Abstract
Sir2 is a NAD+-dependent protein deacetylase that extends lifespan in yeast and worms. This study examines seven human proteins homologous to Sir2 (SIRT1 through SIRT7) for cellular localization, expression profiles, protein deacetylation activity, and effects on human cell lifespan. We found that: 1) three nuclear SIRT proteins (SIRT1, SIRT6, and SIRT7) show different subnuclear localizations: SIRT6 and SIRT7 are associated with heterochromatic regions and nucleoli, respectively, where yeast Sir2 functions; 2) SIRT3, SIRT4, and SIRT5 are localized in mitochondria, an organelle that links aging and energy metabolism; 3) cellular p53 is a major in vivo substrate of SIRT1 deacetylase, but not the other six SIRT proteins; 4) SIRT1, but not the other two nuclear SIRT proteins, shows an in vitro deacetylase activity on histone H4 and p53 peptides; and 5) overexpression of any one of the seven SIRT proteins does not extend cellular replicative lifespan in normal human fibroblasts or prostate epithelial cells. This study supports the notion that multiple human SIRT proteins have evolutionarily conserved and nonconserved functions at different cellular locations and reveals that the lifespan of normal human cells, in contrast to that of lower eukaryotes, cannot be manipulated by increased expression of a single SIRT protein.
INTRODUCTION
Normal human somatic cells in culture have a finite replicative lifespan. After a limited number of cell divisions, the cells eventually enter the state of replicative senescence, in which they show an irreversible growth arrest (Tominaga et al., 2002). Although accumulation of DNA damage (Sedelnikova et al., 2004), response to oxidative stress (Campisi, 2001), and regulation of telomeres (Harrington and Robinson, 2002) are involved in this cellular aging process, current understanding of molecular details of the human cell aging and its possible contribution to in vivo organismal aging in humans is still incomplete. Assuming an evolutionarily conserved mechanism of aging (Tissenbaum and Guarente, 2002), investigation of human proteins similar to the proteins controlling aging in model organisms should be a promising approach.
The silent information regulator 2 (Sir2) is a NAD+-dependent protein deacetylase (Imai et al., 2000) that controls longevity in lower eukaryotes such as Saccharomyces cerevisiae and Caenorhabditis elegans (Kaeberlein et al., 1999; Tissenbaum and Guarente, 2001). An increase in Sir2 activity extends the lifespan in these organisms. In S. cerevisiae, Sir2 extends the replicative lifespan through suppression of formation of extrachromosomal ribosomal DNA circles in the nucleoli (Kaeberlein et al., 1999). The Sir2 protein also plays a critical role in heterochromatic gene silencing through regulation of histone modifications at telomeres, ribosomal DNA clusters, and mating-type loci (Lustig, 1998). The NAD+ dependency of Sir2 activity suggests that the control of lifespan is highly associated with metabolic state. Calorie restriction not only affects the metabolic processes but also extends the lifespan in a wide range of organisms from yeast to mammals (Heilbronn and Ravussin, 2003; Hursting et al., 2003). The insulin/IGF-I signaling pathway, a mediator of aging effects by calorie restriction (Barbieri et al., 2003), has been linked to the expression of a mammalian Sir2 homologue (Cohen et al., 2004). Thus, it is possible that the Sir2-mediated regulation of aging is conserved in higher organisms including humans.
Humans have seven proteins of the sirtuin family (SIRT1 through 7) that share the catalytic domain with Sir2 (Blander and Guarente, 2004; North and Verdin, 2004). SIRT1 is a nuclear protein with the highest sequence similarity to Sir2 and a yeast Sir2-related protein Hst1 (Frye, 2000). SIRT1 can modulate cellular stress response and survival through regulation of p53 (Luo et al., 2001; Vaziri et al., 2001; Langley et al., 2002), NF-κB signaling (Yeung et al., 2004), and FOXO transcription factors (Brunet et al., 2004; Motta et al., 2004). Studies on SIRT1 function (Fulco et al., 2003; Takata and Ishikawa, 2003) and Sirt1 knockout mice (Cheng et al., 2003; McBurney et al., 2003b) suggest its roles in mammalian development and differentiation. SIRT2 is a cytoplasmic protein that deacetylates α-tubulin (North et al., 2003). SIRT3 is localized to mitochondria and becomes activated by the proteolytic processing at N-terminus (Onyango et al., 2002; Schwer et al., 2002). Despite these data on SIRT1, SIRT2, and SIRT3, it is not known whether they regulate the replicative lifespan of human cells. No data have been available for biological functions or cellular localizations of the other four SIRT proteins (SIRT4, 5, 6, and 7). To understand the possible roles of the Sir2 homologues in human aging, a systematic investigation of all seven human SIRT proteins is an important step. In this study, we examine all seven SIRT proteins for their cellular localization, expression profiles, protein deacetylation activity, and effects on cellular lifespan of human cells. Our findings with normal human cells provide the essential set of data toward elucidation of the physiological functions of human SIRT proteins.
MATERIALS AND METHODS
Cell Culture, Treatment, Transfection, and Retroviral Transduction
A normal human fibroblast strain NHF was derived from foreskin (a gift from Dr. Jayne Boyer, University of North Carolina; Sedelnikova et al., 2004). WI-38 was from Coriell Cell Repository (Camden, NJ). HeLa and 293T cells were from American Type Culture Collection (ATCC, Manassas, VA). Human prostate epithelial cells (PrEC) was purchased from Clonetics (San Diego, CA) and maintained according to the supplier's protocol. Plasmid transfection, preparation, and transduction of retroviral vectors were performed according to the Stratagene protocol (http://www.stratagene.com/manuals/217566.pdf). NHF cells and PrEC cells were infected with the retroviral supernatant at 7 population doublings (PDs) and 6 PDs, respectively. To obtain the cells stably overexpressing each SIRT protein, the retroviral vector-infected cells were selected for growth in 400 μg/ml G418 and maintained as described below. The treatment with adriamycin (0.2 μg/ml for 8 h) to induce p53 acetylation followed Sakaguchi et al. (1998). The treatment with bleomycin (10 mU/ml for 20 h) or H2O2 (150 μM for 2 h) to induce premature senescence was previously described (Sedelnikova et al., 2004).
Plasmid Constructs
The full-length cDNA of each SIRT gene was PCR-amplified (PCR primer sequences available upon request) and cloned into the retroviral expression vector pFB-Neo (Stratagene, La Jolla, CA). For constructing the retroviral vectors driving the SIRT proteins tagged with HA (influenza hemagglutinin epitope) at the N-terminus, the corresponding sequences were added to the PCR primers. The full-length cDNA fragments were also cloned into the Gateway Entry vector pENTR/d-TOPO (Invitrogen, Carlsbad, CA), subcloned into pcDNA-DEST47 or pcDNA-DEST40 (Invitrogen) by Gateway LR reaction, and used for expression of cycle 3 GFP or V5-His fusion proteins, respectively. For production of the recombinant SIRT1, SIRT6, and SIRT7 proteins in Hi5 insect cells, the full-length cDNA fragments containing the Gateway signal sequences, and the 5′ cleavage site for Tobacco Etch Virus (TEV) protease were subcloned in the baculovirus expression vector pDest-603 (SIRT1 and 6) or pDest-606 (SIRT7), the Gateway-modified version of pFastBac-Dual vector (Invitrogen). We confirmed by DNA sequencing that all PCR-generated cDNA constructs were free of PCR errors.
Antibodies
The rabbit polyclonal antibodies specifically recognizing SIRT6 and SIRT7 were raised against the following synthetic peptides: SPKRERPTSPAPHRPPKRV for SIRT6 and GWFGRGCTKRTKRKKVT for SIRT7. The affinity-purified antibodies were used in this study. To verify the specificity of the antibodies, RNA interference (RNAi)-mediated knockdown of SIRT6 or SIRT7 expression was performed in HeLa cells by using the pSUPERretro RNAi system (OligoEngine, Seattle, WA). The target sequences were 5′-AAG AAT GTG CCA AGT GTA AGA-3′ for SIRT6 and 5′-TAG CCA TTT GTC CTT GAG GAA-3′ for SIRT7.
The antibody against p53 acetylated at the lysine 382 residue (anti-acetylK382p53) was provided by Drs. Ettore Appella and Shin'ichi Saito (Sakaguchi et al., 1998). The other primary antibodies used were anti-p53 (DO-1 and Pab1801, Santa Cruz Biotechnology, Santa Cruz, CA), anti-SIRT1 (H300, Santa Cruz), anti-β-actin (Abcam, Cambridge, MA), anti-HA (Roche, Indianapolis, IN), anti-nucleoli (Chemicon International, Temecula, CA), anti-V5 (Invitrogen) and anti-cytochrome C (BD Biosciences, San Diego, CA).
Detection of GFP by Fluorescence Microscopy
Forty-eight hours after transfection of the pcDNA-DEST47 vector expressing each SIRT-GFP fusion protein, cells were stained with 100 ng/ml Hoechst 33342 (Molecular Probes, Eugene, OR) and MitoFluor Red 589/594 Dye (Molecular Probes) for 30 min. The microscopic observation (Leica DMIRB, Deerfield, IL) was performed in phenol red-free media.
Indirect Immunofluorescence
For detection of HA-tagged-SIRT1, SIRT6, and SIRT7 proteins, cells were fixed with 4% paraformaldehyde for 15 min, treated with 0.2% Triton X-100 for 15 min, incubated in 10% goat serum at room temperature for 1 h, and incubated overnight at 4°C with anti-HA rat monoclonal antibody (mAb) and anti-nucleoli mouse mAb. The secondary antibodies were Texas Red-X-conjugated goat anti-mouse IgG (H+L) (Molecular Probes) and FITC-conjugated goat anti-rat IgG (H+L; Jackson ImmunoResearch Laboratories, West Grove, PA). HA-SIRT2 was similarly detected using Texas Red-X-conjugated goat anti-rat IgG (H+L; Molecular Probes) as a secondary antibody, in combination with staining with 4′-6-diamidino-2-phenylindole (DAPI; Molecular Probes). The stained cells were examined by Zeiss LSM 510 confocal microscope (Thornwood, NY).
To detect endogenous SIRT proteins and V5-His-tagged SIRT proteins, cell fixation, permeabilization, and preincubation were performed as described above. For the endogenous SIRT6 and SIRT7, anti-SIRT6 or anti-SIRT7 rabbit polyclonal antibody and anti-nucleoli antibody were used, and the secondary antibodies were Texas Red-X-conjugated goat anti-mouse IgG (H+L) and Alexa Fluor 488-conjugated goat anti-rabbit IgG (H+L; Molecular Probes). The V5-His-tagged SIRT proteins were detected using anti-V5 mouse antibody (Invitrogen) and Alexa Fluor 488-conjugated goat anti-mouse IgG (H+L; Molecular Probes). The cells were also stained with DAPI and MitoTracker Orange (Molecular Probes).
Western Blot and Immunoprecipitation
SDS-PAGE and transfer to a PVDF (polyvinylidene difluoride) membrane followed standard procedures. Protein signals were detected using the enhanced chemiluminescence method (ECL, Amersham, Piscataway, NJ). To examine p53 acetylation, immunoprecipitation was carried out as described previously (Sakaguchi et al., 1998). Briefly, after treatment with 0.2 μg/ml adriamycin for 8 h, cells were lysed in lysis buffer (50 mM Tris-HCl at pH 7.5, 5 mM EDTA, 150 mM NaCl, 1% Triton X-100 and protease inhibitor cocktail [Roche, catalogue no. 1836170]) at 4°C for 30 min. Precleared lysates (1 mg of proteins) were incubated with 5 μg of agarose-conjugated DO-1 and 5 μg of agarose-conjugated Pab1801 (Santa Cruz) for 1 h with rotation at 4°C. The immunoprecipitated proteins were analyzed by Western blot using anti-acetylK382p53 or DO-1 antibody.
Preparation of Subcellular Fractions
Subcellular protein fractions were isolated from 293T cells expressing the V5-His-tagged version of SIRT3, SIRT4, or SIRT5 protein by using the Mitochondria Isolation kit (Pierce Biotechnology, Rockford, IL) according to the manufacturer's instructions. The cytoplasmic fraction was concentrated by trichloracetate precipitation. Equal amounts of proteins from nuclear, cytoplasmic, and mitochondrial fractions were analyzed by Western blot using the anti-V5 antibody.
Quantitative Real-time RT-PCR Assay
cDNA synthesis, PCR reaction, real-time detection of PCR products, and quantitative data analysis were carried out as previously described (Sinha-Datta et al., 2004). The sequences of primers and probe for detecting each SIRT gene are available on request, and those for human telomerase reverse transcriptase (hTERT) and β-2-microglobulin were as described (Sinha-Datta et al., 2004). β-2-microglobulin was used as the internal control for normalizing all the data. Human tissue RNA samples were from Human Total RNA Master Panel II (BD Biosciences Clontech, San Jose, CA).
Production of Recombinant Proteins and In Vitro Deacetylation Assays
The recombinant SIRT1, SIRT6, and SIRT7 proteins were produced in the baculovirus vector-infected Hi5 cells. The purification of the recombinant proteins was carried out at the Protein Expression Laboratory, National Cancer Institute/SAIC-Frederick, MD (http://www.ncifcrf.gov/rtp/PEL/), as previously described (Oishi et al., 2005). The N-terminal tags used for affinity purification (His6-glutathione-S-transferase or His6-maltose binding protein) were removed from the recombinant proteins with the TEV protease.
The histone deacetylase assay was performed according to the manufacturer's protocol (Upstate Biotechnology, Lake Placid, NY; protocol no. 17–328, www.upstate.com/img/coa/17-328-22369.pdf). Briefly, the purified SIRT1, SIRT6, or SIRT7 protein (10 μg per reaction) was incubated in HDAC assay buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 10% glycerol) containing the [3H]acetyl histone H4 peptide (amino acid residues 2–24) with or without NAD+ (1 mM) for 2 h at 30°C. The reaction was stopped by acidification and the released tritium was extracted with ethyl acetate. To examine an in vitro deacetylase activity on p53, the SIRT Fluorescent Activity Assay kit (Biomol, Plymouth Meeting, PA; catalogue no. AK-555) was used. The purified proteins (10 μg per reaction) were incubated in the assay buffer (25 mM Tris-HCl, pH 8.0, 2.7 mM KCl, 137 mM NaCl, 1 mM MgCl2, and 1 mg/ml bovine serum albumin) containing the Fluor de Lys-p53 peptide (amino acid residues 379–382 of human p53, Arg-His-Lys-Lys [Ac]) in the presence or absence of NAD+ (500 μM) for 2 h at 37°C. The deacetylase activity was measured in arbitrary fluorescence units at 460 nm.
Examination of Replicative Lifespan and Senescence-associated β-galactosidase Staining
All the retroviral vector-infected cells derived from NHF, WI-38, and PrEC were subcultured at a split ratio of 1:4. The number of cells was counted at each passage, and the number of population doublings (PDs) achieved between passages was determined by log2 (number of cells obtained/number of cells inoculated) (Pereira-Smith and Smith, 1981; Sugawara et al., 1990). To draw the growth curves throughout the replicative lifespan, the cumulative PDs of the cells were calculated and plotted to the days after retroviral infection. When the cells stopped dividing and remained impossible to be further passaged for >2 wk, they were regarded as growth-arrested. Senescence-associated (SA)-β-galactosidase (gal) staining followed the method by Dimri et al. (1995).
RESULTS
Cellular Localization of SIRT-GFP Fusion Proteins and HA-tagged SIRT Proteins
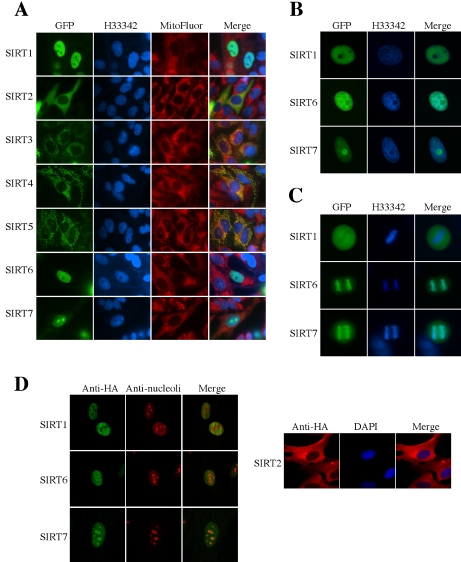
Each of the seven SIRT proteins fused to GFP at their C-terminus was expressed in NHF normal human fibroblasts (Figure 1A). SIRT1-GFP fusion protein was detected in the nuclei as previously reported (Luo et al., 2001; Vaziri et al., 2001), but it appeared to be excluded from the nucleoli (see below for nucleolar costaining) and was not enriched in Hoechst 33342-stained regions, which are thought to contain heterochromatin (see Figure 1B for a magnified nucleus and compare with SIRT6 described below). SIRT2-GFP was distributed in the cytoplasm and SIRT3-GFP colocalized with the mitochondria-specific probe MitoFluor, both consistent with the previous reports (Onyango et al., 2002; Schwer et al., 2002; North et al., 2003). This study for the first time shows the cellular localization of the other four SIRT proteins. Similar to SIRT3-GFP, SIRT4-GFP and SIRT5-GFP fusion proteins colocalized with the MitoFluor (Figure 1A; their mitochondrial localization further verified below). SIRT6-GFP and SIRT7-GFP fusion proteins were in the nuclei but differed in subnuclear localization pattern from SIRT1-GFP and from each other. SIRT6-GFP was distributed throughout the nucleus and, unlike SIRT1-GFP, overlapped significantly with heterochromatic regions stained with Hoechst 33342 (Figure 1, A and B). SIRT7-GFP was concentrated in the nucleolus-like compartments (confirmed as the nucleoli below), in marked contrast to SIRT1-GFP and SIRT6-GFP (Figure 1, A and B). SIRT6-GFP and SIRT7-GFP, but not SIRT1-GFP, were associated with condensed chromosomes in mitosis (Figure 1C).
Figure 1.
Cellular localization of human SIRT proteins. (A) Each SIRT-GFP fusion protein was transiently expressed in NHF normal human fibroblasts. Hoechst 33342 (H33342) and MitoFluor stainings are shown in parallel. The merged pictures of GFP, Hoechst 33342 and MitoFluor are in the most right panels. (B) Magnified nuclei of NHF cells expressing SIRT1-GFP, SIRT6-GFP, or SIRT7-GFP protein. (C) Mitotic NHF cells expressing SIRT1-GFP, SIRT6-GFP, or SIRT7-GFP protein. Condensed chromosomes are strongly stained with Hoechst 33342 in the middle panels. (D) NHF cells expressing HA-tagged SIRT1, SIRT2, SIRT6, or SIRT7 were immunostained with anti-HA antibody, in combination with anti-nucleoli antibody (for SIRT1, SIRT6, and SIRT7) or DAPI staining (for SIRT2).
We also examined the cellular localization of the HA-tagged version of SIRT proteins in NHF. SIRT1, SIRT2, SIRT6, and SIRT7 proteins with the HA tag at their N-terminus showed the similar patterns of localization to their GFP fusion counterparts (Figure 1D). The costaining with anti-nucleoli antibody revealed that HA-SIRT7, but not HA-SIRT1 and HA-SIRT6, was enriched in the nucleoli (Figure 1D, left panel). SIRT3, SIRT4, or SIRT5 with the HA tag at their N-terminus could not be detected (unpublished data) probably because of an N-terminal processing (Schwer et al., 2002).
Production of Anti-SIRT6 and Anti-SIRT7 Antibodies and Cellular Localization of Endogenous SIRT6 and SIRT7 Proteins
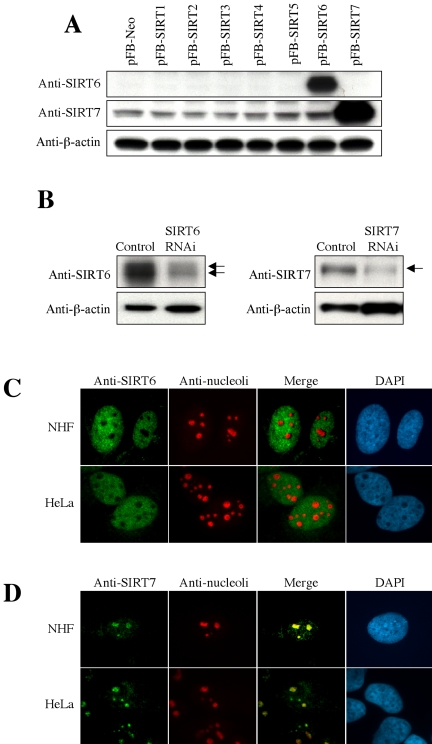
To further examine SIRT6 and SIRT7, which were newly suggested to be nuclear SIRT proteins, we raised the rabbit polyclonal antibodies against them. When NHF cells were infected with the retroviral vector driving each SIRT protein and used in Western blot with these antibodies, they specifically recognized the respective, overexpressed protein (Figure 2A). We further verified the specificity of the antibodies in HeLa cells with the endogenous SIRT6 or SIRT7 expression knocked down via RNAi. The protein signals detected with anti-SIRT6 or anti-SIRT7 antibody were significantly weakened in SIRT6- or SIRT7-knocked-down cells, respectively (Figure 2B). These antibodies were used in indirect immunofluorescence staining to examine cellular localization of endogenous SIRT6 and SIRT7 proteins. As shown in Figure 2, C and D, the endogenous SIRT6 and SIRT7 proteins in NHF and HeLa cells were detected in the nuclei and showed the similar patterns of staining to their GFP fusion and HA-tagged counterparts (Figure 1). In combination with the staining with anti-nucleoli antibody, the exclusion of endogenous SIRT6 from the nucleoli (Figure 2C) and the concentration of endogenous SIRT7 in the nucleoli (Figure 2D) were clearly shown.
Figure 2.
Production of anti-SIRT6 and anti-SIRT7 antibodies and detection of endogenous SIRT6 and SIRT7 proteins. (A) NHF cells were infected with the retroviral vector driving each SIRT (pFB-SIRT1–7) or control vector (pFB-Neo) and used in Western blot with anti-SIRT6 and anti-SIRT7 rabbit polyclonal antibodies. β-actin was a loading control. (B) The endogenous SIRT6 (left) or SIRT7 (right) expression was knocked down by RNA interference (RNAi) in HeLa cells. The arrows indicate the specific signals, which were significantly weakened in the RNAi cells. The nature of two SIRT6 bands is currently unknown. (C and D) Indirect immunofluorescence staining of endogenous SIRT6 (C) and SIRT7 (D) proteins in NHF and HeLa cells. Costaining with anti-nucleoli antibody and DAPI staining are also shown.
SIRT3, SIRT4, and SIRT5 Are Mitochondrial Proteins
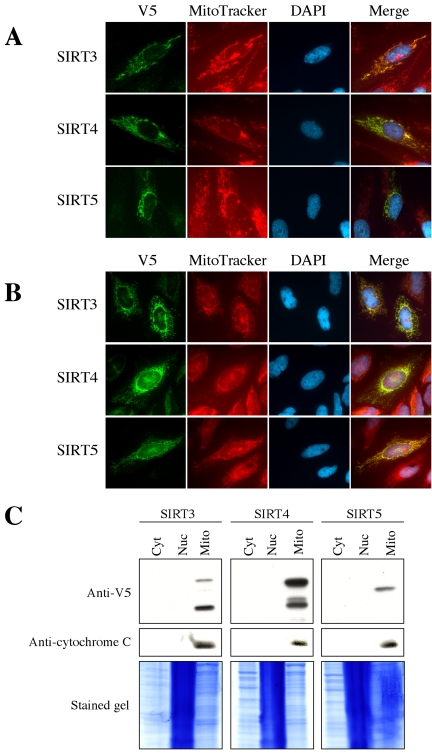
To verify the mitochondrial localization of SIRT4 and SIRT5, the vectors for expression of SIRT3, SIRT4, and SIRT5 with a V5-His tag at their C-terminus were constructed and transfected to NHF and HeLa cells. The indirect immunofluorescence staining with anti-V5 antibody showed that the V5-His-tagged SIRT3, SIRT4, and SIRT5 proteins colocalized with the mitochondrial marker MitoTracker in NHF (Figure 3A) and HeLa cells (Figure 3B), reproducing the observation with the GFP fusion proteins and MitoFluor staining (Figure 1A). In Western blot using subcellular protein fractions from the transfected 293T cells, the mitochondrial fractions exclusively contained the V5-His-tagged SIRT3, SIRT4, and SIRT5 proteins (Figure 3C). These findings strongly suggest that three different SIRT proteins (SIRT3, SIRT4, and SIRT5) are localized in the mitochondria.
Figure 3.
Mitochondrial localization of SIRT3, SIRT4, and SIRT5. (A and B) The V5-His-tagged SIRT3, SIRT4, or SIRT5 was transiently expressed in NHF (A) and HeLa (B). The tagged proteins were detected by immunostaining with anti-V5 antibody. MitoTracker staining and DAPI staining are shown in parallel. (C) Cytoplasmic (Cyt), nuclear (Nuc), and mitochondrial (Mito) proteins were fractionated from 293T cells expressing V5-His-tagged SIRT3 (left), SIRT4 (middle), or SIRT5 (right), and analyzed in Western blot using anti-V5 antibody. Cytochrome C, a well known mitochondrial protein, was detected as a control. The gels stained with SimplyBlue SafeStain (Invitrogen) are also shown. Residual DNA in nuclear fractions gave the stronger staining. The presence of two SIRT3 bands is due to its N-terminal processing (Schwer et al., 2002). The exact nature of multiple SIRT4 bands is not known.
Establishment of Normal Human Cells Overexpressing Each of Human SIRT Proteins
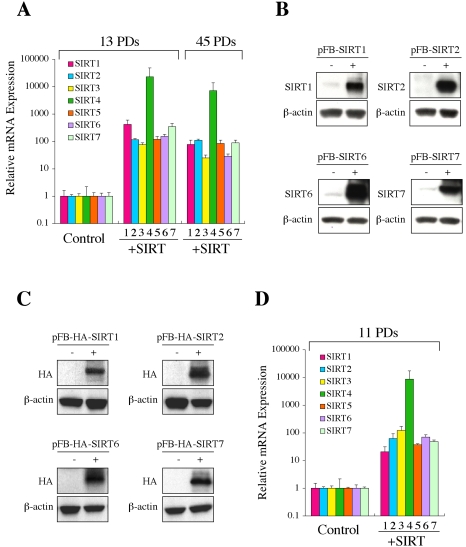
To examine the biological activities of SIRT proteins, we attempted to generate normal human cells stably overexpressing each of them. The retroviral expression vectors (same as those used in Figure 2A) were transduced to NHF cells at 7 PDs and selected for G418 resistance. Neither cell growth retardation nor significant cell death was observed, and at least 95% of the cells were infected and became G418-resistant, excluding a possibility that the cells were selected for an unrelated phenotype (e.g., clonal growth or stress resistance). The quantitative real-time RT-PCR assay showed a significant increase in mRNA expression of the respective SIRT genes in these cells at both early and late PDs (at least ∼25-fold more than the endogenous expression levels in control cells) (Figure 4A). The overexpression of SIRT1, SIRT2, SIRT6, and SIRT7 was also shown in Western blot (Figure 4B). In an independent experiment, we used the retroviral vectors driving the HA-tagged version of the SIRT proteins (same as those described above) and similarly obtained NHF cells overexpressing each HA-tagged SIRT. The overexpression was confirmed by the quantitative real-time RT-PCR assay (for all seven; unpublished data), as well as Western blot and indirect immunofluorescence using anti-HA antibody (for SIRT1, 2, 6, and 7; Figures 4C and 1D). The retroviral vectors (without HA tag) were also transduced to normal human prostate epithelial cells (PrEC) at 6 PDs and the overexpression of each SIRT was confirmed by the quantitative real-time RT-PCR at 11 PDs (Figure 4D).
Figure 4.
Establishment of normal human cells stably overexpressing each human SIRT. (A) NHF cells with the retroviral vector driving each SIRT (+SIRT1–7) at 13 PDs and 45 PDs were examined for mRNA expression of the respective SIRT gene by the real-time quantitative RT-PCR assays. NHF with the empty vector (Control) was also examined for the endogenous expression of each SIRT gene. The data are expressed as relative values to the endogenous expression levels in control cells (defined as 1 for each SIRT gene). (B) Western blot showed the overexpression of SIRT1, SIRT2, SIRT6, or SIRT7 protein in the respective retroviral vector-transduced NHF. The antibodies specific to each SIRT protein were used. (C) Western blot showed the overexpression of HA-SIRT1, HA-SIRT2, HA-SIRT6, or HA-SIRT7 protein in the respective retroviral vector-transduced NHF. Anti-HA antibody was used. (D) PrEC cells with the retroviral vector driving each SIRT (+SIRT1–7) or the empty vector (Control) were examined at 11 PDs as in A.
Cellular p53 Is an In Vivo Substrate for SIRT1 Deacetylase in Normal Human Cells
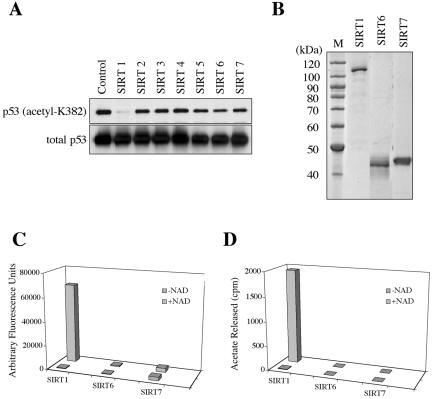
The deacetylation of p53 protein at the lysine residue 382 (K382) by SIRT1 was previously shown in the in vitro studies using the recombinant p53 or p53 peptide and the in vivo assays using cancer or immortalized cell lines (Luo et al., 2001; Vaziri et al., 2001; Langley et al., 2002). No data are available as to the ability of the other human SIRT proteins to deacetylate cellular p53 protein. We examined the effect of each SIRT protein on the K382 acetylation status of cellular, endogenous p53 protein in normal human cells. NHF cells overexpressing each of the SIRT proteins (without HA tag), which were established above, were treated with adriamycin to induce the acetylation of endogenous p53 protein. The cellular protein extracts were immunoprecipitated with anti-p53 antibodies and analyzed by Western blot with anti-acetylK382p53 antibody or anti-p53 antibody (DO-1; Figure 5A). More than 90% reduction in K382-acetylated p53 was reproducibly observed with SIRT1, consistent with the previous findings (Luo et al., 2001; Vaziri et al., 2001; Langley et al., 2002) and first demonstrating that endogenous p53 protein is a major in vivo substrate for human SIRT1 deacetylase in normal human cells. The other six SIRT proteins, including nuclear SIRT6 and SIRT7, had no significant effect on the K382 acetylation status in this in vivo assay.
Figure 5.
Protein deacetylation assays of human SIRT proteins. (A) Cellular p53 protein was immunoprecipitated from control and the SIRT-overexpressing NHF cells, followed by Western blot analysis using anti-acetyl-K382p53 antibody (top panel) and anti-p53 antibody (DO-1, bottom panel). The amounts of K382-acetylated p53 were normalized with those of total p53 protein detected by DO-1. In three independent experiments (a representative result shown here), the SIRT1-overexpressing cells, but not any others, showed a significant decrease in the acetylated p53. (B) Recombinant SIRT1, SIRT6, and SIRT7 proteins were purified from the baculovirus-infected insect cells. One microgram of proteins were analyzed on a SDS-polyacrylamide gel. (C) The purified SIRT1, SIRT6, and SIRT7 proteins were examined for an in vitro deacetylase activity on the synthetic p53 peptide (amino acid residues 379–382). The reactions with and without NAD+ were included. (D) The purified SIRT1, SIRT6, and SIRT7 proteins were examined for an in vitro deacetylase activity on the histone H4 peptide (amino acid residues 2–24) in the presence or absence of NAD+.
SIRT1, But Not SIRT6 and SIRT7, Deacetylates p53 and Histone Peptides In Vitro
To examine in vitro deacetylase activity of three nuclear SIRT proteins, the recombinant SIRT1, SIRT6, and SIRT7 were purified from baculovirus vector-infected Hi5 cells (Figure 5B). When a synthetic p53 peptide containing the acetylated K382 was used as the substrate, SIRT1 showed a strong deacetylase activity in a NAD+-dependent manner, whereas no or little activity was observed with SIRT6 or SIRT7 (Figure 5C). The in vitro deacetylation assay using a histone H4 peptide (containing the acetylated lysine 16 residue) also revealed a NAD+-dependent deacetylase activity of SIRT1, but not SIRT6 or SIRT7 (Figure 5D).
Increased Expression of Human SIRT Proteins Does Not Extend Replicative Lifespan in Normal Human Cells
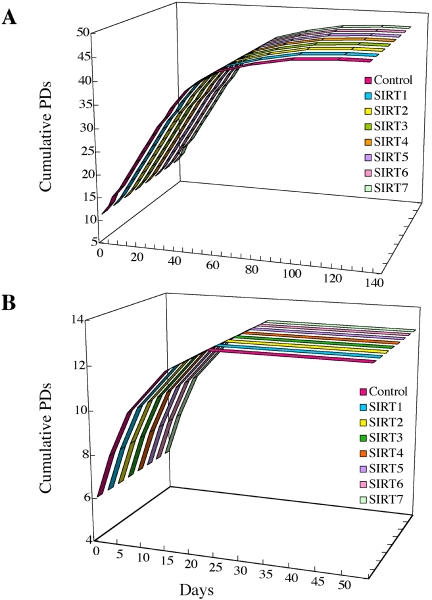
Considering that an increase in yeast Sir2 activity extends the replicative lifespan (Kaeberlein et al., 1999), it is of great interest whether human SIRT proteins regulate replicative lifespan in human cells. We therefore examined the SIRT-overexpressing cells established above for their replicative lifespan in culture. NHF cells overexpressing any one of the SIRT proteins, without or with HA tag, underwent the similar replicative history to control cells infected with the empty vector (Figure 6A and unpublished data). All NHF-derived cells ceased to proliferate with enlarged and flattened cell morphology and the positive SA-β-gal staining at 47 PDs, when untransduced NHF cells also senesce. As shown in Figure 4A (see 45 PDs), the cells maintained high levels of the respective SIRT expression even at the end stage of the replicative lifespan. We also examined the effects of the SIRT proteins on replicative lifespan in normal human cells of epithelial origin. All SIRT-overexpressing PrEC cells, as well as control cells with the empty vector, became senescent at 13 PDs 3 wk after retroviral infection (Figure 6B). Similarly, the replicative lifespan of WI-38 cells, another normal human fibroblast strain, was not extended by any of the SIRT proteins (unpublished data). Thus, we conclude that an increased expression of any single SIRT protein by itself does not extend cellular replicative lifespan in normal human cells under standard culture conditions.
Figure 6.
Replicative lifespan of NHF (A) and PrEC (B) overexpressing human SIRT proteins. The replicative histories of control and SIRT-overexpressing cells are shown as the growth curves, where the cumulative PDs are plotted to the days after retroviral transduction. All NHF-derived and PrEC-derived cells ceased to proliferate at 47 and at 13 PDs, respectively, with the characteristic features of cellular senescence such as enlarged and flat cell morphology and positive SA-β-gal staining.
The ectopic expression of the human telomerase reverse transcriptase (hTERT) extends replicative lifespan in normal human cells (Bodnar et al., 1998). The quantitative real-time RT-PCR assay showed no activation of the hTERT gene in any of the SIRT-overexpressing NHF cells (unpublished data), consistent with no lifespan extension in these cells. We also tested the SIRT-overexpressing cells for sensitivity to premature stress-induced senescence. After oxidative stress or DNA damage (150 μM of H2O2 for 2 h or 10 mU/ml bleomycin for 20 h; Sedelnikova et al., 2004), all SIRT-overexpressing NHF cells, as well as vector control cells and untransduced NHF cells, underwent senescent growth arrest (unpublished data).
Expression of Endogenous SIRT1 Protein Decreases during Replicative Aging in Normal Human Fibroblasts
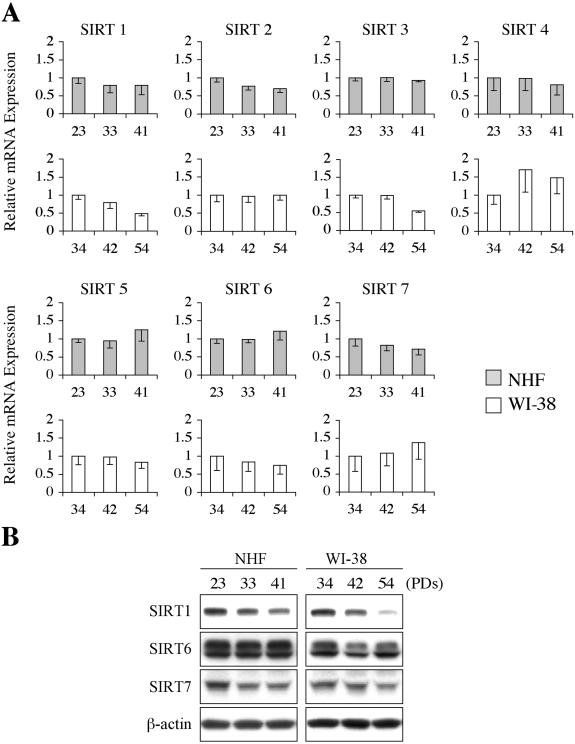
The expression levels of endogenous SIRT genes were examined during replicative lifespan in NHF and WI-38 fibroblasts. The real-time RT-PCR assays for quantitatively detecting each SIRT mRNA did not find a significant change common in NHF and WI-38 (Figure 7A). For example, the gradual decrease in SIRT1 mRNA was observed in WI-38, but not in NHF. However, Western blot showed that SIRT1 expression at the protein level decreased as a function of PDs in both NHF and WI-38 (Figure 7B). Slight changes, if any, in SIRT6 or SIRT7 protein expression were not strictly associated with PDs the cells underwent.
Figure 7.
Endogenous SIRT expression during replicative aging in normal human fibroblasts. (A) The real-time RT-PCR assays quantitated mRNA expression of each SIRT gene in NHF (at 23, 33, and 41 PDs) and WI-38 (at 34 PDs, 42 PDs, and 54 PDs). (B) The same set of cells as in (A) was examined in Western blot for SIRT1, SIRT6, and SIRT7 protein expression. β-actin was a loading control.
Expression Profiles of SIRT Genes in Human Organs
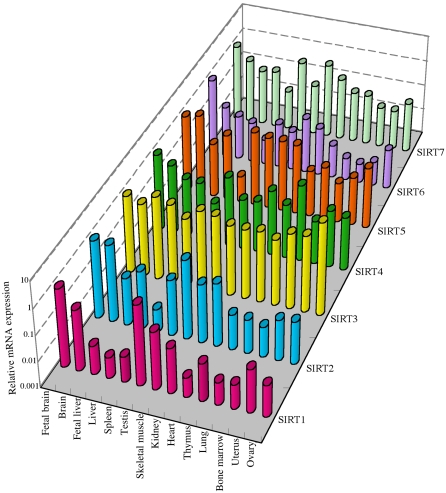
The ubiquitous mRNA expression of five SIRT genes (SIRT1 through 5) were previously reported (Frye, 1999). However, the previous data were not quantitative and did not include SIRT6 and SIRT7. We quantitatively examined the expression levels of all seven SIRT genes in human organs (Figure 8). The results not only confirmed their ubiquitous expression in a wide variety of tissues but also revealed the expression profiles unique to each SIRT gene. For all SIRT genes, brain and testis were among the adult organs where the high levels of mRNA expression were observed. For all but SIRT2 and SIRT5, fetal brain showed the higher expression than adult brain, possibly suggesting their roles in the developmental processes.
Figure 8.
Expression profiles of SIRT genes in human organs. Twelve adult organs, fetal brain, and fetal liver were examined for the expression of each SIRT gene by the quantitative real-time RT-PCR assay. For each SIRT, the expression level in fetal brain was defined as 1, and the expression levels in the other organs were expressed as the relative values to fetal brain.
DISCUSSION
This study determined the cellular localization of each SIRT protein. We confirmed the previously reported localization of SIRT1 (nucleus), SIRT2 (cytoplasm), and SIRT3 (mitochondria). Like SIRT3, two of the newly characterized SIRT proteins (SIRT4 and SIRT5) were shown to be in the mitochondria by colocalization of the GFP fusion proteins (Figure 1A) and the V5-His-tagged proteins (Figure 3, A and B) with the mitochondria-specific probes, as well as by the presence of the V5-His-tagged proteins in isolated mitochondria (Figure 3C). Given that mitochondria is an organelle involved in both aging and energy metabolism (Merry, 2004) and that the sirtuin family of proteins function in a NAD+-dependent manner (Imai et al., 2000; Blander and Guarente, 2004; North and Verdin, 2004), an interesting possibility is that one or more of the mitochondrial SIRT proteins may function in linking metabolic and aging processes in humans.
In addition to SIRT1, SIRT6 and SIRT7 were identified as nuclear proteins in this study. Importantly, these three nuclear SIRT proteins are different in subnuclear localization from one another. Consistent with the previous reports (Langley et al., 2002; McBurney et al., 2003a, 2003b), SIRT1 is not concentrated in the nucleoli (Figure 1). It is not significantly associated with Hoechst 33342-stained regions, in agreement with the euchromatic localization of mouse Sirt1 (McBurney et al., 2003b). Our determination of SIRT6 and SIRT7 localization was based on four independent experiments, i.e., detection of the GFP fusion proteins (C-terminal fusion; Figure 1, A and B), indirect immunofluorescence staining of the HA-tagged proteins (N-terminal tag; Figure 1D) and the V5-His-tagged proteins (C-terminal tag; unpublished data), and most convincingly, indirect immunofluorescence staining of the endogenous proteins with the SIRT6- and SIRT7-specific polyclonal antibodies (Figure 2, C and D). Our data reveal that SIRT6 is excluded from the nucleoli and highly associated with the heterochromatic regions and that SIRT7 is concentrated in the nucleoli. Although SIRT6 and SIRT7 belong to the different classes of the sirtuin family from yeast Sir2 and SIRT1 (Frye, 2000), they are similar in cellular localization to yeast Sir2, which functions in the nucleoli and heterochromatic regions (Gotta et al., 1997; Lustig, 1998; Blander and Guarente, 2004; North and Verdin, 2004).
The results of the in vitro deacetylation assay using a histone substrate may suggest an evolutionary complexity of the sirtuin family. Despite their similarity to yeast Sir2 in cellular localization, SIRT6 and SIRT7 failed to deacetylate the histone H4 peptide containing the acetylated lysine 16 residue, a known target site of yeast Sir2 deacetylase (Imai et al., 2000; Shankaranarayana et al., 2003), whereas SIRT1 showed a strong deacetylase activity in a NAD+-dependent manner (Figure 5D). This is consistent with the previous finding in a different in vitro assay system (North et al., 2003) and a recent report that mouse Sirt6 and Sirt7 have no in vitro deacetylase activity (Liszt et al., 2005). Vaquero et al. (2004) suggested that SIRT1 could promote the formation of facultative heterochromatin at an experimentally recruited euchromatic site through deacetylation of the H4 lysine 16. Thus, mammalian SIRT1 might have evolved to control the transcription of a specific set of single-copy genes through its evolutionarily conserved deacetylase activity (Fulco et al., 2003; Takata and Ishikawa, 2003; Picard et al., 2004), rather than to regulate the chromatin structure and transcription at ribosomal DNA clusters or constitutive heterochromatin. On the other hand, it is likely that the heterochromatin-associated SIRT6 and nucleolar SIRT7 proteins function at the evolutionarily conserved sites either through deacetylating an unidentified substrate(s) or in a deacetylase activity-independent manner. The association of SIRT6 and SIRT7, but not SIRT1, with condensed chromosomes at mitosis (Figure 1C) may suggest their roles in the dynamic regulation of chromosome structure and function. It will be important to identify the interacting proteins or physiological substrates. It is also interesting to examine whether and how an ADP-ribosyltransferase activity (Frye, 1999; Liszt et al., 2005) is involved in their functions.
The in vivo deacetylation assay demonstrated a strong ability of SIRT1 to deacetylate cellular p53 protein at the K382 residue in normal human cells (Figure 5A). In contrast, any of the other six SIRT proteins did not show a significant effect on the p53 acetylation status, suggesting that p53 is not a major in vivo substrate for these SIRT proteins. These findings agree with the results of in vitro deacetylation assay, in which SIRT1, but not SIRT6 or SIRT7, showed the deacetylase activity on a synthetic p53 peptide (Figure 5C). However, our in vivo assay might not be able to detect an effect of nonnuclear SIRT proteins on a small fraction of cellular p53 protein located in the cytoplasm or mitochondria (Baptiste and Prives, 2004; Murphy et al., 2004). SIRT2, SIRT3, SIRT4, and SIRT5 therefore deserve further examination for p53 deacetylase activity.
This study for the first time examined the effects of human SIRT proteins on replicative lifespan of human cells. For this purpose, we have successfully established normal human cells of both fibroblastic and epithelial origins that stably express an increased amount of each SIRT protein (Figure 4). Neither human fibroblasts (NHF and WI-38) nor epithelial cells (PrEC) overexpressing any of the SIRT proteins showed a change in replicative lifespan under standard culture conditions (Figure 6, A and B). Although the deacetylation of p53 may reduce its activity (Brooks and Gu, 2003) and the loss of p53 activity extends cellular replicative lifespan (Hara et al., 1991; Morris et al., 2002), the strong inhibition of DNA damage-induced p53 acetylation by SIRT1 (Figure 5A) was not associated with an extension of the replicative lifespan. Although SIRT1 represses p53-dependent apoptotic cell death induced by high-dose irradiation or etoposide (Luo et al., 2001; Vaziri et al., 2001), the SIRT1-overexpressing NHF was as sensitive to milder DNA damage and oxidative stress, which induce senescent growth arrest rather than immediate cell death (Sedelnikova et al., 2004), as control cells and cells overexpressing the other SIRT proteins. These findings suggest that the regulation of p53 acetylation by SIRT1 does not primarily control replicative senescence and stress-induced senescence in normal human cells.
We conclude that the replicative lifespan of normal human cells cannot be manipulated by increased expression of a single SIRT protein. Our data present a striking contrast to the lifespan extension in lower eukaryotes by Sir2 and its homologues (Kaeberlein et al., 1999; Tissenbaum and Guarente, 2001; Rogina and Helfand, 2004) and suggest complex layers of regulation of replicative aging in human cells. Nonetheless, we do not exclude a role of human SIRT proteins in cellular or organismal aging processes. This study presents a finding that implies a link between cellular aging and a human SIRT protein: the expression of endogenous SIRT1 protein progressively decreased as normal human fibroblasts replicatively aged in culture (Figure 7B). An important future study will be RNAi-mediated knockdown of endogenous SIRT proteins in normal human cells. Finally, the expression profiles of SIRT genes in various organs (Figure 8) provide important information for transgenic and knockout mouse studies. This is evidenced by the phenotypes of mice deficient for Sirt1. Their major defects were abnormal development of retina and impaired spermatogenesis (Cheng et al., 2003; McBurney et al., 2003b), which agree with the abundant expression of SIRT1 in brain and testis.
Acknowledgments
We thank Dr. Paul Goldsmith for expertise in antibody production; Drs. Shin'ichi Saito and Ettore Appella for anti-acetyl-Lys382p53 antibody; Drs. Dominic Esposito, Ralph Hopkins, and William Gillette (Protein Expression Laboratory, NCI/SAIC-Frederick) for purification of recombinant SIRT proteins; Dr. Jayne Boyer for NHF; and Drs. Frederick Alt and Katrin Chua for helpful discussion. This research was supported by the Intramural Research Program of the NIH, NCI. E.M. is supported by JSPS Research Fellowship for Japanese Biomedical and Behavioral Researchers at the NIH. J.Y.P and J.M.B. were NIH Summer Interns.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-01-0033) on August 3, 2005.
Abbreviations used: NAD+, nicotinamide adenine dinucleotide; GFP, green fluorescent protein; HA, influenza hemagglutinin epitope; PDs, population doublings; PrEC, prostate epithelial cells.
References
- Baptiste, N., and Prives, C. (2004). p53 in the cytoplasm: a question of overkill? Cell 116, 487–489. [DOI] [PubMed] [Google Scholar]
- Barbieri, M., Bonafe, M., Franceschi, C., and Paolisso, G. (2003). Insulin/IGF-I-signaling pathway: an evolutionarily conserved mechanism of longevity from yeast to humans. Am. J. Physiol. Endocrinol. Metab. 285, E1064–E1071. [DOI] [PubMed] [Google Scholar]
- Blander, G., and Guarente, L. (2004). The sir2 family of protein deacetylases. Annu. Rev. Biochem. 73, 417–435. [DOI] [PubMed] [Google Scholar]
- Bodnar, A. G., Ouellette, M., Frolkis, M., Holt, S. E., Chiu, C. P., Morin, G. B., Harley, C. B., Shay, J. W., Lichtsteiner, S., and Wright, W. E. (1998). Extension of life-span by introduction of telomerase into normal human cells. Science 279, 349–352. [DOI] [PubMed] [Google Scholar]
- Brooks, C. L., and Gu, W. (2003). Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr. Opin. Cell Biol. 15, 164–171. [DOI] [PubMed] [Google Scholar]
- Brunet, A. et al. (2004). Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303, 2011–2015. [DOI] [PubMed] [Google Scholar]
- Campisi, J. (2001). From cells to organisms: can we learn about aging from cells in culture? Exp. Gerontol. 36, 607–618. [DOI] [PubMed] [Google Scholar]
- Cheng, H. L., Mostoslavsky, R., Saito, S., Manis, J. P., Gu, Y., Patel, P., Bronson, R., Appella, E., Alt, F. W., and Chua, K. F. (2003). Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. USA 100, 10794–10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, H. Y., Miller, C., Bitterman, K. J., Wall, N. R., Hekking, B., Kessler, B., Howitz, K. T., Gorospe, M., De Cabo, R., and Sinclair, D. A. (2004). Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305, 390–392. [DOI] [PubMed] [Google Scholar]
- Dimri, G. P., et al. (1995). A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92, 9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye, R. A. (1999). Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 260, 273–279. [DOI] [PubMed] [Google Scholar]
- Frye, R. A. (2000). Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273, 793–798. [DOI] [PubMed] [Google Scholar]
- Fulco, M., Schiltz, R. L., Iezzi, S., King, M. T., Zhao, P., Kashiwaya, Y., Hoffman, E., Veech, R. L., and Sartorelli, V. (2003). Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell 12, 51–62. [DOI] [PubMed] [Google Scholar]
- Gotta, M., Strahl-Bolsinger, S., Renauld, H., Laroche, T., Kennedy, B. K., Grunstein, M., and Gasser, S. M. (1997). Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 16, 3243–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara, E., Tsurui, H., Shinozaki, A., Nakada, S., and Oda, K. (1991). Cooperative effect of antisense-Rb and antisense-p53 oligomers on the extension of life span in human diploid fibroblasts, TIG-1. Biochem. Biophys. Res. Commun. 179, 528–534. [DOI] [PubMed] [Google Scholar]
- Harrington, L., and Robinson, M. O. (2002). Telomere dysfunction: multiple paths to the same end. Oncogene 21, 592–597. [DOI] [PubMed] [Google Scholar]
- Heilbronn, L. K., and Ravussin, E. (2003). Calorie restriction and aging: review of the literature and implications for studies in humans. Am. J. Clin. Nutr. 78, 361–369. [DOI] [PubMed] [Google Scholar]
- Hursting, S. D., Lavigne, J. A., Berrigan, D., Perkins, S. N., and Barrett, J. C. (2003). Calorie restriction, aging, and cancer prevention: mechanisms of action and applicability to humans. Annu. Rev. Med. 54, 131–152. [DOI] [PubMed] [Google Scholar]
- Imai, S., Armstrong, C. M., Kaeberlein, M., and Guarente, L. (2000). Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800. [DOI] [PubMed] [Google Scholar]
- Kaeberlein, M., McVey, M., and Guarente, L. (1999). The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13, 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley, E., Pearson, M., Faretta, M., Bauer, U. M., Frye, R. A., Minucci, S., Pelicci, P. G., and Kouzarides, T. (2002). Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 21, 2383–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszt, G., Ford, E., Kurtev, M., and Guarente, L. (2005). Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 280, 21313–21320. [DOI] [PubMed] [Google Scholar]
- Luo, J., Nikolaev, A. Y., Imai, S., Chen, D., Su, F., Shiloh, A., Guarente, L., and Gu, W. (2001). Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107, 137–148. [DOI] [PubMed] [Google Scholar]
- Lustig, A. J. (1998). Mechanisms of silencing in Saccharomyces cerevisiae. Curr. Opin. Genet. Dev. 8, 233–239. [DOI] [PubMed] [Google Scholar]
- McBurney, M. W., Yang, X., Jardine, K., Bieman, M., Th'ng, J., and Lemieux, M. (2003a). The absence of SIR2alpha protein has no effect on global gene silencing in mouse embryonic stem cells. Mol. Cancer Res. 1, 402–409. [PubMed] [Google Scholar]
- McBurney, M. W., Yang, X., Jardine, K., Hixon, M., Boekelheide, K., Webb, J. R., Lansdorp, P. M., and Lemieux, M. (2003b). The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol. Cell. Biol. 23, 38–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merry, B. J. (2004). Oxidative stress and mitochondrial function with aging— the effects of calorie restriction. Aging Cell 3, 7–12. [DOI] [PubMed] [Google Scholar]
- Morris, M., Hepburn, P., and Wynford-Thomas, D. (2002). Sequential extension of proliferative lifespan in human fibroblasts induced by over-expression of CDK4 or 6 and loss of p53 function. Oncogene 21, 4277–4288. [DOI] [PubMed] [Google Scholar]
- Motta, M. C., Divecha, N., Lemieux, M., Kamel, C., Chen, D., Gu, W., Bultsma, Y., McBurney, M., and Guarente, L. (2004). Mammalian SIRT1 represses forkhead transcription factors. Cell 116, 551–563. [DOI] [PubMed] [Google Scholar]
- Murphy, M. E., Leu, J. I., and George, D. L. (2004). p53 moves to mitochondria: a turn on the path to apoptosis. Cell Cycle 3, 836–839. [PubMed] [Google Scholar]
- North, B. J., Marshall, B. L., Borra, M. T., Denu, J. M., and Verdin, E. (2003). The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11, 437–444. [DOI] [PubMed] [Google Scholar]
- North, B. J., and Verdin, E. (2004). Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 5, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi, S. et al. (2005). Evaluation of macrocyclic Grb2 SH2 domain-binding peptide mimetics prepared by ring-closing metathesis of C-terminal allylglycines with an N-terminal beta-vinyl-substituted phosphotyrosyl mimetic. Bioorg. Med. Chem. 13, 2431–2438. [DOI] [PubMed] [Google Scholar]
- Onyango, P., Celic, I., McCaffery, J. M., Boeke, J. D., and Feinberg, A. P. (2002). SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc. Natl. Acad. Sci. USA 99, 13653–13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Smith, O. M., and Smith, J. R. (1981). Expression of SV40 T antigen in finite life-span hybrids of normal and SV40-transformed fibroblasts. Somatic Cell Genet. 7, 411–421. [DOI] [PubMed] [Google Scholar]
- Picard, F., Kurtev, M., Chung, N., Topark-Ngarm, A., Senawong, T., Machado De Oliveira, R., Leid, M., McBurney, M. W., and Guarente, L. (2004). Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 429, 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogina, B., and Helfand, S. L. (2004). Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. USA 101, 15998–16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi, K., Herrera, J. E., Saito, S., Miki, T., Bustin, M., Vassilev, A., Anderson, C. W., and Appella, E. (1998). DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12, 2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer, B., North, B. J., Frye, R. A., Ott, M., and Verdin, E. (2002). The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 158, 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedelnikova, O. A., Horikawa, I., Zimonjic, D. B., Popescu, N. C., Bonner, W. M., and Barrett, J. C. (2004). Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat. Cell Biol. 6, 168–170. [DOI] [PubMed] [Google Scholar]
- Shankaranarayana, G. D., Motamedi, M. R., Moazed, D., and Grewal, S. I. (2003). Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr. Biol. 13, 1240–1246. [DOI] [PubMed] [Google Scholar]
- Sinha-Datta, U., Horikawa, I., Michishita, E., Datta, A., Sigler-Nicot, J.C., Brown, M., Kazanji, M., Barrett, J. C., and Nicot, C. (2004). Transcriptional activation of hTERT through the NF-kappaB pathway in HTLV-I-transformed cells. Blood 104, 2523–2531. [DOI] [PubMed] [Google Scholar]
- Sugawara, O., Oshimura, M., Koi, M., Annab, L. A., and Barrett, J. C. (1990). Induction of cellular senescence in immortalized cells by human chromosome 1. Science 247, 707–710. [DOI] [PubMed] [Google Scholar]
- Takata, T., and Ishikawa, F. (2003). Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem. Biophys. Res. Commun. 301, 250–257. [DOI] [PubMed] [Google Scholar]
- Tissenbaum, H. A., and Guarente, L. (2001). Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410, 227–230. [DOI] [PubMed] [Google Scholar]
- Tissenbaum, H. A., and Guarente, L. (2002). Model organisms as a guide to mammalian aging. Dev. Cell 2, 9–19. [DOI] [PubMed] [Google Scholar]
- Tominaga, K., Olgun, A., Smith, J. R., and Pereira-Smith, O. M. (2002). Genetics of cellular senescence. Mech. Ageing Dev. 123, 927–936. [DOI] [PubMed] [Google Scholar]
- Vaquero, A., Scher, M., Lee, D., Erdjument-Brumage, H., Tempst, P., and Reinberg, D. (2004). Human SirT1 interacts with histone H1 and promotes formation of a facultative heterochromatin. Mol. Cell 16, 93–105. [DOI] [PubMed] [Google Scholar]
- Vaziri, H., Dessain, S. K., Ng Eaton, E., Imai, S. I., Frye, R. A., Pandita, T. K., Guarente, L., and Weinberg, R. A. (2001). hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107, 149–159. [DOI] [PubMed] [Google Scholar]
- Yeung, F., Hoberg, J. E., Ramsey, C. S., Keller, M. D., Jones, D. R., Frye, R. A., and Mayo, M. W. (2004). Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23, 2369–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]