Abstract
The virus–host interactions that influence hepatitis C virus (HCV) replication are largely unknown but are thought to involve those that disrupt components of the innate intracellular antiviral response. Here we examined cellular antiviral pathways that are triggered during HCV RNA replication. We report that (i) RNA replication of HCV subgenomic replicons stimulated double-stranded RNA (dsRNA) signaling pathways within cultured human hepatoma cells, and (ii) viral RNA replication efficiency corresponded with an ability to block a key cellular antiviral effector pathway that is triggered by dsRNA and includes IFN regulatory factor-1 (IRF-1) and protein kinase R (PKR). The block to dsRNA signaling was mapped to the viral nonstructural 5A (NS5A) protein, which colocalized with PKR and suppressed the dsRNA activation of PKR during HCV RNA replication. NS5A alone was sufficient to block both the activation of IRF-1 and the induction of an IRF-1-dependent cellular promoter by dsRNA. Mutations that clustered in or adjacent to the PKR-binding domain of NS5A relieved the blockade to this IRF-1 regulatory pathway, resulting in induction of IRF-1-dependent antiviral effector genes and the concomitant reduction in HCV RNA replication efficiency. Our results provide further evidence to support a role for PKR in dsRNA signaling processes that activate IRF-1 during virus infection and suggest that NS5A may influence HCV persistence by blocking IRF-1 activation and disrupting a host antiviral pathway that plays a role in suppressing virus replication.
Virus infection of mammalian cells triggers an innate intracellular antiviral response, in part through the accumulation of viral double-stranded RNA (dsRNA) replication products (1). dsRNA provides an immediate signal for the activation of specific transcription factors, including NFκB, IFN regulatory factor (IRF)-1, and IRF-3, which stimulate the expression of host antiviral and antiproliferative gene products (2). The activation of IRF-1 is proposed to proceed through a protein kinase R (PKR)-dependent pathway, in which PKR indirectly signals the modification of IRF-1 and activation of its DNA-binding activity (3). Active IRF-1 binds to the IRF-E motif within the IFN-stimulated response promoter element (ISRE) and participates in the induction of cellular genes that impact virus replication either directly or through stimulation of the adaptive immune response (4).
Infection with hepatitis C virus (HCV) frequently results in a persistent infection, suggesting that it has evolved efficient mechanism(s) for blocking the host cell's innate antiviral response. Two HCV proteins in particular, NS5A and E2, have been implicated in the ability of the virus to regulate the host response to dsRNA. Both proteins have been shown to bind and inhibit PKR (5, 6). Moreover, some molecular epidemiological studies have correlated the sequence of the PKR-interacting region of E2 or NS5A with HCV persistence and the widespread resistance to the current IFN-based therapy for HCV infection (7, 8). This finding suggests that E2 and NS5A may contribute to viral persistence by inhibiting the intracellular antiviral response that is triggered by dsRNA and signaled by PKR (5, 9). In this study we examined the impact of HCV RNA replication and protein expression on the innate antiviral response induced by dsRNA in human cells. We report that dsRNA-responsive antiviral pathways are triggered during HCV RNA replication, and that NS5A regulation of dsRNA-signaling events may alter the host environment sufficiently to favor persistent virus replication.
Methods
Preparation of the HCV Replicon and cDNA Expression Constructs.
The HCV replicon cDNA, pHCVneo17, was assembled by using overlapping oligonucleotides according to the sequence of the HCV genotype 1b replicon I377/NS3–3′ (10). The assembled cDNA was cloned into a pUC-derived recipient plasmid adjacent to a T7 RNA polymerase-promoter site to yield pHCVNeo17. Human IRF-1 was cloned from IFN-treated UHCV11 cells by reverse transcription–PCR (RT-PCR) using Moloney murine leukemia virus reverse transcriptase (Promega) and an oligo(dT) primer as directed by the manufacturer. Primer pairs specific for IRF-1 were used to PCR amplify IRF-1, and the product was cloned into pCR2.1 (Invitrogen) to yield pCR2.1-IRF-1. pFLAG-CMV, pFLAG-NS5A 1B-1 (9), pCDNA1neo, pcDNA1neo-PKRK296R, pcDNA1neo PKR wild type (wt; ref. 11), pIFNβ-luc (12), and pGbp2-luc (13) have been described. The NS5A-coding region of replicon clone 10A was amplified by RT-PCR from total RNA recovered from replicon cell line 10A. The resulting cDNA was cloned into the HindIII site of pFLAG CMV to yield pFLAG-NS5A 10A. The assembly of the HCV replicon cDNA and the cloning of each expression construct was verified by nucleotide sequence analysis. The sequences of oligonucleotide primers used in this study are available on request.
Cell Culture and Transfection.
All cells were propagated in DMEM supplemented with 10% FBS, 200 μM l-glutamine, penicillin, and streptomycin. Parental Huh7 cells and primary replicon cell isolates were prepared by M. Giovani and R. DeFrancesco (Instituto di Ricerche di Biologia Moleculare P. Angeletti SPA/Merck). In vitro transcription of pHCVneo17, RNA transfection, and recovery of Huh7 cells harboring the HCV replicon RNA were conducted exactly as described (10). From this process, Huh7 cells, termed 10A and H27, which harbor distinct HCV RNA replicon quasispecies, were isolated in 400 μg/ml G418 and characterized. The absence of pHCVNeo17 DNA in 10A and H27 cells was confirmed by PCR analysis, and authentic HCV RNA replication was confirmed by RT-PCR for the viral negative-strand replicative intermediate. The complete nucleotide and deduced amino acid sequence of the H27 and 10A replicons were determined directly from purified RT-PCR products. Passaged replicon cell populations were generated by transfecting fresh Huh7 cells with 200 ng of total RNA isolated from the primary 10A and H27 replicon cells. Cells harboring the passaged replicon RNA were selected for 14 days in 400 μg/ml G418. For tetracycline-regulated protein expression, the indicated cell lines were cultured in DMEM ± 1 μg/ml tetracycline. wt and Pkr-null mouse embryo fibroblasts (MEFs) were cultured as described (14).
For dsRNA treatment, cell culture was conducted in the presence of 40 μg/ml poly inosine:cytosine (pIC) as described (13). Cells were then incubated for 2 h and harvested, and extracts were prepared for protein analysis. Where indicated, dsRNA treatment was accompanied by a simultaneous incubation with 100 units/ml IFN-α. Superfect (Qiagen) was used to transfect cells with plasmid DNA. For luciferase assays, cells were harvested 2 h after dsRNA treatment and processed by using the Luciferase Assay Kit (Promega). Transfection efficiencies were monitored by cotransfection with pSVβgal (Promega) and determination of β-galactosidase activity in cell lysates.
Protein Analysis.
In vitro translation products were synthesized by using the TNT system (Promega). Immunoblot and immunoprecipitation analyses were conducted exactly as described (9). For electrophoretic mobility-shift assayss (EMSAs), nuclear extracts were isolated and subject to EMSA exactly as described (12), using 32P-labeled double-stranded oligonucleotide probes corresponding to the ISRE of ISG15, ISG54, or the synthetic IRF-E/C1 oligomer (15). PKR in vitro protein kinase assay was performed on anti-PKR immunocomplexes (16) in the presence of 20 ng of recombinant eukaryotic initiation factor 2α. Immunofluorescence analysis of PKR and NS5A in HCV replicon cells was conducted as described (16), using a 1:100 dilution of anti-NS5A rabbit serum and 1:200 dilution of anti-PKR 71/10 mAb, followed by incubation with 1:1,000 dilutions of donkey anti-rabbit Texas red and goat anti-mouse FITC Abs. Cells were visualized by fluorescence microscopy.
mRNA Expression and HCV RNA Quantitation.
Expression of Gbp2, Lmp2, 2′-5′oligoadenylate synthetase, and actin was assessed from control and HCV replicon cells by RT-PCR from total RNA, an oligo(dT) primer, and Moloney murine leukemia virus reverse transcriptase. Then 2 μl of product from each reaction was subjected to gene-specific PCR for 22 cycles. Quantitation of viral RNA from replicon cultures was performed by real-time RT-PCR procedure (17) by using the Applied Biosystems Prism 7700 sequence detection system.
Results
Characterization of Distinct Quasispecies of the HCV Subgenomic Replicon.
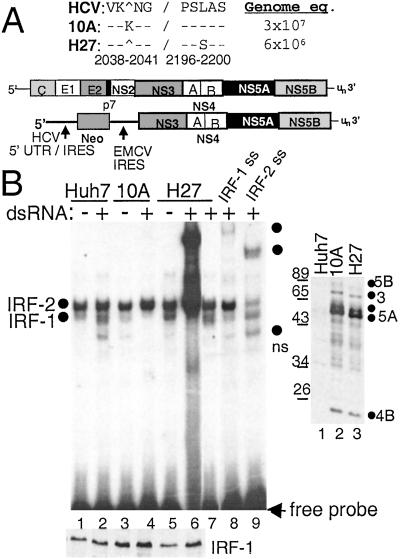
The recent development of the subgenomic HCV RNA replicon system by Bartenschlager and coworkers (10) has established the first suitable cell culture/HCV replication system for the examination of virus–host interactions. An HCV genotype 1B replicon was therefore constructed based on this system. Huh7 cell lines, termed 10A and H27, were isolated that harbored replicons encoding distinct quasispecies of the HCV subgenome (Fig. 1A) and supported the persistent replication of the HCV RNA.
Figure 1.
Viral RNA replication efficiency and IRF-1 regulation of HCV replicon quasispecies. (A) Organization of the HCV genome and subgenomic replicon, with NS5A sequence variation between replicon clones 10A and H27. The amino acid positions are numbered according to the HCV 1b reference sequence, GenBank accession no. D11168. Replicon copy number is expressed as genome equivalents per micrograms of total cellular RNA. (B Right) Immunoblot analysis of HCV proteins in parental Huh7 cells (lane 1) or cells harboring replicon 10A (lane 2) or H27 (lane 3). The blot was probed with anti-HCV patient serum, and the positions of individual HCV proteins were identified by parallel comparison to identical blots that were probed separately with HCV protein-specific antisera. (B Left) Parental Huh-7 cells (lanes 1, 2, 8, and 9) or cells harboring replicon 10A (lanes 3 and 4) or H27 (lanes 5–7) were cultured for 2 h in the presence (+) or absence (−) of dsRNA. Nuclear extract (2 μg) was subjected to EMSA (Upper) or immunoblot analysis for IRF-1 protein levels (Lower). EMSA was conducted by using a probe encoding the ISRE of ISG54. Complexes were identified as IRF-1 or IRF-2 by supershift (ss) analysis of extracts from dsRNA-treated Huh7 cells by using Abs (Santa Cruz Biotechnology) to IRF-1 (lane 8) or IRF-2 (lane 9). Lane 7 shows the resolved complexes within a 10-fold dilution of the extract shown in lane 6. Dots indicate the probe-bound complexes. ns denotes a nonspecific complex not reproducibly observed. In B Upper entire gel is shown; all subsequent EMSA panels show only the region of the gel with probe-bound complexes.
Direct nucleotide sequencing of the full-length replicon RT-PCR product from RNA derived from 10A and H27 cells revealed the presence of nucleotide substitutions scattered throughout the HCV-coding region. However, nonsynonymous mutations were identified only in the NS5A-coding region of each replicon and were limited to a single codon each. Replicon 10A encoded an in-frame K insertion at amino acid position 2040, located near the amino terminus of NS5A. The H27 replicon encoded a single amino acid substitution, L2198S, located within the major phosphorylation region of NS5A and just adjacent to the amino-terminal end of the PKR-binding domain (18).
In exponentially growing cultures, replicon 10A replicated to >5-fold higher levels than replicon H27, but we did not observe a difference in steady-state protein expression levels between the two replicons (Fig. 1). Thus, the K2040 insertion provides a greater level of efficiency to viral RNA replication than does the L2198S mutation of the H27 variant, implying that these mutations confer different functional properties to NS5A. Previous studies have identified the NS5A-coding region as a major site for mutations that confer cell culture adaptation to HCV subgenomic replicons (19, 20), suggesting important roles for NS5A in supporting HCV RNA replication. Our results support this notion and identify two additional sites within NS5A that affect HCV RNA replication.
Differential Regulation of IRF-1 by HCV Replicon Quasispecies.
We hypothesized that the differences in the level and efficiency of HCV RNA replication may depend, in part, on the ability of NS5A to antagonize the dsRNA-responsive antiviral pathways of the host cell. We therefore used EMSA to characterize dsRNA signaling to the ISRE in parental Huh7 cells and cells harboring the 10A or H27 replicon quasispecies. As seen in Fig. 1B, we identified IRF-1 and IRF-2 as factors in Huh7 cells that bind the ISRE. All cell lines exhibited the constitutive DNA-binding activity of IRF-2 (21), whereas cells harboring the H27 replicon exhibited a basal level of active, DNA-bound IRF-1 that was not apparent in extracts from the control Huh7 or replicon 10A cells. Treatment with dsRNA stimulated the DNA-binding activity of IRF-1 in Huh7 and H27 cells but not in cells harboring the 10A replicon. Moreover, H27 cells were hyperactive to exogenous dsRNA and exhibited a superresponse of an ≈10-fold excess of IRF-1 binding over their basal level and over dsRNA-treated parental Huh7 cells (Fig. 1B, see lanes 2, 5, 6, and 7). Importantly, IRF-1 was expressed to similar levels in all cell lines (Fig. 1B, Lower). These results demonstrate that (i) dsRNA-induced antiviral pathways are intact and can be activated in Huh7 and replicon H27 cells, and (ii) replication of clone 10A renders a specific block in antiviral-signaling pathways that activate IRF-1 in response to dsRNA. The basal level of active IRF-1 within replicon H27 cells and the hypersensitivity of these cells to exogenous dsRNA implies that HCV RNA replication may stimulate or “prime” host dsRNA-signaling pathways, perhaps through dsRNA structures encoded within the HCV genome and replication intermediates (22).
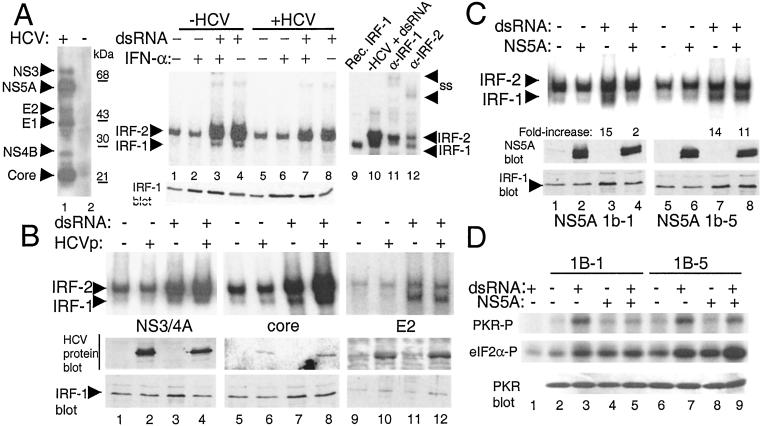
Characterization of HCV Proteins Involved in the Regulation of IRF-1.
To identify the HCV-encoded protein(s) that influence the dsRNA-induced activation of IRF-1, we conducted comparative EMSA on extracts prepared from human cell lines expressing either the entire HCV ORF or subgenomic fragments of HCV. In the absence of HCV protein expression, dsRNA stimulated the activation of IRF-1 (Fig. 2A, lanes 3 and 4). In contrast, dsRNA failed to activate IRF-1 upon expression of the full HCV ORF (Fig. 2A, lanes 7 and 8), confirming that HCV genome expression can render a block in the IRF-1 activation pathway induced by dsRNA. Importantly, the block to IRF-1 activation could not be overcome by cotreatment of cells with IFN-α, which resulted in increased levels of IRF-1 expression (Fig. 2A Lower).
Figure 2.
IRF-1 regulation by HCV proteins. (A Left) Immunoblot of HCV proteins in UHCV11 cells, harboring the ORF from HCV 1A (35), cultured to induce (+HCV; lane 1) or repress (−HCV; lane 2) ORF expression. The blot was probed with anti-HCV patient serum. (A Right) HCV ORF expression in UHCV11 cells was repressed (lanes 1–4; −HCV) or induced (lanes 5–8; +HCV), and cells were left untreated or treated with IFN-α and/or dsRNA as indicated. Nuclear extracts were subjected to EMSA by using a labeled ISG15 ISRE probe. Lanes 9–12 show the positive identification (indicated by arrows) of IRF-1 or IRF-2 complexes by using recombinant. in vitro-translated IRF-1 (lane 9), or anti-IRF-1 (lane 11) or anti-IRF-2 Abs (lane 12) to generate supershift (SS) complexes derived from the extract represented in lane 10. (A Lower) An immunoblot of IRF-1 protein levels within the extract from each corresponding lane. (B and C Upper) EMSA of nuclear extracts from UNS3/4A cells (36) (lanes 1–4; expressing NS3 and NS4A), UTH⋅28 cells (37) (lanes 5–8; expressing core), or HE2 cells (lanes 9–12; expressing the HCV 1A E2 protein) (J.P. and M.G., unpublished data) (B), and HNS5A 1b-1 or HNS5A 1b-5 cells (9) (C) cultured to control the tetracycline-regulated expression of the respective HCV proteins. Above each lane − and + indicate expression of the HCV proteins (HCVp) or treatment with dsRNA. EMSA was conducted with a labeled C1 probe. The positions of the IRF-1 and IRF-2 complexes are indicated. (C) The level of dsRNA-induced activation of IRF-1 DNA-binding activity was quantitated by PhosphorImager analysis. Numbers beneath the corresponding lanes indicate the fold increase of IRF-1 DNA-binding activity after treatment of cells with dsRNA, either in the absence (lane 3 and 7) or presence (lanes 4 and 8) of NS5A expression. (Lower) Immunoblot analysis of HCV or IRF-1 protein levels. (D) PKR activity in HNS5A 1b-1 and HNS5A 1b-5 cells cultured to induce or repress NS5A expression. Where indicated, cells were treated with dsRNA for 2 h. Extracts (200 μg) were subjected to immunoprecipitation and PKR assay by using nonimmune serum (lane 1) or anti-PKR polyclonal serum (lanes 2–9). Panels show phosphorylation of PKR (PKR-P; Top) and the eukaryotic initiation factor 2α substrate (eIF2α-P; Middle). (Bottom) Input PKR levels for each corresponding lane.
We also examined how cell lines expressing E2, NS3/NS4A proteins, or the HCV core protein responded to dsRNA. The DNA-binding activity of IRF-1 was efficiently activated by dsRNA in both the presence and absence of these HCV proteins (Fig. 2B). Thus, the HCV E2, NS3, NS4A, and core proteins do not impact dsRNA-induced antiviral signaling to IRF-1.
We next determined whether the HCV NS5A protein could influence IRF-1 activation. To ascertain the potential role of NS5A as a PKR inhibitor in the regulation of IRF-1, we examined the activity of PKR and IRF-1 in cells that expressed isogenic NS5A quasispecies capable of inhibiting PKR (NS5A 1b-1) or that harbored mutations within the PKR-binding domain that disrupted PKR-regulatory function (NS5A 1b-5) (9). In the absence of NS5A expression, dsRNA induced a ≥14-fold increase in IRF-1 DNA-binding function (Fig. 2C), which was not significantly affected by expression of NS5A 1b-5. In contrast, expression of NS5A 1b-1 rendered cells refractory to dsRNA signaling and blocked the induction of the active IRF-1/DNA complex (Fig. 2C, compare lanes 3 and 4). Thus, NS5A was sufficient to block the dsRNA-induced activation of IRF-1, and this block was relieved by mutations that disrupt PKR-regulatory function. Analysis of PKR activity in NS5A cell lines revealed that NS5A 1b-1 expression inhibited PKR and suppressed the dsRNA induction of PKR activity, but no inhibitory effects on PKR were observed in cells expressing NS5A 1b-5 (Fig. 2D). Taken together, these results suggest that the dsRNA-induced activation of IRF-1 is signaled through a PKR-dependent pathway that is blocked by NS5A, and that this block is relieved by mutations in NS5A that disrupt its PKR regulatory properties.
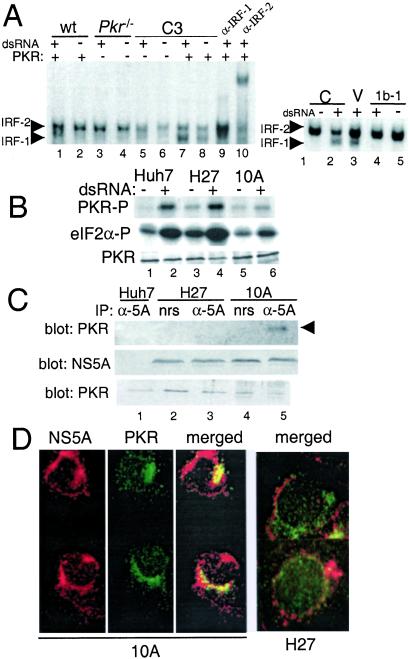
NS5A Blocks a PKR-Dependent Pathway to IRF-1 Activation.
To determine whether NS5A was indeed disrupting IRF-1 activation through a PKR-dependent mechanism, we examined the dsRNA activation of IRF-1 in wt MEFs or MEFs nullizygous for a functional Pkr gene (14). dsRNA treatment of wt MEFs induced the formation of an active IRF-1/DNA complex (Fig. 3A Left, see lanes 1–4). As previously demonstrated (13, 23), Pkr-null MEFs were refractory to dsRNA and failed to activate IRF-1 upon dsRNA treatment. However, we found that this deficit could be complemented by the tetracycline-regulated expression of wt PKR (Fig. 3A Left, lanes 5–8). In contrast, the transient expression of NS5A 1b-1 disrupted the dsRNA-induced activation of IRF-1 in wt MEFs and conferred an IRF-1 phenotype comparable to Pkr-null MEFs (Fig. 3A Right). These results confirm that NS5A is sufficient to disrupt the PKR-dependent activation of IRF-1 that is signaled by dsRNA. NS5A may therefore influence IRF-1 function by regulating PKR activity during HCV RNA replication.
Figure 3.
NS5A regulation of a PKR-dependent IRF-1 activation pathway. (A Left) EMSA of extracts from wt MEFs (lanes 1 and 2), Pkr-null MEFs (lanes 3 and 4), and C3 MEFs cultured to suppress (lanes 5 and 6) or induce (lanes 7 and 8) wt PKR expression, respectively. Lanes 9 and 10 show supershift of extracts from dsRNA-treated wt MEFs. (A Right) EMSA of extracts from wt MEFs, either untransfected (controls; lanes 1 and 2) or after transfection of vector alone (lane 3) or a plasmid encoding NS5A 1b-1 (lanes 4 and 5). Cells were cultured for 2 h in the presence or absence of dsRNA. EMSAs used the C1 probe. (B) Parental Huh7 cells (lanes 1 and 2) and cells harboring replicon H27 (lanes 3 and 4) or 10A (lanes 5 and 6) were cultured in the presence or absence of dsRNA for 2 h, and extracts were subjected to PKR protein kinase assay as described in Fig. 2D. PKR autophosphorylation (Top), phosphorylation of eukaryotic initiation factor 2α substrate (Middle), and PKR input levels (Bottom). (C) Extracts (250 μg) from parental Huh7 cells (lane 1) or cells harboring replicon H27 (lanes 2 and 3) or 10A (lanes 4 and 5) were subject to immunoprecipitation with anti-NS5A polyclonal serum (lanes 1, 3, and 5) or nonimmune serum (lanes 2 and 4), and the products were analyzed by immunoblot using anti-PKR mAb (Top). (Middle and Bottom) Amount of NS5A and PKR present in 25 μg of the input extracts. Arrow points to PKR. (D) Protein distribution of NS5A (red) and PKR (green) in replicon 10A and H27 cells was determined by immunocytochemical staining and two-color digital microscopy. (Left) Individual and merged images for replicon 10A. (Right) Replicon H27 merged images. Yellow indicates protein codistribution.
HCV RNA Replicons Have Distinct PKR Regulatory Properties.
Our results raised the possibility that the more efficient replication of the 10A replicon may involve an NS5A-imposed block in the antiviral response of the host cell that is otherwise triggered by dsRNA and signaled by PKR. To investigate this idea, we assessed the impact of HCV RNA replication on endogenous PKR activity. In vitro kinase assay of anti-PKR immunoprecipitation products revealed a higher basal level of active PKR in replicon H27 cells compared to parental Huh7 cells, and the specific activity of PKR increased in both cell lines after their exposure to dsRNA (Fig. 3B). Cells harboring replicon 10A exhibited a very low basal level of PKR activity and showed virtually no increase in activity in response to dsRNA. In parallel experiments, we were able to recover PKR from anti-NS5A immunoprecipitation reactions of extracts prepared from replicon 10A cells but not from H27 or Huh7 cells (Fig. 3C). In addition, immunocytochemical staining was used to examine the subcellular distribution of NS5A and PKR in cells harboring either the 10A or H27 replicon. Cells harboring the 10A replicon exhibited considerable overlap between the cellular distribution of NS5A and PKR (Fig. 3D), whereas no evidence for codistribution of the two proteins was observed in cells harboring the H27 replicon. Taken together, these results demonstrate that the NS5A protein encoded within replicon 10A binds to PKR and suppresses kinase activity during HCV RNA replication. It should be noted that both replicon quasispecies possess identical sequences within their 64-aa PKR-binding domain. The lack of any apparent impact on PKR activity in H27 cultures suggests that the L2198S mutation or possibly other aa substitutions that cluster outside, but proximal to the PKR-binding domain (24), may alter the ability of NS5A to bind and regulate PKR.
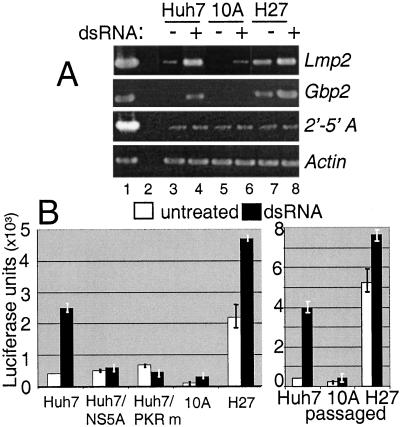
Regulation of IRF-1-Dependent Gene Expression During HCV RNA Replication.
To determine whether the regulation of IRF-1 affected host cell gene expression during HCV RNA replication, we measured the dsRNA induction of Gbp2 and Lmp2, both of which require functional IRF-1 for inducible expression (25, 26). As controls, we included an assessment of 2′-5′oligoadenylate synthetase and β-actin, whose levels are not regulated by dsRNA (27). As seen in Fig. 4A, a basal level of Gbp2 and Lmp2 expression in H27 cells correlated with the high basal level of active IRF-1 induced by the H27 replicon (compare with Fig. 1). Moreover, dsRNA treatment of Huh7 and H27 replicon cells induced the expression of both genes. In contrast, a basal level of Gbp2 or Lmp2 expression was not apparent in cells harboring the 10A replicon, and the mRNAs were not induced by dsRNA treatment. Expression of β-actin and 2′-5′oligoadenylate synthetase were similar between cells. These results suggest that the block to IRF-1 activation by the 10A replicon was sufficient to disrupt the induction of IRF-1-dependent genes during HCV RNA replication. For Gbp2, we confirmed that the effect of HCV RNA replication on gene expression was occurring at the level of promoter induction, by examining the expression of a transfected Gbp2 promoter/enhancer reporter construct (Gbp2-luc; Fig. 4B). The basal and dsRNA-induced expression of Gbp2-luc paralleled IRF-1 activity and Gbp2 mRNA levels in Huh7, 10A, and H27 cells. To further assess the role of NS5A and PKR in the regulation of this IRF-1-dependent promoter, Huh7 cells were cotransfected with dominant-negative mutant PKR or an NS5A expression construct derived from the 10A replicon. Gbp2-luc induction by dsRNA was blocked by expression of mutant PKR. Moreover, expression of the 10A NS5A protein alone was sufficient to repress the dsRNA-induced activation of Gbp2-luc in Huh7 cells to similar levels conferred by the 10A replicon (Fig. 4B). Taken together, our results confirm that the basal activation of IRF-1 by the H27 replicon conferred increased levels of IRF-1-dependent gene expression and that the corresponding deficiency in gene expression in cells harboring the 10A replicon was mediated through NS5A abrogation of PKR function.
Figure 4.
Regulation of IRF-1-dependent gene expression. (A) Expression levels of Lmp2, Gbp2, 2′-5′oligoadenylate synthetase, and actin were assessed by RT-PCR from total RNA isolated from Huh7 (lanes 3 and 4), replicon 10A cells (lanes 5 and 6), or replicon H27 cells (lanes 7 and 8) cultured in the presence or absence of dsRNA for 2 h. Lane 1: + plasmid PCR control product. Lane 2: − control using the RNA shown in lane 8 but omitting the RT step. (B) The level of dsRNA-induced stimulation of the Gbp2 promoter-luc reporter construct (average units of luc activity and SE for four experiments) was determined by luciferase assay of extracts from cells cultured in the absence of dsRNA (empty bars) or for 2 h in the presence of dsRNA (filled bars). Cells were cotransfected with pSVβgal and vector alone, or for Huh7 cells, an expression plasmid encoding the 10A NS5A sequence or dominant-negative PKR (PKR m). (Right) Gbp2-luc activity was assessed from parental Huh7 cells and those harboring the passaged replicon 10A or H27 RNA.
We sought to confirm that the regulation of IRF-1-dependent gene expression was directed by the HCV RNA replicons rather than a host cell phenotype. We transfected new Huh7 cells with total RNA isolated from the primary cell isolates harboring the 10A or H27 replicons and selected new G418-resistant 10A and H27 replicon populations. The Gbp2-luc-promoter regulation phenotype associated with each replicon was maintained in these new replicon cultures (Fig. 4B Right). We therefore attribute the differential regulation of IRF-1 and IRF-1-dependent gene expression to properties encoded within the HCV replicon quasispecies and not to host cell differences.
Discussion
Our results demonstrate that HCV RNA replication can trigger dsRNA-induced antiviral pathways of the host cell, leading to activation of PKR and IRF-1. Many, if not all, animal viruses activate or regulate PKR during infection (28). Our studies demonstrate that the NS5A protein can inhibit PKR during HCV RNA replication. NS5A inhibition of PKR correlated with a block in IRF-1 activation and subsequently the dsRNA-induced stimulation of IRF-1-dependent gene expression. Moreover, expression of NS5A alone was sufficient to block induction of an IRF-1-dependent promoter by dsRNA. Thus, HCV may disrupt IRF-1 activation, and affect host gene expression, by targeting PKR.
The precise pathway of IRF-1 activation during virus infection has not been conclusively determined. Various studies indicate that IRF-1 DNA-binding activity and transactivation functions are signaled by PKR and are directly regulated by phosphorylation. Hiscott and coworkers (29) identified two clusters of regulatory phosphorylation sites within IRF-1 that dramatically influenced transactivation function. More recently, Rahat et al. (30) demonstrated that phosphorylation induced the DNA binding of IRF-1 and its transactivation of the HLA-DRα locus. In this report we confirmed that PKR is indeed an upstream transducer of IRF-1 activation signals. Taken together, these results support a model during virus infection in which PKR initiates a dsRNA-induced-signaling cascade that results in the phosphorylation of IRF-1 and activation of its DNA-binding properties. Preliminary work from our laboratory suggests that PKR indirectly signals the hyperphosphorylation of IRF-1, and that NS5A can block this phosphorylation pathway (J.P. and M.G., unpublished results). It is notable that expression of the E2 protein, which has been described as a PKR inhibitor (6), did not influence IRF-1 activation. Consistent with this finding, we found that E2 did not block the dsRNA-induced activation of PKR in vivo (data not shown). It is possible that E2 retention in the endoplasmic reticulum lumen (31) or differential glycosylation of E2 may have precluded interaction with PKR, or that E2 may regulate PKR independently of dsRNA and IRF-1.
NS5A is normally localized in a perinuclear/cytoplasmic context (32). The 10A NS5A protein was similarly localized, and we found that its distribution overlapped with PKR and supported NS5A-PKR complex formation. In contrast, the H27 NS5A protein did not interact with PKR but exhibited a perinuclear and cytoplasmic-punctate pattern independent of PKR distribution. These results suggest that K2040 insertion in the 10A species may alter the subcellular localization of NS5A sufficiently to support interaction with PKR. Alternatively, the L2198S mutation in H27 NS5A may interfere with PKR binding or colocalization by directly influencing the confirmation of the PKR-binding domain. Further structure-function analyses of these NS5A domains will be necessary to determine their role in regulation of NS5A localization and function. Overall, our results suggest that efficient HCV RNA replication may involve a block in PKR-dependent signaling. These findings indicate that regulation of PKR may provide a replicative advantage to those viral quasispecies that can block the dsRNA-induced host antiviral response to effect suppression of antiviral genes. In support of this idea, a previous study has demonstrated that mutations that clustered in or around the PKR-binding domain of NS5A dramatically influenced the efficiency of replication of culture-adapted HCV RNA replicons (19). In the context of our results, this finding suggests that disruption of the PKR-binding domain may compromise NS5A function and relieve the block upon the IRF-1 activation pathway, thereby placing limitations upon HCV RNA replication. Such a model is supported by studies of NS5A sequences isolated from HCV-infected patients, in which sequence complexity has been associated with reduced viral load, and increased sensitivity to the host antiviral response (33).
Our results demonstrate that HCV RNA replication can trigger the promoter activation and expression of cellular genes that depend on IRF-1, and that NS5A, through its ability to inhibit PKR, can suppress IRF-1-dependent gene expression. Recent studies demonstrate that IRF-1 cooperates with IRF-3 and IRF-7 in the regulation of a range of antiviral genes, including the IFN-α and IFN-β genes (12, 34). HCV regulation of IRF-1 may therefore impact other IRF pathways to influence host gene expression on a more global scale. Such regulation may create a more hospitable cellular environment for persistent HCV replication.
Acknowledgments
We thank R. Defrancesco and M. Giovani for primary replicon isolates, W. Lee, S. Kimball, I. Julkunnen, A. Hovanessian, B. Williams, and D. Moradpour for cell lines, Abs, and reagents, W. Breshnahan for critical review of this manuscript, and W. Milling for technical assistance. We thank Dr. M. Katze for reagents and for helpful discussions. This study was supported by National Institutes of Health Grants AI48235 and AI35522 to M.G. and D.S., respectively. M.G. is the Nancy C. and Jeffery A. Marcus Endowed Scholar in Medical Research, in honor of Dr. Bill S. Vowell.
Abbreviations
- dsRNA
double-stranded RNA
- elf2α
eukaryotic initiation factor 2-α
- EMSA
electrophoretic mobility-shift assay
- HCV
hepatitis C virus
- Gbp2
guanylate-binding protein-2
- IFN
interferon
- IRF
IFN regulatory factor
- ISRE
IFN-stimulated response element
- MEF
mouse embryo fibroblast
- NS5A
nonstructural 5A
- RT-PCR
reverse transcription–PCR
- PKR
protein kinase R
- wt
wild type
References
- 1.Biron C A. Curr Opin Microbiol. 1999;2:374–381. doi: 10.1016/s1369-5274(99)80066-6. [DOI] [PubMed] [Google Scholar]
- 2.Stark G R, Kerr I M, Williams B R G, Silverman R H, Schreiber R D. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 3.Williams B R G. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 4.Taniguchi T, Lamphier M S, Tanaka N. Biochim Biophys Acta. 1997;1333:9–17. doi: 10.1016/s0304-419x(97)00014-0. [DOI] [PubMed] [Google Scholar]
- 5.Gale M, Jr, Korth M J, Tang N M, Tan S-L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 6.Taylor D R, Shi S T, Romano P R, Barber G N, Lai M M C. Science. 1999;285:107–110. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- 7.Witherell G W, Beibeke P. J Med Virol. 2000;63:8–16. [PubMed] [Google Scholar]
- 8.Berg T, Mas Marques A, Hohne M, Wiedenmann B, Hopf U, Schreier E. Hepatology. 2000;32:1386–1395. doi: 10.1053/jhep.2000.20527. [DOI] [PubMed] [Google Scholar]
- 9.Gale M, Jr, Kwieciszewski B, Dossett M, Nakao H, Katze M G. J Virol. 1999;73:6506–6516. doi: 10.1128/jvi.73.8.6506-6516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lohmann V, Korner F, Kock J-O, Theilmann L, Bartenschlager R. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 11.Katze M G, Wambach M, Wong M-L, Garfinkel M, Meurs E, Chong K, Williams B R G, Hovanessian A G, Barber G N. Mol Cell Biol. 1991;11:5497–5505. doi: 10.1128/mcb.11.11.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fredericksen, B., Akkaraju, G., Foy, E., Wang, C., Pflugheber, J., Chen, Z. J. & Gale, M., Jr. (2002) Viral Immunol., in press. [DOI] [PubMed]
- 13.Kumar A, Yang Y-L, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, Williams B R G. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y-L, Reis L F L, Pavlovic J, Aguzzi A, Schäfer R, Kumar A, Williams B R G, Aguet M, Weissmann C. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada H, Taniguchi T, Tanaka N. Biochimie. 1998;80:641–650. doi: 10.1016/s0300-9084(99)80017-0. [DOI] [PubMed] [Google Scholar]
- 16.Gale M, Jr, Katze M G. Methods Companion Methods Enzymol. 1997;11:383–401. doi: 10.1006/meth.1996.0436. [DOI] [PubMed] [Google Scholar]
- 17.Martell M, Gomez J, Esteban J, Sauleda S, Quer J, Cabot B, Esteban R, Guardia J. J Clin Microbiol. 1999;37:327–332. doi: 10.1128/jcm.37.2.327-332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan S-L, Katze M G. Virology. 2001;284:1–12. doi: 10.1006/viro.2001.0885. [DOI] [PubMed] [Google Scholar]
- 19.Blight K J, Kolykhalov A A, Rice C M. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 20.Kreiger N, Lohmann V, Bartenschlager R. J Virol. 2001;75:4614–4654. doi: 10.1128/JVI.75.10.4614-4624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka N, Kawakami T, Taniguchi T. Mol Cell Biol. 1993;13:4531–4538. doi: 10.1128/mcb.13.8.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Francesco R. J Hepatol. 1999;31:47–53. doi: 10.1016/s0168-8278(99)80374-2. [DOI] [PubMed] [Google Scholar]
- 23.Der S D, Yang Y-L, Weissman C, Williams B R G. Proc Natl Acad Sci USA. 1997;94:3279–3283. doi: 10.1073/pnas.94.7.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nousbaum J-B, Polyak S J, Ray S C, Sullivan D G, Larson A M, Carithers R L, Gretch D R. J Virol. 2000;74:9028–9038. doi: 10.1128/jvi.74.19.9028-9038.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White L C, Wright K L, Felix N J, Ruffner H, Reis L F, Pine R, Ting J P. Immunity. 1996;5:365–376. doi: 10.1016/s1074-7613(00)80262-9. [DOI] [PubMed] [Google Scholar]
- 26.Briken V, Ruffner H, Schwarz A, Reis L, Strehlow I, Decker T, Staeheli P. Mol Cell Biol. 1995;15:975–982. doi: 10.1128/mcb.15.2.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geiss G K, Jin G, Guo J, Bumgarner R E, Katze M G, Sen G C. J Biol Chem. 2001;276:30178–30182. doi: 10.1074/jbc.c100137200. [DOI] [PubMed] [Google Scholar]
- 28.Gale M, Jr, Katze M G. Pharmacol Ther. 1998;78:29–46. doi: 10.1016/s0163-7258(97)00165-4. [DOI] [PubMed] [Google Scholar]
- 29.Lin R, Hiscott J. Mol Cell Biochem. 1999;191:169–180. [PubMed] [Google Scholar]
- 30.Rahat M A, Chernichovski I, Lahat N. Int Immunol. 2001;13:1423–1432. doi: 10.1093/intimm/13.11.1423. [DOI] [PubMed] [Google Scholar]
- 31.Reed K E, Rice C M. In: Molecular Characterization of Hepatitis C Virus. Reesink H W, editor. Basel: Karger; 1998. pp. 1–37. [DOI] [PubMed] [Google Scholar]
- 32.Polyak S J, Paschal D M, Mcardle S, Gale M J, Jr, Moradpour D, Gretch D R. Hepatology. 1999;29:1262–1271. doi: 10.1002/hep.510290438. [DOI] [PubMed] [Google Scholar]
- 33.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Maruno F, Sato C. N Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 34.Au W C, Pitha P M. J Biol Chem. 2001;276:41629–41637. doi: 10.1074/jbc.M105121200. [DOI] [PubMed] [Google Scholar]
- 35.Moradpour D, Kary P, Rice C M, Blum H E. Hepatology. 1998;28:192–201. doi: 10.1002/hep.510280125. [DOI] [PubMed] [Google Scholar]
- 36.Wolk B, Sansonno D, Krausslich H, Dammacco F, Rice C, Blum H, Moradpour D. J Virol. 2000;74:2293–2304. doi: 10.1128/jvi.74.5.2293-2304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moradpour D, Englert C, Wakita T, Wands J R. Virology. 1996;222:51–63. doi: 10.1006/viro.1996.0397. [DOI] [PubMed] [Google Scholar]