Abstract
Aconitase and isocitrate dehydrogenase (IDH) enzyme activities were detected in anaerobically prepared cell extracts of the obligate anaerobe Bacteroides fragilis. The aconitase gene was located upstream of the genes encoding the other two components of the oxidative branch of the Krebs cycle, IDH and citrate synthase. Mutational analysis indicates that these genes are cotranscribed. A nonpolar in-frame deletion of the acnA gene that encodes the aconitase prevented growth in glucose minimal medium unless heme or succinate was added to the medium. These results imply that B. fragilis has two pathways for α-ketoglutarate biosynthesis—one from isocitrate and the other from succinate. Homology searches indicated that the B. fragilis aconitase is most closely related to aconitases of two other Cytophaga–Flavobacterium–Bacteroides (CFB) group bacteria, Cytophaga hutchinsonii and Fibrobacter succinogenes. Phylogenetic analysis indicates that the CFB group aconitases are most closely related to mitochondrial aconitases. In addition, the IDH of C. hutchinsonii was found to be most closely related to the mitochondrial/cytosolic IDH-2 group of eukaryotic organisms. These data suggest a common origin for these Krebs cycle enzymes in mitochondria and CFB group bacteria.
Early studies of central metabolism in the obligate anaerobe Bacteroides fragilis suggested that heme is important for the generation of ATP from glucose (reviewed in ref. 1). Through these studies it was shown that ATP biosynthesis is linked to a cytochrome b-dependent electron transport system in which fumarate serves as the terminal electron acceptor (2, 3). Because B. fragilis is incapable of de novo heme biosynthesis, an exogenous source of heme is required for cytochrome assembly (4). Molar growth yield studies indicate that B. fragilis cultivated in the presence of heme generates 4.5 mol of ATP per mole of glucose, as opposed to 1.7 mol of ATP per mole of glucose in the absence of heme (2). Because the theoretical yield of ATP from anaerobically grown Escherichia coli is only 2.5 mol of ATP per mole of glucose, it has been proposed that this electron transport system gives B. fragilis a growth advantage in the anaerobic environment of the colon (2).
Studies with radioactively labeled succinate indicate that B. fragilis is capable of synthesizing α-ketoglutarate by the reductive carboxylation of succinyl-CoA (5). Because heme is essential for the reduction of fumarate to succinate (6, 7), in the step preceding the formation of succinyl-CoA, α-ketoglutarate biosynthesis by this pathway is predicted to be heme-dependent. This observation, coupled with the observation that some Bacteroides species lack isocitrate dehydrogenase (IDH) activity (8), has led to the conclusion that these bacteria lack the oxidative branch of the Krebs cycle and rely on the reductive branch for α-ketoglutarate synthesis. However, a report of redox-sensitive enzymes in a related bacterium, Bacteroides thetaiotaomicron, indicates the presence of aconitase activity in anaerobically prepared cell extracts (9). Furthermore, B. fragilis is capable of growth in the absence of heme, albeit at a slower rate (2, 10). Therefore, this bacterium may synthesize trace amounts of heme to maintain fumarate reductase activity, may reduce fumarate in a heme-independent fashion, as seen in E. coli (11), or may generate α-ketoglutarate via a succinyl-CoA-independent pathway.
Under aerobic conditions, many nonautotrophic organisms use the Krebs cycle for (i) the generation of metabolic energy in the form of ATP and GTP; (ii) the reduction of adenine dinucleotides, such as NAD+ and NADP+, for use as electron donors; and (iii) the generation of biosynthetic precursors such as oxaloacetate, succinyl-CoA, and α-ketoglutarate (12). When cells experience anaerobiosis, the rate of NADH and NAD(P)H oxidation decreases because of the limited availability of external terminal electron acceptors. During anaerobiosis some facultative anaerobes, such as E. coli, split the Krebs cycle into two opposing half-cycles termed the oxidative branch and the reductive branch (12). Under such conditions, the NADH and NAD(P)H produced during glycolysis and by the oxidative branch can be reoxidized via the reductive branch, allowing maintenance of the cytoplasmic redox potential. In addition, the reductive branch functions to provide the cell with oxaloacetate and succinyl-CoA, and the oxidative branch functions to provide the cell with α-ketoglutarate.
We show here that in addition to a heme-dependent pathway for α-ketoglutarate biosynthesis, B. fragilis also has a heme-independent pathway for α-ketoglutarate biosynthesis. This heme-independent pathway uses the enzymes of the oxidative branch of the Krebs cycle—citrate synthase, aconitase, and IDH. Until this study, to our knowledge, all reported bacterial aconitases have been found to belong to either the aconitase A or the aconitase B group. Phylogenetic analysis of the A group has provided great insight into the origin of the aconitase-like iron regulatory proteins (IRPs) of eukaryotes. However, because these groups are quite distinct from the mitochondrial aconitases, the origin of the mitochondrial aconitase has remained unclear.
It is well accepted that the mitochondrion is derived from an ancestral α-proteobacterium. It is curious to note, however, that the aconitases encoded by the α-proteobacterial group are most similar to aconitases of the A group. Unexpectedly, it was discovered that the aconitase of B. fragilis is most similar to aconitases of the mitochondrial group. This finding provides a potential link to a better understanding of the origin of mitochondrial Krebs cycle components, and perhaps to the origin of the mitochondrion. We speculate that some of these genes were contributed by an ancestral Cytophaga–Flavobacterium–Bacteroides (CFB) group bacterium.
Materials and Methods
Strains, Media, and Growth Conditions.
The bacterial strains used in this study are described in Table 1. B. fragilis strains were grown in an anaerobic chamber (Coy Laboratory Products, Ann Arbor, MI) at 37°C with an atmosphere of 5% CO2, 10% H2, and 85% N2 (Airgas Northeast, Radnor, PA) by using supplemented brain heart-infusion medum (BHIS) (13) or anaerobic minimal medium with 0.5% glucose (AMMgluc). AMMgluc is based on the formulation described in ref. 10 with the following changes: resazurin, VFA solution, B-vitamin solution, and casitone were omitted; vitamins B12 (0.5 μg/ml) and K3 (1 μg/ml) were added; medium was buffered with 0.1 M potassium phosphate (pH 7.2); and 50 μg/ml of thymine was added for growth of thy− strains. Unless otherwise stated, all AMMgluc media were supplemented with 5 μg/ml of hematin as the sole source of heme. Hematin stock solutions were prepared by dissolving 5 g of hemin chloride in 1 liter of 0.1 M NaOH. Glassware used for hematin-restricted cultures was baked at 200°C for 5 h to destroy trace heme contamination.
Table 1.
Bacterial strains and plasmids used in this study
| Name | Relevant characteristics | Source or ref. no. |
|---|---|---|
| B. fragilis | ||
| TM4000 | Wild-type, RifR, TpS | 23 |
| ADB77 | TM4000 ΔthyA1, TpR | Lab stock |
| ADB237 | ADB77 acnA∷pADB237, TetR | This work |
| ADB245 | ADB77 ΔacnA245 | This work |
| E. coli | ||
| DH5α | ΔlacZ M15 | 14 |
| DW1030 | Host strain for pRK2317 | 42 |
| Plasmids | ||
| pCR2.1TOPO | TA cloning vector, lacZα, AmpR | Invitrogen |
| pRK231.7 | RP4 derivative, TetR, Tra+, AmpS | 15 |
| pYT102 | p15A ori, CmR, RP4 oriT; B. fragilis suicide vector, thyA+, TetR | 17 |
| pADB237 | pYT102 with acnA internal fragment | This work |
| pADB245 | pYT102 ΔacnA245 | This work |
| pADB250 | pCR2.1TOPO containing acnA | This work |
E. coli strain DH5α (14) was used for cloning, and strain DW1030/RK2317 (15) was used for mobilization of plasmids from DH5α to B. fragilis. E. coli strains were grown in L medium (Difco) at 37°C. Competent cells were prepared by the RbCl method and transformed as described (16). Chloramphenicol (25 μg/ml), ampicillin (100 μg/ml), tetracycline (2 μg/ml), rifampicin (50 μg/ml), gentamicin (50 μg/ml), and trimethoprim (100 μg/ml) were used as indicated.
acnA Sequencing and Analysis.
Oligonucleotide primers used in this study are described in Table 2. The acnA gene was amplified from the B. fragilis TM4000 chromosome by PCR, using Taq DNA polymerase (GIBCO/BRL) and the primers acon03 and acon06. Primer sequences were based on sequence data obtained from the B. fragilis ATCC 25285 preliminary genome sequence produced by the Pathogen Sequencing Group at the Sanger Institute (http://www.sanger.ac.uk). The TM4000 acnA fragment was cloned in pCR2.1TOPO (Invitrogen), following the manufacturer's protocol, to create the plasmid pADB250. Plasmid and PCR product purifications were performed by using QIAprep spin columns (Qiagen, Chatsworth, CA). Sequencing of the insert in pADB250 was performed with an ABI 3100 sequencing apparatus (Perkin–Elmer) by the Department of Physiology at Tufts University, using the M13- and acnA-specific primers described in Table 2.
Table 2.
Oligonucleotide primers used in this study
| Primer | Sequence or source |
|---|---|
| acon01 | 5′-GATACAAGCTTATCGGGGGGCTGAACG-3′ |
| acon02 | 5′-GTTCGGATCCCGGATTCACAATCAGC-3′ |
| acon03 | 5′-TCCAAAGCTTCGAACATGGTACGATAGGAGG-3′ |
| acon04 | 5′-AAGGCCATGGGAACATGCTCCATCTTCCC-3′ |
| acon05 | 5′-AAAGCCATGGGTAAGTGTACGACAGACC-3′ |
| acon06 | 5′-CGTTGGATCCGGTATTGTCGGTACATCCG-3′ |
| 61RAB | 5′-GGCGCGCCGTAAGGAAAGTGGCTCTCAG-3′ |
| 1842 | 5′-CAAGGCGACAAGGTGCTGATGC-3′ |
| M13 Reverse | Invitrogen (product no. N53002) |
| M13 Forward | Invitrogen (product no. N52002) |
Preliminary Fibrobacter succinogenes sequence data were obtained from The Institute for Genomic Research (http://www.tigr.org). Preliminary Cytophaga hutchinsonii sequence data were obtained from the Department of Energy Joint Genome Institute (http://www.jgi.doe.gov) through the Pedant web site (http://pedant.gsf.de). All other sequence data discussed in this report were obtained from the National Center for Biotechnology Information (NCBI) web site (http://www.ncbi.nlm.nih.gov). Sequences were analyzed by using DNA STRIDER 1.2, editseq, and megalign (DNAstar, Madison, WI), WISCONSIN PACKAGE VERSION 10.1 (GCG), and MACVECTOR 7.0 (Oxford Molecular, Madison, WI).
Disruption and Deletion of acnA.
The B. fragilis aconitase-deficient strain ADB237 was constructed by a single crossover recombination between the suicide vector pADB237 and the B. fragilis ADB77 chromosome at the acnA locus. pADB237 was constructed by cloning a PCR-amplified internal acnA fragment in the suicide vector pYT102 (17). Amplification was performed by using primers acon01 and acon02. HindIII and BamHI restriction sites used for cloning of the PCR product in pYT102 are underlined in the acon01 and acon02 primer sequences. pADB237 was mobilized from DH5α to B. fragilis ADB77 by using the broad-host range plasmid RK2317 as the mobilizer (15). Cointegrants were selected on BHIS medium containing gentamicin, rifampicin, and tetracycline. Cointegrants with pADB237 recombined at the acnA locus were identified in a colony PCR-based screen in which the vector-specific primer 61RAB and the external chromosomal primer acon06 directed amplification of a 1.7-kbp chromosome/vector junction fragment.
An in-frame deletion of the acnA gene in the ADB77 chromosome was constructed by using a two-step double-crossover technique (17). The allelic exchange plasmid used was constructed as follows. A fragment consisting of 715 bp of acnA 5′ noncoding sequence and 67 bp of amino-terminal coding sequence was amplified by using primers acon03 and acon04. A fragment consisting of 557 bp of carboxyl-terminal acnA coding sequence and 227 bp of 3′ sequence was amplified by primers acon05 and acon06. These fragments were digested with NcoI, which cuts at the sites underlined in the acon04 and acon05 primer sequences. After digestion, the fragments were ligated to form the acnA245 allele. acnA245 was digested sequentially with HindIII and BamHI for ligation into pYT102 to generate pADB245. HindIII and BamHI sites used for cloning are underlined in the acon03 and acon06 primer sequences, respectively.
The pADB245 suicide vector was mobilized into ADB77 as described above. Tetracycline-resistant isolates were screened by colony PCR using primers acon03 and the vector primer 1842, as well as with primers 61RAB and acon06. Isolates that were positive for recombination of the suicide vector at the acnA locus were chosen for further use. As the suicide vector pADB245 contains the B. fragilis thyA gene, recombination at acnA results in thymine prototrophy and trimethoprim sensitivity. Cointegrants were grown overnight from single colonies in BHIS with thymine. Overnight cultures were plated on glucose minimal medium containing thymine and trimethoprim to select for isolates that had experienced the second recombination event. Trimethoprim-resistant colonies were screened for tetracycline sensitivity on BHIS plates containing tetracycline. Tetracycline-sensitive isolates were screened by colony PCR using primers acon03 and acon06 to distinguish between wild-type acnA resolution products and acnA deletion resolution products. By using this primer pair, a 3.2-kb fragment was amplified from the wild-type acnA locus, whereas a 1.6-kb fragment was amplified from the acnA245 locus.
Enzyme Assays.
Enzyme assays were performed by using crude extracts from anaerobically grown cells prepared by sonication in the anaerobic chamber (9). Exponential phase cultures were harvested in screw-cap conical tubes by centrifugation at 2,300 × g at 4°C in a Sorvall RC2-B centrifuge. Cells were resuspended in 1 ml of chilled prereduced 100 mM Tris⋅HCl (pH 8) and transferred to 2-ml microcentrifuge tubes. Samples were centrifuged at 2,000 × g in a microcentrifuge at 4°C. Cells were resuspended in 1 ml of 100 mM Tris⋅HCl (pH 8) and disrupted by sonication for 2 min with 50% output on a 50% duty cycle with a Branson Sonifier 250. During sonication, samples were maintained at 4°C by using an ArcticIce block (USA Scientific, Ocala, FL). Sonicates were clarified by centrifugation at 2,000 × g in a microcentrifuge at 4°C. Clarified samples were transferred to 2-ml microcentrifuge tubes and maintained at 4°C. Aconitase activity was measured by following the conversion of isocitrate to cis-aconitate as indicated by increase in absorbance at 240 nm (18). One unit is defined as the amount of enzyme required to increase absorbance by 0.0033 units in 1 min in the presence of 5.2 mg/ml of d,l-isocitrate (Sigma). IDH activity was measured by following isocitrate-dependent conversion of NAD+ to NADH as indicated by increase in absorbance at 340 nm (19). One unit is defined as the amount of enzyme necessary to reduce 1 μmol of NAD+ to NADH, in 1 min, in the presence of 5.2 mg/ml of d,l-isocitrate. Fumarase activity was measured by following the increase in absorbance at 240 nm in the presence of 10 mM malate (20). One unit is defined as the amount of enzyme required to increase absorbance 0.01 units in 1 min. All assays were performed by using a reaction buffer of 100 mM Tris⋅HCl, pH 8. Reactions were started by addition of cell extract to the reaction mixture. All solutions were prepared with preboiled distilled water to ensure minimal O2 contamination.
Nucleotide Sequence Accession Nos.
The nucleotide sequence of the B. fragilis TM4000 acnA gene has been deposited in GenBank under the accession no. AF434843.
Results
B. fragilis Has Aconitase and IDH Activity.
Crude cell extracts from B. fragilis ADB77 grown in AMMgluc were assayed for the presence of aconitase and IDH activity. Because studies of enzymes of central metabolism in B. thetaiotaomicron reveal that some of these proteins are highly sensitive to oxidative inactivation (9), B. fragilis cell extracts were prepared and maintained under anaerobic conditions. B. fragilis was found to express the activities of both of these key oxidative branch enzymes (Table 3).
Table 3.
Aconitase, IDH, and fumarase activities in crude cell extracts of B. fragilis ADB77 (wild-type) and the aconitase mutants ADB237 and ADB245
| Strain | Specific activity [unit (mg protein)−1]*
|
||
|---|---|---|---|
| Aconitase | IDH | Fumarase | |
| ADB77 | 89 ± 10 | 6.0 ± 0.8 | 810 ± 7 |
| ADB237 | <0.4 | <0.3 | 820 ± 40 |
| ADB245 | <0.2 | 6.0 ± 0.3 | 680 ± 30 |
Values are the mean ± SD from three determinations.
B. fragilis Encodes an Aconitase Homologue.
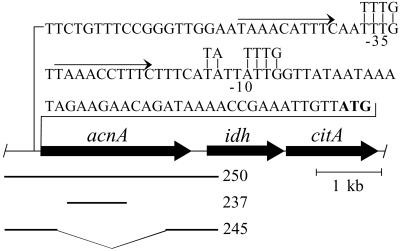
By using the Bacillus subtilis aconitase sequence (21) as the query sequence in a tblastn search of the unfinished B. fragilis ATCC 25285 genome, a B. fragilis aconitase homologue was identified. The gene encoding this aconitase homologue, termed acnA, was found to be located upstream of the genes encoding homologues of the other two components of the oxidative branch of the Krebs cycle, IDH and citrate synthase (Fig. 1). The coding sequences of acnA and idh are separated by 162 bp, whereas the coding sequences of idh and citrate synthase (citA) are separated by 12 bp, suggesting these genes may constitute an operon. Putative B. fragilis -35 and -10 promoter elements (22) were identified 63 and 39 bp, respectively, upstream of the acnA coding sequence. A 10-bp imperfect direct repeat was found flanking the acnA -35 region. Putative -35 and -10 sequences were also identified 174 and 125 bp, respectively, upstream of the idh coding sequence (data not shown), suggesting that idh and citA transcription may occur independently of acnA transcription.
Figure 1.
Physical map of the B. fragilis acnA region. The genes encoding aconitase (acnA), IDH (idh), and citrate synthase (citA) are depicted by large arrows. The sequence shown above the map illustrates the putative -35 and -10 promoter elements (underlined); the B. fragilis consensus -35 and -10 sequences are aligned above (22). The predicted AcnA translational start codon is marked in bold. The small arrows indicate direct repeats flanking the -35 sequence. The bar labeled 250 represents the region that was amplified from the B. fragilis TM4000 chromosome and cloned into pCR2.1-TOPO to generate pADB250. The bar labeled 237 represents the 0.88-kb fragment that was amplified from the TM4000 chromosome and cloned into pYT102 to generate pADB237. The bars labeled 245 represent the 0.8-kb fragments that were amplified from the TM4000 chromosome and cloned into pYT102 to generate pADB245.
Because B. fragilis ATCC 25285 is not identical to B. fragilis TM4000 (23), our reference strain for genetic studies, oligonucleotides were designed for PCR amplification and sequencing of the acnA region from TM4000. A 3,183-bp PCR product containing acnA was cloned in pCR2.1-TOPO, generating the plasmid pADB250. Two independent clones were fully sequenced on both strands. Sequence comparison of the ATCC 25285 and TM4000 acnA gene and gene product revealed 99.6% nucleotide identity and 99.6% amino acid identity (data not shown).
The acnA Gene Is Required for Aconitase Activity in B. fragilis.
To determine whether acnA is essential for aconitase activity in B. fragilis, the acnA locus was disrupted by using the suicide vector pADB237. Because pADB237 contains a fragment corresponding to nucleotides 295–1,181 of the acnA coding sequence (Fig. 1), a single-crossover recombination between the suicide vector and the acnA locus should disrupt expression of acnA by uncoupling the 3′ end of the gene from its promoter. The structure of the acnA locus of the cointegrant strain ADB237 was verified by using colony PCR (data not shown).
The aconitase locus of ADB237 is predicted to encode the amino-terminal 394 residues of the B. fragilis aconitase. Because this truncated peptide lacks 8 of the 17 residues conserved in the active site of all enzymatically functional aconitases (24), this peptide is predicted to be nonfunctional. Cell extracts were prepared from this strain and assayed for aconitase and IDH activity, as described above. As expected, ADB237 showed no detectable aconitase activity (Table 3). In addition, IDH activity was undetectable in this extract. It is important to note that the fumarase activity of the ADB237 extract was comparable to that of the wild-type extract, indicating that the ADB237 extract was not inactivated during preparation. The concomitant loss of aconitase activity and IDH activity in ADB237 is consistent with the idea that acnA and idh are members of an operon. However, because aconitase is a known posttranscriptional regulator (25), it is possible that aconitase is required for IDH expression. It is also possible that IDH is a component of a multisubunit metabolon (26), which requires aconitase for activity. In either case, the loss of aconitase would result in the loss of IDH activity in the cell extracts.
To determine whether the presence of aconitase is necessary for IDH activity, strain ADB245 was constructed by allelic exchange. ADB245 harbors a 1,617-bp in-frame deletion of the acnA coding sequence from nucleotides 68 to 1,684 (Fig. 1). The acnA deletion was verified by using a colony PCR (data not shown).
The 1.6-kb acnA allele of ADB245 has the potential to encode a 210-aa peptide. Because this peptide lacks 13 of the 17 conserved active site residues, it is predicted to be nonfunctional. Consistent with this prediction, extracts of ADB245 showed no detectable aconitase activity (Table 3). However, the ADB245 extract showed wild-type levels of IDH activity, indicating that the lack of IDH activity in ADB237 is not because of the lack of a functional aconitase, but is more likely the result of polar effects from the aconitase gene disruption.
acnA Is Essential for Growth in the Absence of Heme.
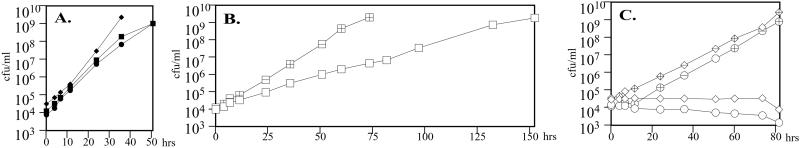
To determine whether aconitase has a functional role in B. fragilis central metabolism, the growth of strains ADB237 and ADB245 was assessed in AMMgluc containing 7.7 μM heme; the growth rates of both acnA mutants were indistinguishable from that of the wild-type ADB77 (Fig. 2A). Because ADB237 and ADB245 have no detectable aconitase activity but are not auxotrophic for α-ketoglutarate or for the amino acids derived from this key metabolic intermediate, B. fragilis must have an aconitase-independent pathway for the synthesis of α-ketoglutarate. A potential explanation for this observation comes from the report that B. fragilis has the capacity for the reductive carboxylation of succinyl-CoA (5).
Figure 2.
The B. fragilis aconitase mutants require succinate for growth in minimal medium. (A) Growth of the wild-type (ADB77, solid squares) and aconitase mutants (ADB237, solid diamonds; ADB245, solid circles) in AMMgluc with 7.7 μM heme. (B) Growth of the wild-type in the absence of heme (ADB77, open squares) with the addition of 18 mM succinate (ADB77, crossed squares) to the growth medium. (C) Stasis of the aconitase mutants in the absence of heme (ADB237, open diamonds; ADB245, open circles) is overcome by the addition of succinate (ADB237, crossed diamonds; ADB245, crossed circles).
Because B. fragilis requires exogenous heme for the reduction of fumarate to succinate (6, 7), the precursor of succinyl-CoA, it is reasonable to predict that aconitase-independent α-ketoglutarate biosynthesis is heme-dependent. In validation of this prediction, the acnA mutant strains failed to grow in AMMgluc when heme was omitted from the growth medium (Fig. 2C). It is important to note that the doubling time of the wild-type strain increased from 2.4 h in the presence of heme to 8.4 h in the absence of heme (Fig. 2 A and B). It has been suggested that this change in growth rate is because of a decreased capacity to generate ATP (2). However, in the presence of succinate (18 mM) the doubling time of the wild-type heme-restricted culture was reduced to ≈4.5 h (Fig. 2B). This observation suggests that, under these growth conditions, the primary role of heme is in succinate biosynthesis. Consistent with the observation that B. fragilis can use succinyl-CoA as a precursor in α-ketoglutarate biosynthesis, the growth rate of the heme-restricted aconitase mutants was restored to that of the wild-type strain by the addition of succinate to the growth medium (Fig. 2C). Taken together, these data indicate that B. fragilis has two separate pathways to generate α-ketoglutarate, either of which is sufficient for growth.
B. fragilis Encodes a Mitochondrion-Like Aconitase.
blast analysis was used to gain insight into the relatedness of the B. fragilis AcnA (AcnA-Bf) to aconitase and aconitase-like proteins of other bacteria and eukaryotes. When the AcnA-Bf was used as the query sequence in tblastn searches against the NCBI microbial genome database and against the genome sequences available at the Pedant web site, the peptide showed 62% identity and 75% similarity to the putative aconitase of F. succinogenes, and 64% identity and 77% similarity to the putative aconitase of C. hutchinsonii. Although these bacteria are quite diverse in habitat, extensive phylogenetic analyses have demonstrated that they are all members of the broad group of CFB eubacteria (27, 28). The sequence conservation observed between these aconitase homologues is further evidence of the common ancestry shared by these seemingly dissimilar bacteria.
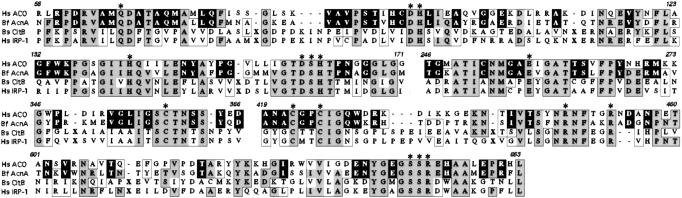
When the AcnA-Bf was used as the query in a blastp search against the nonredundant database at the NCBI, the peptide showed 59% identity and 74% similarity to the mitochondrial aconitase of the red algae Gracilaria gracilis. Within the nonredundant database the closest bacterial homologue to AcnA-Bf was the aconitase of Aquifex aoelicus (37% identity, 51% similarity). Akin to the mitochondrial aconitases, AcnA-Bf lacks the numerous insertions that are characteristic of the bacterial aconitase-A/IRP group (Fig. 3).
Figure 3.
Partial amino acid sequence alignment of the B. fragilis aconitase (Bf) with the mitochondrial aconitase of Homo sapiens (Hs ACO; GenBank accession no. Q99798), IRP-1 of H. sapiens (Hs; GenBank accession no. NP_002188), and aconitase-A of B. subtilis (Bs; GenBank accession no. G69599). Alignments were generated by using the CLUSTAL W algorithm with the MACVECTOR default settings. Numbers above the alignment refer to residues of the B. fragilis AcnA sequence. Residues that are similar to the B. fragilis sequence are boxed, those which are identical are shaded. Identical residues unique to the B. fragilis AcnA and the human mitochondrial ACO are boxed in black. Conserved active-site residues are marked by asterisks. Gaps in the alignment of 4 to 27 residues have been replaced by an X.
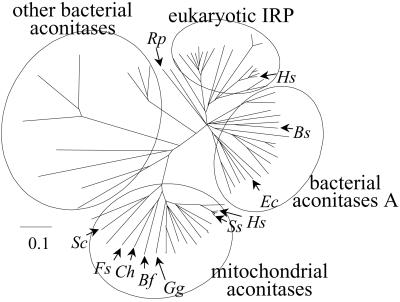
A multiple sequence alignment among the CFB aconitases, mitochondrial aconitases, IRP-aconitases A, and other bacterial aconitases was used to reconstruct a phylogenetic tree (Fig. 4). From this analysis, it is clear that the CFB aconitases belong to the mitochondrial aconitase group. It is noteworthy that the α-proteobacterium Rickettsia prowazekii, which is thought to share close ancestry with the mitochondrion (29–32), encodes an aconitase that falls within the bacterial subgroup of the IRP-aconitase A group (31, 32). These observations indicate that the genes encoding the mitochondrial and CFB aconitases are derived from a common source distant from that which gave rise to the cytosolic and bacterial aconitases.
Figure 4.
Phylogenetic tree of aconitase and IRP amino acid sequences from bacteria and eukaryotes. Trees were generated by calculating neighbor-joining distance with PAUPSEARCH (GCG) and visualized with TREEVIEWPPC 1.6.5 (43). Rp, R. prowazekii; Hs, H. sapiens; Bs, B. subtilis; Ec, E. coli; Ss, S. scrofa; Gg, G. gracilis; Bf, B. fragilis; Ch, C. hutchinsonii; Fs, F. succinogenes; Sc, Saccharomyces cerevisiae. See Fig. 6A for a fully annotated version of this tree.
When a phylogenetic tree was reconstructed from a multiple sequence alignment of eukaryotic and eubacterial IDH sequences, a different pattern of relatedness was observed with respect to the CFB representatives. First, although IDH activity has been observed in F. succinogenes cell extracts (8), the genome does not seem to encode any close homologue of IDH. Next, the B. fragilis IDH is clearly a member of the bacterial IDH-1 group (see Fig. 6B, which is published as supporting information on the PNAS web site, www.pnas.org). Finally, the IDH of C. hutchinsonii is one of three eubacterial IDHs that fall into the cytosolic/mitochondrial IDH-2 group. This observation is consistent with the previous suggestion that the eukaryotic IDH-2 group is of eubacterial origin (33).
Discussion
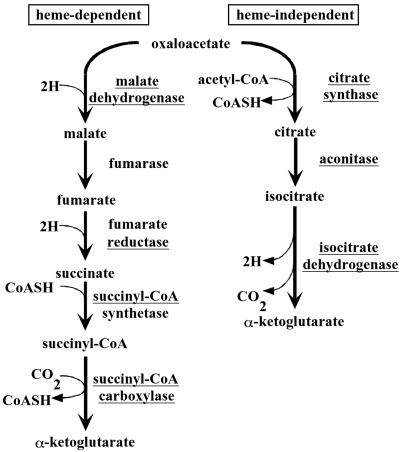
Early studies on central metabolism in Bacteroides species indicated that some of these bacteria synthesize α-ketoglutarate only via the reductive branch of the Krebs cycle (8). However, the presence of aconitase activity in the related bacterium B. thetaiotaomicron has recently been documented (9). In this study, we show that B. fragilis encodes the activities of the oxidative branch of the Krebs cycle. Through mutational analysis we have demonstrated that the genes coding for these activities seem to form an operon and are required for α-ketoglutarate biosynthesis in the absence of heme. However, these genes seem to be dispensable for α-ketoglutarate biosynthesis in the presence of heme or succinate. Based on these data, we present a model for the bimodal synthesis of α-ketoglutarate in B. fragilis (Fig. 5). The intriguing possibility that these activities are regulated by environmental conditions such as nutrient deprivation and/or oxidative stress is under investigation.
Figure 5.
Model for the dual pathways of α-ketoglutarate biosynthesis in B. fragilis. The left pathway is heme-dependent at the reduction of fumarate to succinate. The right pathway is heme-independent and involves a mitochondrion-like aconitase for the conversion of citrate to isocitrate. Pathways are based on results of this study and on refs. 1 and 5. The enzymes that catalyze these pathways are underlined.
Unlike other eubacteria that code for an aconitase similar to the cytoplasmic IRP, members of the CFB group contain an aconitase with an extraordinary resemblance to aconitases of the mitochondrial group. Although it is likely that this gene is of CFB ancestry, we cannot exclude the possibility that an ancestral CFB group bacterium acquired this gene from a lower eukaryote, such as G. gracilis. However, in addition to the presence of a mitochondrion-like aconitase, C. hutchinsonii also codes for an IDH that bears striking similarity to proteins of the IDH-2 group. In eukaryotes, proteins in this group show localization to both the cytosol and the mitochondrion. Previous reports of the Sphingomonas yanoikuyae IDH indicate that it is also a member of the IDH-2 group (34). Consistent with our results, an extensive phylogenetic analysis of the IDH-2 group supports the notion that this group has a eubacterial root (33). Taken together, these data are indicative of a common origin of the oxidative branch of the Krebs cycle in mitochondria and CFB group bacteria.
Numerous phylogenetic analyses indicate that the mitochondrial genome is of α-proteobacterial origin (29–32). However, mitochondrial proteins are encoded by both the mitochondrial genome and the nuclear genome (32). The nuclear-encoded mitochondrial genes seem to be of both bacterial and eukaryotic ancestry (32, 35). In yeast, approximately one-half of such genes seem to be of bacterial origin, whereas only one-third is predicted to be of eukaryotic origin (32, 35). During mitochondrial evolution, numerous genes may have been transferred from an ancestral mitochondrial genome to the nuclear genome (36, 37). However, it is unlikely that α-proteobacteria are the sole prokaryotic source of nuclear-encoded mitochondrial proteins (35). In fact, few of the nuclear-encoded mitochondrial genes are of α-proteobacterial origin, whereas a large number of these genes seem to be derived from other bacterial origins (35).
One model that offers an explanation for the polyphyletic origin of the nuclear genes encoding the mitochondrial proteome is the serial endosymbiont theory (38, 39). Like the endosymbiont theory, this theory suggests that the intimate association between a eukaryotic host and an endosymbiont permitted the translocation of several endosymbiont genes to the host genome. The serial endosymbiont theory suggests that these events have occurred successively throughout the history of the eukaryote (40). Some of these events are made obvious by the presence of genomous organelles, whereas other events are marked by genes left behind in the nuclear genome. In support of the idea that a CFB group bacterium may have been a participant in such a relationship, present-day endosymbionts of some Acanthamoebae spp. can be unambiguously assigned to the CFB group of eubacteria (41).
In further support for the idea that an ancestral CFB group bacterium has contributed to the nuclear-encoded mitochondrial gene repertoire, we have identified additional examples of B. fragilis genes whose products bear greater homology to nuclear-encoded mitochondrial proteins than to their eubacterial counterparts. For example, the B. fragilis carbamoyl phosphate synthetase is 55% identical and 72% similar to the mitochondrial carbamoyl phosphate synthetase III of the rainbow trout, Oncorhynchus mykiss. The closest bacterial homologue to the B. fragilis carbamoyl phosphate synthetase is the CarB of B. subtilis (43% identical, 62% similar). It is important to note that not all nuclear-encoded mitochondrial proteins show this extreme conservation with proteins of the CFB group bacteria, consistent with a model in which multiple bacteria contributed to the nuclear-encoded mitochondrial genes. From this study, it is evident that further analysis of metabolism in the CFB group bacteria will provide insight into the evolution of metabolic systems in mitochondria.
Supplementary Material
Acknowledgments
We thank A. L. Sonenshein, D. W. Lazinski, and A. Camilli for comments on the manuscript; A. L. Sonenshein and A. W. Serio for technical advice on the aconitase assays; B. O'Malley for assistance with bioinformatics; and B. Belitsky for his continued interest and valuable suggestions pertaining to this study. This study was supported by Public Health Service Grant AI-19497 from the National Institutes of Health.
Abbreviations
- IDH
isocitrate dehydrogenase
- IRP
iron regulatory protein
- CFB
Cytophaga–Flavobacterium–Bacteroides
- AMMgluc
anaerobic minimal medium with 0.5% glucose
- BHIS
supplemented brain heart-infusion medium
Footnotes
References
- 1.Macy J M, Probst I. Annu Rev Microbiol. 1979;33:561–594. doi: 10.1146/annurev.mi.33.100179.003021. [DOI] [PubMed] [Google Scholar]
- 2.Macy J M, Probst I, Gottschalk G. J Bacteriol. 1975;123:436–442. doi: 10.1128/jb.123.2.436-442.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris M A, Reddy C A. J Bacteriol. 1977;131:922–928. doi: 10.1128/jb.131.3.922-928.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuller M D, Caldwell D R. Can J Microbiol. 1982;28:1304–1310. doi: 10.1139/m82-195. [DOI] [PubMed] [Google Scholar]
- 5.Allison M J, Robinson I M, Baetz A L. J Bacteriol. 1979;140:980–986. doi: 10.1128/jb.140.3.980-986.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macy J M, Ljungdahl L G, Gottschalk G. J Bacteriol. 1978;134:84–91. doi: 10.1128/jb.134.1.84-91.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen M, Wolin M J. J Bacteriol. 1981;145:466–471. doi: 10.1128/jb.145.1.466-471.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joyner A E, Baldwin R L. J Bacteriol. 1966;92:1321–1330. doi: 10.1128/jb.92.5.1321-1330.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan N, Imlay J A. Mol Microbiol. 2001;39:1562–1571. doi: 10.1046/j.1365-2958.2001.02343.x. [DOI] [PubMed] [Google Scholar]
- 10.Varel V H, Bryant M P. Appl Microbiol. 1974;18:251–257. doi: 10.1128/am.28.2.251-257.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iverson T M, Luna-Chavez C, Cecchini G, Rees D C. Science. 1999;284:1961–1966. doi: 10.1126/science.284.5422.1961. [DOI] [PubMed] [Google Scholar]
- 12.Liao J C, Chao Y P, Patnaik R. Ann NY Acad Sci. 1994;745:21–34. doi: 10.1111/j.1749-6632.1994.tb44361.x. [DOI] [PubMed] [Google Scholar]
- 13.Thompson J T, Malamy M H. J Bacteriol. 1990;172:2584–2593. doi: 10.1128/jb.172.5.2584-2593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodcock D M, Crowther P J, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith S S, Michael M Z, Graham M W. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guiney D G, Hasegawa P, Davis C E. Proc Natl Acad Sci USA. 1984;83:7203–7206. doi: 10.1073/pnas.81.22.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan D, Jessee J, Bloom F R. Methods Enzymol. 1991;204:63–113. doi: 10.1016/0076-6879(91)04006-a. [DOI] [PubMed] [Google Scholar]
- 17.Tang Y. Ph.D. thesis. Boston: Tufts University; 2000. [Google Scholar]
- 18.Craig J E, Ford M J, Blaydon D C, Sonenshein A L. J Bacteriol. 1997;179:7351–7359. doi: 10.1128/jb.179.23.7351-7359.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kornberg A. Methods Enzymol. 1955;1:707–709. [Google Scholar]
- 20.Massey V. Methods Enzymol. 1955;1:729–735. [Google Scholar]
- 21.Dingman D W, Sonenshein A L. J Bacteriol. 1987;169:3062–3067. doi: 10.1128/jb.169.7.3062-3067.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bayley D P, Rocha E R, Smith C J. FEMS Microbiol Lett. 2000;193:149–154. doi: 10.1111/j.1574-6968.2000.tb09417.x. [DOI] [PubMed] [Google Scholar]
- 23.Tally F P, Snydman D R, Shimell M J, Malamy M H. J Bacteriol. 1982;151:686–691. doi: 10.1128/jb.151.2.686-691.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gruer M J, Artymiuk P J, Guest J R. Trends Biochem Sci. 1997;22:3–6. doi: 10.1016/s0968-0004(96)10069-4. [DOI] [PubMed] [Google Scholar]
- 25.Tang Y, Guest J R. Microbiology. 1999;145:3069–3079. doi: 10.1099/00221287-145-11-3069. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell C G. Biochem J. 1996;313:769–774. doi: 10.1042/bj3130769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffiths E, Gupta R S. Microbiology. 2001;147:2611–2622. doi: 10.1099/00221287-147-9-2611. [DOI] [PubMed] [Google Scholar]
- 28.Amann R, Ludwig W, Schleifer K H. J Gen Microbiol. 1988;134:2815–2821. doi: 10.1099/00221287-134-10-2815. [DOI] [PubMed] [Google Scholar]
- 29.Andersson S G E, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. Nature (London) 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 30.Lang B F, Gray M W, Burger G. Annu Rev Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- 31.Kurland C G, Andersson S G E. Microbiol Mol Biol Rev. 2000;64:786–820. doi: 10.1128/mmbr.64.4.786-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karlberg O, Canbäck B, Kurland C G, Andersson S G E. Yeast. 2000;17:170–187. doi: 10.1002/1097-0061(20000930)17:3<170::AID-YEA25>3.0.CO;2-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nekrutenko A, Hillis D M, Patton J C, Bradley R D, Baker R J. Mol Biol Evol. 1998;15:1674–1684. doi: 10.1093/oxfordjournals.molbev.a025894. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Lau P C K. Gene. 1996;168:15–21. doi: 10.1016/0378-1119(95)00732-6. [DOI] [PubMed] [Google Scholar]
- 35.Gray M W, Burger G, Lang B F. Genome Biol. 2001;2:1018.1–1018.5. doi: 10.1186/gb-2001-2-6-reviews1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurland C G. BioEssays. 1992;14:709–714. doi: 10.1002/bies.950141013. [DOI] [PubMed] [Google Scholar]
- 37.Berg O G, Kurland C G. Mol Biol Evol. 2000;17:951–961. doi: 10.1093/oxfordjournals.molbev.a026376. [DOI] [PubMed] [Google Scholar]
- 38.Gray M W, Burger G, Lang B F. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 39.Sogin M L. Curr Opin Genet Dev. 1997;7:792–799. doi: 10.1016/s0959-437x(97)80042-1. [DOI] [PubMed] [Google Scholar]
- 40.Heddi A, Grenier A, Khatchadourian C, Charles H, Nardon P. Proc Natl Acad Sci USA. 1999;96:6814–6819. doi: 10.1073/pnas.96.12.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horn M, Harzenetter M D, Linner T, Schmid E N, Muller K D, Michel R, Wagner M. Environ Microbiol. 2001;3:440–449. doi: 10.1046/j.1462-2920.2001.00210.x. [DOI] [PubMed] [Google Scholar]
- 42.Robillard N J, Tally F P, Malamy M H. J Bacteriol. 1985;164:1248–1255. doi: 10.1128/jb.164.3.1248-1255.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Page R D. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.