decreases after 13.5 days, and is barely detected at 18.5 days. In mouse brain, Insm1 is strongly expressed for 2 weeks after birth but shows little or no expression thereafter. Transfection of cells with GFP-tagged INSM1 revealed that INSM1 is expressed exclusively in the nucleus. We identified proteins that interacted with INSM1 by the yeast two-hybrid system and the binding of one of them, Cbl-associated protein (CAP), to INSM1 was confirmed by in vitro pull-down experiments, nuclear colocalization, and co-immunoprecipitation assays. Further studies showed that both INSM1 and CAP proteins were present in the nucleus of insulinoma cells and that endogenous INSM1 protein was co-precipitated with antibody to CAP. These findings raise the possibility that during embryo development CAP may enter the nucleus through its own nuclear localization signal or by binding to INSM1.
Keywords: zinc fingers, nuclear protein, mouse chromosome 2, intronless gene, embryo development, CAP
Introduction
INSM1 (IA-1) cDNA was isolated from a human insulinoma subtraction library [1,2]. It encodes a 510-amino-acid protein consisting of both a SNAG motif [3] and an alanine/proline-rich region at the amino terminus and five zinc-finger motifs at the carboxyl terminus. INSM1 is located on chromosome 20p11.2 and is expressed in fetal pancreas, brain, and tumors of neuroendocrine origin (for example, pheochromocytoma, medullary thyroid carcinoma, insulinoma, pituitary tumors, and small cell lung carcinoma), but is not expressed in normal adult tissues or most non-neuroendocrine tumors [2,4]. Clinical studies on a panel of human lung cancer cells revealed that INSM1 mRNA is expressed in 97% of small cell lung cancers and 13% of non-small-cell lung cancers (which have neuroendocrine features) [4]. The 5′-upstream region (2090 bp) of INSM1 contains several tissue-specific regulatory elements, which might account for its unique tumor expression pattern [5]. Functional studies revealed that INSM1 possesses transcriptional repressor activity and that the DNA-binding domain interacts and regulates the 5′-flanking region of mouse Neurod1 [6]. Differentiation studies showed that Insm1 expression is closely associated with induced differentiation of the rat pancreatic acinar cell line (AR42J) into insulin-positive cells and expression of the islet-specific transcription factor genes Pdx1, Neurod1, and Nkx6-1 [7]. These findings suggest that INSM1 functions as a developmentally regulated transcription factor in neuroendocrine cells.
To further elucidate the functional role and temporal expression pattern of Insm1, we cloned and characterized a mouse Insm1 homolog that is expressed in early mouse embryo development. The data presented here show that mouse Insm1 is highly conserved and intronless. To understand how the INSM1 protein functions, we used a yeast two-hybrid system to identify interacting proteins. We found that the Cbl-associated protein, CAP [8–13], interacts with the INSM1 protein and that both INSM1 and CAP are present in the nucleus.
Results
Chromosomal Localization of Mouse Insm1
We screened a phagemid library derived from mouse βTC-1 cells with radiolabeled human INSM1. Two positive clones with inserts of 1.4 and 2.5 kb were obtained. The total sequence of the two overlapping clones was 2909 bases and contained a single open reading frame (ORF) encoding a protein of 521 amino acids (Figs. 1A and 1B). The initial ATG codon was flanked by a Kozak translation sequence [14] and a SNAG motif was founded at amino acids 1–7 [3]. The 3′-UTR contained two polyadenylation consensus sequences (AATAAA) [15]. The N terminus contained two proline-rich regions (amino acids 43–57 and 75–85), whereas the C terminus contained a single proline-rich region (amino acids 405–414) and five zinc-finger motifs of the Cys2-His2 type (residues 274–294, 302–322, 375–395, 454–475, and 482–503). Sequence analysis revealed a nuclear localization signal (NLS; residues 350–367) immediately before the third zinc-finger motif. The degree of relatedness of mouse INSM1 to human INSM1 is high, with 86% identity at the protein level (Table 1). Comparison with INSM1 homologs in zebrafish (partial sequence), Drosophila melanogaster [16], and Caenorhabditis elegans showed 63%, 31%, and 26% identity, respectively. Two-point maximum-likelihood analysis yielded a significant lod (log of the odds ratio) score for linkage of mouse Insm1 to the marker D2Mit109 in the chromosome 2 radiation-hybrid map of RB04.02 RH panel (4.81 cR from D2Mit109, LOD > 3.0). Its localization is in the order D2mit491-D2Mit109-Insm1-D2mit492 on mouse chromosome 2 (Fig. 2A).
FIG. 1.

Mouse Insm1 cDNA. (A) Analysis of the deduced amino acid sequence of mouse INSM1. The longest clone was 2909 bp and contained an ORF encoding a protein of 521 amino acids. The deduced protein sequence of mouse INSM1 is characterized by a SNAG motif (blue, amino acids 1–7), three proline-rich sequences (green, amino acids 43–57, amino acids 75–85, and amino acids 405–414) and five C2H2 zinc-finger motifs (yellow). A NLS sequence (amino acids 350–367) is shown in red. Two polyadenylation consensus sequences (dashed lines) are seen in the 3′-untranslated region. (B) Diagrammatic representation of mouse INSM1. SNAG motif, proline-rich regions, five zinc-finger motifs, and an NLS are shown.
TABLE 1.
Chromosomal localization and degree of relatedness to human INSM1
| Relationship (%) to human INSM1 | |||||
|---|---|---|---|---|---|
| Species | Chromosome locus | Amino acid residues | I | S | GenBank acc. no. |
| Human | 20p11.2 | 510 | 100 | 100 | NM_002196 |
| Mouse | 2 | 521 | 86 | 87 | NM_016889 |
| Zebrafish | LG17 | 129* | 63 | 69 | AF411143 |
| Drosophila | 3L-61E | 496 | 31 | 37 | AF203690 |
| C. elegans | V | 286 | 26 | 31 | AF283983 |
Partial sequence. (I) Identity; (S) Similarity; Zebrafish, LG7, linkage group 17.
FIG. 2.

Intronless gene and chromosomal mapping of mouse Insm1. (A) Mouse Insm1 is tightly linked to the marker D2Mit109, which localizes mouse Insm1 between 80 and 82 cM on the integrated map of chromosome 2. (B) The size of PCR products amplified from mouse Insm1 cDNA and mouse Insm1 genomic DNA is the same, indicating that Insm1 is an intronless gene.
Mouse Insm1 Is an Intronless Gene
We determined the genomic structure of mouse Insm1 by PCR analysis and sequencing. A pair of primers flanking the ORF of mouse Insm1 was used to PCR amplify both mouse cDNA and mouse genomic DNA. Both mouse cDNA and mouse genomic DNA templates produced a distinct PCR band of 1.6 kb (Fig. 2B). This argues that mouse Insm1 is intron-less and similar to human INSM1. This was further supported by sequencing of a 6.0-kb EcoRI genomic clone, which failed to show any introns.
Dominant Expression of Mouse Insm1 in the Early Mouse Embryo
The expression pattern of human INSM1 indicated that it was restricted to fetal pancreas, brain, and tumors of neuroendocrine origins [1,7]. In this study, we examined the temporal expression of mouse Insm1 at various stages of mouse embryogenesis. A 3.4-kb band corresponding to mouse Insm1 mRNA was first seen in embryos at 10.5 days, remained strongly expressed through 13.5 days, and then gradually decreased in intensity (Fig. 3A). Northern blots on postpartum brain showed a strong 3.4-kb band for up to 2 weeks with little or no expression thereafter (Fig. 3B). Insm1 was not detected in the brain, liver, heart, spleen, lung, skeletal muscle, kidney, and testis of adult mice (data not shown).
FIG. 3.
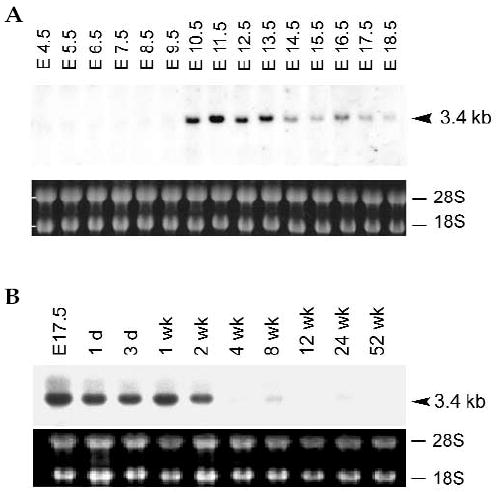
Northern blot analysis of mouse Insm1 expression during mouse embryo development. Total RNA obtained from embryos and post-partum brains was hybridized with a mouse Insm1 probe. A 3.4-kb band from embryos (A) and post-partum brains (B) revealed positive signals from day 10.5 until 2 weeks post-partum.
Expression and Nuclear Localization of Mouse Insm1
Because mouse Insm1 encodes a zinc-finger DNA-binding protein, it is likely to be expressed in the nuclear compartment. We determined the distribution of mouse INSM1 protein in the subcellular fractions of the βTC-1 cell line by differential centrifugation. Equal amount of protein from each fraction was separated on SDS-PAGE, transferred onto nitro-cellulose membranes and blotted with mouse INSM1 antibodies. We showed by western blot analysis that the INSM1 protein is concentrated in whole cell lysate and the nuclear fraction as a 58-kDa band (Fig. 4). This band was not found in the mitochondria, microsome, or cytosol fractions. To confirm the cellular fractionation data, green fluorescent protein (GFP) fused to mouse INSM1 and the GFP vector alone were transiently transfected into HeLa and Calu-6 cells. The Insm1-transfected cells showed nuclear fluorescence with little or no fluorescence in the cytoplasm (Figs. 5A–5D).
FIG. 4.
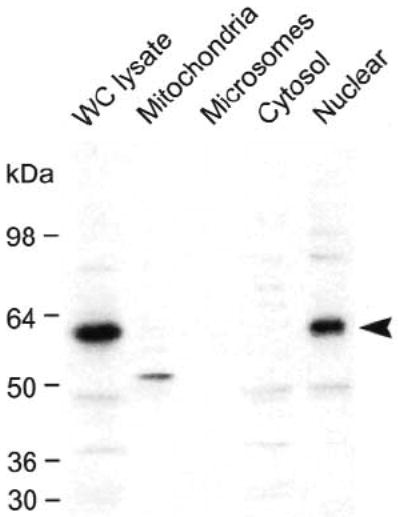
Cellular fractionation of mouse INSM1 protein in βTC-1 cells. Approximately 15 μg of proteins isolated from whole cell (WC) lysate, mitochondria, microsome, cytosol, and nuclear extract fractions were separated on 12% SDS-PAGE, transferred to a nitrocellulose membrane, and blotted with affinity purified rabbit polyclonal INSM1 antibody. The mouse INSM1 protein, approximately 58 kDa in size, is found in the whole cell lysate and the nuclear fraction. Nonspecific minor bands are observed in several of the fractions.
FIG. 5.
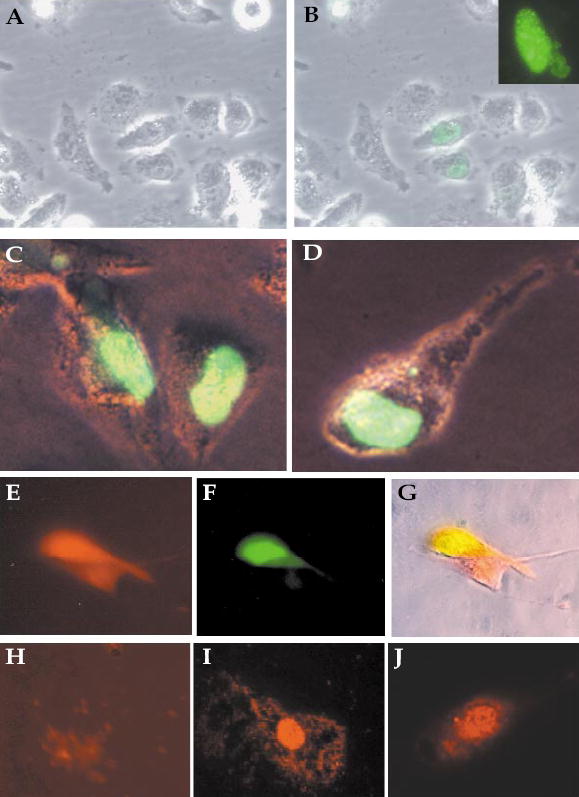
Nuclear localization of mouse INSM1 by immunofluores-cence staining. Untransfected Calu-6 cells (A), Calu-6 cells (B), and HeLa cells (C, D) were transfected with the GFP–INSM1 constructs. The dot-like distribution of GFP–INSM1 in the nucleus was observed by confocal microscopy (inset). (E–G) Calu-6 cells cotransfected with plasmids expressing RFP–CAP (E) and GFP–INSM1 (F). (G) Merge of the two fields. Endogenous INSM1 (I) and CAP (J) proteins detected in the nucleus of HIT-T15 cells stained with antibodies to INSM1 or CAP, respectively. (H) HIT-T15 cells incubated with preimmune serum.
Identification of CAP as a Mouse INSM1 Interacting Protein by the Yeast Two-Hybrid System
Using full-length INSM1 as the bait, we screened βTC-1 and mouse brain libraries for interacting proteins. Positive clones were retested for interaction with INSM1 by triple-nutrient selection. We isolated and sequenced 20 clones that survived nutrient selection and were LacZ positive. Two of them contained an overlapping 1.4-kb insert with an ORF of 1113 bp encoding a polypeptide of 371 amino acids. Sequence analysis of this INSM1 interacting polypeptide (designated A13) revealed that it contained the three SH3 domains (amino acids 486–724) of CAP (GenBank acc. no. AF078666). A search of the GenBank database uncovered a 168-bp insert in A13 representing an alternative splice variant of CAP [12,17]. This 56-amino-acid insert is located immediately after amino acid 483 of mouse CAP and, based on the human genomic DNA sequence (GenBank acc. no. AL160288), corresponds to exon 25 [17]. Both the mouse and human isoforms of CAP contain a potential NLS located immediately before the three SH3 domains of CAP.
Mouse INSM1 Interacts with CAP in a GST Pull-Down Assay
To see whether CAP interacted with INSM1 in vitro, we carried out GST pull-down protein-interaction assays. A partial-length CAP isoform (amino acids 435–740) was produced as a GST fusion protein (Fig. 6A) and immobilized on glutathione Sepharose beads. [35S]Cystine/methionine-labeled INSM1 was produced by in vitro transcription-coupled translation as a 58-kDa protein (Fig. 6B, lane 1) similar in size to the endogenous INSM1 protein detected in the nuclear fraction of βTC-1 cells. For pull-down experiments, the GST–CAP fusion protein that had been immobilized on glutathione Sepharose beads was incubated with [35S]cystine/methionine-labeled INSM1, eluted, and separated on a 12% SDS-PAGE gel. INSM1 was readily pulled down by GST–CAP (Fig. 6B, lane 3), but not by GST alone (Fig. 6B, lane 2).
FIG. 6.
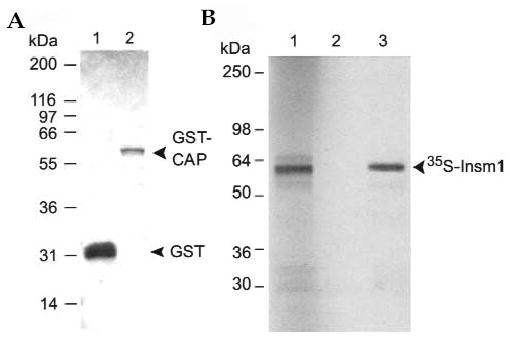
Mouse INSM1 interacts with CAP in vitro. (A) Expressed and purified GST (lane 1) and GST–CAP (lane 2) fusion proteins were electrophoresed on a 12% SDS-PAGE and stained by Coomassie blue. (B) The GST–CAP fusion protein was immobilized on glutathione Sepharose beads and then incubated with 35S-labeled INSM1. After extensive washing, bound [35S]INSM1 was eluted and separated on a 12% SDS-PAGE. Lane 3 shows that the 58-kDa 35S-labeled INSM1 protein interacted with and was pulled down by GST–CAP, but not by GST alone (lane 2). The first lane, loaded with 35S-labeled INSM1 protein, served as a size control.
Mouse INSM1 Co-immunoprecipitates with CAP in Mammalian Cells
The strong expression of c-myc tagged CAP and GFP tagged INSM1 fusion proteins in transfected Calu-6 and HeLa cells is seen in Fig. 7A. For co-immunoprecipitation assays, we cotransfected Calu-6 and HeLa cells with c-myc–CAP and GFP–INSM1. Cell lysates were immunoprecipitated with antibody to c-myc, electrophoresed on 12% SDS-PAGE, transferred to nitrocellulose, and then western blotted with antibody to either c-myc or GFP. Immunoprecipitation of the cell lysate with antibody to c-myc pulled down both c-myc–CAP (lanes 1 and 2) and GFP–INSM1 (lanes 4 and 5) suggesting that INSM1 and CAP interacted with each other in transfected cells (Fig. 7B). Furthermore, immunoprecipitation of c-myc–CAP transfected Calu-6 cells with antibody to c-myc co-precipitated endogenous INSM1 (Fig. 7C).
FIG. 7.
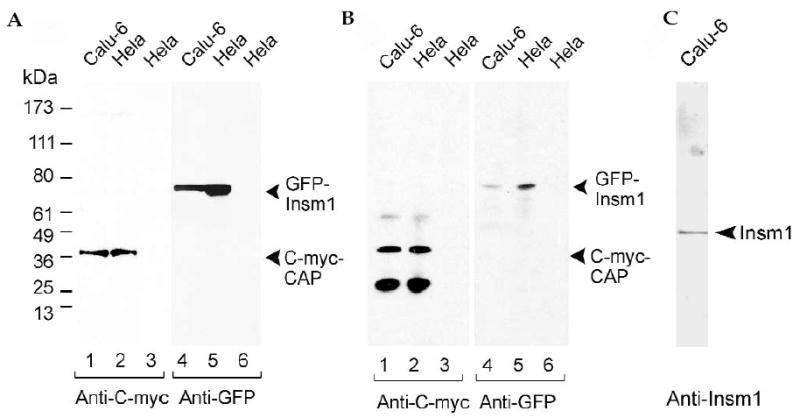
Co-immunoprecipitation of mouse INSM1 and CAP in cultured cells. (A) Expression of CAP tagged with c-myc (lanes 1 and 2) and mouse INSM1 tagged with GFP (lanes 4 and 5) in Calu-6 (lanes 1 and 4) and HeLa (lanes 2 and 5) cells. Lanes 3 and 6 represent untransfected cells. Cell extracts were separated on SDS-PAGE, transferred to a nitrocellulose membrane, and stained with antibody to c-myc or GFP. The 37-kDa and 78-kDa bands, respectively, show that CAP and INSM1 were strongly expressed in Calu-6 and HeLa cells. (B) Calu-6 and HeLa cells were co-transfected with c-myc–CAP and INSM1–GFP. Cell lysates were immunoprecipitated with antibody to c-myc coupled to Sepharose beads (or Sepharose beads alone), transferred to a nitrocellulose membrane, and stained with antibody to c-myc or GFP. Lanes 1 and 2 show that immunoprecipitation with antibody to c-myc pulled down c-myc–CAP and lanes 4 and 5 show that GFP–INSM1 was co-precipitated with c-myc–CAP. Lanes 3 and 6 represent untransfected cells. The 61- and 27-kDa bands in lanes 1 and 2 are the heavy and light chains of the antibody to c-myc used for immunoprecipitation. (C) Calu-6 cells that express endogenous INSM1 were transfected with c-myc–CAP. Cell lysates were immunoprecipitated with antibody to c-myc that had been coupled to Sepharose beads, electrophoresed on SDS-PAGE, transferred to a nitrocel-lulose membrane, and stained with antibody to INSM1. Endogenous INSM1 co-precipitated in the c-myc–CAP pull-down.
Mouse INSM1 and CAP Colocalize in the Nucleus of Transfected and Untransfected Cells
We cotransfected Calu-6 cells with plasmids expressing GFP–INSM1 and RFP–CAP. Both INSM1 and CAP were expressed in the nucleus (Figs. 5E–5G). Endogenous INSM1 and CAP proteins also were found in the nucleus of the hamster insulinoma cell line, HIT-T15, by staining with flu-orescein-labeled antibodies to INSM1 and CAP (Figs. 5H–5J).
Discussion
We have shown here that mouse Insm1 is an intron-less gene that encodes a protein of 521 amino acids and is located on chromosome 2. It is 11 amino acids longer than and 86% identical to its human counterpart. The C-terminal domain of mouse INSM1 has five putative zinc-finger DNA binding motifs of the form Cys-X2-4-Cys-Xl2-His-X3-4-His. The five zinc-finger motifs are identical in mice and humans. However, the first zinc finger of both human and mouse INSM1 has an arginine in place of the second histidine. The N-terminal domain of mouse INSM1 is proline-rich and contains a SNAG motif [3]. The SNAG motif of INSM1 differs from other members of the SNAG family in that a glycine replaces a serine at position four. Mouse INSM1 also has a nuclear localization signal (NLS) before the third zinc finger. Cellular fractionation, direct fluorescence microscopy with mouse INSM1 tagged with GFP, as well as immunostaining with antibody to mouse INSM1 showed that mouse INSM1 is located in the nucleus.
In terms of its function, recent studies showed that the zinc-finger domain of the INSM1 protein binds to a consensus DNA sequence of TG/TC/TC/TT/AGGGGG/TCG/A and functions as a transcriptional repressor [6]. Target sequence analysis revealed that the INSM1 protein regulates the mouse Neurod1 5′-upstream sequence. As Neurod1 has a crucial role in the development of the pancreas and nervous system, identification of INSM1 as a regulator of Neurod1 places INSM1 in the complex cascade of factors important for neuroendocrine development.
INSM1 has three proline-rich regions: two in the N-terminal region (amino acids 43–57 and 75–85) and one in the C-terminal region (amino acids 405–414), the latter being located between the second and third zinc-finger motifs. The function of these proline-rich regions is still not clear, but because proline-rich regions are known to have repressor activity by interfering with the initiation and/or assembly of transcriptional machinery [18,19], the C-terminal proline-rich region may contribute to the repressor activity of the INSM1 molecule. In some cases crucial protein–protein interactions involve only a very small sequence within a larger functional domain [20–22]. As proline-rich regions also are known to serve as binding sites for SH3 domains [23–25], it is likely that the two proline-rich regions at the N terminus of INSM1 serve as the binding site for CAP. In fact, ongoing studies indicate that CAP binds to the 180-amino-acid N terminus of INSM1, which contains the two proline-rich regions.
The finding that mouse Insm1 is transiently expressed during embryo development from day 10 to 2 weeks post-partum, but minimally or not at all thereafter, adds support to the idea that Insm1 has a role in development and differentiation. The expression of INSM1 mRNA in certain tumors [4] suggests that INSM1 also may be involved in tumorogenesis. Analysis of the protein database showed that both mouse and human INSM1 belong to a family of genes with a SNAG repressor motif at the N terminus and a zinc-finger transcriptional motif in the C-terminal half of the molecule. The SNAG transcriptional repressor family consists of at least 20 proteins and can be divided into three subfamilies [26]: Snail/Slug proteins, which are involved in organ development [27]; GFI1 (growth factor independence) proteins, which are involved in apoptosis and cell cycle arrest [3]; and INSM1 proteins, which seem to be involved in transcription regulation and cell differentiation [6,7]. In addition to mouse and human INSM1, the INSM1 family includes mouse MLT-1 (methylated in liver tumor) and a human homolog [26]. Mouse MLT-1 is 50% identical to human INSM1 and is thought to be a tumor suppressor gene that can be silenced by methylation. Human MLT-1 (INSM2), is 53% identical to human INSM1. Homologs of mouse and human INSM1 also are found in zebrafish, Drosophila, and C. elegans.
In addition to the interactions that can take place through the SNAG and zinc-finger motifs of INSM1, the present study shows that INSM1 can interact with a splice variant of CAP which contains an NLS. This was demonstrated by co-immunoprecipitation of either transfected GFP–INSM1 or endogenous INSM1 with anti-c-myc antibody to the c-myc tagged CAP fusion protein. CAP is found primarily in the cytoplasm of cells [10], but recently it also was found in the nucleus [12]. In spinocerebellar ataxia 7, CAP colocalized with ataxin-7 in neuronal intranuclear inclusions. In our studies, the presence of an NLS in both INSM1 and the CAP splice variant and the strong nuclear staining of both INSM1 and CAP proteins suggest that the INSM1–CAP interaction takes place in the nuclear compartment. CAP splice variants with a NLS are now known to exist in brain, retina, and skeletal muscle [12].
Besides its interaction with c-cbl, the C-terminal SH3 domain of CAP is known to interact and form complexes with a variety of other signaling molecules including focal adhesion kinase (FAK), son of sevenless (SOS), and the insulin receptor [9,11,28]. These interactions, in turn, can trigger a broad cascade of biological events. In this context it is interesting to speculate that CAP complexes might be recruited into the nucleus either through the binding of CAP to INSM1 and or through CAP’s own splice variant NLS. These CAP complexes then might affect the transcriptional machinery of INSM1-targeted downstream gene expression. The fact that mouse Insm1 is expressed during embryo development and in neuroendocrine tumors suggests that mouse Insm1 also may have a role in rapid tissue growth and differentiation. That this role may not be trivial is supported by the fact that some members of the SNAG family are tumor suppressors [26] and that homologs of Insm1 can be found in a variety of species going back to C. elegans and Drosophila.
Materials and Methods
Mouse Insm1 cDNA and genomic sequence
A phagemid cDNA library was constructed from βTC-1 cells with insert sizes ranging from 0.4 to 5 kb. The library was screened by standard filter hybridization methods using a full-length human INSM1 cDNA 32P-labeled probe. Positive clones were re-screened and two overlapping clones with insert sizes of 1.4 and 2.5 kb, were isolated and sequenced. To isolate Insm1 genomic DNA, a mouse 129/SVJ genomic DNA library (Stratagene, La Jolla, CA) was hybridized with [32P]-labeled full-length mouse Insm1 cDNA. Two positive clones with insert sizes of 12 and 21 kb were isolated and database searches performed.
Chromosomal mapping
The chromosomal location of mouse Insm1 was determined using the RB04.02 chromosome mapping panel (Research Genetics, Huntsville, AL). A 258-bp fragment was PCR-amplified with primers at the 3′-UTR of mouse Insm1 (sense, 5′-TTCGCTTCGGTGGAGCATGAC-3′; anti-sense, 5′-CAGAGATTGGTAGGCAGAGC-3′). The PCR results were used to query the RHserver for mapping results. Chromosomal mapping data was then integrated as centimorgans (cM) by querying the Mouse Genome Institute (MGI). In addition, zebrafish chromosomal mapping was carried out using the LN54 radiation hybrid panel [29], and two paired-primers were designed from the 3′-UTR region of zebra fish Insm1 (sense, 5′-CCTGTGTCACAT-GTTGCAGTCG-3′; antisense, 5′-TCGAGTAGACAACACGGACGCAC-3′). The PCR conditions were as described [30].
Northern blot analysis
Blots of mouse whole embryos and postnatal brain tissues (Seegene, Inc., Seoul, Korea) were probed with a 32P-labeled XbaI fragment (587 bp) from the 3′-UTR of mouse Insm1 cDNA. The blots were hybridized (PerfectHyb Plus Hybridization Buffer, Sigma-Aldrich, Inc., St Louis, MO), washed according to manufacturer’s specifications, and subjected to autoradiography at −80°C for 3 days.
Plasmid constructs
To express proteins in E. coli, mouse Insm1 (amino acids 1–521) and CAP (amino acids 435–740) were PCR amplified and ligated into a pGEX-4T-1 vector (Amersham Pharmacia, Arlington Heights, IL). For mammalian cell expression, a GFP–INSM1 fusion protein was constructed by ligating the mouse Insm1 cDNA into the eukaryotic expression vector pEGFP-C2 (Clontech, Palo Alto, CA). c-myc–CAP fusion protein was constructed by ligating the C terminus of the mouse CAP gene (amino acids 435–740) into a pCMV-Tag3 vector (Stratagene). All constructs generated by PCR were sequenced by an ABI Prism 377 automatic sequencer. To make a bait plasmid construct for two-hybrid screening, the Insm1 coding region was PCR amplified and subcloned into p-GAL4-BD Cam phagemid (Stratagene) and designated pGal4-BD-Insm1.
Construction of βTC-1 two-hybrid library
Total RNA was extracted from mouse βTC-1 cells and purified on an oligo-dT affinity column (Stratagene). Approximately 5 μg mRNA was used to construct a Matchmaker cDNA library with Stratagene HybriZAP Two-Hybrid cDNA Gigapack Cloning Kit according to the manufacturer’s instructions. The final phage library contained approximately 1.4 × 106 clones. The phage library was amplified in XL-1 blue MRF bacterial host cells. Excision of the pAD-Gal4 phagemid vector from the HybriZAP vector was performed with ExAssist helper phage. A pre-made mouse brain cDNA Matchmaker library was purchased from Clontech.
Yeast transformation and two-hybrid library screening
INSM1 bait (pGaL4-BD-Insm1) and an equal amount of a cDNA library (GAL4-AD-library) were used to transform YRG-2 yeast cells according to the manufacturer’s protocol (Stratagene). Yeast DNA (containing both bait and target phagemids) was prepared and used to transform E. coli, XL-1 blue. The phagemid clones containing the target DNA resistant to ampicillin treatment were selected for further confirmation and sequence analysis.
Antibodies and immunostaining
New Zealand white rabbits were immunized with purified recombinant mouse INSM1 and subjected to affinity column purification. Polyclonal anti-CAP peptide antibodies (amino acids CEKRAKDDSRRVVKST) were obtained from Upstate Biotechnology (Lake Placid, NY). All fluorescein-conjugated secondary antibodies were obtained from Jackson Immunoresearch Lab, West Grove, PA.
Cell lines and transfection
HeLa S-3 (human cervix adenocarcinoma), Calu-6 (human lung anaplastic carcinoma), and HIT-T15 (Syrian hamster insuli-noma) cell lines were obtained from ATCC (Manassas, VA). βTC-1 cells (a mouse insulinoma cell line) was kindly provided by Edward Leiter (The Jackson Laboratory). Column-purified plasmid DNA (Qiagen, Santa Clarita, CA) was used to transiently transfect mammalian cells using GenePORTER2 Transfection Reagent (Gene Therapy Systems, Inc., San Diego, CA). Cells were plated to 50–70% confluence and transfected with 10 μg of DNA (100 mm dish). Stable HeLa S-3 and Calu-6 cell lines containing c-myc–CAP were treated with G418 (1 mg/ml) for 3–4 weeks for selection. Of 10 G418-resistant colonies, 8 were confirmed to express c-myc–CAP protein using anti-c-myc antibody in western blot analysis.
Cellular fractionation
βTC-1 cells (2 × 108) were sonicated in PBS buffer and centrifuged at 500g for 10 minutes. The resuspended pellet was homogenized, pelleted, washed 3 times, resuspended in PBS buffer, and cleared by centrifugation (20,000g for 30 minutes). The first post-nuclear supernatant was used to purify mitochondria by the digitonin method [31]. All non-mitochondria fractions were pooled and centrifuged at 30,000g for 30 minutes. The resulting pellet was designated the microsome fraction and the supernatant was designated the cytosol fraction.
Direct fluorescence and immunofluorescence microscopy
Transfected cells were grown on a 2-well chamber slide (Nalge Nunc International Corp., Naperville, IL) for 48 hours and examined for expression of GFP–INSM1 fusion protein. For confocal studies, cells were fixed with 2% paraformaldehyde for 10 minutes, and then examined using a TCS4D confocal laser scanning microscope (Leica Microsystems, Heidelberg, Germany). For immunofluorescence studies, cells were fixed with 4% paraformaldehyde for 15 minutes, rinsed three times, incubated for 15 minutes in 50 mM NH4Cl/PBS, washed, and then permeabilized with 0.1% Triton X-100 for 10 minutes. Slides then were incubated with appropriate primary and secondary antibodies.
Protein expression and pull-down assay
GST fusion proteins were expressed in E. coli strain BL21 by induction with 1 mM isopropyl-β-d-thiogalactoside (IPTG) at 37°C for 3 hours. Cells then were disrupted by sonication in PBS containing 0.5% Triton X-100 and 1 mM PMSF. GST fusion proteins were purified by passage through a glutathione agarose affinity column. INSM1 protein was excised from GST–INSM1 by thrombin digestion. Trace amounts of thrombin were removed from INSM1 by Glutathione Sepharose 4B (Amersham Pharmacia Biotech) absorption. Purified GST–CAP fusion proteins were used directly in protein–protein interaction assays. Purified plasmid pBluescript-Insm1 was used as template for in vitro transcription-coupled translation (Promega, Madison, WI) with [35S]cystine/methionine (Amersham Pharmacia Biotech).
GST pull-down protein-interaction experiments were performed by immobilizing GST or GST-fusion proteins on the glutathione Sepharose beads. [35S]INSM1 was added to the beads in binding buffer (10 mM Tris-HCl, pH 7.5, 10% glycerol, 0.1 mM EGTA, 100 mM KCL, 0.1% NP40, 0.1% Triton X-100, 1 mM DTT). Unbound protein was removed and the beads were washed eight times with buffer. The immobilized GST target proteins and [35S]INSM1 that bound to the beads were eluted by boiling for 5 minutes in 13 Laemmli SDS-PAGE sample buffer. Samples were analyzed by SDS-PAGE. Gels were stained with Coomassie blue for GST and GST-tagged proteins and radioautographed for detection of INSM1 protein.
Protein isolation and western blot
Cell lysates were prepared with M-Per Mammalian Protein Extraction Reagent (PIERCE, Rockford, IL). Aliquots of the supernatant containing 50 μg of protein were boiled in 1× NuPAGE LDS sample buffer, electrophoresed on 4–15% NuPAGE Bis-Tris gel and transferred electrophoretically to nitrocellulose membranes (Invitrogen, Carlsbad, CA). c-myc–CAP and GFP–INSM1 expression were detected using the Western-light Chemiluminescent Detection System (Tropix, Inc., Bedford, MA).
Co-immunoprecipitation
Cultured cell lysates (500 μl), cleared with formalin-fixed (4%) Staphylococcus aureus crude cell suspension (Sigma-Aldrich), were mixed with 20 μl of Sepharose beads, conjugated with mouse anti-c-myc antibody (2 μg/μl, Clone 9E10; Research Diagnostics, Inc., Flanders, NJ), or 20 μl Sepharose beads (control), and then mixed overnight at 4°C. Protein complexes which bound to the Sepharose beads conjugated with anti-c-myc antibody were collected by centrifugation. After washing four times with a reaction buffer (20 mM Tris, pH 8.0, 150 mM NaCl, and 0.5% Triton X-100, and 0.05% sodium azide), protein complexes were resuspended in 50 μl 1× NuPAGE LDS sample buffer. Proteins then were eluted and analyzed by western blot.
Acknowledgments
We thank M. Tsang for the chromosomal mapping of Insm1 in zebrafish, and Charles Wohlenberg for technical help.
Footnotes
Sequence data from this article have been deposited in the DDBJ/EMBL/GenBank Data Libraries under accession number NM_016889 (Insm1).
References
- 1.Goto Y, et al. A novel human insulinoma-associated cDNA, IA-1, encodes a protein with “zinc-finger” DNA-binding motifs. J Biol Chem. 1992;267:15252–15257. [PubMed] [Google Scholar]
- 2.Lan MS, Li Q, Lu J, Modi WS, Notkins AL. Genomic organization, 5’-upstream sequence, and chromosomal localization of an insulinoma-associated intron-less gene, IA-1. J Biol Chem. 1994;269:14170–14174. [PubMed] [Google Scholar]
- 3.Grimes HL, Chan TO, Zweidler-McKay PA, Tong B, Tsichlis PN. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol Cell Biol. 1996;16:6263–6272. doi: 10.1128/mcb.16.11.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lan MS, Russell EK, Lu J, Johnson BE, Notkins AL. IA-1, a new marker for neuroendocrine differentiation in human lung cancer cell lines. Cancer Res. 1993;53:4169–4171. [PubMed] [Google Scholar]
- 5.Li Q, Notkins AL, Lan MS. Molecular characterization of the promoter region of a neuroendocrine tumor marker, IA-1. Biochem Biophys Res Commun. 1997;236:776–781. doi: 10.1006/bbrc.1997.7054. [DOI] [PubMed] [Google Scholar]
- 6.Breslin MB, Zhu M, Notkins AL, Lan MS. Neuroendocrine differentiation factor, IA-1, is a transcriptional repressor and contains a specific DNA-binding domain: identification of consensus IA-1 binding sequence. Nucleic Acids Res. 2002;30:1038–1045. doi: 10.1093/nar/30.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu M, Breslin MB, Lan MS. Expression of a novel zinc-finger cDNA, IA-1, is associated with rat AR42J cells differentiation into insulin-positive cells. Pancreas. 2002;24:139–145. doi: 10.1097/00006676-200203000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Sparks AB, Hoffman NG, McConnell SJ, Fowlkes DM, Kay BK. Cloning of ligand targets: systematic isolation of SH3 domain-containing proteins. Nat Biotechnol. 1996;14:741–744. doi: 10.1038/nbt0696-741. [DOI] [PubMed] [Google Scholar]
- 9.Ribon V, Printen JA, Hoffman NG, Kay BK, Saltiel AR. A novel, multifuntional c-Cbl binding protein in insulin receptor signaling in 3T3-L1 adipocytes. Mol Cell Biol. 1998;18:872–879. doi: 10.1128/mcb.18.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandai K, et al. Ponsin/SH3P12: an l-afadin- and vinculin-binding protein localized at cell-cell and cell-matrix adherens junctions. J Cell Biol. 1999;144:1001–1017. doi: 10.1083/jcb.144.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baumann CA, et al. CAP defines a second signalling pathway required for insulin-stimulated glucose transport. Nature. 2000;407:202–207. doi: 10.1038/35025089. [DOI] [PubMed] [Google Scholar]
- 12.Lebre AS, et al. Ataxin-7 interacts with a Cbl-associated protein that it recruits into neuronal intranuclear inclusions. Hum Mol Genet. 2001;10:1201–1213. doi: 10.1093/hmg/10.11.1201. [DOI] [PubMed] [Google Scholar]
- 13.Lin WH, et al. Molecular scanning of the human sorbin and SH3-domain-containing-1 (SORBS1) gene: positive association of the T228A polymorphism with obesity and type 2 diabetes. Hum Mol Genet. 2001;10:1753–1760. doi: 10.1093/hmg/10.17.1753. [DOI] [PubMed] [Google Scholar]
- 14.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birnstiel ML, Busslinger M, Strub K. Transcription termination and 3’ processing: the end is in site! Cell. 1985;41:349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- 16.Stivers C, Brody T, Kuzin A, Odenwald WF. Nerfin-1 and -2, novel Drosophila Zn-finger transcription factor genes expressed in the developing nervous system. Mech Dev. 2000;97:205–210. doi: 10.1016/s0925-4773(00)00409-3. [DOI] [PubMed] [Google Scholar]
- 17.Lin WH, et al. Cloning, mapping, and characterization of the human sorbin and SH3 domain containing 1 (SORBS1) gene: a protein associated with c-Abl during insulin signaling in the hepatoma cell line Hep3B. Genomics. 2001;74:12–20. doi: 10.1006/geno.2001.6541. [DOI] [PubMed] [Google Scholar]
- 18.Guiral M, Bess K, Goodwin G, Jayaraman PS. PRH represses transcription in hematopoietic cells by at least two independent mechanisms. J Biol Chem. 2001;276:2961–2970. doi: 10.1074/jbc.M004948200. [DOI] [PubMed] [Google Scholar]
- 19.Bonni S, et al. TGF-β induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat Cell Biol. 2001;3:587–595. doi: 10.1038/35078562. [DOI] [PubMed] [Google Scholar]
- 20.Madden SL, Cook DM, Rauscher FJ., 3rd A structure-function analysis of transcriptional repression mediated by the WT1, Wilms’ tumor suppressor protein. Oncogene. 1993;8:1713–1720. [PubMed] [Google Scholar]
- 21.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 22.Fisher AL, Ohsako S, Caudy M. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol Cell Biol. 1996;16:2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu H, et al. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 24.Musacchio A, Saraste M, Wilmanns M. High-resolution crystal structures of tyrosine kinase SH3 domains complexed with proline-rich peptides. Nat Struct Biol. 1994;1:546–551. doi: 10.1038/nsb0894-546. [DOI] [PubMed] [Google Scholar]
- 25.Goudreau N, et al. NMR structure of the N-terminal SH3 domain of GRB2 and its complex with a proline-rich peptide from Sos. Nat Struct Biol. 1994;1:898–907. doi: 10.1038/nsb1294-898. [DOI] [PubMed] [Google Scholar]
- 26.Tateno M, et al. Identification of a novel member of the snail/Gfi-1 repressor family, mlt 1, which is methylated and silenced in liver tumors of SV40 T antigen trans-genic mice. Cancer Res. 2001;61:1144–1153. [PubMed] [Google Scholar]
- 27.Hemavathy K, Ashraf SI, Ip YT. Snail/slug family of repressors: slowly going into the fast lane of development and cancer. Gene. 2000;257:1–12. doi: 10.1016/s0378-1119(00)00371-1. [DOI] [PubMed] [Google Scholar]
- 28.Kurakin A, Hoffman NG, Kay BK. Molecular recognition properties of the C-terminal Sh3 domain of the Cbl associated protein, Cap. J Pept Res. 1998;52:331–337. doi: 10.1111/j.1399-3011.1998.tb00657.x. [DOI] [PubMed] [Google Scholar]
- 29.Hukriede NA, et al. Radiation hybrid mapping of the zebrafish genome. Proc Natl Acad Sci USA. 1999;96:9745–9750. doi: 10.1073/pnas.96.17.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai T, Krause MW, Odenwald WF, Toyama R, Notkins AL. The IA-2 gene family: homologs in Caenorhabditis elegans, Drosophila and zebrafish. Diabetologia. 2001;44:81–88. doi: 10.1007/s001250051583. [DOI] [PubMed] [Google Scholar]
- 31.Moreadith RW, Fiskum G. Isolation of mitochondria from ascites tumor cells permeabilized with digitonin. Anal Biochem. 1984;137:360–367. doi: 10.1016/0003-2697(84)90098-8. [DOI] [PubMed] [Google Scholar]


