Abstract
We screened a custom-made candidate gene cDNA array comprising 300 genes. Genes chosen have either been implicated in schizophrenia, make conceptual sense in the light of the current understanding of the disease, or are located on high-susceptibility chromosome locations. The array screen using prefrontal cortex tissue from 10 schizophrenia and 10 control brains revealed robust up-regulation of apolipoprotein L1 (apo L1) by 2.6-fold. The finding was cross-validated in a blinded quantitative PCR study using prefrontal cortex tissue from the Stanley Foundation brain collection, Bethesda, MD. This collection consists of 15 schizophrenia, 15 bipolar disorder, 15 major depression, and 15 control individuals, all 60 brains being well-matched on conventional parameters, with antipsychotic drug exposure in the schizophrenia and bipolar disorder groups. Significant up-regulation of apo L1 gene expression in schizophrenia was confirmed. Using quantitative PCR, expression profiles of other members of the apo L family (apo L2–L6) were investigated, showing that apo L2 and L4 were highly significantly up-regulated in schizophrenia. Results were then confirmed in an independent set of 20 schizophrenia and 20 control brains from Japan and New Zealand. Apo L proteins belong to the group of high density lipoproteins, with all six apo L genes located in close proximity to each other on chromosome 22q12, a confirmed high-susceptibility locus for schizophrenia and close to the region associated with velocardiofacial syndrome that includes symptoms of schizophrenia.
Schizophrenia is a major public health problem. Typically, psychosis begins in early adulthood and interferes with education, employment, and social functioning. A hereditary contribution has been established beyond doubt, and neuroimaging and neuropathological studies show subtle brain abnormalities (1–3). The dorsal prefrontal cortex has been particularly implicated in the pathophysiological dysfunction in schizophrenia (4, 5).
The sequencing of the human genome undoubtedly will open new exciting avenues for research into complex disorders such as schizophrenia. Furthermore, the availability of novel molecular profiling techniques such as microarrays and differential display–PCR now allow a global approach to screening for abnormalities in gene expression. Such approaches have the benefit that no assumption has to be made as to whether pathology is caused primarily by genetic and/or environmental factors, as any causative effect will inevitably lead to changes in gene expression.
For this study, we designed a candidate gene cDNA array to investigate changes in gene expression in the prefrontal cortex in schizophrenia. We found robust and reliable changes in expression of apolipoprotein (apo) L1, L2, and L4. Lipoproteins provide cells with lipids that are of vital importance to normal cell functioning. Lipids are used to generate energy, are building blocks for biomembranes, are essential for the synthesis of numerous lipophilic molecules (e.g., steroid hormones and vitamin E), and play an important role in cell signaling and antioxidative mechanisms (6, 7). Most lipoproteins are expressed in many tissues and cell types, including the brain and cerebrospinal fluid (6). Apo L belongs to the high density lipoprotein family that plays a central role in cholesterol transport (8). The cholesterol content of membranes is important in cellular processes such as modulating gene transcription and signal transduction both in the adult brain and during neurodevelopment (7).
Methods
Tissue Collection.
Fresh-frozen prefrontal cortex tissue was obtained from the Stanley Foundation brain collection and neuropathology consortium, Bethesda, MD, as well as from a collection of schizophrenia and control brains from Japan (Department of Psychiatry, Tokyo Metropolitan Matsuzawa Hospital) and from New Zealand (New Zealand Neurological Foundation Human Brain Bank, University of Auckland, School of Medicine). The Stanley Foundation brain collection consists of well-matched prefrontal cortex tissue derived from 15 schizophrenia, 15 bipolar disorder, 15 major depression, and 15 control individuals (9). The New Zealand/Japan collection consists of corresponding prefrontal cortex tissue from 20 schizophrenia and 20 matched controls. For all cases, tissue was collected from patients who had previously been diagnosed according to DSM-IIIR (33) or DSM-IV (34). The Stanley Foundation brain collection is well-matched for age, pH, sex, race, side of the brain, and postmortem interval, the details of which have been described by Torrey et al. (9). In brief, the mean age in the schizophrenia group was 44.2 (range 25–62) years, 42.3 (range 25–61) years for bipolar disorder, 46.4 (range 30–65) for major depression, and 48.1 (range 29–68) for normal controls. The sex ratio was nine males and six females in all groups, and the average postmortem interval for the four groups ranged between 23.7 and 33.7 h (see ref. 9 for more details). All 60 tissue samples from the Stanley Foundation were analyzed blind to diagnostic status with the code being retained by the foundation until analysis was completed. The New Zealand/Japan brain collection was matched for age, sex, and postmortem interval. Whereas brains from the Stanley Foundation collection were predominantly young to middle-aged, the New Zealand/Japan collection consists of mainly elderly and chronically (mostly lifelong) hospitalized individuals. For the schizophrenia group the mean age was 65.3 years (range 43–85 years) and for controls 62.7 years (range 42–84 years); the sex ratio was 15 males and five females for the schizophrenia group and 17 males and three females for controls. Postmortem interval averages were 8.1 h (range 2–18 h) for schizophrenia and 12.6 h (range 2–25 h) for controls.
RNA Extraction and cDNA Synthesis.
Total RNA was extracted from postmortem prefrontal cortices of schizophrenia, bipolar, depressed, and control brains by using the Tri-reagent RNA extraction protocol (Sigma) in conjunction with a mechanical homogenizer (Thermohybaid, Middlesex, U.K.). Typical yields were 1 μg of total RNA per 1 mg of tissue. RNA sample concentrations were determined by UV spectrophotometry (average OD260/280 >1.8). RNA integrity was confirmed by direct visualization of 18S and 28S rRNA bands after agarose gel electrophoresis. RNA samples (8 μg) were incubated with 10 units of DNase I (Roche Diagnostics) at 37°C for 20 min to remove residual DNA followed by inactivation at 65°C for 10 min. RNA samples were further purified by using a CHROMA SPIN column (CLONTECH) according to the manufacturer's instructions. RNA samples (0.5 μg) were reverse-transcribed with avian myeloblastosis virus reverse transcriptase (Roche Diagnostics) using random hexamers.
High-Density Filter Array Procedure.
Gene expression profiles of 10 control and 10 schizophrenia brain samples (selected from the New Zealand/Japan collection) were analyzed by hybridization screening of nylon membrane cDNA arrays with radioactively labeled first-strand cDNAs. The arrays contained 10 control probes and 300 IMAGE cDNAs complementary to mRNAs of noted or implicated importance in schizophrenia. The clones were selected from the United Kingdom Human Genome Mapping Project Resource Centre HuGen2 gene index and by sequence comparison versus the dbEST database (National Center for Biotechnology Information, Bethesda, MD). Each cDNA was selected on the basis that it represented the most 3′ end of a mRNA and lacked repeat sequences. cDNAs were amplified by PCR using the primers IMAGE-M13F (5′-GTTTTCCCAGTCACGACGTTG-3′) and IMAGE-M13R (5′-TGAGCGGATAACAATTTCACACAG-3′). The identities of the cDNAs were verified by dye terminator cycle sequencing (Perkin–Elmer). Control cDNAs included Arabidopsis thaliana cytochrome c554 gene (used for hybridization signal normalization); A. thaliana poly(A) sequences of 50, 60, and 90 bp and the vector pT7T3D (negative controls); and housekeeping genes actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). PCR-amplified cDNAs were purified (Milipore) and diluted to a concentration of 0.5 μg/μl. cDNAs were denatured by the addition of NaOH to 0.2 M and robotically spotted (Flexys, Genomic Solutions, Huntingdon, U.K.) in quadruplicate onto Hybond-N+ membranes (Amersham Pharmacia). cDNAs were fixed onto membranes by UV crosslinking (70,000 μJ/cm2). Twenty five micrograms of total RNA from each sample was reverse-transcribed by 200 units of Superscript II-RT (Life Technologies, Gaithersburg, MD) extending an oligo(dT25)VN (8.3 μM) primer in the presence of 0.55 μM [33P]dATP. Each RNA sample was spiked with 2 ng of A. thaliana cytochrome c554 mRNA. RNA was destroyed by alkaline hydrolysis and labeled cDNAs were purified (QIAquick PCR purification columns, Qiagen, Chatsworth, CA). Before hybridization, labeled cDNAs were incubated with 17 μg of oligo(dA80) for 2.5 h at 42°C in an attempt to prevent hybridization of poly(T) sequences to poly(A) tails of immobilized cDNAs. Hybridization was for 48 h at 42°C in 9.5 ml of hybridization buffer (Ultrahyb, Ambion). Arrays were washed sequentially in 2× SSC, 0.1% SDS at room temperature (2 × 5 min), 2× SSC, 0.1% (wt/vol) SDS at 42oC (2 × 5 min), and 0.1× SSC, 0.1% (wt/vol) SDS at 42oC (2 × 15 min) and exposed before scanning with the Typhoon 8600 variable mode imager (Amersham Pharmacia). To normalize for variation in robotic spotting each membrane was stripped and rehybridized with 5 ng of 33P-dATP-labeled pBluescript II SK(−) vector. After image acquisition, hybridization signals were quantified by using ArrayVision (Image Research, St. Catherines, Canada). Signal processing was subsequently performed by using Bluescript vector data to correct for variation in the relative amount of DNA present at equivalent positions on different arrays. Variations in labeling, hybridization, washing, and duration of phosphor screen exposure for different array hybridizations were normalized against plant spike expression profiles (A. thaliana cytochrome c554). Quantified data were expressed as absolute gene expression levels and were comparable within all RNA samples.
Real-Time Quantitative PCR.
Real-time PCR was performed by using an Applied Biosystems Prism 7700 sequence detection system. Reactions were performed in a 25-μl volume including diluted cDNA sample, 0.5 μM primers, 2 mM MgCl2, nucleotides, TaqDNA polymerase, and buffer included in the SYBR Green I Mastermix (Applied Biosystems). Diluted cDNA samples produced from 12.5 ng total RNA were added to each well. A typical protocol took 2 h to complete, allowing for the detection of multiple transcripts and replicates within a 96-well plate format. PCR cycling conditions were 50°C for 2 min, 95°C for 10 min, and 40 cycles of 94°C for 15 s, 60°C for 1 min. SYBR Green I reverse transcriptase–PCR data were collected by using sequence detector software (version 1.6, Applied Biosystems). Amplification plots and predicted Ct values from the exponential phase of the PCR were exported directly into Microsoft excel worksheets for analysis.
Primers, Control, and Standard Curve Generation.
Several primer sets for each gene were designed to intron/exon boundaries (eliminating contaminating genomic DNA signals). This procedure has allowed us to compare independently the mRNA transcript level detected for the same gene. Primer sequences were designed by using primer express software (version I, Applied Biosystems). In general, amplicons were between 70 and 120 nt. Melting curve analysis and agarose gel electrophoresis allowed us to verify the specificity of PCR amplifications (data not shown). Standard curve assays were performed by measuring mRNA transcript levels obtained with specific primer sets from a control cDNA sample diluted at 2-fold intervals. A similar standard curve was performed for the housekeeping gene GAPDH. Therefore we could normalize the mRNA transcript level against GAPDH at each dilution (ΔCt). The relationship between ΔCt and log input mRNA concentration was linear for all primer sets analyzed. The slope of the standard curve should be ≤0.1 for accurate mRNA transcript level determination. All standard curve and sample assays were performed in duplicate to improve the accuracy of mRNA transcript detection. No-template control assays were performed for each primer set used, which produced negligible signal detection, typically 38–40 Cts in value. RNA quality and quantity of each sample was assessed by normalizing data with respect to the GAPDH mRNA transcipt level determined. All primer sets used for analyzing diluted cDNA samples typically produced 20–30 Cts on average, whereas the GAPDH value was 18 Cts within the main detection window, 0–40 Cts in value i.e., within the accepted detection window.
In Situ Hybridization.
In situ hybridization was carried out as described by Herrero et al. (10). In brief, tissue sections were fixed in 4% paraformaldehyde, rinsed in 0.1 M PBS, and dehydrated through 70%, 80%, 90%, and 100% ethanol. Sections were then incubated with 2 ng 35S-labeled antisense oligonucleotides in 100 μl hybridization buffer (20% SSC/10% dextrane sulfate/1× Denhardt's solution/50% deionized formamide/400 μg/ml denatured salmon testes DNA/0.3% 2-mercaptoethanol) at 37°C for 16–18 h. To demonstrate the specificity of the hybridization, control sections were hybridized with 2 ng 35S-labeled probe plus 200 ng (100-fold) unlabeled probe. After hybridization, slides were passed through three brief rinses in 1× SSC, followed by three 30-min stringent washes in 1× SSC at 55°C, with a final wash in 1× SSC at room temperature for 1 h. All washes contained 0.0001% 2-mercaptoethanol. Sections were rinsed in 300 mM ammonium acetate/70% ethanol and allowed to air dry. Dried sections were exposed to BioMax MR film (Kodak) for 7 days.
For cellular transcript localization slides were coated in Ilford K-5 nuclear tract emulsion. After exposure for 3 weeks at 4°C in light-tight boxes, sections were processed in Ilford phenisol developer for 5 min, washed in 2% chrome alum/2% sodium metabisuphite for 5 min, fixed in 30% (wt/vol) sodium thiosulphate for 10 min, and finally rinsed extensively in dH2O. Sections were then dried before being counterstained with methylene blue and coverslipped.
Statistical Analysis.
An unpaired Student's t test (two-tailed) was used for both cDNA array and real-time PCR data to detect significant changes in gene expression. Data transformation (log 10) was performed if the individual subject group data exhibited a non-normal distribution or the SD between groups were significantly different as detected by F test statistics.
Analysis of covariance was used to investigate the possible confounding effects of a number of characteristics on the relationship between gene expression and the main effect of clinical diagnosis.
Results
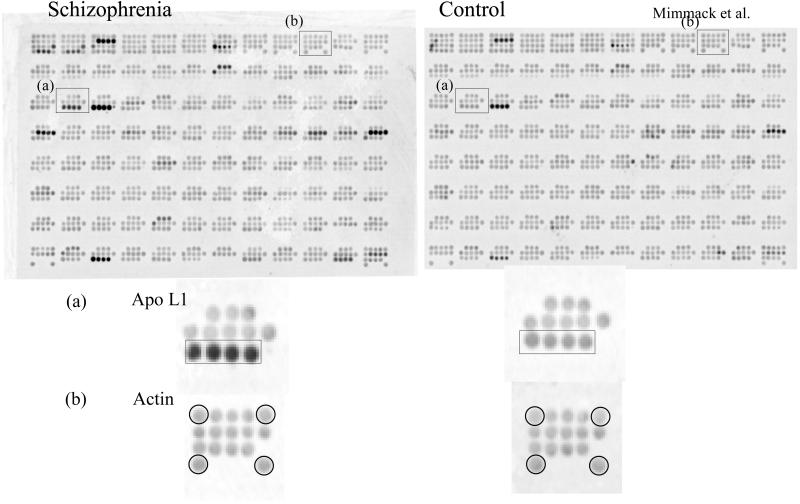
A custom-made cDNA array consisting of more than 300 schizophrenia candidate genes was screened with cDNA probes derived from the prefrontal cortex of 10 schizophrenia and 10 control samples taken from the New Zealand/Japan postmortem brain collection (see Methods). In total we identified 12 genes that showed a statistically significant change in expression above a cut-off set at ±1.4-fold. Among those, the apo L1 gene showed a robust up-regulation of 2.6-fold overall in schizophrenia (P = 0.03) (Fig. 1).
Figure 1.
Results of hybridization screening of nylon membrane cDNA arrays with radioactively labeled first-strand cDNA derived from control and schizophrenia prefrontal cortex RNA. The expression of apo L1 (a) and the housekeeping gene actin (b) are highlighted. Additional differentially expressed genes were detected after normalization procedures.
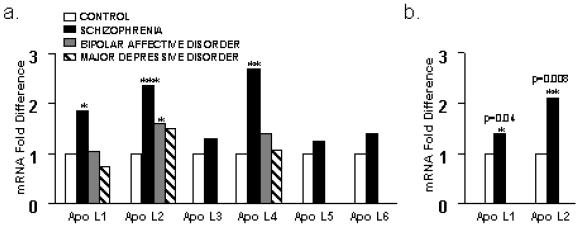
We went on to cross-validate this finding on an additional independent set of brains by using an independent method. Apo L1 expression was validated blind by using real-time PCR on the Stanley Foundation brain collection consisting of 60 brain samples comprising 15 samples in each group of schizophrenia, bipolar disorder, depression, and normal controls (see Methods). The breaking of the blind code by the Stanley Foundation showed that apo L1 was increased in the schizophrenia group by an average of 1.85-fold, which was statistically significant (P = 0.01) (Fig. 2a, Tables 1 and 2). In a next step we validated apo L1 expression on an additional 20 schizophrenia and 20 control samples from the New Zealand/Japan brain collection (including the 10 schizophrenia and 10 control samples from the array screen). This brain collection is less well matched and in contrast to the Stanley brain collection consists of predominantly elderly, chronically hospitalized patients (see Methods). Nevertheless, we could confirm a significant 1.4-fold increase of apo L1 expression in schizophrenia (P = 0.04); however, real-time PCR values showed overall more variability, particularly in the oldest individuals (Fig. 2b).
Figure 2.
(a) Relative mRNA fold differences in apo L gene expression (apo L1–L6) in the prefrontal cortex of control, schizophrenia, bipolar, and major depression patients as determined by real-time PCR on the Stanley Foundation brain collection. Note that the bipolar group shows up-regulation of apo L1, L2, and L4 genes, which reaches significance for apo L2 gene expression. Significant differences are indicated by * as determined by unpaired Student's t test (two-tailed). See Tables 1 and 2 for details. (b) Relative mRNA fold differences in gene expression in the prefrontal cortex of apo L1 and L2 determined in 20 schizophrenia and 20 control samples (Japan, New Zealand). Significant differences are indicated by P values and * as determined by unpaired Student's t test (two-tailed).
Table 1.
Fold differences of apo L gene expression in the Stanley Foundation brain collection: Comparison of psychiatric subject groups with normal controls
| Gene | Schizophrenia
|
Bipolar disorder
|
Major depressive disorder
|
|||
|---|---|---|---|---|---|---|
| Fold | P value | Fold | P value | Fold | P value | |
| ApoL1 | 1.85 | 0.13 | 1.07 | 0.69 | −1.07 | 0.69 |
| ApoL2 | 2.37 | 0.00004 | 1.60 | 0.05 | 1.53 | 0.17 |
| ApoL3 | 1.30 | 0.12 | NA | NA | ||
| ApoL4 | 2.70 | 0.009 | 1.40 | 0.39 | 1.08 | 0.77 |
| ApoL5 | 1.25 | 0.24 | NA | NA | ||
| ApoL6 | 1.41 | 0.12 | NA | NA | ||
The Stanley Foundation brain collection comprising 15 well-matched schizophrenia, bipolar, depression, and control samples was used to determine the mean percentage differences for each apo L gene (apo L1–apo L6). P values are shown, and P values < 0.05 were considered significant for unpaired Student's t tests (two-tailed). NA denotes subject groups not analyzed.
Table 2.
Real-time PCR analysis of gene expression for apo L1 and apo L2 in individual control and schizophrenic subjects (2)
| Control group | ΔCt | mRNA level (×1,000) | Schizophrenia group | ΔCt | mRNA level (×1,000) |
|---|---|---|---|---|---|
| 1 | 10.49 | 0.69 | 1 | 9.02 | 1.92* |
| 2 | 10.76 | 0.57 | 2 | 10.03 | 0.95 |
| 3 | 10.23 | 0.83 | 3 | 8.64 | 2.51 |
| 4 | 10.82 | 0.55 | 4 | 9.71 | 1.19 |
| 5 | 11.46 | 0.35 | 5 | 9.35 | 1.53 |
| 6 | 10.45 | 0.71 | 6 | 8.92 | 2.06* |
| 7 | 11.84 | 1.27 | 7 | 9.25 | 1.64 |
| 8 | 8.96 | 2.01 | 8 | 8.88 | 2.12* |
| 9 | 9.71 | 1.19 | 9 | 8.33 | 3.11 |
| 10 | 9.16 | 1.74 | 10 | 8.67 | 2.45 |
| 11 | 9.52 | 1.36 | 11 | 8.59 | 2.59 |
| 12 | 10.60 | 0.64 | 12 | 8.68 | 2.43 |
| 13 | 10.30 | 0.79 | 13 | 9.47 | 1.41 |
| 14 | 11.06 | 0.47 | 14 | 9.96 | 1.00 |
| 15 | 9.35 | 1.53 | 15 | 8.51 | 2.74 |
| Av | 10.31 | 0.91 | Av | 9.06 | 1.97 |
The ΔCt value was derived by normalizing mRNA transcript levels against the housekeeping gene GAPDH. Each ΔCt was produced by the mean values taken for two independent replicates. Note that a higher ΔCt value indicates lower expression levels (a ΔCt decrease of 1 corresponds to a 2-fold increase in gene expression). The mRNA level value was obtained by the equation 2−ΔCt.
denotes schizophrenia patients who were not treated with antipsychotics at time of death.
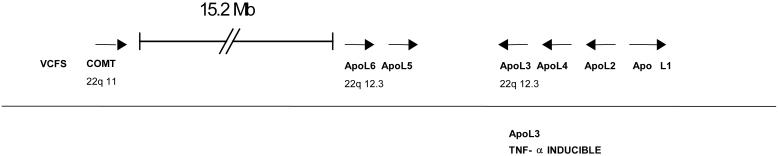
The apo L1 gene is part of a related gene family that is arranged in a cluster on chromosome 22q12.3 (Fig. 3) (11). To establish expression levels of the other five apo L genes (apo L2–L6), we performed real-time PCR assays on the Stanley Foundation samples, comparing expression levels in the 15 schizophrenia brains with the normal controls. Interestingly, apo L2 and L4 were also highly significantly up-regulated at 2.4-fold (P = 0.0004) and 2.8-fold (P = 0.009), respectively (Fig. 2a, Tables 1 and 2). The extremely significant up-regulation of apo L2 was validated on the New Zealand/Japan brain collection of 20 schizophrenia and 20 control brains, giving a 2.1-fold increase of apo L2 (P = 0.008) (Fig. 2b). It is worth noting that the apo L1 and L2 genes are highly homologous (82% sequence and 67% amino acid identity, respectively) and are expressed at high levels (compared with GAPDH), whereas apo L4 transcripts are relatively less abundant in the prefrontal cortex.
Figure 3.
Schematic diagram showing the genomic organization of members of the apo L gene family (apo L1–L6) on human chromosome 22q12.3. Linkage of schizophrenia to chromosome 22q11–22q13 has been reported and was replicated in several independent studies. Note the proximity of the apo L cluster to the loci for velocardiofacial syndrome (VCFS) and catechol-o-methyltransferase (COMT) (see text).
Analysis of covariance showed no correlations with potential confounds for the observed gene up-regulation for either apo L1 and L2 in both schizophrenia and bipolar disorder. All F statistics for covariates fell well short of statistical significance: in particular, lifetime drug exposure (F = 0.24 for apo L1 and F = 0.14 for apo L2), number of years of antipsychotic treatment (F = 0.04 for apo L1 and F = 0.07 for apo L2), and type of antipsychotic (typical versus atypical antipsychotics) at time of death (F = 0.11 for apo L1 and F = 0.12 for apo L2). We also considered postmortem delay, age, sex, brain pH, alcohol, and illicit drug use (past and at time of death) as covariates. Again, there was no significant correlations with potential confounds for the observed gene up-regulation for apo L1 and L2.
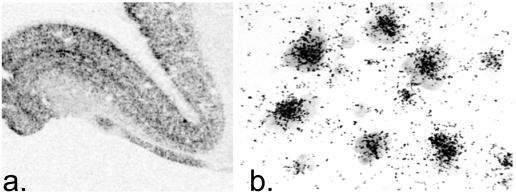
It has previously been reported that some of the apo L genes are expressed in the brain; however, the cellular distribution has not been investigated so far. We therefore examined the expression patterns of apo L1 and L2 by using in situ hybridization on control tissue showing a pan-neuronal expression in the human prefrontal cortex (Fig. 4).
Figure 4.
Autoradiogram (a) and low-power photomicrograph (b) illustrating the expression of apo L1 transcripts in the prefrontal cortex of a control individual. Apo L1 transcripts appear to be expressed in all neurons at high levels. The apo L2 gene also showed a neuronal distribution of expression (not shown).
Discussion
We report up-regulation of three members of the apo L gene family (apo L1, L2, and L4) that was consistent in schizophrenia compared with controls, within and between groups. Independent analysis with cDNA arrays was followed by cross-validation with quantitative real-time PCR on tissue taken from the prefrontal cortex of two independent tissue collections obtained from three countries. The sample size comprising altogether 35 schizophrenia and 35 control brains as well as 15 bipolar disorder and 15 depression brains demonstrated a reproducible up-regulation of apo L1 and apo L2 in patients with schizophrenia.
We cannot completely rule out drug treatment as a confounding factor for changes in gene expression. However, several lines of evidence suggest that our observations are more likely to be associated with the disease process itself, rather than being secondary to antipsychotic treatment. First, the three individuals with schizophrenia who had received no medication by the time of death showed an even greater increase of apo L1 and L2 gene expression than did drug-treated schizophrenia patients (Table 2). Second, bipolar disorder patients who had been treated with antipsychotic drugs showed no significant increase in apo L1 and apo L4 expression and only marginally significant up-regulation of apo L2 (Fig. 3). Finally, analysis of covariance showed no relationship between gene expression and several summary measures of drug exposure, namely lifetime drug exposure, number of years of antipsychotic, and type of antipsychotic (typical versus atypical antipsychotics) at time of death.
Interestingly, the apo L genes are clustered on chromosome 22q12 (Fig. 3). Several reports have suggested linkage of this chromosome region to schizophrenia as well as, although to a lesser degree, bipolar disorder (12–14). Furthermore, this chromosome region is of interest because the velocardiofacial syndrome (VCFS) is located close by on chromosome 22q11 (Fig. 3) (15). About one-third of VCFS cases experience schizophrenia-like psychoses. A recent study also reported a significant association between catechol-o-methyltransferase (COMT) genotype and abnormal prefrontal cortical function; COMT is also located on chromosome 22q11 (Fig. 3) (16).
The function of apo L proteins in the brain is unknown. Using in situ hybridization we found that apo L1 and L2 show pan-neuronal expression in the prefrontal cortex (Fig. 4). It is known that apo L proteins are functionally associated with apo A1, the major protein of high density lipoprotein and of importance in cholesterol transport (8). The central nervous system contains almost a quarter of the unesterified cholesterol present in the whole body. These sterols are largely located in two pools, namely the membranes of glial cells and neurons and in myelin sheaths. The cholesterol in the brain is almost exclusively synthesized in situ (7). Abnormalities in cholesterol turnover have been associated with a number of neurodegenerative disorders such as Alzheimer's disease and Niemann–Pick disease (6, 17). Numerous previous studies have reported alterations in genes and proteins associated with lipid metabolism in schizophrenia, for example, changes in phospholipid content, fatty acid content, and cholesteryl esters (18). Elevation of apo D in the prefrontal cortex in both schizophrenia and bipolar disorder has been observed (19). Furthermore, a recent microarray study found that several myelin-related genes are abnormally expressed in the prefrontal cortex of schizophrenia, and it is feasible that our observations fit into the picture of myelin dysregulation in schizophrenia (20). Such abnormalities could account for the longitudinal aspect of the schizophrenia phenotype, which manifests with multiple abnormalities long before the onset of actual psychosis (21).
However, lipoproteins have a role in a wide variety of other biological functions. High density lipoprotein has antiviral activity and has been shown to inhibit HIV infectivity and virus-induced syncytia formation. Apo A1 has also been found to inhibit herpes simplex virus-induced cell fusion at physiological concentrations and reduces viral spread (22, 23). Apos are engaged in the regulation of protease activity and calcium signaling and in intercellular signaling during embryonic development (6, 24, 25). Some lipoprotein receptors are known to act as receptors for Reelin, a protein that plays an important role in cortical lamination and has been shown to be down-regulated in the prefrontal cortex in schizophrenia (26). Lipoproteins also play an important role in axonal growth. Posse de Chaves et al. (27) recently demonstrated that distal axons of cultured sympathetic neurons internalize proteins and lipids from lipoproteins and transport them retrogradely to the cell body.
Some evidence for the potential role of apo L proteins comes from differential display studies investigating cytokine-induced gene expression changes in endothelial cells (28). The authors found that apo L3 expression was up-regulated by either monocyte-conditioned medium or tumor necrosis factor α (TNF-α), suggesting a role for apo L in inflammatory processes. The dysregulation of the inflammatory response system has been linked to the pathophysiology of schizophrenia (29). TNF-α is a proinflammatory cytokine that can exert neurotrophic and neurotoxic effects (30). The TNF-α gene is located on chromosome 6p21, another high-susceptibility locus for schizophrenia (31). A recent study found the TNF2(A) allele significantly increased in schizophrenia patients (32). Further investigations are required to establish whether other apo L genes are regulated by TNF-α directly or by other proteins in the TNF-α signal transduction and transcription pathway.
In summary, we have shown that apo L1, L2, and L4 are significantly up-regulated in the prefrontal cortex of schizophrenia patients. The very large sample size of 100 brains in total, the consistency of our findings within disease groups, across independent brain collections and between independent techniques, as well as the lack of association with antipsychotic drug exposure lead us to be confident in suggesting that abnormalities in the expression of these genes may be involved in the genesis of schizophrenia.
Acknowledgments
We thank Drs. John Coadwell and Simon Andrews for generous and invaluable bioinformatics support. Thanks to Dr. Eurof Walters for his support with statistical data analysis; Wendy Brooks for assistance in array preparation; Prof. Ed Bullmore and Drs. Lawrence Wilkinson and Nick Allen for useful comments and discussions; and Dr. Nigel Miller for help in figure preparation. Thanks to the New Zealand Neurological Foundation Human Brain Bank (supported by grants to R.L.M.F. from the New Zealand Neurological Foundation and the Health Research Council of New Zealand) for providing high-quality brain tissue. This research was supported by the Theodore and Vada Stanley Foundation and the donations of the Stanley Foundation Brain Collection courtesy of Drs. Michael B. Knable, E. Fuller Torrey, Maree J. Webster, and Robert H. Yolken, who also provided much encouragement and valued discussions; the Biotechnology and Biological Sciences Research Council, and the National Alliance for Research on Schizophrenia and Depression. P.C.E. is the 2001 Lattner Investigator (National Alliance for Research on Schizophrenia and Depression). The Department of Psychiatry, Univ. of Cambridge gratefully acknowledges center support from the Stanley Foundation.
Abbreviations
- apo
apolipoprotein
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- TNF-α
tumor necrosis factor α
References
- 1.Bullmore E T, Brammer M J, Rabe-Hesketh S, Curtis V A, Morris R G, Williams S C, Sharma T, McGuire P K. Hum Brain Mapp. 1999;7:38–48. doi: 10.1002/(SICI)1097-0193(1999)7:1<38::AID-HBM4>3.0.CO;2-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benes F M, Davidson J, Bird E D. Arch Gen Psychiatry. 1986;43:31–35. doi: 10.1001/archpsyc.1986.01800010033004. [DOI] [PubMed] [Google Scholar]
- 3.Benes F M, Vincent S L, Alsterberg G, Bird E D, SanGiovanni J P. J Neurosci. 1992;12:924–929. doi: 10.1523/JNEUROSCI.12-03-00924.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinberger D R, Berman K F, Zec R F. Arch Gen Psychiatry. 1986;43:114–124. doi: 10.1001/archpsyc.1986.01800020020004. [DOI] [PubMed] [Google Scholar]
- 5.Bertolino A, Esposito G, Callicott J H, Mattay V S, Van Horn J D, Frank J A, Berman K F, Weinberger D R. Am J Psychiatry. 2000;157:26–33. doi: 10.1176/ajp.157.1.26. [DOI] [PubMed] [Google Scholar]
- 6.Danik M, Champagne D, Petit-Turcotte C, Beffert U, Poirier J. Crit Rev Neurobiol. 1999;13:357–407. doi: 10.1615/critrevneurobiol.v13.i4.20. [DOI] [PubMed] [Google Scholar]
- 7.Dietschy J M, Turley S D. Curr Opin Lipidol. 2001;12:105–112. doi: 10.1097/00041433-200104000-00003. [DOI] [PubMed] [Google Scholar]
- 8.Duchateau P N, Pullinger C R, Orellana R E, Kunitake S T, Naya-Vigne J, O'Connor P M, Malloy M J, Kane J P. J Biol Chem. 1997;272:25576–25582. doi: 10.1074/jbc.272.41.25576. [DOI] [PubMed] [Google Scholar]
- 9.Torrey E F, Webster M, Knable M, Johnston N, Yolken R H. Schizophr Res. 2000;44:151–155. doi: 10.1016/S0920-9964(99)00192-9. [DOI] [PubMed] [Google Scholar]
- 10.Herrero M T, Augood S J, Asensi H, Hirsch E C, Agid Y, Obeso J A, Emson P C. Brain Res Mol Brain Res. 1996;42:149–155. doi: 10.1016/s0169-328x(96)00157-x. [DOI] [PubMed] [Google Scholar]
- 11.Page N M, Butlin D J, Lomthaisong K, Lowry P J. Genomics. 2001;74:71–78. doi: 10.1006/geno.2001.6534. [DOI] [PubMed] [Google Scholar]
- 12.Schwab S G, Lerer B, Albus M, Maier W, Hallmayer J, Fimmers R, Lichtermann D, Minges J, Bondy B, Ackenheil M, et al. Am J Med Genet. 1995;60:436–443. doi: 10.1002/ajmg.1320600515. [DOI] [PubMed] [Google Scholar]
- 13.Myles-Worsley M, Coon H, McDowell J, Brenner C, Hoff M, Lind B, Bennett P, Freedman R, Clementz B, Byerley W. Am J Med Genet. 1999;88:544–550. [PubMed] [Google Scholar]
- 14.Kelsoe J R, Spence M A, Loetscher E, Foguet M, Sadovnick A D, Remick R A, Flodman P, Khristich J, Mroczkowski-Parker Z, Brown J L, et al. Proc Natl Acad Sci USA. 2001;98:585–590. doi: 10.1073/pnas.011358498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy K C, Jones L A, Owen M J. Arch Gen Psychiatry. 1999;56:940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- 16.Egan M F, Goldberg T E, Kolachana B S, Callicott J H, Mazzanti C M, Straub R E, Goldman D, Weinberger D R. Proc Natl Acad Sci USA. 2001;98:6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holtzman D M, Bales K R, Tenkova T, Fagan A M, Parsadanian M, Sartorius L J, Mackey B, Olney J, McKeel D, Wozniak D, Paul S M. Proc Natl Acad Sci USA. 2000;97:2892–2897. doi: 10.1073/pnas.050004797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horrobin D F, Bennett C N. Prostaglandins Leukotrienes Essent Fatty Acids. 1999;60:141–167. doi: 10.1054/plef.1999.0027. [DOI] [PubMed] [Google Scholar]
- 19.Thomas E A, Dean B, Pavey G, Sutcliffe J G. Proc Natl Acad Sci USA. 2001;98:4066–4071. doi: 10.1073/pnas.071056198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hakak Y, Walker J R, Li C, Wong W H, Davis K L, Buxbaum J D, Haroutunian V, Fienberg A A. Proc Natl Acad Sci USA. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones P, Rodgers B, Murray R, Marmot M. Lancet. 1994;344:1398–1402. doi: 10.1016/s0140-6736(94)90569-x. [DOI] [PubMed] [Google Scholar]
- 22.Srinivas R V, Birkedal B, Owens R J, Anantharamaiah G M, Segrest J P, Compans R W. Virology. 1990;176:48–57. doi: 10.1016/0042-6822(90)90229-k. [DOI] [PubMed] [Google Scholar]
- 23.Martin I, Dubois M C, Saermark T, Ruysschaert J M. Biochem Biophys Res Commun. 1992;186:95–101. doi: 10.1016/s0006-291x(05)80780-6. [DOI] [PubMed] [Google Scholar]
- 24.Herz J. Neuron. 2001;29:571–581. doi: 10.1016/s0896-6273(01)00234-3. [DOI] [PubMed] [Google Scholar]
- 25.Gotthardt M, Trommsdorff M, Nevitt M F, Shelton J, Richardson J A, Stockinger W, Nimpf J, Herz J. J Biol Chem. 2000;275:25616–25624. doi: 10.1074/jbc.M000955200. [DOI] [PubMed] [Google Scholar]
- 26.Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer R E, Richardson J A, Herz J. Cell. 1999;97:689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 27.Posse De Chaves E I, Vance D E, Campenot R B, Kiss R S, Vance J E. J Biol Chem. 2000;275:19883–19890. doi: 10.1074/jbc.275.26.19883. [DOI] [PubMed] [Google Scholar]
- 28.Horrevoets A J, Fontijn R D, van Zonneveld A J, de Vries C J, ten Cate J W, Pannekoek H. Blood. 1999;93:3418–3431. [PubMed] [Google Scholar]
- 29.Muller N, Riedel M, Ackenheil M, Schwarz M J. Eur Arch Psychiatry Clin Neurosci. 1999;249:62–68. doi: 10.1007/pl00014187. [DOI] [PubMed] [Google Scholar]
- 30.Rothwell N J, Hopkins S J. Trends Neurosci. 1995;18:130–136. doi: 10.1016/0166-2236(95)93890-a. [DOI] [PubMed] [Google Scholar]
- 31.Peltonen L. Nature (London) 1995;378:665–666. doi: 10.1038/378665a0. [DOI] [PubMed] [Google Scholar]
- 32.Boin F, Zanardini R, Pioli R, Altamura C A, Maes M, Gennarelli M. Mol Psychiatry. 2001;6:79–82. doi: 10.1038/sj.mp.4000815. [DOI] [PubMed] [Google Scholar]
- 33.American Psychiatric Association. Diagnostic and Statistical Manual. 3rd Ed. Washington, DC: Am. Psychiatric Assoc.; 1987. revised. [Google Scholar]
- 34.American Psychiatric Association. Diagnostic and Statistical Manual. 4th Ed. Washington, DC: Am. Psychiatric Assoc.; 1994. [Google Scholar]