Abstract
Spa2p is a nonessential protein that regulates yeast cell polarity. It localizes early to the presumptive bud site and remains at sites of growth throughout the cell cycle. To understand how Spa2p localization is regulated and to gain insight into its molecular function in cell polarity, we used a coimmunoprecipitation strategy followed by tandem mass spectrometry analysis to identify proteins that associate with Spa2p in vivo. We identified Myo1p, Myo2p, Pan1p, and the protein encoded by YFR016c as proteins that interact with Spa2p. Strikingly, all of these proteins are involved in cell polarity and/or actin function. Here we focus on the functional significance of the interactions of Spa2p with Myo2p and Myo1p. We find that localization of Spa2GFP to sites of polarized growth depends on functional Myo2p but not on Myo1p. We also find that Spa2p, like Myo2p, cosediments with F-actin in an ATP-sensitive manner. We hypothesize that Spa2p associates with actin via a direct or indirect interaction with Myo2p and that Spa2p may be involved in mediating polarized localization of polarity proteins via Myo2p. In addition, we observe an enhanced cell-separation defect in a myo1spa2 strain at 37°C. This provides further evidence that Spa2p is involved in cytokinesis and cell wall morphogenesis.
INTRODUCTION
Polarized cell growth is of fundamental importance for numerous cellular functions including differentiation, proliferation, and morphogenesis. Despite differences in the specific molecular mechanisms, essentially all cells follow a general scheme in their development of cell polarity (Drubin and Nelson, 1996). Cells first recognize an external cue (e.g., cell-cell contact or chemical gradients) or an internal cue that establishes the site of polarization at the cell surface. Next, specific proteins are recruited to the site to interpret the cue. These proteins stimulate the assembly of a polarized cytoskeleton, which mediates delivery of proteins, cell wall constituents, organelles, and other factors necessary for the initiation and maintenance of polarized cell surface growth.
Proper cell polarity in the budding yeast Saccharomyces cerevisiae is critical for its ability to divide and mate. During the vegetative cycle, budding yeast cells respond to an internal molecular cue and polarize their growth toward a specific site on the cell surface to form a bud. The bud site selection proteins dictate the position of the new bud and determine the axis of polarity (for review, see Chant, 1999). In contrast, during mating, yeast cells respond to an external cue, that of peptide pheromone secreted by cells of the opposite mating type, which orients polarized growth in the direction of their mating partner.
Many of the components that influence cell polarity in yeast are necessary for both budding and mating processes and are localized to sites of polarized growth: the presumptive bud site, bud tip, mother-bud neck, and the projection tip of mating cells. Such factors include cytoskeletal elements, motor proteins, G-proteins, and a group of proteins that includes Spa2p, Pea2p, Bud6p, and Bni1p. How these proteins are localized to and maintained at sites of polarized growth is unclear.
Spa2p is a nonessential protein that regulates yeast cell polarity but whose precise molecular function is unknown. Previous studies have shown that mutants lacking Spa2p exhibit several polarity-related phenotypes. First, the precise spatial pattern of cell division of wild-type a/α cells is altered in an a/α spa2/spa2 strain. Wild-type diploid a/α cells divide in a bipolar budding pattern, in which new buds are formed either near the previous bud site or at the opposite end of the cell. By contrast, diploid a/α spa2/spa2 cells bud in a random pattern (Snyder, 1989; Zahner et al., 1996). Second, a/α spa2/spa2 cells exhibit a rounder cell morphology than that of wild-type cells, suggesting that they exhibit reduced apical growth (Sheu et al., 2000). Several other spa2 polarity defects are manifested in haploid cells and include altered morphology in response to mating pheromone (broad mating projection instead of pointed), inefficient mating to an enfeebled mating partner (Chenevert et al., 1994), and a defect in cell fusion during mating (Gammie et al., 1998). Finally, spa2 mutants display a cytokinesis defect, most evident in a/α diploid cells (Snyder et al., 1991).
Consistent with its role in cell polarity, Spa2p localizes to sites of polarized growth in both vegetatively growing and mating cells. Spa2p localizes to the presumptive bud site, to the tips of small buds, to the mother-bud neck before cytokinesis, and to the tips of mating factor-induced projections (Snyder, 1989; Gehrung and Snyder, 1990; Arkowitz and Lowe, 1997). Furthermore, Spa2p interacts with a number of proteins involved in cell polarity and signaling. First, Spa2p interacts with the cell polarity proteins Pea2p, Bud6/Aip3p, and Bni1p. Both coimmunoprecipitation studies and two-hybrid assays have demonstrated that Spa2p interacts with Pea2p (Sheu et al., 1998) and Bni1p (Fujiwara et al., 1998). In addition, Spa2p interacts with Bud6p/Aip3p by two-hybrid assay, and Spa2p, Pea2p, and Bud6p/Aip3p cosediment in sucrose gradients as a 12S complex (Sheu et al., 1998). Of note, Spa2p, Pea2p, Bud6/Aip3p, and Bni1p have similar localization patterns throughout the cell cycle, and mutants lacking these proteins exhibit similar phenotypes (Snyder, 1989; Valtz and Herskowitz, 1996; Zahner et al., 1996; Amberg et al., 1997; Evangelista et al., 1997; Shih, 2001). Thus, it has been suggested that these proteins function together in regulating cell polarity in yeast (Sheu et al., 1998).
Spa2p also interacts with several signaling proteins of the pheromone-response, protein kinase C (PKC; cell-integrity) MAPK, and high osmolarity growth (HOG) pathways. Spa2p interacts with Ste11p and Ste7p (components of the pheromone-response MAPK module) and Mkk1p and Mkk2p (MEKs of the PKC pathway) by the two-hybrid assay (Sheu et al., 1998). Furthermore, Mpk1p and Mkk1p localize to sites of polarized growth in a Spa2p-dependent manner and membrane-bound Spa2p is sufficient to recruit Mkk1 and Mpk1p but not other MAP kinases to the cell cortex. These results suggest that Spa2 may function as a scaffold-like protein to recruit the Mpk1p-MAP kinase module to sites of polarized growth (van Drogen and Peter, 2002). Spa2p also coimmunoprecipitates with Ssk2p (MAPKKK of the HOG pathway) in osmotically stressed cells and efficient localization of Ssk2p at the bud tip depends on Spa2p (Yuzyuk and Amberg 2003).
Until recently, with the proposal that Spa2 may function as a scaffold protein for the cell wall integrity pathway during polarized growth (van Drogen and Peter, 2002), the role of Spa2p in determining cell polarity remained unclear despite the long list of interacting proteins reviewed above. To further elucidate its function, we used a coimmunoprecipitation method coupled with tandem mass spectrometry analysis to identify potential in vivo binding partners of Spa2p. Here, we identify Myo1p, Myo2p, Pan1p, and the protein encoded by YFR016c as new binding partners of Spa2p. Significantly, all of these proteins are involved in cell polarity and/or actin function (Watts et al., 1987; Johnston et al., 1991; Lillie and Brown, 1994; Govindan et al., 1995; Brown, 1997; Duncan et al., 2001; Ho et al., 2002). We show that Spa2p, like Myo2p, cosediments with actin in an ATP-sensitive manner and that localization of Spa2GFP to sites of polarized growth depends on Myo2p. We also explore the relationship of MYO1 and SPA2 and provide functional links between Spa2p and cell separation and cytokinesis.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
Yeast strains used in this study are described in Table 1. Standard yeast growth conditions and genetic manipulations are described in Rose et al. (1990). Cells were grown in YEPD medium at 30°C unless otherwise noted.
Table 1.
Yeast strains used in this study
| Name | Genotype | Source |
|---|---|---|
| JSY162 | MATapep4::HIS3 prb1Δ1.6R ura3-52 his3Δ 200 trp1 lys2-801 leu2Δ1 can1 gal | This study |
| JSY163 | MATaSPA2-zz:Kl URA3 pep4::HIS3 prb1Δ1.6R ura3-52 his3Δ 200 trp1 lys2-801 leu2Δ1 can1 gal | This studya |
| JSY214 | MATaSPA2-HA:TRP1 pep4::HIS3 prb1Δ1.6R ura3-52 his3Δ 200 trp1 lys2-801 leu2Δ1 can1 gal | This studya |
| JSY261 | MATaMYO2-myc:Kan pep4::HIS3 prb1Δ1.6R ura3-52 his 3Δ 200 trp1 lys2-801 leu2Δ1 can1 gal | This studya |
| JSY262 | MATaMYO2-myc:Kan spa2::Kl URA3 pep4::HIS3 prb1Δ1.6R ura3-52 his3Δ 200 trp1 lys2-801 leu2Δ1 can1 gal | This studya |
| YEF473A | MATatrp1Δ63 leu2-Δ1 ura3-52 his3-Δ200 lys2-801 | J. Pringle |
| YEF1681 | MATaMYO1-GFP:Kan trp1Δ63 leu2-Δ1 ura3-52 his3-Δ200 lys2-801 | E. Bi |
| JSY212 | MATaMYO1-GFP:Kan spa2::Kl URA3 trp1Δ63 leu2-Δ1 ura3-52 his3-Δ200 lys2-801 | This studyb |
| YEF2056 | MATamyo1::HIS3 trp1Δ63 leu2-Δ1 ura3-52 his3-Δ200 lys2-801 +YCp50-MYO1 | E. Bi |
| YEF1813 | MATamyo1::HIS3 trp1Δ63 leu2-Δ1 ura3-52 his3-Δ200 lys2-801 | E. Bi |
| JSY120 | MATaspa2::Cg HIS3 trp1Δ63 leu2-Δ1 ura3-52 his3-Δ200 lys2-801 | This studyb |
| JSY235 | MATamyo1::HIS3 spa2::Cg TRP1 trp1Δ63 leu2-Δ1 ura3-52 his3-Δ200 lys2-801 | This studyb |
| JSY154 | MATaSPA2-GFP:TRP1 trp1Δ63 leu2-Δ1 ura3-52 his3-Δ200 lys2-801 | This studyb |
| JSY213 | MATaSPA2-GFP:TRP1 myo1::HIS3 trp1Δ63 leu2-Δ1 ura3-52 his3-Δ200 lys2-801 | This studyb |
| JSY244 | MATaMYO2-GFP:Kan trp1Δ63 leu2-Δ1 ura3-52 his3-Δ200 lys2-801 | This studyb |
| JSY245 | MATaMYO2-GFP:Kan spa2::Cg HIS3 trp1Δ63 leu2-Δ1 ura3-52 his3-Δ200 lys2-801 | This studyb |
| NY580 | MATaPEP4::URA3 ura3-52 leu2-3112 | P. Novick |
| JSY278 | MATaSPA2-HA:Kan PEP4::URA3 ura3-52 leu2-3112 | This studyc |
| NY1125 | MATα myo2-66 PEP4::URA3 ura3-52 his4-619 | P. Novick |
| JSY279 | MATα myo2-66 SPA2-HA:Kan PEP4::URA3 ura3-52 his4-619 | This studyc |
| ABY531 | MATα MYO2:HIS3 ade2-101 his3Δ200 leu2-3112 lys2-801 | A. Bretscher |
| JSY237 | MATaMYO2:HIS3 ade2-101 his3Δ200 leu2-3112 lys2-801 | This studyd |
| JSY231 | MATα MYO2:HIS3 SPA2GFP:Kan ade2-101 his3Δ200 leu2-3112 lys2-801 | This studyd |
| JSY257 | MATaMYO2:HIS3 SPA2GFP:Kan ade2-101 his3Δ200 leu2-3112 lys2-801 | This studyd |
| ABY533 | MATα myo2-13:HIS3 ade2-101 his3Δ200 leu2-3112 lys2-801 | A. Bretscher |
| JSY232 | MATα myo2-13:HIS3 SPA2GFP:Kan ade2-101 his3Δ200 leu2-3112 lys2-801 | This studyd |
| ABY535 | MATα myo2-66:HIS3 ade2-101 his3Δ200 leu2-3112 lys2-801 | A. Bretscher |
| JSY234 | MATα myo2-66:HIS3 SPA2GFP:Kan ade2-101 his3Δ200 leu2-3112 lys2-801 | This studyd |
| JSY254 | MATamyo2-66:HIS3 SPA2GFP:Kan ade2-101 his3Δ200 leu2-3112 lys2-801 | This studyd |
| ABY536 | MATα myo2-16:HIS3 ade2-101 his3Δ200 leu2-3112 lys2-801 | A. Bretscher |
| JSY233 | MATα myo2-16:HIS3 SPA2GFP:Kan ade2-101 his3Δ200 leu2-3112 lys2-801 | This studyd |
| JSY255 | MATamyo2-16:HIS3 SPA2GFP:Kan ade2-101 his3Δ200 leu2-3112 lys2-801 | This studyd |
| LWY2598 | MATaura3-52 leu2-3112 trp1Δ90 lys2-801 ade8::HIS3 | L. Weisman |
| LWY2599 | MATamyo2-2 ura3-52 leu2-3112 trp1Δ90 lys2-801 ade8::HIS3 | L. Weisman |
| JSY276 | MATaSPA2GFP:Kan ura3-52 leu2-3112 trp1Δ90 lys2-801 ade8::HIS3 | This studye |
| JSY277 | MATamyo2-2 SPA2GFP:Kan ura3-52 leu2-3112 trp1Δ90 lys2-801 ade8::HIS3 | This studye |
| JCY452 | MATα trp1 leu2 ura3 his3 lys2 suc2 | B. Wendland |
| JCY455 | MATα pan1-20 trp1 leu2 ura3 his3 lys2 suc2 | B. Wendland |
| IH4203 | MATα pan1-20 SPA2GFP:HIS3 trp1 leu2 ura3 his3 lys2 suc2 | This studyf |
These strains are derived from JSY162.
These strains are derived from YEF473A (Bi and Pringle, 1996).
These strains are isogenic with NY580 (Reck-Peterson et al., 2001).
These strains are isogenic with ABY531 (Schott et al., 1999).
These strains are isogenic with LWY2598 and LWY2599 (Catlett and Weisman, 1998).
This strain is derived from JCY452 (Wendland and Emr, 1998).
Strain Construction
C-terminal tagging with protein A (Spa2-zz) was performed as described by Puig et al. (1998). Briefly, a PCR product was generated that contained the coding information for a protein A Ura3p cassette flanked by the final 48 nucleotides of the Spa2p coding region and 48 nucleotides downstream of the stop codon. The PCR product was transformed into JSY162 by the lithium acetate method, essentially as described (Ito et al., 1983) to create JSY163. Correct integration into the genome was verified by PCR. Expression of the fusion protein was confirmed by Western blot.
Similarly, C-terminal tagging with GFP, 3HA and 13Myc was performed as described by Longtine et al. (1998). PCR products were generated that contained a GFP Kan, 3HA Trp1p, 3HA Kan, or 13Myc Kan coding cassette flanked by Spa2p coding sequence. These amplified products were transformed into yeast and correct substitution verified by PCR. Expression of the fusion protein was confirmed by Western blot. The Spa2GFP Kan construct was transformed into ABY531, ABY533, ABY535, and ABY536 to create JSY231, JSY232, JSY234, and JSY233, respectively. JSY231, JSY233, and JSY234 were crossed to JSY237 to generate diploids, which were then used to obtain JSY257, JSY256, and JSY254. The Spa2GFP Trp1 construct was transformed into YEF1813 to generate JSY213. The Myo1GFP construct was transformed into YEF473A to create JSY212. The Spa2HA Kan construct was transformed into NY580 and NY1125 to create JSY278 and JSY279, respectively. JSY261 was created by transforming the Myo2-myc Kan construct into JSY162.
Gene deletions were constructed as described by Puig et al. (1998) or Kitada et al. (1995). Briefly, a PCR product was generated that contained a Kluyveromyces lactis URA3 (or Candida glabrata HIS3 or TRP1) cassette flanked by sequences immediately upstream and downstream of the SPA2 open reading frame. The amplified fragment was transformed into yeast. Correct integration was verified by PCR and phenotypic analysis. The SPA2 knock-out construct (K. lactis URA3-based) was transformed into YEF1681 to create JSY212 and transformed into JSY261 to create JSY262. Similarly, C. glabrata-based SPA2 knock-out constructs were transformed into JSY244, YEF473A, and YEF2056 to create JSY245, JSY120, and JSY235, respectively. JSY235 transformants were later cured of the YCp50-MYO1 plasmid by selection for growth on 5-FOA plates.
Immunoprecipitation of Proteins Associated with Spa2-zz
Log-phase cells from a 250-ml culture of JSY162 or JSY163 were harvested and washed with 50 ml H2O. The pellet was resuspended in 1.5 ml IgG buffer containing 50 mM Tris, pH 7.5, 1 mM EDTA, 5 mM MgCl2, 0.1 mM dithiothreitol (DTT), 5% glycerol, 1% NP40, 150 mM NaCl, and a Complete mini protease inhibitor tablet (1 tablet/10 ml IgG buffer stock, Boehringer Mannheim, Indianapolis, IN). Cells were lysed with 0.5-mm glass beads (Biospec, Bartlesville, OK) for 10 s at 4°C, repeated 10 times, with 1-min resting periods on ice, using a Mini-Bead Beater (Biospec). Lysates were cleared by spinning in a microfuge at 14,000 rpm for 10 min at 4°C. Clarified lysates were diluted fivefold before incubation with IgG-Sepharose (Amersham Pharmacia, Piscataway, NJ). Fifteen Eppendorf tubes with 500-μl aliquots of diluted lysate were incubated with 25 μl IgG-Sepharose beads for 90 min at 4°C. The resin was washed extensively with IgG buffer, and bound proteins were eluted with IgG buffer containing 1 M MgCl2 and concentrated with chloroformmethanol precipitation (Wessel and Flugge, 1984). Bound proteins from the 15 Eppendorf tubes were pooled and resolved by SDS-PAGE (6.5% SDS-Gel) and visualized by standard Coomassie Blue staining (Ausubel, 1991).
In-Gel Digestion of Protein Excised from SDS-PAGE
The dominant bands observed by Coomassie Blue staining were excised and minced. Gel pieces were dehydrated in acetonitrile for 10 min, centrifuged, and the supernatant was aspirated. This step was repeated and followed by evaporation of the gel pieces under vacuum. Fifty microliters milli-Q water was added, and the gel slices were allowed to re-swell for 15 min. Excess water was removed and the gel pieces were again dehydrated in acetonitrile for 15 min. Excess acetonitrile was removed and the gel pieces were then dried under vacuum.
In-gel protein digestion and peptide extraction were performed according to the procedures of Shevchenko et al. (1996). Gel pieces were rehydrated at 4°C in a digestion buffer containing 50 mM NH4HCO3 and 12.5 ng/μl modified porcine trypsin (Promega, Madison, WI; sequencing grade). After 45 min, the tubes were topped up with milli-Q water to just cover the gel pieces and transferred to 37°C for overnight incubation.
Analysis of Peptides by Microelectrospray LC-MS/MS
Microelectrospray columns were constructed from 360 μm od × 75 μm id fused silica capillary with the column tip tapered to a 5–10-μm opening. The columns were packed with 200 Å 5-μm C18 beads (Michrom BioResources, Auburn, CA), a reverse-phase packing material, to a length of 10–12 cm. The flow-through of the column was split precolumn to achieve a flow rate of 200 nl/min. The mobile phase used for gradient elution consisted of 1) 0.4% acetic acid, 0.005% heptafluorobutyric acid, and 5% acetonitrile and 2) 0.4% acetic acid and 0.005% heptafluorobutyric acid in acetonitrile. The gradient was linear from 0.5–45% in the first mobile phase solution in 35 min followed by 45–65% in 5 min in the second solution.
Tandem mass spectra were recorded on an LCQ ion trap mass spectrometer (Thermoquest, San Jose, CA) equipped with an in-house microelectrospray ionization source. Needle voltage was set at 1.6 kV. Nonredundant yeast protein sequence databases were searched directly with the tandem mass spectra using the computer algorithm, SEQUEST, described previously (Eng et al., 1994; Yates et al., 1995).
Cell Fractionation
For crude fractionations, log-phase cells from a 100-ml culture of JSY214 were harvested, washed with 20 ml H2O, and the pellet resuspended in 1 ml 50 mM Tris, pH 7.5, 1 mM EGTA, 0.1 mM DTT, 150 mM KCl, and a Complete mini protease inhibitor tablet (1 tablet/10 ml buffer stock, Boehringer Mannheim). The cells were lysed with 0.5-mm glass beads (Biospec) for 45 s at 4°C, repeated three times, with 1-min resting periods on ice, using a Mini-Bead-Beater-8 (Biospec). The cell extract was cleared by centrifugation at 2000 × g for 5 min at 4°C in a Beckman TLA100.2 rotor/Beckman Optima ultracentrifuge (Beckman Intruments, Palo Alto, CA) and designated S1 (supernatant 1). The S1 fraction was then centrifuged at 30,000 × g for 30 min at 4°C (Beckman TLA100.2 rotor) to create fraction S2 (supernatant 2) and P2 (pellet 2). Finally, the S2 fraction was centrifuged at 100,000 × g for 1 h at 4°C (Beckman TLA100.2 rotor) to create S3 (supernatant 3) and P3 (pellet 3). Samples from each fraction were separated by SDS-PAGE and transferred to nitrocellulose, and immunoblots were performed with anti-HA monoclonal antibody (1:1000, Covance), anti-Myo2p antibody (1:2000, Reck-Peterson et al., 1999), anti-Snc1p antibody (1:1000, generous gift of Pat Brennwald, Cornell University Medical College), or anti-Pea2p antibody (1:200, Valtz and Herskowitz, 1996).
Actin Cosedimentation Assays
Log-phase cells from 100 ml of NY580, NY1125, JSY278, JSY279, JSY261, or JSY262 were harvested and lysed as described above in buffer A: 20 mM imidazole, 75 mM KCl, 1 mM EGTA, 2.5 mM MgCl2, 2 mM DTT, and a Complete mini-protease inhibitor tablet (1 tablet/10 ml buffer A stock, Boehringer Mannheim) or a protease inhibitor cocktail (1 mM Pefabloc-sc [Boehringer], 10 mM pepstatin A, 10 mM leupeptin, 1 mM benzamidine, pH 7.2). JSY278 and JSY279 were grown to log phase at 25°C and then shifted to 37°C for 2.5 h before harvesting and cell lysis.
The lysate was spun at 2000 × g for 5 min at 4°C (Beckman TLA100.2 rotor/Beckman Optima ultracentrifuge) and the resulting supernatant spun at 257,000 × g for 20 min at 4°C (Beckman TLA100.2 rotor). The supernatant (S3) and P3 fractions from this spin were used for actin cosedimentation assays.
To monitor actin binding of soluble Myo2p, S3 (supernatant after a 257,000 × g spin for 20 min) was mixed with 7 μM phalloidin-stabilized F-actin purified from chicken skeletal muscle (Spudich and Watt, 1971) or 10 μM phalloidin-stabilized F-actin purified from Acanthamoeba (generously provided by D. Mullins, UCSF) in buffer A with or without 4 mM ATP. The reaction mixture was incubated on ice for 10 min and then centrifuged at 175,000 × g for 20 min at 4°C (Beckman TLA100 rotor) to pellet the F-actin.
To monitor actin binding of particle-associated Myo2p in P3, 7 μM F-actin was mixed with resuspended P3 in the presence of 20 mM MgCl2 (to form paracrystalline actin bundles that pellet at low speeds), 25 μM phalloidin (Boehringer-Mannheim) in buffer A with or without 4 mM ATP. The reaction mixture was incubated on ice for 10 min and then spun at 21,000 × g for 15 min at 4°C (Beckman TLA100 rotor) to pellet the F-actin bundles.
The resulting supernatant and pellet fractions were separated by SDS-PAGE. The lower half of the gel was stained with Coomassie Blue to visualize actin. The top half of the gel was transferred to nitrocellulose, and immunoblots were performed with anti-HA antibody, anti-Myo2p tail antibody, or anti-Pea2p antibody.
Microscopy
Microscopic analysis was performed using an Olympus BX60 with a 100X UplanApo objective (Lake Success, NY). Cells were visualized by differential interference contrast (DIC) or epifluorescence. Images were captured with a SPOT2e camera (Diagnostic Instruments, Sterling Heights, MI) and down-loaded directly into Adobe Photoshop (Adobe Systems, San Jose, CA).
Two scoring strategies were used to quantify Spa2GFP localization phenotype. In one scheme, the percentage of cells with some Spa2-GFP in the correct location (bud site, bud cortex, or neck cortex) was determined. This assay was performed three times, scoring >200 cells for each strain. In the other scheme, cells that properly localized Spa2GFP were classified into three groups: localization at presumptive bud site/tips of small buds, localization to mediumsized buds, and localization at the neck in large-budded cells.
RESULTS
Identification of Proteins That Associate with Spa2p
To understand how Spa2p localization to sites of growth is regulated and gain insight into its function in yeast cell polarity, a coimmunoprecipitation method coupled with tandem mass spectrometry was used to identify proteins that physically associate with Spa2p. We constructed a strain that expressed a fusion protein (Spa2-zz) consisting of Spa2 and two Protein A z-domains (zz) to immunoprecipitate Spa2p and its associated proteins. Cells expressing Spa2-zz as their only source of Spa2 protein exhibited a Spa2+ phenotype with respect to shmoo morphology and bud-site selection in an a/α diploid strain (Chenevert et al., 1994; Zahner et al., 1996), demonstrating that the fusion protein was functional.
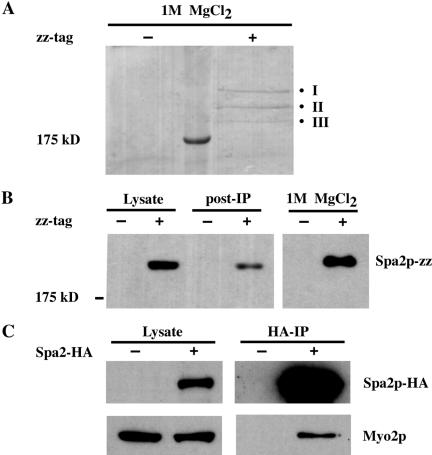
IgG Sepharose was added to yeast whole-cell extracts derived from wild-type or Spa2-zz strains to precipitate Spa2-zz and associated proteins. The resin was washed, and bound proteins were eluted with 1 M MgCl2, separated by SDS-PAGE, and visualized with Coomassie Blue (see Materials and Methods). We found three protein bands present specifically in the Spa2-zz immunoprecipitate and excised them individually from the gel for analysis by tandem mass spectrometry (Figure 1, A and B). Eight different tryptic peptides of the protein encoded by YFR016c were identified from Band I; 18 different peptides of Spa2p and eight different peptides of Myo1p were identified from Band II; 14 different peptides of Myo2p and 7 different peptides of Pan1p were identified from Band III (Table 2). Thus, we identified the protein encoded by YFR016c, Myo1p, Myo2p, and Pan1p as proteins that potentially can complex with Spa2p.
Figure 1.
Immunoprecipitation of proteins that interact with Spa2-zz. (A) Lysates derived from untagged (JSY162) or Spa2-zz (JSY163) strains were incubated with IgG-Sepharose beads for 90 min at 4°C. The resin was washed, and bound proteins were eluted with 1 M MgCl2. Eluted proteins were concentrated, separated on 6.5% SDS-PAGE, and visualized by Coomassie Blue staining. Three bands (I, II, and III) were present specifically in the Spa2-zz immunoprecipitate and individually excised for tandem mass spectrometry analysis. (B) Samples of the lysates before and after (post-IP) incubation with IgG-Sepharose beads and the 1 M MgCl2 eluant were separated by SDS-PAGE, transferred to nitrocellose, and visualized by immunoblotting with preimmune serum. (C) Myo2p coimmunoprecipitates with Spa2p-HA. Proteins prepared from untagged (JSY162) and Spa2p-HA (JSY214) strains were immunoprecipitated with HA-Sepharose beads for 2 h at 4°C. Total lysates and immunoprecipitates were analyzed on immunoblots and probed with anti-HA or anti-Myo2p antibodies.
Table 2.
Peptides identified by LC-MS/MS
| Excised band | Protein name | Position | Peptide identifieda |
|---|---|---|---|
| I | YFR016c | 171-184 | ESTGIEVGNSPITR |
| 239-251 | VNIVQDEPVNVEK | ||
| 529-542 | SSIIEIEGSANSAK | ||
| 578-588 | DDVEIVEAVEK | ||
| 637-647 | QEGTAELSNEK | ||
| 730-742 | VQISTEQAETTQK (5) | ||
| 1093-1109 | IVDDSELNALLQSLDAK (2) | ||
| 1205-1224 | GHNDLIGNWEEIEEANEDYK (2) | ||
| II | Spa2 | 1-13 | MGTSSEVSLAHHR |
| 71-84 | IGEDANQPDYLLPK | ||
| 151-175 | TSTNSSSVTQVAPNVSVQPSLVIPK | ||
| 523-537 | SDSNGESTTSNEGNR | ||
| 596-615 | AINSPIIRPSSSNGVPTTSR | ||
| 634-638 639-643 644-649 | NSSHK EDNDK YVSPIK | ||
| 650-667 | AVTSASNSASSNISEIPK | ||
| 676-691 | IGTVIPPSENQVPNIK | ||
| 873-889 | NFQEPLGNVESPDMTQK (2) | ||
| 899-907 908-918 | AVGPESDSR VESPGMTGQIK (2) | ||
| 969-976 977-981 | LASSGEVDK IESPR | ||
| 985-1003 | ESESLEAVGNTIPSNMTVK | ||
| 1060-1067 | TPSSATLK | ||
| 1069-1085 | SGLPEPNSQIVSPELAK (2) | ||
| 1100-1117 | ETNKPHTETITSVEPTNK (2) | ||
| 1125-1130 | DADLNR | ||
| 1431-1440 | LAGIAFDVAK (2) | ||
| Myo1 | 850-862 | IKPLLTSSNDMTR | |
| 1013-1018 | LENEIK | ||
| 1063-1074 | LQSLVTENSDLR | ||
| 1337-1351 | AHYDAENAISALHSK | ||
| 1450-1459 | EALLSEQLDR | ||
| 1695-1711 | TDALQISNAALSSSTQK | ||
| 1761-1773 | NIDLYEENQTLQK | ||
| 1916-1925 | NIDSNNAQSK | ||
| III | Myo2 | 180-203 | YFASVEEENSATVQHQVEMSETEQ |
| 205-219 | ILATNPIMEAFGNAK | ||
| 354-366 | NDASLSADEPNLK | ||
| 395-408 | IVSNLNYSQALVAK | ||
| 514-525 | LGILSLLDEESR (2) | ||
| 595-610 | ASTNETLINILEGLEK (2) | ||
| 621-629 | LELEQAGSK | ||
| 735-746 | ETTEEDIISVVK | ||
| 947-957 | VIELTQNLASK | ||
| 1009-1022 | TIENNLQSTEQTLK | ||
| 1055-1071 | TLVEYQTLNGDLQNEVK (2) | ||
| 1075-1079 | EEIAR | ||
| 1474-1493 | LISQYQVADYESPIPQEILR | ||
| 1561-1574 | IVDLVAQQVVQDGH | ||
| Pan1 | 581-595 | SNFNNNLIDNSSQDK | |
| 617-629 | GLLDSPTAVEIFR | ||
| 740-752 | NEEQSSFSSPSAK | ||
| 930-940 | SSISDISASLK | ||
| 981-1000 | SVTESSPFVPSSTPTPVDDR (2) | ||
| 1055-1070 | EQPQQIAGSSNLVEPR | ||
| 1248-1260 | DASASSTSTFDAR |
The numbers in parentheses indicate the number of times a peptide was identified if it was identified more than once.
Using a different C-terminal tag, we further confirmed the interaction between Spa2p and Myo2p by coimmunoprecipitation of Myo2p with Spa2-HA. Anti-HA immunoprecipitates from yeast extracts derived from wild-type or Spa2-HA strains were loaded on SDS-PAGE, blotted onto nitrocellulose membrane, and probed with Myo2p antibody. Figure 1C shows that Myo2p was readily detected in the Spa2HA immunocomplex.
Subcellular Fractionation of Spa2p Is Similar to That of Myo2p
Myo2p is a yeast class V myosin that is believed to be a motor protein that brings secretory vesicles into the bud tip along actin cables. Subcellular fractionation studies of Myo2p indicate that it is present in both plasma membrane and microsomal fractions (Reck-Peterson et al., 1999). Similar studies have shown that Bud6p/Aip3p, which interacts by two-hybrid assay and cosediments with Spa2p (Sheu et al., 1998), is also present in the microsomal fraction (Jin and Amberg, 2000). Given that Spa2p interacts with Myo2p and Bud6p/Aip3p, we hypothesized that Spa2p would also be present in both plasma membrane and microsomal fractions. We characterized the subcellular fractionation of Spa2p in wild-type cells using differential centrifugation, following the scheme described by Reck-Peterson et al. (1999) and Jin and Amberg (2000).
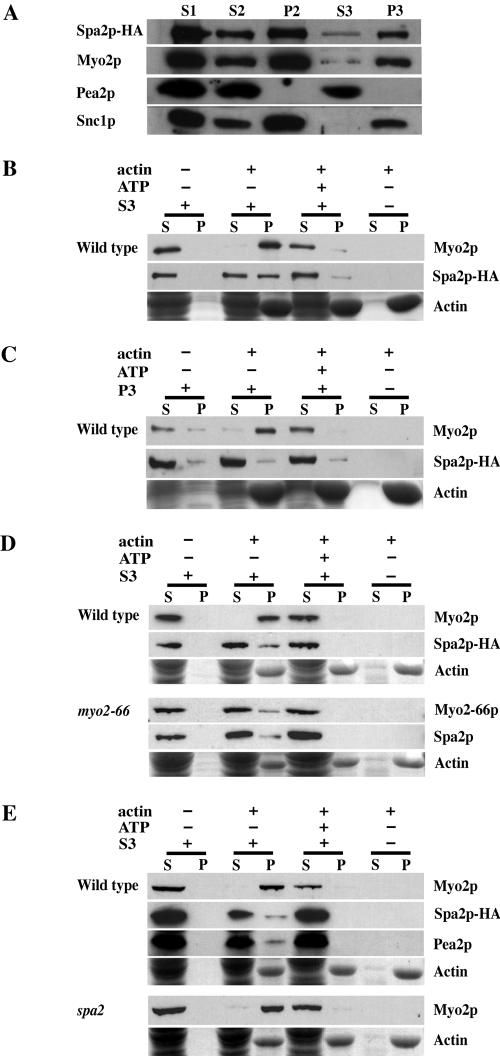
Cell extracts obtained by mechanical lysis were clarified by centrifugation at 2000 × g to remove cell debris (S1). S1 was first fractionated at 30,000 × g for 30 min to create supernatant 2 (S2) and pellet 2 (P2). S2 was then fractionated at 100,000 × g for 1 h to create supernatant 3 (S3) and pellet 3 (P3). Samples from each fraction were separated by SDS-PAGE, transferred to nitrocellulose, and probed with anti-HA or anti-Myo2 antibodies or antibodies for other markers. As seen in Figure 2A, Spa2p was present in every fraction, exhibiting a pattern remarkably similar to that of Myo2p. Interestingly, we found Pea2p, a protein involved in cell polarity that interacts tightly with Spa2p (Sheu et al., 1998; Valtz and Herskowitz, 1996) to be present only in the S2 and the S3 fractions and not in the P2 and P3 fractions (Figure 2A).
Figure 2.
(A) Spa2p cofractionates with Myo2p. Lysate from logphase cells of the Spa2-HA strain (JSY214) was spun at 2000 × g to generate supernatant 1 (S1). S1 was spun at 30,000 × g, resulting in S2 and pellet 2 (P2). S2 was spun at 100,000 × g to generate S3 and P3. Gel samples were prepared using volumetric stoichiometry and equal volumes of each sample were loaded per lane, separated by SDS-PAGE, and transferred to nitrocellulose. Blots were then probed with anti-HA, anti-Myo2p, anti-Pea2p, or anti-Snc1p antibodies. P2 is the plasma membrane fraction, as Pma1p, a marker of the plasma membrane in yeast, is found primarily in P2 (Reck-Peterson et al., 1999). P3 is the microsomal fraction and is enriched in late secretory vesicles (Goud et al., 1988). Snc1p, a late vesicleborne, integral membrane protein (Protopopov et al., 1993), is present primarily in the P3 fraction. (B–E) Actin cosedimentation assays. S3 supernatant or P3 pellet fractions derived from lysates of wild-type strains, NY580 (B and C), JSY278 (D) or JSY261 (E); myo2-66 strain, JSY279, (D); or spa2Δ strain, JSY262 (E) were used for actin cosedimentation assays. Wild-type and myo2-66 strains in D were grown at 25°C and shifted to 37°C for 2.5 h before harvesting and cell lysis. Myo2p, Myo2-66p, Spa2p-HA, and Pea2p present in the S3 (B, D, and E) or Myo2p and Spa2p-HA present in the P3 (C) were incubated with F-actin in the presence or absence of 4 mM ATP. After centrifugation to pellet the F-actin, the supernatant and pellet fractions were immunoblotted with antibodies to Myo2p, HA or Pea2p. Actin was visualized by Coomassie Blue staining. A fraction of Spa2p and Pea2p (present in S3) cosediment with actin in an ATP-sensitive manner similar to that of Myo2p.
Spa2p and Myo2p Cosediment with Actin
Like most myosins, yeast Myo2p can cosediment with F-actin only in the absence, but not in presence, of 4 mM ATP (Reck-Peterson et al., 2001). We wanted to determine whether Spa2p could associate and cosediment with actin through its interaction with Myo2p. First, Myo2 and Spa2 proteins present in the S3 or P3 fractions were assayed for their ability to cosediment with chicken skeletal muscle F-actin. As seen in Figure 2B, Myo2p from the S3 fraction cosedimented with actin in the absence but not in the presence of ATP. A pool of Spa2 protein from the S3 fraction also cosedimented with actin in a similar ATP-sensitive manner, presumably via its interaction with Myo2p. By contrast, Spa2p present in the P3 fraction did not exhibit this behavior, although Myo2p in the same P3 fraction was able to cosediment with actin in an ATP-sensitive manner (Figure 2C).
To determine if association of Spa2p with actin is dependent on Myo2p, we assayed sedimentation of Spa2p in the myo2-66 background. Myo2-66 protein from myo2-66 cells shifted to nonpermissive conditions showed impaired binding to actin in the absence of ATP, consistent with the presence of a point mutation in the predicted actin-binding face of the motor domain in this mutant (Figure 2D and Reck-Peterson et al., 2001). In the myo2-66 background, a pool of Spa2 protein continued to cosediment with F-actin, similar to its behavior in the wild-type background. Given these results, no conclusions can be made regarding the dependence of Spa2p cosedimentation with actin upon Myo2p.
Because Pea2p has been shown to interact tightly with Spa2p (Sheu et al., 1998), we also wanted to determine if Pea2p might also cosediment with actin via its interaction with Spa2p. Figure 2E shows that a pool of Pea2p cosedimented with actin similarly to Spa2p. We were unable to determine if the pool of Pea2p that associates with actin is dependent on Spa2p because Pea2p is not stable in the spa2 background (Valtz and Herskowitz, 1996 and unpublished data).
Although we were unable to draw any conclusions regarding the dependence of Spa2p cosedimentation with actin upon Myo2p or the dependence of Pea2p cosedimentation with actin upon Spa2p, our results suggest that Spa2p, Pea2p, and Myo2p exist together in a complex that associates with actin in an ATP-sensitive manner.
Spa2p Localization Depends on Myo2p
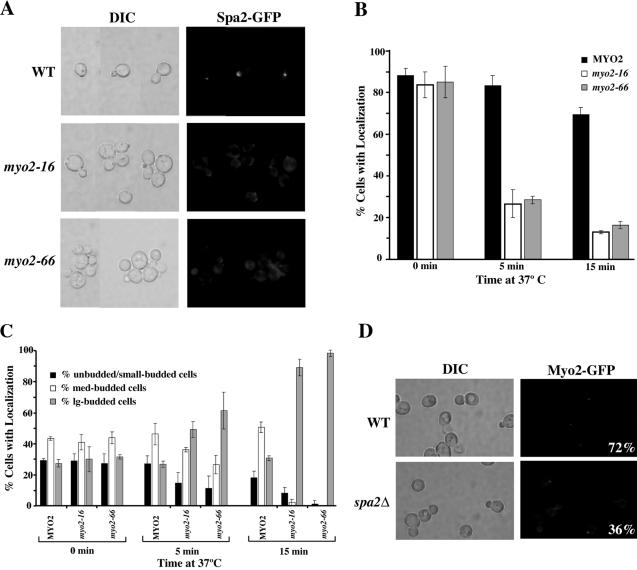
In wild-type cells, Myo2p localizes to sites of polarized growth in a pattern identical to that of Spa2p (Snyder, 1989; Gehrung and Snyder, 1990; Lillie and Brown, 1994; Arkowitz and Lowe, 1997). Colocalization of these proteins led us to investigate the dependence of polarized localization of each protein on the other. We constructed Spa2GFP fusions in several myo2 mutant backgrounds to determine the dependence of Spa2p localization on Myo2p function. Current models for myosin function propose that the motor domain moves along actin filaments, and the tail domain binds cargo. We first examined the dependence of Spa2GFP localization on Myo2p motor function in budding cells. myo2-66 encodes a protein with a mutation in the actin-interacting motor domain (Lillie and Brown, 1994). The strain carrying this allele exhibited severe defects in Spa2GFP localization within 5 min after a temperature shift to nonpermissive temperature, 37°C (Figure 3A); only 28% of myo2-66 cells exhibited polarized localization of Spa2p compared with 84% of wild-type cells (Figure 3B).
Figure 3.
(A) Localization of Spa2GFP depends on functional Myo2p. Exponentially growing wild-type (JSY257), myo2-16 (JSY255), and myo2-66 (JSY254) cells expressing Spa2-GFP were shifted to 37°C for 5 min. Cells were visualized by DIC microscopy (left panels). Spa2-GFP localization in the same cells was visualized by fluorescence microscopy (right panels). All cells are shown at the same magnification; all images were captured with the same exposure time. (B and C) Quantitative analysis of Spa2GFP localization in myo2 mutants. (B) Percentage of cells exhibiting polarized localization of Spa2GFP at different cell stages was determined in wild-type (JSY257), myo2-16 (JSY255), and myo2-66 (JSY254) strains after a 5- or 15-min shift to 37°C. This assay was performed three times, scoring >200 cells for each strain. (C) Distribution of cells exhibiting polarized localization of Spa2GFP after 5- or 15-min shift to 37°C. Cells were classified into three groups: unbudded/small budded (representing cells that exhibit apical growth), medium budded (representing cells that exhibit isotropic growth), and large budded (representing cells that exhibit repolarization to the mother-bud neck). This assay was performed three times, scoring at least 100 cells for each strain. (D) Localization of Myo2GFP partially depends on Spa2p. Exponentially growing wild-type (JSY244) or spa2Δ(JSY245) cells expressing Myo2-GFP were examined by DIC and fluorescence microscopy for proper polarized localization of Myo2GFP. 72% of wild-type cells exhibit polarized Myo2-GFP localization compared with 36% of spa2Δ cells. (n = 400)
We next examined the dependence of Spa2GFP localization on Myo2p tail domain function in budding cells. The tail domain of Myo2p contains distinct regions involved in the movement of different cargoes (Schott et al., 1999; Catlett et al., 2000; Yin et al., 2000). Several alleles, including conditional lethal mutations affecting the tail domain of Myo2 (myo2-13 and myo2-16), affect maintenance of the polarized distribution of secretory vesicles (Schott et al., 1999) but not vacuolar inheritance (Schott et al., 1999; Catlett et al., 2000). Strains carrying the myo2-16 allele exhibited loss of Spa2GFP localization within 5 min after a shift to 37°C (Figure 3, A and B). Similar observations were made for myo2-13 (unpublished data). By contrast, Spa2GFP remained polarized at sites of growth in another tail-domain mutant, myo2-2 (unpublished data), which causes defects in vacuolar inheritance but is not involved in the essential functions of Myo2p (Catlett and Weisman, 1998).
In the quantitative analysis of Spa2GFP localization in the myo2 strains described above, we observed that Spa2GFP was more readily lost at the presumptive bud site and at bud tips than at the mother-bud neck after the mutant strains were shifted to nonpermissive conditions. Figure 3C shows the distribution of cells exhibiting polarized localization of Spa2GFP after MYO2, myo2-16, and myo2-66 strains were shifted to nonpermissive conditions for 0, 5, or 15 min. The distribution of cells (with respect to cell cycle stage based on bud size) that properly localized Spa2GFP before shifting to 37°C was similar for each of the strain backgrounds. Twenty-seven to 29% of the cells that properly localized Spa2GFP were unbudded or small-budded cells, 40–43% were medium-budded cells, and 27–32% were large-budded cells. After a 5-min shift to 37°C, wild-type cells that properly localized Spa2GFP continued to exhibit a similar cell distribution pattern (27% unbudded or small-budded, 46% medium-budded, 26% large-budded cells). By contrast, myo2-16 cells that properly localized Spa2GFP after a 5-min shift to 37°C exhibited a different distribution: 14% of these cells were unbudded or small-budded cells, 36% were medium-budded cells, and 49% were large-budded cells. myo2-66 cells that properly localized Spa2GFP after a 5-min shift to 37°C exhibited a cell distribution similar to that of myo2-16 cells: Eleven percent were unbudded or small-budded cells, 26% were medium-budded cells, and 61% were large-budded cells. This shift in the distribution of cells that properly localized Spa2GFP toward the large-budded cells in both myo2-16 and myo2-66 cells was more pronounced after 15 min at 37°C.
Taken together, these results indicate that efficient localization of Spa2p to sites of polarized growth in vegetative cells requires functional Myo2 head and tail function. The fact that a small population of Myo2-deficient cells is able to localize Spa2GFP to the mother-bud neck suggests that there may be a redundant pathway involved in localizing Spa2p to the neck of large-budded cells.
Because both Spa2p and Myo2p colocalize at the tips of mating cell projections (shmoo tips), we also examined the localization dependence of Spa2GFP on Myo2p in mating cells. We observed that Spa2GFP is not polarized in the shmoo tips of both myo2-66 and myo2-16 cells observed at 5 min after shift to nonpermissive temperature (unpublished data).
We also examined the localization dependence of Myo2p on Spa2p in vegetative cells and observed that Myo2p exhibited some dependence on Spa2p for its localization to sites of growth (Figure 3D). Whereas 72% of wild-type cells properly localized Myo2GFP to sites of polarized growth, only 36% of spa2 cells were able to localize Myo2GFP. Interestingly, loss of Myo2GFP localization in spa2 cells did not show preference for any particular cell stage, in contrast to localization of Spa2GFP in the absence of functional Myo2p.
Localization of Spa2p Does Not Depend on Myo1p, Pan1p, or the Protein Encoded by YFR016c
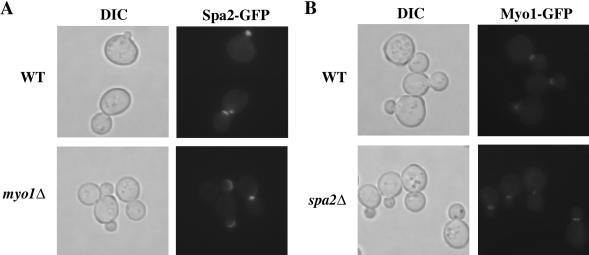
Because both Spa2p and Myo1p, a yeast class II myosin, localize to the bud neck during part of the cell cycle (Snyder, 1989; Gehrung and Snyder, 1990; Arkowitz and Lowe, 1997; Bi et al., 1998; Lippincott and Li, 1998;), we next analyzed the localization interdependence of Spa2p and Myo1p. Figure 4 shows that Spa2p and Myo1p are independent of each other for their proper localization. Spa2GFP localized to all sites of growth in the absence of Myo1p, and Myo1p localized as a ring to the presumptive bud site and to the bud neck before contracting to a point and disappearing in the absence of Spa2p. These results indicate that the localization codependence of Spa2 and Myo2 is specific and not a general property of yeast myosins.
Figure 4.
Polarized localization of Spa2p and Myo1p are independent of each other. Exponentially growing wild-type (JSY154) and myo1Δ(JSY213) cells expressing Spa2GFP (A) or wild-type (YEF1681) and spa2Δ(JSY212) cells expressing Myo1GFP were examined under DIC (left side) or fluorescence (right side) microscopy.
We also examined the dependence of Spa2GFP localization on the other Spa2p-interacting proteins that we identified: Pan1p and the protein encoded by YFR016c. Pan1p is involved in organization of the actin cytoskeleton and in endocytosis and has been shown to interact with the Arp2/3 complex and activate actin nucleation (Duncan et al., 2001). We observed that Spa2GFP localization was not disrupted in pan1-20 cells (Wendland and Emr, 1998) after a 5-min shift to 37°C (unpublished data). The protein encoded by YFR016c has 22% similarity over a segment of 108 amino acids residues to Uso1p, a protein involved in ER-vesicle transport. The null mutant is reported to be viable (Winzeler et al., 1999). Recent work by Ho et al. (2002) identified actin and actin-associated proteins as other binding partners of the protein encoded by YFR016c. We constructed the mutant strain lacking YFR016c and found that it is not required for localization of Spa2GFP to sites of growth (unpublished data). Furthermore, we found that the yfr016c mutant was not defective for bud-site selection in a haploid or in a/α diploid strains and was not defective for shmoo morphology in the YEF473A background (unpublished data).
Genetic Interaction of Spa2p with Myo1p
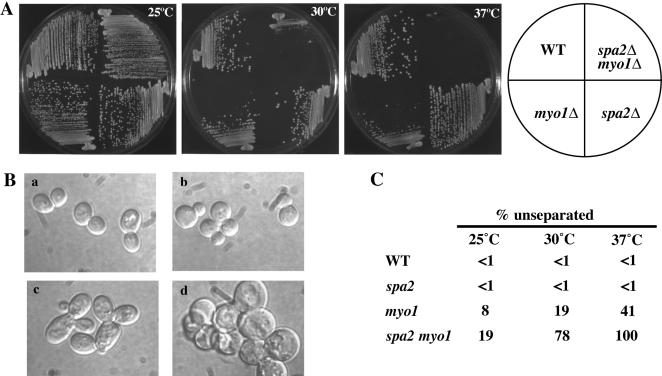
Although Spa2p and Myo1p do not depend on each other for localization, they colocalize to the mother-bud neck during part of the cell cycle and share similar mutant phenotypes with respect to cytokinesis. Thus, we examined the genetic relationship between SPA2 and MYO1. We constructed a spa2 myo1 double mutant and assayed its growth and morphology at different temperatures (Figure 5, A and B). At 25 and 30°C, spa2 and myo1 strains were both viable and grew at a rate similar to wild type. Although the spa2 myo1 strain grew at a rate indistinguishable from wild type at 25°C, it exhibited a slow-growth phenotype at 30°C. At 37°C, the spa2 myo1 strain was inviable, in contrast to myo1, which exhibited a slow-growth phenotype, and spa2, which grew at a similar rate as wild-type cells.
Figure 5.
Synthetic lethality between spa2Δ and myo1Δ. (A) Wild-type (YEF473A), myo1Δ(YEF1813), spa2Δ(JSY120), and spa2Δ myo1Δ(JSY235) strains were grown on YEPD plates at 25, 30, or 37°C. Plates were photographed after 3–4 d. At 30°C, only the spa2Δ myo1Δ strain exhibited slow growth compared with wild type. At 37°C, the myo1Δ strain exhibited a slow-growth phenotype compared with wild type, and the spa2Δ myo1Δ strain was not viable. (B) spa2Δ exacerbates the cell separation defect of myo1Δ. Log-phase (a) wild-type, (b) spa2Δ, (c) myo1Δ, and (d) spa2Δ myo1Δ cells grown in YEPD culture at 25°C were shifted to 30°C for 3.5 h and then examined under DIC microscopy. (C) The percentage of cells displaying three or more cells connected together was determined for each strain at 25°C, after shift to 30°C for 3.5 h, and after shift to 37°C for 3.5 h (after mild sonication). The assay was done three times and at least 200 cells were scored for each strain. Less than 1% of wild-type and spa2Δ cells exhibited a cell-separation defect after 3.5 h at 30°C compared with 19% of myo1Δ cells and 78% of spa2Δ myo1Δ cells. This defect was enhanced in both myo1Δ and spa2Δ myo1Δ strains after a shift to 37°C for 3.5 h.
The morphological characteristics of myo1 mutants include formation of attached cells, abnormal distribution of cell wall chitin, and formation of an abnormally thickened chitinous junction between mother and daughter cells (Rodriguez and Paterson, 1990; Bi et al., 1998; Lippincott and Li, 1998; Hales et al., 1999). We found that after shifting cells to the nonpermissive temperature for 3.5 h, all spa2 myo1 cells were in chains (n = 600; defined as three or more cell bodies connected together after mild sonication). In contrast, 41% of myo1 cells and <1% of spa2 cells were found in chains after 3.5 h of growth at 37°C (Figure 5, B and C).
DISCUSSION
We have discovered new physical associations with Spa2p and validated some of these interactions in vivo. Our results reveal that Spa2p associates with several proteins involved in actin function and cell polarity, providing new insight into the molecular function of Spa2p in yeast cell polarity. The existence of a physical interaction between Spa2p and Myo2p combined with protein localization codependence suggests a number of models. In one model, Spa2p is a cargo protein of Myo2p, either alone or as part of a multiprotein complex. Whether the interaction between Spa2p and Myo2p is required for the establishment of polarized localization of Spa2p remains unclear. However, the Spa2p-Myo2p interaction may be important for the maintenance of Spa2p and other polarity proteins at sites of growth. In addition, a genetic interaction between spa2 and myo1 provides further evidence that Spa2p is involved in cytokinesis and cell wall morphogenesis.
Spa2p Interacts with Proteins Involved in Cell Polarity and Actin Function
Using a coimmunoprecipitation strategy coupled with tandem mass spectrometry analysis we identified Myo1p, Myo2p, Pan1p, and the protein encoded by YFR016c as proteins that interact with Spa2p. We have further confirmed the interaction between Spa2p and Myo2p by coimmunoprecipitation of Myo2p with Spa2-HA. Although we do not know if the interaction between Spa2p and Myo2p is direct, we believe that it is functionally significant based on subcellular fractionation studies, actin cosedimentation assays, and localization dependence studies.
Differential centrifugation of cell extracts showed that Spa2p was present in every fraction in which Myo2p was present. We believe this to be a significant finding because not all proteins exhibited a similar pattern of fractionation by this method. For example, Pea2p, a protein involved in cell polarity that interacts tightly with Spa2p (Valtz and Herskowitz, 1996; Sheu et al., 1998) was present only in the S2 and the S3 fractions and not in the P2 and P3 fractions (Figure 2A). The observation that Spa2p was present in some cell fractions without Pea2p indicates that Spa2p and Pea2p are not obligate heteromultimers.
Myo2p has been shown to associate with actin in an ATP-sensitive manner (Reck-Peterson et al., 2001). We showed that Spa2p also exhibited this same property. Because Spa2p lacks an obvious actin-binding domain, we hypothesize that the ATP-sensitive association between Spa2p and actin is most likely a consequence of the interaction between Spa2p and Myo2p. Interestingly, we also found that a pool of Pea2p cosedimented with actin similarly to Spa2p (Figure 2E) and we hypothesize that this is likely via the interaction of Pea2p with Spa2p, which further interacts with Myo2p and actin. We were unable, however, to determine if the pool of Pea2p that associates with actin is dependent on Spa2p because Pea2p is not stable in the spa2 background (unpublished data; Valtz and Herskowitz, 1996).
If the interaction between Spa2p and actin occurs via Myo2p, we expect that the Spa2-actin association would be disrupted in the myo2-66 mutant, which carries a temperature-sensitive mutation in the Myo2p actin-binding domain. Our results, however, neither confirmed nor disproved this prediction because it is apparent that the myo2-66 mutant retained some functional protein, which was able to cosediment with actin (Figure 2D; + actin, – ATP, + S3; pellet fraction; bottom panel). This population of functional Myo2-66p may have been adequate to bind Spa2p and could be an explanation for the presence of Spa2p in the pellet fraction along with actin and Myo2-66p (Figure 2D; + actin, – ATP, + S3; bottom panel). Alternatively, Myo2-66p may be stabilized by association with Spa2p or a Spa2p complex. In this case, the subpopulation of Spa2p-Myo2-66p complexes would remain associated with F-actin despite the temperature shift. In any case, the fact that Myo2-66p itself associated with actin in these experiments, we cannot draw any conclusions about the requirement of Myo2p for the association of Spa2p and actin.
Another approach by which to address the requirement of Myo2p for the Spa2p-actin association would be to examine the Spa2p-actin association in a myo2 mutant background in which the Spa2p-Myo2p interaction is disrupted. If we believe that Spa2p is a cargo protein of Myo2p, a cargo-specific allele such as myo2-13 or myo2-16 may be an appropriate background by which to test this hypothesis in future studies.
An alternative explanation for our results is the possibility that Myo1p may be redundant with Myo2p in binding actin, and thus, the fraction of Spa2p associating with actin in the myo2-66 mutant may be a consequence of its interaction with Myo1p. Alternatively, Spa2p may bind to actin in an ATP-sensitive manner via other Spa2-interacting proteins which also bind actin, such as Bud6p/Aip3p or Bni1p. In any case, Spa2p, Pea2p, and Myo2p likely form a stable complex that associates with actin in an ATP-sensitive manner to mediate cell polarity and/or actin function.
Spa2p Depends on Myo2p for Localization
In further support of a functionally significant relationship between Spa2p and Myo2p, we found that proper maintenance of Spa2p localization to sites of polarized growth depends on functional Myo2p motor and tail domains. We observed depolarization of Spa2GFP to occur within 5 min at 37°C in temperature-sensitive myo2-66 mutant, which suggests that the Myo2p motor domain has an important role in targeting Spa2p to sites of growth.
Our observation that polarized localization of Spa2GFP was also affected in the myo2-16 strain within 5 min at 37°C suggests that proper localization of Spa2p may be dependent on an interaction between Spa2p and the region of the Myo2p tail that is required for the essential function carried out by Myo2p. This hypothesis is supported by our observation that polarized Spa2GFP distribution was not disrupted by the tail domain mutation myo2-2, which causes defects in vacuolar inheritance but does not affect the Myo2 essential function (Catlett and Weisman, 1998).
Now that we have a link between Spa2p and Myo2p, the next goal is to understand the nature of their interaction. Future studies which address whether the interaction between Spa2p and Myo2p is direct or dependent upon other polarity proteins, as well as to identify the interaction domains for these two proteins would further our understanding of Spa2p function in cell polarity. The tail domain of Myo2p is thought to be the cargo-binding domain and has been shown to be important for vacuolar and vesicular movement as well as spindle pole orientation (Catlett and Weisman, 1998; Schott et al., 1999; Catlett et al., 2000; Yin et al., 2000). Thus, if the interaction is direct, we predict that Spa2p also associates with the Myo2p tail, perhaps mediated by the Spa2 box, the region within Spa2p that is necessary and sufficient for localization of Spa2p to sites of growth (Arkowitz and Lowe, 1997).
Possible Functional Role of the Myo2p-Spa2p Interaction
Myo2p is a yeast class V myosin that is believed to be a motor protein that brings secretory vesicles into the bud tip along actin cables. This hypothesis is supported by the fact that myo2 mutants accumulate post-Golgi vesicles and grow isotropically at the restrictive temperature, indicating that secretion continues but is not polarized (Johnston et al., 1991; Govindan et al., 1995). In addition, myo2 mutations genetically interact with several post-Golgi sec mutations (Govindan et al., 1995), and Myo2p localizes to sites of polarized growth (Snyder, 1989; Lillie and Brown, 1994). Furthermore, maintenance of the polarized distribution of Sec4p, the vesicle-associated Rab protein, depends on Myo2p. Depolarization of Sec4p has previously been shown to occur at the nonpermissive temperature within 5 min, whereas actin organization remains generally polarized in both the myo2-66 and myo2-16 mutants (Schott et al., 1999).
Based on the current understanding of Myo2p function and our findings that Spa2p associates with Myo2p and that the maintenance of polarized Spa2p localization depends on Myo2p, we propose three models for the functional role of the Spa2p-Myo2p interaction. First, Spa2p, either directly or as part of a multiprotein complex, functions as a bridge between the Myo2p tail and secretory vesicles to facilitate Myo2p transport of its cargo along actin cables. This model is plausible since Bud6p/Aip3p has previously been shown to associate with post-Golgi vesicles and depend on Myo2p for maintenance of polarized localization (Jin and Amberg, 2000) as well as associate with Spa2p (Sheu et al., 1998). It is, therefore, possible that the complex of Spa2p-Pea2p-Bud6p/Aip3p mediates the interaction between secretory vesicles and Myo2p.
A second model for the functional role of the Spa2p-Myo2p interaction is that Myo2p is involved in transporting Spa2p and its associated polarity related proteins to sites of polarized growth. This might occur via an interaction between Spa2p and Myo2p-associated secretory vesicles, either directly or as part of a multiprotein complex. The mechanism by which Spa2p is localized to sites of polarized growth has not yet been fully elucidated so this possibility is intriguing.
The two models described above assume either direct or indirect interaction of Spa2p with secretory vesicles and transport of Spa2p by Myo2p along actin cables. Both models would thus predict loss of Spa2p tip localization in a late secretory mutant. Given that Bud6p/Aip3p has been shown to depend on the secretory pathway (Jin and Amberg 2000) for its localization, it would be informative to assess whether the localization of Spa2p and Bud6p/Aip3p is affected differentially by the secretory pathway. Our preliminary results indicate that Spa2p is at least partially dependent on the secretory pathway for its localization to sites of polarized growth (unpublished data). However, more careful studies are necessary to determine if the dependence on the secretory pathway is related to the initial localization of Spa2p to the presumptive bud site and/or to the maintenance of Spa2p at the chosen site.
This preliminary finding might suggest a third model for the role of the Spa2p-Myo2p interaction, in which the initial localization of Spa2p to the presumptive bud site is independent of F-actin (Ayscough et al., 1997) and the secretory pathway, but maintenance of Spa2p at the chosen site requires interaction with Myo2p. In this model, Spa2p marks the anchor site for secretory vesicles and interaction between Spa2p and Myo2p (and/or Bud6p/Aip3p) would restrict the secretory vesicles to the site marked by Spa2p.
A connection between Spa2p and late secretory vesicles is conceivable and is supported by the recent observation that spa2 cells exhibit diffuse localization of Sec4p at the bud tip during apical growth and at the division site during repolarization just before cytokinesis (Sheu et al., 2000). The failure to concentrate Sec4p at polarized sites is also observed in mutants defective for other polarity proteins, Pea2p, Bud6p/Aip3p, and Bni1p (Sheu et al., 2000), which physically interact with Spa2p (Fujiwara et al., 1998; Sheu et al., 1998). Furthermore, cell-surface expansion in all of these mutants is not confined to the distal tip of the bud (Sheu et al., 2000).
The observation that Myo2p partially depends on Spa2p for polarized localization suggests that Spa2p may also have a role in regulating efficient Myo2p targeting and/or maintenance at sites of growth. If we suppose that Myo2p localization represents a balance between rapid translocation of Myo2p along actin cables to growth sites and slower release or diffusion from these sites, as suggested by Schott et al. (1999), it is conceivable that Spa2p could have a role in either or both of these processes. The observation that Sec4p is diffusely localized at polarized sites in spa2 cells is consistent with either possibility (Sheu et al., 2000).
Spa2p Involvement in Cytokinesis, Cell Separation, and Cell Wall Morphogenesis
Previous observations indicate that Spa2p may have a role in cytokinesis and cell wall morphogenesis. First, Spa2p localizes to the mother-bud neck prior to cytokinesis (Snyder, 1989; Snyder et al., 1991; Arkowitz and Lowe, 1997). Second, spa2-7 mutants exhibit a mild defect in cell separation (Snyder, 1989; Snyder et al., 1991). Several groups have also observed that the mother/bud necks in spa2Δ cells are wider than those in wild-type cells (Zahner et al., 1996; Sheu et al., 2000; Yuzyuk and Amberg 2003). Third, genetic interactions between SPA2 and other genes involved in cytokinesis and cell wall morphology, including CDC10 and CHS5, have been identified. For example, cdc10-10 strains require Spa2p for growth (Flescher et al., 1993), and the spa2 chs5 double null mutant exhibits a severe growth defect at 37°C (Santos and Snyder, 2000). It is notable that the septin protein, Cdc10p, is a GTP-binding protein that is involved in cytokinesis and, like Spa2p, localizes to the bud neck (reviewed in Longtine et al., 1996). Chs5p is a regulatory subunit for Chitin Synthase III, which is responsible for chitin synthesis and deposition in both budding and mating cells. Finally, Spa2p interacts with another member of the septin family of proteins, Shs1p/Sep7p, by two-hybrid assay (Mino et al., 1998).
The synthetic lethal genetic interaction that we observed between spa2 and myo1 at 37°C, in which the myo1 cell separation and cytokinesis defect at nonpermissive temperature is enhanced in the myo1 spa2 strain, provides further indication that Spa2p is involved in cytokinesis, cell separation, and cell wall morphogenesis. The physical association between Spa2p and Myo1p as well as the genetic relationship identified here sheds light on how Spa2p may participate in these processes and provides direction for future experiments.
Cytokinesis in budding yeast is accomplished by the concerted action of the actomyosin contractile ring and the formation of the septum, which presumably requires targeted secretion to the bud neck. Thus, knowing whether a septum is formed in the spa2 myo1 mutant strain at the nonpermissive temperature would be important for understanding the function of Spa2p in cytokinesis. Examination of the septum structure by electron microscopy would allow us to assess the presence or absence of vesicle accumulation near the region of the bud neck and determine if the cytoplasm has been cleaved in the single and double mutant strains at nonpermissive temperature.
Our finding that Spa2p localization to the bud neck does not depend on Myo2p suggests that there may be a redundant mechanism for localizing Spa2p to the bud neck. Because Spa2p has been shown to interact with the septin Shs1p/Sep7p by two-hybrid (Mino et al., 1998), it would be interesting to see if Spa2p localization to the bud neck is dependent on Shs1p/Sep7p. This result may also provide further insight into the role of Spa2p in cytokinesis
Protein Complexes Involved in Cell Polarity
Cell polarity in yeast requires a number of components that function to govern the establishment and maintenance of a polarized actin cytoskeleton necessary for polarized secretion. Several independent studies aimed toward understanding the molecular function of many of these components have already identified numerous protein-protein associations important for the development of cell polarity using the two-hybrid system or by coimmunoprecipitation methods. Drees et al. (2001) used a large-scale, array-based two-hybrid approach, which identified new links in the network of protein-protein associations controlling polarity development.
Prior coimmunoprecipitation studies indicate that Spa2p interacts with Pea2p and Bni1p (Fujiwara et al., 1998; Sheu et al., 1998). Spa2p also interacts with Bud6p/Aip3p by two-hybrid assay, and Spa2p, Pea2p, and Bud6p/Aip3p cosediment in sucrose gradients (Sheu et al., 1998). Thus, it is likely that Spa2p functions in a complex with Pea2p, Bud6p, and Bni1p (Sheu et al., 1998). In addition, Spa2p interacts with components of the pheromone-response MAPK module (Ste11p and Ste7p) and MEKs of the PKC cell-integrity pathway (Mkk1p and Mkk2p) by the two-hybrid assay (Sheu et al., 1998). We have found that Spa2p interacts with Myo1p, Myo2p, Pan1p, and the protein product encoded by YFR016c. Thus, Spa2 interacts with numerous proteins, which together form a network that regulates actin function and polarity development (Figure 6). It is likely that the protein-protein associations in this network change over time and space. Understanding the role of Spa2p will likely require a definition of these complexes during the cell cycle.
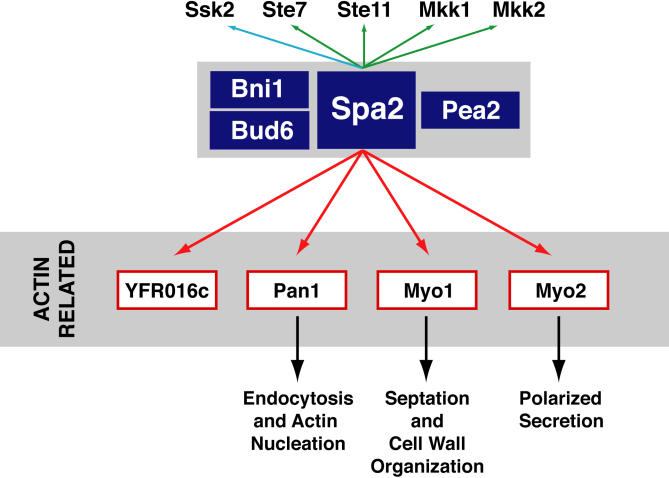
Figure 6.
Summary of interactions among proteins involved in cell polarity pathways. Spa2p, Pea2p, Bni1p, and Bud6p are thought to comprise a complex involved in regulating cell polarity (Sheu et al., 1998). Green arrows indicate interactions previously demonstrated by two-hybrid (Sheu et al., 1998); a blue arrow between Spa2p and Ssk2p represents the interaction previously demonstrated by coimmunoprecipitation (Yuzyuk and Amberg, 2003); red arrows indicate interactions identified in this study by coimmunoprecipitation; black arrows indicate a biological pathway. (Arrows do not necessarily represent direct interactions in this protein interaction map.) Actin and actin-associated proteins have been identified as other binding partners of the protein encoded by YFR016c (Ho et al., 2002; not shown in figure). Thus, the new binding partners of Spa2p (in red boxes) are all involved in actin function and cell polarity.
Acknowledgments
We thank D. Mullins for providing purified F-actin; P. Brennwald for anti-Snc1p antibody; and E. Bi, J. Pringle, P. Novick, A. Bretscher, L. Weisman, and B. Wendland for yeast strains. We also thank A. Sil, L. Huang, S. Shaham, R. Tabtiang, and one reviewer for critical discussions and comments on this work. This work was supported by National Institutes of Health Grants DK-25387 (M.S.M.), DK-55389 (M.S.M.), and GM-48052 (I.H.). J.L.S. was supported by the Medical Scientist Training Program.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–02–0108) on July 19, 2005.
References
- Amberg, D. C., Zahner, J. E., Mulholland, J. W., Pringle, J. R., and Botstein, D. (1997). Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol. Biol. Cell 8, 729–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkowitz, R. A., and Lowe, N. (1997). A small conserved domain in the yeast Spa2p is necessary and sufficient for its polarized localization. J. Cell Biol. 138, 17–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F. M. (1991). Current Protocols in Molecular Biology, Vol. 2, New York: John Wiley & Sons, 10.6.1. [Google Scholar]
- Ayscough, K. R., Stryker, J., Pokala, N., Sanders, M., Crews, P., Drubin, D. G. (1997). High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137, 399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, E., Maddox, P., Lew, D. J., Salmon, E. D., McMillan, J. N., Yeh, E., and Pringle, J. R. (1998). Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 142, 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, E., and Pringle, J. R. (1996). ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell Biol. 16, 5264–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, S. S. (1997). Myosins in yeast. Curr. Opin. Cell Biol. 9, 44–48. [DOI] [PubMed] [Google Scholar]
- Catlett, N. L., Duex, J. E., Tang, F., and Weisman, L. S. (2000). Two distinct regions in a yeast myosin-V tail domain are required for the movement of different cargoes. J. Cell Biol. 150, 513–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett, N. L., and Weisman, L. S. (1998). The terminal tail region of a yeast myosin-V mediates its attachment to vacuole membranes and sites of polarized growth. Proc. Natl. Acad. Sci. USA 95, 14799–14804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant, J. (1999). Cell polarity in yeast. Annu. Rev. Cell Dev. Biol. 15, 365–391. [DOI] [PubMed] [Google Scholar]
- Chenevert, J., Valtz, N., and Herskowitz, I. (1994). Identification of genes required for normal pheromone-induced cell polarization in Saccharomyces cerevisiae. Genetics 136, 1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees, B. L. et al. (2001). A protein interaction map for cell polarity development. J. Cell Biol. 154, 549–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin, D. G., and Nelson, W. J. (1996). Origins of cell polarity. Cell 84, 335–344. [DOI] [PubMed] [Google Scholar]
- Duncan, M. C., Cope, M.J.T.V, Goode, B. L., Wendland, B., and Drubin, D. G. (2001). Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat. Cell Biol. 3, 687–690. [DOI] [PubMed] [Google Scholar]
- Eng, J. K., McCormack, A. L., and Yates, J. R., III. (1994). An approach to correlate MS/MS data to amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., Blundell, K., Longtine, M. S., Chow, C. J., Adames, N., Pringle, J. R., Peter, M., and Boone, C. (1997). Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276, 118–122. [DOI] [PubMed] [Google Scholar]
- Flescher, E. G., Madden, K., and Snyder, M. (1993). Components required for cytokinesis are important for bud site selection in yeast. J. Cell Biol. 122, 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, T., Tanaka, K., Mino, A., Kikyo, M., Takahashi, K., Shimizu, K., and Takai, Y. (1998). Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell 9, 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie, A. E., Brizzio, V., and Rose, M. D. (1998). Distinct morphological phenotypes of cell fusion mutants. Mol. Biol. Cell 9, 1395–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrung, S., and Snyder, M. (1990). The SPA2 gene of Saccharomyces cerevisiae is important for pheromone-induced morphogenesis and efficient mating. J. Cell Biol. 111, 1451–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goud, B., Salminen, A., Walworth, N. C., and Novick, P. J. (1988). A GTP-binding protein required for secretion rapidly associates with secretory vesicles and the plasma membrane in yeast. Cell 53, 753–768. [DOI] [PubMed] [Google Scholar]
- Govindan, B., Bowser, R., and Novick, P. (1995). The role of Myo2, a yeast class V myosin, in vesicular transport. J. Cell Biol. 128, 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales, K. G., Bi, E., Wu, J. Q., Adam, J. C., Yu, I. C., and Pringle, J. R. (1999). Cytokinesis: an emerging unified theory for eukaryotes? [see comments]. Curr. Opin. Cell Biol. 11, 717–725. [DOI] [PubMed] [Google Scholar]
- Ho, Y. et al. (2002). Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183. [DOI] [PubMed] [Google Scholar]
- Ito, H., Fukuda, Y., Murata, K., and Kimura, A. (1983). Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H., and Amberg, D. C. (2000). The secretory pathway mediates localization of the cell polarity regulator Aip3p/Bud6p. Mol. Biol. Cell 11, 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, G. C., Prendergast, J. A., and Singer, R. A. (1991). The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J. Cell Biol. 113, 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada, K., Yamaguchi, E., and Arisawa, M. (1995). Cloning of the Candida glabrata TRP1 and HIS3 genes, and construction of their disruptant strains by sequential integrative transformation. Gene 165, 203–206. [DOI] [PubMed] [Google Scholar]
- Lillie, S. H., and Brown, S. S. (1994). Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. J. Cell Biol. 125, 825–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott, J., and Li, R. (1998). Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol. 140, 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., DeMarini, D. J., Valencik, M. L., Al-Awar, O. S., Fares, H., De Virgilio, C., and Pringle, J. R. (1996). The septins: roles in cytokinesis and other processes. Curr. Opin. Cell Biol. 8, 106–119. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., McKenzie, A., 3rd, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998). Additional modules for versatile and economical P.C.R-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Mino, A., Tanaka, K., Kamei, T., Umikawa, M., Fujiwara, T., and Takai, Y. (1998). Shs1p: a novel member of septin that interacts with Spa2p, involved in polarized growth in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 251, 732–736. [DOI] [PubMed] [Google Scholar]
- Protopopov, V., Govindan, B., Novick, P., and Gerst, J. E. (1993). Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. Cell 74, 855–861. [DOI] [PubMed] [Google Scholar]
- Puig, O., Rutz, B., Luukkonen, B. G., Kandels-Lewis, S., Bragado-Nilsson, E., and Séraphin, B. (1998). New constructs and strategies for efficient PCR-based gene manipulations in yeast. Yeast 14, 1139–1146. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson, S. L., Novick, P. J., and Mooseker, M. S. (1999). The tail of a yeast class V myosin, myo2p, functions as a localization domain. Mol. Biol. Cell 10, 1001–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson, S. L., Tyska, M. J., Novick, P. J., and Mooseker, M. S. (2001). The yeast class v myosins, myo2p and myo4p, are nonprocessive actin-based motors. J. Cell Biol. 153, 1121–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, J. R., and Paterson, B. M. (1990). Yeast myosin heavy chain mutant: maintenance of the cell type specific budding pattern and the normal deposition of chitin and cell wall components requires an intact myosin heavy chain gene. Cell Motil. Cytoskelet. 17, 301–308. [DOI] [PubMed] [Google Scholar]
- Rose, M. D., Winston, F. and Heiter, P. 1990. Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 123 pp.
- Santos, B., and Snyder, M. (2000). Sbe2p and sbe22p, two homologous Golgi proteins involved in yeast cell wall formation. Mol. Biol. Cell 11, 435–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott, D., Ho, J., Pruyne, D., and Bretscher, A. (1999). The COOH-terminal domain of Myo2p, a yeast myosin V, has a direct role in secretory vesicle targeting. J. Cell Biol. 147, 791–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu, Y. J., Barral, Y., and Snyder, M. (2000). Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol. Cell Biol. 20, 5235–5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu, Y. J., Santos, B., Fortin, N., Costigan, C., and Snyder, M. (1998). Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell. Biol. 18, 4053–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Shih, J. L. (2001). Analysis of cell polarity determinants in Saccharomyces cerevisiae. Ph.D. Dissertation. San Francisco: University of California, San Francisco.
- Snyder, M. (1989). The SPA2 protein of yeast localizes to sites of cell growth. J. Cell Biol. 108, 1419–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder, M., Gehrung, S., and Page, B. D. (1991). Studies concerning the temporal and genetic control of cell polarity in Saccharomyces cerevisiae. J. Cell Biol. 114, 515–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich, J. A., and Watt, S. (1971). The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosintroponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871. [PubMed] [Google Scholar]
- Valtz, N., and Herskowitz, I. (1996). Pea2 protein of yeast is localized to sites of polarized growth and is required for efficient mating and bipolar budding. J. Cell Biol. 135, 725–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Drogen, F., and Peter, M. (2002). Spa2p functions as a scaffold-like protein to recruit the Mpk1p MAP kinase module to sites of polarized growth. Curr. Biol. 12, 1698–1703. [DOI] [PubMed] [Google Scholar]
- Watts, F. Z., Shiels, G., and Orr, E. (1987). The yeast MYO1 gene encoding a myosin-like protein required for cell division. EMBO J. 6, 3499–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland, B., and Emr, S. D. (1998). Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J. Cell Biol. 141, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel, D., and Flugge, U. I. (1984). A method for the quantitative recovery of protein. Anal. Biochem. 138, 141–143. [DOI] [PubMed] [Google Scholar]
- Winzeler, E. A. et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Yates, J. R. Dithiothreitol., Eng, J. K., McCormack, A. L., and Schieltz, D. (1995). Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67, 1426–1436. [DOI] [PubMed] [Google Scholar]
- Yin, H., Pruyne, D., Huffaker, T. C., and Bretscher, A. (2000). Myosin V orientates the mitotic spindle in yeast. Nature 406, 1013–1015. [DOI] [PubMed] [Google Scholar]
- Yuzyuk, T., and Amberg, D. C. (2003). Actin recovery and bud emergence in osmotically stressed cells requires the conserved actin interacting mitogen-activated protein kinase kinase kinase Ssk2p/MTK1 and the scaffold;Protein Spa2p. Mol. Biol. Cell 14, 3013–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner, J. E., Harkins, H. A., and Pringle, J. R. (1996). Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 16, 1857–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]