Abstract
The initial steps of spliceosomal small nuclear ribonucleoprotein (snRNP) maturation take place in the cytoplasm. After formation of an Sm-core and a trimethylguanosine (TMG) cap, the RNPs are transported into the nucleus via the import adaptor snurportin1 (SPN) and the import receptor importin-β. To better understand this process, we identified SPN residues that are required to mediate interactions with TMG caps, importin-β, and the export receptor, exportin1 (Xpo1/Crm1). Mutation of a single arginine residue within the importin-β binding domain (IBB) disrupted the interaction with importin-β, but preserved the ability of SPN to bind Xpo1 or TMG caps. Nuclear transport assays showed that this IBB mutant is deficient for snRNP import but that import can be rescued by addition of purified survival of motor neurons (SMN) protein complexes. Conserved tryptophan residues outside of the IBB are required for TMG binding. However, SPN can be imported into the nucleus without cargo. Interestingly, SPN targets to Cajal bodies when U2 but not U1 snRNPs are imported as cargo. SPN also relocalizes to Cajal bodies upon treatment with leptomycin B. Finally, we uncovered an interaction between the N- and C-terminal domains of SPN, suggesting an autoregulatory function similar to that of importin-α.
INTRODUCTION
A key feature of all eukaryotic cells is their ability to regulate the flow of macromolecules between various subcellular compartments. The nuclear envelope is one of the best examples of this type of cellular partitioning, because the nuclear pore complexes (NPCs) embedded within this structure allow for the selective transport of specific RNA and protein cargoes (reviewed in Rout and Aitchison, 2001; Suntharalingam and Wente, 2003; Pante, 2004). Individual cargoes contain nuclear localization signals (NLSs) and/or nuclear export signals (NESs), which are recognized by nuclear transport receptors collectively called karyopherins (reviewed in Fried and Kutay, 2003). Karyopherins can be divided into two subfamilies, called importins and exportins, depending on the direction of cargo transport (reviewed in Mosammaparast and Pemberton, 2004).
Despite their opposing directionalities, most importins and exportins are structurally related to importin-β (reviewed in Harel and Forbes, 2004). Importin-β family members are characterized by an N-terminal Ran binding domain and a series of HEAT repeats (reviewed in Andrade et al., 2001). The HEAT repeats interact with the FG-rich motifs present in most nucleoporins and allow for passage of cargo through the NPC. The direction of cargo transport is regulated by a small GTPase called Ran (Izaurralde et al., 1997). In the nucleus, Ran exists primarily in the GTP-bound state, whereas cytoplasmic Ran is predominantly GDP bound. Nuclear RanGTP promotes dissociation of importins from their cargoes and association of exportins with their substrates, thereby conferring directionality to the system (Görlich and Mattaj, 1996).
An additional group of adaptor proteins mediates cellular transport in cooperation with the importin-β superfamily. These adaptors facilitate transport of cargoes that cannot bind directly to a given receptor protein. For example, importin-α forms the bridge between most “classical” NLS motifs and importin-β (Adam and Gerace, 1991; Adam and Adam, 1994; Moroianu et al., 1995; Weis et al., 1995). The N-terminal region of importin-α contains an importin-β binding (IBB) motif, whereas the C-terminal domain mediates recognition of the NLS-containing cargoes (Görlich et al., 1996; Conti et al., 1998). Interestingly, the N-terminal IBB domain also contains a weak NLS that is thought to perform an autoregulatory function (Conti et al., 1998; Kobe, 1999). Thus, adaptor proteins such as importin-α must shuttle between the nucleus and the cytoplasm, binding cargo in one compartment and releasing it in the other. However, transport proteins are not the only factors known to shuttle.
Certain cargo proteins (e.g., cyclins, heterogeneous nuclear ribonucleoprotein [RNP] proteins) are known to contain both NLSs and NESs (reviewed in Dreyfuss et al., 2002), and these factors also shuttle between the nucleus and cytoplasm. Sm-RNPs represent a unique category of cargoes, because they are one of the few factors known to make two “one-way” trips, traveling from the nucleus to the cytoplasm and back again, albeit with significant remodeling on each leg of the circuit (reviewed in Will and Lührmann, 2001; Kiss, 2004). Interestingly, the RNA component of the RNP forms an integral part of the signals used for these transport events. Export of small nuclear (sn)RNA transcripts from the nucleus to the cytoplasm is mediated by specific factors that recognize the RNA pol II-encoded 7-methylguanosine (m7G) cap structure (Jarmolowski et al., 1994; Ohno et al., 2000; Masuyama et al., 2004). Once in the cytoplasm, snRNAs are assembled with core factors, called Sm proteins, forming a stable RNP. This process is mediated by the activity of the survival of motor neurons (SMN) protein complex (Meister et al., 2002; Yong et al., 2004). After Sm-core formation, the m7G cap is hypermethylated by an enzyme called Tgs1 to create a 2,2,7-trimethylguanosine (TMG) cap (Mouaikel et al., 2002, 2003; Verheggen et al., 2002). The TMG cap and the Sm core form two separable NLSs through which two independent import adaptors use the same import receptor, importin-β (Fischer et al., 1993; Marshallsay and Lührmann, 1994; Palacios et al., 1997). Snurportin1 (SPN) is the adaptor protein for the TMG cap-dependent pathway (Huber et al., 1998; Huber et al., 2002), whereas the SMN complex is required for the Sm-core pathway (Narayanan et al., 2004). Subsequently, importin-β exits the nucleus in a complex with RanGTP (Izaurralde et al., 1997; Hieda et al., 1999); whether components of the SMN complex are exported from the nucleus is unknown. Recycling of SPN is carried out by the export receptor exportin1 (Xpo1/Crm1; Paraskeva et al., 1999).
Human SPN is a 45-kDa protein that contains three known functional domains, consisting of an N-terminal IBB motif, a centrally located TMG cap binding domain, and an ill-defined region responsible for binding to Xpo1 (Figure 1A). The SPN N terminus shares significant similarity with the IBB domain of importin-α, but the TMG-binding domain is completely novel, with no obvious similarity to other RNA-binding proteins (Huber et al., 1998). Despite the fact that SPN binds to Xpo1 with high affinity, the protein lacks a discernible leucine-rich NES (Paraskeva et al., 1999). To better define the motifs within SPN that are important for its function, we undertook a mutational analysis of the protein. Using a combination of in vivo localization, in vitro binding, and nuclear transport assays, we identified specific residues within both the IBB and TMG domains that are required for proper SPN function, found evidence for trafficking of SPN to Cajal bodies, and identified a potential autoinhibitory interaction. Together, these studies provide important insight into role of SPN in the biogenesis of small nuclear RNPs.
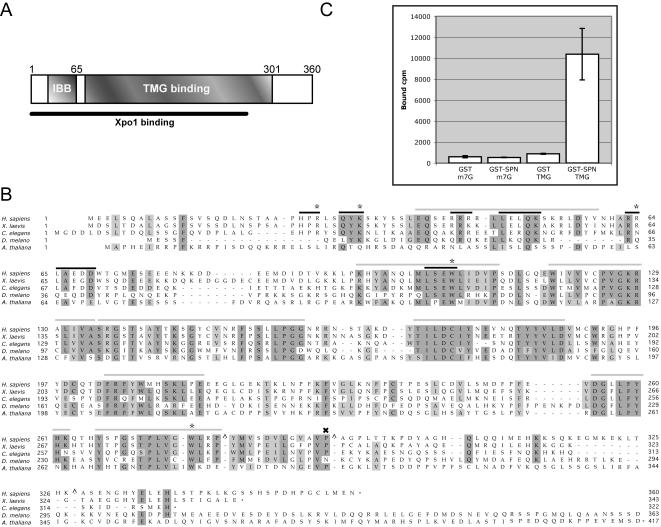
Figure 1.
Schematic of SPN, alignment of SPN orthologues, and TMG cap binding assay. (A) Cartoon of SPN illustrating the TMG cap, Xpo1, and IBB. The IBB of SPN is defined as amino acid residues 26–65, based on similarity with the IBB of importin-α (Huber et al., 1998). The region of SPN responsible for Xpo1 binding activity has not been mapped and may not be a modular domain (Huber et al., 1998; Paraskeva et al., 1999). Based on proteolytic cleavage of SPN and UV cross-linking studies, the TMG-binding domain is thought to span residues 79–301 (Strasser et al., 2004). (B) Alignment of SPN orthologues. Human, frog, worm, fly, and plant SPN proteins are aligned, with identities shaded dark and similarities shaded light. A subset of the mutations used in this study is illustrated. Asterisks indicate alanine point substitutions and include R27, K32, R64, W107, and W276. Black bars indicate block alanine substitutions and include 25–27, 30–32, 43–45, 48–52, 63–64, 65–69, and 104–107. Gray bars indicate residues that were deleted and replaced with an amino acid linker consisting of IVAGS and include 39–52, 96–112, 119–134, 135–159, 170–187, 203–214, 255–262, and 266–279. The X indicates residue P291 that was mutated to leucine. Note that this alignment does not include the predicted C terminus of the Drosophila melanogaster orthologue. Carats (^) mark sites where one or more amino acids were excluded to facilitate the alignment. (C) Recombinant SPN can distinguish between m7G- and TMG-capped RNA. GST pulldowns were conducted using GST or GST-SPN and radiolabeled m7G- or TMG-capped U2 snRNA. After a 1-h incubation at 4°C, complexes were washed and bound counts determined.
MATERIALS AND METHODS
Plasmid Construction and Mutant Generation
All deletions, single and block amino acid mutations as well as truncations were generated using the QuikChange mutagensis kit (Stratagene, La Jolla, CA); primer sequences are available on request. Deletions involved removal of the indicated amino acids along with insertion of an in-frame five-residue linker (5′-ATCGTCGCAGGATCC-3′) that includes a novel BamH I restriction site used for identification of positive clones. All constructs were subsequently sequenced throughout the entire SPN open reading frame. Primers containing BamH I and Not I restriction sites were used to PCR amplify human Xpo1 from Myc-Xpo1, and this fragment was subsequently cloned into pET 24b (Novagen, Madison, WI).
Protein Purification
Glutathione S-transferase (GST)- and His-tagged proteins were expressed in the Escherichia coli strain BL-21 Star (DE3) (Invitrogen, Carlsbad, CA). Cells were grown at 37°C to an optical density at 600 nm of 0.6, followed by induction with 1 mM isopropyl β-d-thiogalactoside (Sigma-Aldrich, St. Louis, MO). Cells were induced at 30°C for 2 h except for cells expressing RanQ69L (gift from K. Weis, Department of Molecular and Cell Biology, University of California, Berkeley, CA), which were induced at 25°C for 4 h. GST- and His-tagged constructs were purified using either glutathione beads (GE Healthcare, Piscataway, NJ) or Ni-NTA agarose beads (QIAGEN, Valencia, CA) as per the manufacturers' instructions. RanQ69L was purified as described previously (Klebe et al., 1993; Nilsson et al., 2001) and loaded with GTP as described previously (Askjaer et al., 1998).
Generation of Radiolabeled RNA
A plasmid containing an Ascaris U2 snRNA gene driven by a T3 promoter (gift of T. Nilsen, Center for RNA Molecular Biology, Case Western Reserve University, Cleveland, OH) was linearized with Sma I. Linearized DNA was then purified by phenol/chloroform extraction, resuspended in TE buffer, and used to generate single-stranded RNA. In vitro transcription using the Riboprobe system (Promega, Madison, WI) was then conducted in the presence of radiolabeled UTP, and m7G- or TMG-cap analogs (as directed), and resulting RNA was purified using Bio-Spin Tris columns (Bio-Rad, Hercules, CA). One microgram of GST or GST-tagged protein was then incubated with 1.6 × 106 counts of RNA for 1 h at 4°C. Beads were then washed four times with mRIPA (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1% NP-40, 1 mM EDTA) containing 2 mM dithiothreitol (DTT) plus protease inhibitor cocktail tablets (Roche Diagnostics, Indianapolis, IN) and bound counts determined by an LS6500 scintillation counter (Beckman Coulter, Fullerton, CA).
GST-Pulldown Assays
E. coli lysates containing GST, GST-SPN, or mutant SPN were incubated with glutathione beads for 1 h at 4°C and washed two times with 1× phosphate-buffered saline (PBS). All pulldowns used 1 μg of GST-SPN except for experiments involving Xpo1, which used 2 μg. Glutathione-bead captured GST, GST-SPN, or mutant SPN was incubated with 1 μg of importin-β for 1 h at 4°C in 800 μl of mRIPA buffer. Pulldowns using Xpo1 involved incubation of GST, GST-SPN, or mutant SPN with 150 μl of E. coli lysate expressing Xpo1-His for 3 h in the presence of 30 μg of RanQ69L-GTP. Leptomycin B (LMB; Calbiochem, San Diego, CA) was added at 20 nM to E. coli lysate 1 h preceding the addition to glutathione-bead captured GST-SPN. Reactions were incubated with gentle inversion for 1 h at 4°C and washed four times with 1 ml of mRIPA, resuspended in 10 μl of 5× SDS loading buffer, boiled, and analyzed by SDS-PAGE. After transfer to nitrocellulose, membranes were probed with the appropriate primary and secondary antibodies before chemiluminescence detection (Pierce Chemical, Rockford, IL). The assay shown in Figure 2B was conducted as described above except that a buffer described in Paraskeva et al. (1999) was used. This reaction was incubated and washed in 50 mM HEPES-KOH, pH 7.5, 200 mM NaCl, 5 mM Mg(OAc)2, and 0.005% digitonin.
Figure 2.
SPN mutants defective in TMG-cap binding also fail to interact with Xpo1. (A) Mutation of residue W107 or residues 104–107 abolish TMG binding. GST pulldowns were conducted using GST alone, GST-SPN, and the following GST-tagged SPN mutants: R27A, W107A, 104–107A, Δ119-134, Δ203-214, and P291L in the presence of radiolabeled, TMG-capped U2 snRNA. After incubation and washes, bound counts were determined using a scintillation counter. (B) SPN binding to Xpo1 is extremely sensitive to mutation. GST pulldowns were conducted using GST, GST-SPN (± LMB) and the following GST-tagged SPN mutants: 25-27A, R27A, 104-107A, W107A, Δ119-134, Δ203-214, W276A, and P291L in the presence of lysate expressing recombinant Xpo1-His and containing RanQ69L-GTP. Western blot analysis was conducted using anti-Xpo1 and anti-GST antibodies (loading control). Input shows 5% of the total lysate used in the pulldown.
Immunochemical Methods
HeLa-ATCC cells were cultured in DMEM (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum and penicillin/streptomycin (Invitrogen) to 70% confluence. Cells were harvested and electorporated using a GenePulser Xcell electroporator (Bio-Rad) as directed using 2 μg of DNA. Cells were then seeded on slides (Nalge Nunc International, Rochester, NY) for 16 h, fixed in 4% paraformaldehyde, and permeabilized in 0.5% Triton X-100 as described previously (Frey and Matera, 1995). Incubation in 10% normal goat serum preceded antibody detection. LMB at 20 nM was added to cell culture media for 1 h before cell fixation.
Solid Phase Binding Assays
Solid phase binding assays were performed essentially as described in Bednenko et al. (2003), with the following modifications. Two hundred nanograms of importin-β was adsorbed to each well and GST-SPN binding reactions were performed in PBS containing 0.2% NP-40 and 1% bovine serum albumin (BSA). GST-SPN was detected using an anti-GST antibody (GE Healthcare).
Import Assays
HeLa-ATCC cells were grown to 50% confluence on slides (Nalge Nunc International) and washed once with P buffer [50 mM HEPES-KOH, pH 7.5, 50 mM KOAc, 8 mM Mg(OAc)2, 2 mM EGTA, 1 mM DTT, and 1 μg/ml each aprotinin, leupeptin, and pepstatin]. Cells were permeabilized with digitonin in the presence of an ATP regenerating system (0.2 mg/ml BSA, 1 mM ATP, 10 mM creatine phosphate, 50 μg/ml creatine phosphokinase; Roche Diagnostics) plus 0.2 mM GTP for 5 min at 26°C. Cells were then washed twice and incubated in P buffer for 15 min at 26°C to remove endogenous transport factors. After two washes with P buffer, cells were transferred to T buffer [20 mM HEPES-KOH, pH 7.5, 80 mM KOAc, 4 mM Mg(OAc)2, 1 mM DTT, and 1 μg/ml each of aprotinin, leupeptin, and pepstatin] before performing the import assay. Import reactions were incubated at 26°C for 30–40 min. Unless specified otherwise, each reaction contained 0.2 mg/ml tRNA, 0.2 mg/ml BSA, 1 mM ATP, 10 mM creatine phosphate, 50 μg/ml creatine phosphokinase (Roche Diagnostics), and 40 nM Cy3-labeled U1 or U2 snRNPs (kind gift from R. Lührmann, Department of Cellular Biochemistry, Max-Planck-Institute of Biophysical Chemistry, Göttingen, Germany; Sumpter et al., 1992; Segault et al., 1995; Huber et al., 1998) and 800 ng each of green fluorescent protein (GFP)-SPN and importin-β. Purified SMN or control complexes (Pellizzoni et al., 2002; Narayanan et al., 2004) were a gift from G. Dreyfuss (Howard Hughes Medical Institute, Department of Biochemistry and Biophysics, University of Pennsylvania, Philadelphia, PA) and were used at 400 ng/assay. After incubating, cells were washed in transport buffer and then fixed in 4% paraformaldehyde for 10 min at room temperature and permeabilized with 0.5% Triton for 5 min. Cells were visualized by a Zeiss Axioplan upright epifluorescent microscope (100× objective). Digital images were taken with a Hamamstsu ORCA-ER C4742–95 charge-coupled device camera and Open Lab software (Improvision, Lexington, MA).
Antibodies
A rabbit polyclonal anti-coilin antibody (R124) was generated (Covance Research Products, Berkley, CA) using a His-tagged fragment of coilin consisting of the C-terminal 214 amino acids of human coilin. Mouse monoclonal anti-Xpo1 (BD Biosciences, San Diego, CA), rabbit polyclonal anti-Myc, and mouse monoclonal anti-GST (Santa Cruz Biotechnology, Santa Cruz, CA) were used at 1:5000, whereas (R124) was used at 1:600. A His-probe (Pierce Chemical) was used at 1:5000 to detect His-tagged proteins as per the manufacturer's instructions. Secondary antibodies used were goat anti-mouse- and goat anti-rabbit-conjugated horseradish peroxidase at 1:5000 (Pierce Chemical) and goat anti-rabbit-conjugated Texas Red (Vector Laboratories, Burlingame, CA).
RESULTS
Mutational Analyses Identify Residues Important for TMG Binding
SPN contains a small N-terminal IBB domain and a large centrally located TMG cap binding domain (Figure 1A). Although the central region of the protein is conserved among higher eukaryotes (Figure 1B), it does not share significant similarity with other known RNA binding domains. To gain insight into the process of snRNP import, we set out to define sequences that are critical for SPN function. As a first step in our analysis, we tested wild-type recombinant GST-SPN for its ability to bind to TMG capped snRNA. Radiolabeled U2 snRNAs were transcribed in vitro in the presence of either m7G or TMG cap dinucleotide triphosphates. As shown in Figure 1C, GST-SPN specifically recovered TMG- but not m7G-capped RNA, whereas only background levels of U2 RNA bound to GST alone. We conclude that, at least by this criterion, recombinant GST-SPN is a functional protein.
Previous studies used truncation mutants in attempts to map the various domains of SPN (Huber et al., 1998). Therefore, we generated a large battery of block substitution and internal deletion mutants in various conserved regions throughout the length of the SPN molecule and tested them for their ability to bind to TMG caps in vitro. The entire data set is summarized in Table 1. For comparison, the sequence conservation of the mutated regions is illustrated in Figure 1B. Because tryptophan and other aromatic residues are known to play important roles in binding to m7G caps (reviewed in Quiocho et al., 2000; Fechter and Brownlee, 2005), we paid special attention to conserved motifs containing such residues. Surprisingly, we found that nearly all of the deletion mutants abolished TMG binding (Figure 2A and Table 1). We therefore made a number of point and block substitution mutations in these conserved motifs (e.g., W107A, 104-107A, 203-207A, and W276A) and found that these also significantly reduced binding to TMG capped snRNAs (Figure 2A and Table 1). Two mutations bordering the TMG domain (Δ1-65 and P291L) disrupted TMG binding only slightly (Figure 2A and Table 1). Together with previous findings (Huber et al., 1998; Strasser et al., 2004), these results identify a minimal TMG binding domain, located between residues 100 and 280.
Table 1.
In vitro binding and in vivo localization studies
| SPN construct | Imp β | TMG | Xpo1 | Localization |
|---|---|---|---|---|
| Wild type | + | + | + | Cytoplasmic |
| (1-65) | +++ | n.d. | –a | Nucleoplasmic |
| (1-280) | ++ | n.d. | +/–b | Nuc+Cyto |
| R27A | – | +/– | + | Cytoplasmic |
| 25-27A | – | +/– | +/– | Cytoplasmic |
| 30-32A | + | n.d. | n.d. | n.d. |
| 43-45A | + | n.d. | n.d. | n.d. |
| 48-52A | ++ | n.d. | – | Nucleoplasmic |
| 63-64A | + | n.d. | n.d. | n.d. |
| 65-69A | + | n.d. | n.d. | n.d. |
| 104-107A | ++ | – | +/– | Nucleoplasmic |
| W107A | +c | – | +/– | Cytoplasmic |
| W107,276A | +c | – | +/– | Nuc+Cyto |
| 203-207A | +a | – | –a | Nucleoplasmic |
| W276A | +c | – | +/– | Cytoplasmic |
| P291L | ++ | +/– | + | Cytoplasmic |
| Δ1-65 | – | +b | –b | Nucleoplasmic |
| Δ39-52 | +/– | n.d. | +/– | Nuc+Cyto |
| Δ96-112 | ++ | – | – | Nucleoplasmic |
| Δ119-134 | + | – | +/– | Nucleoplasmic |
| Δ135-159 | + | n.d. | +/– | Nucleoplasmic |
| Δ170-187 | + | n.d. | – | Nucleoplasmic |
| Δ203-214 | + | – | +/– | Nucleoplasmic |
| Δ255-262 | + | n.d. | – | Nucleoplasmic |
| Δ266-279 | + | – | – | Nucleoplasmic |
Point Mutants in the TMG Domain Can Interact with Xpo1 In Vivo
After import of newly assembled snRNPs, SPN must be recycled to the cytoplasm to facilitate additional rounds of snRNP import. Recycling depends on the ability of SPN to interact with its export receptor, Xpo1 (Paraskeva et al., 1999). Despite the lack of a discernible NES, Xpo1 binds to SPN with 50-fold greater affinity than it does to leucine-rich NES-containing proteins such as HIV Rev (Paraskeva et al., 1999). We therefore tested whether the Xpo1 interaction with SPN was sensitive to LMB (Fornerod et al., 1997; Ossareh-Nazari et al., 1997; Kudo et al., 1998). Treatment with 20 nM LMB significantly reduced Xpo1 binding to GST-SPN (Figure 2B), suggesting that SPN binding uses the typical NES docking site on Xpo1. Using a similar pulldown assay, we also tested various mutant GST-SPN constructs for their ability to form a ternary export complex with Xpo1 and RanGTP in vitro. As shown in Figure 2B, the deletion mutants we tested were dramatically reduced for binding to Xpo1, although faint bands could be detected upon long exposures. Two of the TMG point substitution mutants also bound Xpo1 to a lesser degree; W276A was moderately impaired relative to wild-type SPN, whereas W107A displayed significantly reduced binding (Figure 2B).
To characterize the recycling capacities of these TMG domain mutants in vivo, we analyzed the steady-state subcellular distributions of various GFP-tagged constructs in the presence or absence of LMB. As expected, wild-type GFP-SPN localized to the cytoplasm and redistributed to the nucleoplasm upon treatment with LMB (Figure 3A). Despite their reduced capacities for Xpo1 binding in vitro, we found that the W107A and W276A constructs localized to the cytoplasm in untreated cells and relocalized to the nucleoplasm upon LMB treatment (Figure 3A). We therefore conclude that W107A and W276A functionally interact with Xpo1 in vivo. In contrast, block substitution or deletion mutations within the TMG domain resulted in proteins that did not bind Xpo1 in vitro and localized to the nucleus under steady-state conditions in vivo (Figure 3A and Table 1). The results suggest that each of the TMG domain mutants described above can bind to importin-β, because they were either nuclear in untreated cells or they relocalized to the nucleus after treatment with LMB (Table 1).
Figure 3.
In vivo localization of SPN to Cajal bodies depends upon TMG binding but not Xpo1 binding. (A) SPN point mutants reduced for Xpo1 binding in vitro can interact with Xpo1 in vivo. The subcellular localization of GFP-SPN as well as mutant constructs W107A, and W276A were studied after transient transfection of HeLa cells in the presence or absence of LMB. GFP-tagged constructs bearing deletions or block substitutions (Δ119-134, 104-107A, and Δ203-214) were found to be nucleoplasmic in the absence of LMB treatment. (B) Xpo1 and TMG binding mutants fail to accumulate in Cajal bodies. HeLa cells were transiently transfected with wild-type GFP-SPN, -SPN(W107A), or -SPNΔ203-214 and treated with 20 nM LMB for 1 h. Immunofluorescence was then conducted with anti-coilin antibodies to localize Cajal bodies. Arrows mark Cajal bodies in the wild-type panel. (C) Xpo1 is enriched in Cajal bodies in the absence of LMB and is depleted from these nuclear bodies upon treatment. HeLa cells were transiently transfected with Xpo1-GFP and treated with 20 nM LMB for 1 h. Immunofluorescence was then conducted with anti-coilin antibodies to localize Cajal bodies. Bar, 10 μm.
Cargo Binding Is Not a Requirement for SPN Nuclear Import
The fact that the TMG domain deletion mutants were nuclear suggested that SPN can be imported into the nucleus in the absence of RNA cargo. We decided to test this hypothesis by using the digitonin-permeabilized HeLa cell system (Adam et al., 1990). As shown in Figure 4, wild-type GFP-SPN and Cy3-labeled U1 snRNPs were efficiently imported into the nucleus when incubated with recombinant importin-β at 26°C. GFP-SPN also was imported in the absence of the labeled U1 snRNPs (Figure 4), although the level of nucleoplasmic signal was somewhat variable and localization to the nuclear rim was more pronounced (see Discussion). Thus, SPN can be imported into the nucleus in the absence of exogenous cargo. Because we cannot exclude that the protein was imported along with endogenous factors present in the permeabilized HeLa cells, we tested a TMG domain mutant in this assay. GFP-SPN(104-107A) was used for these studies, because this construct can bind neither TMG caps (Figure 2A) nor Xpo1 (Figure 2B) and is nuclear upon transfection into HeLa cells (Figure 3A). This substitution mutant was therefore used in a nuclear transport assay by incubating it in the presence of importin-β and Cy3-U1 snRNPs at 26°C. As shown in Figure 4, GFP-SPN(104-107A) was imported into the nucleus (albeit with a pronounced accumulation at the nuclear envelope), but the construct was completely defective in transporting snRNPs. These results not only demonstrate that TMG domain mutants are incapable of importing snRNPs but also reveal that SPN does not require an RNA cargo to access to the nucleus.
Figure 4.
SPN import does not require bound cargo. The ability of GFP-SPN or -SPN(104-107A) to mediate U snRNP import was examined using an in vitro nuclear transport assay. Recombinant importin-β and purified Cy3-labeled U1 snRNPs were incubated with either wild-type or mutant His-GFP-SPN constructs and digitonin-permeabilized HeLa cells. Top, GFP-SPN and U1 snRNPs were efficiently imported. Bottom, GFP-SPN(104-107A) was imported into the nucleus (with pronounced enrichment at the nuclear rim). However, the mutant construct failed to mediate U1 import. Middle, GFP-SPN was imported in the absence of added U snRNPs, showing variable degrees of nuclear accumulation; some cells displayed prominent rim staining, whereas others were more uniformly labeled. Import reactions were incubated at 26°C for 35 min. Bar, 10 μm.
Leptomycin B Causes SPN to Localize in Nuclear Cajal Bodies
As described above, short treatments with LMB resulted in a dramatic relocalization of GFP-SPN from the cytoplasm to the nucleus (Figure 3A). Most of the protein was distributed throughout the nucleoplasm, excluding nucleoli. However, we were surprised to find that wild-type SPN also accumulated in nucleoplasmic foci. Costaining with anti-coilin antibodies revealed that these foci were, in fact, Cajal bodies (Figure 3B). Interestingly, the LMB-induced accumulation of a given construct in Cajal bodies correlated with its ability to bind TMG-capped RNA. For example, the steady-state distribution of GFP-SPN(W107A) is primarily cytoplasmic in untreated cells, but the protein relocalizes to the nucleus after LMB treatment (Figure 3B). Consistent with its inability to bind TMG caps (Figure 2A), W107A did not accumulate in the Cajal bodies of LMB-treated cells. Likewise, W276A and all of the TMG-domain deletion mutants we tested were negative for Cajal body accumulation upon LMB treatment (Figure 3, A and B, and Table 1). Thus, only constructs that were capable of binding to TMG caps accumulated in Cajal bodies.
Previously, Boulon et al. (2004) showed that Xpo1 can be detected in Cajal bodies under steady-state conditions. Because SPN interacts with Xpo1, it was therefore possible that this interaction was responsible for tethering SPN to Cajal bodies during LMB treatment. We tested this idea by treating cells with LMB and then staining for Xpo1 and coilin. As shown in Figure 3C, Xpo1 localizes to Cajal bodies in untreated cells but fails to accumulate in them after LMB treatment. We therefore conclude that Xpo1 is not the factor that anchors SPN to Cajal bodies after inhibition of export.
SPN Accumulates in Cajal Bodies after Nuclear Import of U2 snRNPs
Based on the localization of newly synthesized GFP-tagged Sm proteins, snRNPs are thought to be imported into the nucleus and to transiently localize in Cajal bodies before proceeding on to speckles (Sleeman et al., 1999). The data in Figure 4 would seem to contradict this interpretation, because neither Cy3-labeled U1 snRNPs nor GFP-SPN seemed to accumulate in nuclear foci after the transport assay. However, Sleeman et al. (1999) also showed that postmitotic Sm proteins (snRNPs) bypass the Cajal body step and localize directly to nuclear speckles. Because the Cy3-labeled U1 snRNPs we used in our assays were purified from HeLa nuclei, they are more similar to postmitotic U1 snRNPs. Recent work strongly suggests that the final steps of U2 snRNP assembly take place in the Cajal body, involving addition of SF3a and SF3b to the maturing RNP and conversion from a 12S to a 17S particle (Will et al., 2002; Nesic et al., 2004; Tanackovic and Krämer, 2005). We therefore hypothesized that RNPs corresponding to 12S preU2 snRNPs might target to Cajal bodies. As shown in Figure 5, when Cy3-labeled U2 snRNPs (12S form) were used in the nuclear transport assay, localization of both SPN and U2 could clearly be detected in Cajal bodies. As expected, U1 snRNPs were imported into the nucleus but did not accumulate in Cajal bodies (Figure 5). These results demonstrate that SPN localizes to Cajal bodies after nuclear import of U2 snRNPs.
Figure 5.
SPN accumulates in Cajal bodies after import of U2 but not U1 snRNPs. The localizations of GFP-SPN and U snRNPs after snRNP import were determined using an in vitro nuclear transport assay. Recombinant importin-β and purified Cy3-labeled U1 or U2 snRNPs were incubated with wild-type His-GFP-SPN and digitonin-permeabilized HeLa cells. Cajal bodies were localized by immunofluorescence using an anti-coilin antibody. Bar, 10 μm.
A Conserved Arginine Residue Is Required for Binding to Importin-β
As part of our mutational analysis, various SPN constructs also were tested for their ability to bind importin-β. We found that neither the deletion nor the substitution mutations within the TMG binding domain had an effect on importin-β binding, with the exception of SPN(104-107A) and SPNΔ96-112, which bound slightly better than wild type (Supplemental Figure S1 and Table 1). We therefore concentrated our efforts within the SPN IBB domain and found that, as expected, deletion of the entire N-terminal domain (Δ1-65) abolished the interaction with importin-β (Figure 6A). However, a smaller deletion in the IBB (Δ39-52) had only a modest effect (Table 1). Intriguingly, certain alanine-scanning mutations of conserved regions within the IBB disrupted binding to importin-β (e.g., 25-27A), whereas others (e.g., 48-52A) enhanced the binding (Figure 6A and Table 1). The molecular implications of the SPN(48-52A) mutation will be discussed below. Given that SPN(25-27A) failed to bind to importin-β, the results suggest that this motif contains residue(s) necessary for the interaction.
Figure 6.
Mutants incapable of binding importin-β in vitro are efficiently imported in vivo. (A) Mutation of residue R27 disrupts SPN binding to importin-β. GST-pulldown assays were conducted using GST (negative control), GST-SPN, and the following SPN mutants: Δ1-65 (N-terminal deletion of 65 amino acids), R27A, 48-52A or P291L, along with recombinant His-myc-importin-β. Western analysis was conducted using anti-myc and anti-GST (loading control) antibodies. Input shows 10% of the total used in the pulldown. (B) Alignment of N-terminal regions of human importin-α and various SPN orthologues (human, Xenopus, and worm). Residues R27, K32, and R64 are marked with asterisks, regions 25–27, 30–32, 48–52, and 63–65 are overlined with bars. (C) Mutants deficient in importin-β binding are imported in vivo. HeLa cells were transiently transfected with wild-type GFP-SPN, or GFP-tagged SPN mutants 25-27A and R27A and treated with 20 nM LMB for 1 h. Immunofluorescence was then conducted with anti-coilin antibodies to localize Cajal bodies. Arrows indicate CBs in untreated cells. Bar, 10 μm.
Because the crystal structure of the IBB of importin-α complexed with importin-β has been solved (Cingolani et al., 1999), we compared the IBB domains of SPN and importin-α (Figure 6B). Notably, in the α/β cocrystal, three importin-α residues that make direct contacts with importin-β are conserved in human SPN (Figure 6B, asterisks). Mutation of only one of these regions (R27) disrupted binding; substitutions within motifs containing the other two residues (K32 and R64) had no effect (Table 1 and Figure 6A). We tested the GST-SPN(R27A) mutant and found that it fails to interact with importin-β in vitro (Figure 6A). To measure the apparent binding affinities (i.e., the relative Kd values) of the IBB mutants, we used a solid phase binding assay (Bednenko et al., 2003). In agreement with our qualitative analysis (Figure 6A), we found that both the (R27A) and (25-27A) mutations decreased the affinity of the IBB motif by roughly 20-fold (Supplemental Figure S2). The apparent affinities of importin-β for wild-type, (R27A) and (25-27A) SPN constructs were 4.7 ± 1.0, 92.5 ± 18.0, and 132.9 ± 28.5 nM, respectively.
Importantly, the (R27A) mutation had little effect on SPN′s ability to bind either TMG caps (Figure 2A) or Xpo1 (Figure 2B), suggesting that these functions were unperturbed. We therefore analyzed the subcellular localization of the two IBB mutants (25-27A and R27A) and found that they were similar to wild type (Figure 6C). Likewise, treatment with LMB demonstrated that the constructs were imported into the nucleus (presumably by an SMN-mediated pathway, see below) and subsequently exported to the cytoplasm by Xpo1 in vivo (Figure 6C). This latter finding was interesting because the 25-27A mutant bound poorly to Xpo1 in vitro (Figure 2B). We also noted that each of the IBB mutant constructs accumulated in Cajal bodies upon LMB treatment (Figure 6C), as predicted by their ability to bind TMG-capped RNAs in vitro (Figure 2A and Table 1). Having successfully identified a mutant that interacts with Xpo1 but is incapable of binding to importin-β, we next tested the GFP-SPN(R27A) construct in a nuclear transport assay.
SPN(R27A) Is Defective in snRNP Import
Together with purified Cy3-labeled U1 snRNPs, GFP-tagged SPN constructs were assayed for import using recombinant importin-β and digitonin-permeabilized HeLa cells. As shown in Figure 7, the import of both GFP-SPN and Cy3-U1 was robust when cells were incubated at 26°C for 35 min. Strikingly, GFP-SPN(R27A) was incapable of supporting U1 import (Figure 7). Thus, despite the fact that SPN(R27A) can bind TMG-capped snRNAs, the mutant was defective for snRNP import in vitro. Significantly, we found that import of both snRNPs and SPN(R27A) could be rescued by addition of purified SMN complexes (Figure 7). When control protein complexes were used, or if importin-β was left out of the reaction, neither SPN(R27A) nor snRNPs were imported (Figure 7). These studies provide an explanation for the nuclear localization of SPN(R27A) upon LMB treatment in vivo (Figure 6C), demonstrating that SPN binding to the TMG cap does not interfere with the SMN-mediated, cap-independent snRNP import pathway (Narayanan et al., 2004). Furthermore, the results indicate that the interaction between SPN and importin-β is not required to stabilize binding of the SMN complex to importin-β.
Figure 7.
SPN binding to the TMG cap does not interfere with cap-independent import. Digitonin-permeabilized HeLa cells were incubated at 26°C for 35 min with purified Cy3-labeled U1 snRNPs, recombinant importin-β and either GFP-SPN or GFP-SPN(R27A) in the presence or absence of purified SMN complexes. Where indicated importin-β was omitted from the import reaction. Note that neither GFP-SPN(R27A) nor Cy3-U1 snRNPs were imported in the absence of added SMN complexes. Both were imported when 400 ng of purified SMN complexes were added along with T buffer and an ATP regenerating system (see Materials and Methods). Bar, 10 μm.
SPN Subdomains Form an Intramolecular Interaction
During the course of our importin-β binding studies, we recovered two SPN mutations that actually increased importin-β binding, relative to wild-type (Figure 6A). One such mutation was within the IBB (48-52A), whereas the other was in the C terminus (P291L). One possibility suggested by these observations is that the C terminus of the protein adopts a conformation that partially sequesters the N terminus, thereby reducing access to importin-β. We therefore truncated the C-terminus of the protein and assayed binding to importin-β relative to wild type. We generated two constructs, one truncating the entire C terminus SPN(1-65), whereas the other removed the last 80 aa, SPN(1-280). Notably, both constructs (Figure 8A and Supplemental Figure S1) bound importin-β to a greater extent than either wild-type or the P291L mutant.
Figure 8.
SPN N- and C-terminal domains interact. (A) The binding of SPN to importin-β is increased upon mutation or truncation of the C terminus. Pulldown analysis was conducted using GST, GST-SPN, GST-SPN(1-65), or GST-SPN(P291L) and recombinant importin-β. The pulldowns were analyzed by Western blot and probed with anti-myc antibody or anti-GST as the loading control. Input shows 10% of the total used in the pulldown. (B) The N-terminal IBB domain and C terminus of SPN interact. GST pulldown assays were conducted using GST (negative control), GST-SPN(1-65), or GST-SPN(1-65) containing the 48-52 mutation, referred to as GST(1-65,48-52A). Lysates containing recombinant His-SPN(65-360), referred to as SPN-Cter(wt), or His-SPN(65-360) containing the P291L mutation, referred to as SPN-Cter(P291L) were used in the binding experiments. Western blot analysis was conducted and the blot probed with a His-probe or anti-GST antibody (loading control). Inputs show 1% of the total lysate used in the pulldown.
To directly assay for cross-talk between SPN subdomains, we generated differentially tagged N- and C-terminal SPN fragments. Recombinant His-SPN(65-360) was incubated with GST-SPN(1-65). Western analysis demonstrated that these two fragments interacted in trans (Figure 8B, lane 3). Control assays with GST-alone were negative (Figure 8B, lanes 1 and 2). We reasoned that in the context of the full-length protein, substitution mutations that increased binding to importin-β in cis (i.e., 48-52A and P291L) did so by disrupting a putative intramolecular interaction between elements located in the N and C termini. Such a disruption might allow the SPN IBB domain to adopt a more “open” conformation. These substitution mutations should also disrupt the ability of isolated N and C termini to interact in trans. We therefore tested this prediction by introducing these substitution mutations within the N- and C-terminal fragment backbones and found that they abolished the interaction (Figure 8B, lanes 4 and 5). Thus, mutations that stimulate importin-β binding in the context of full-length SPN disrupted association between the N- and C-terminal domains of SPN supplied in trans. These data indicate that the C terminus of the protein can attenuate the affinity of SPN for importin-β by sequestering the SPN IBB domain.
DISCUSSION
In vitro, Sm-class snRNPs can be imported into the nucleus via two separate importin-β-dependent pathways (Fischer et al., 1993; Marshallsay and Lührmann, 1994; Palacios et al., 1997). One pathway depends on the presence of a TMG cap and is mediated by SPN (Huber et al., 1998, 2002), whereas the other is cap-independent and relies upon the SMN complex (Narayanan et al., 2004). Previously, we showed that truncation of the entire IBB domain of SPN resulted in a protein that localizes primarily to the nucleus (Table 1; Narayanan et al., 2002). Thus, in vivo, the domain through which SPN is thought to be imported is actually dispensable for import. However, the N terminus of SPN also is required for binding to Xpo1 in vitro (Table 1; Paraskeva et al., 1999), suggesting that the IBB truncation construct (GFP-SPNΔ1-65) is able to access an alternative import pathway. Because SPNΔ1-65 retains the ability to bind TMG caps (Huber et al., 1998), we theorized that import of the mutant protein was facilitated by an import signal present on newly assembled snRNPs (Narayanan et al., 2002). We tested this hypothesis directly, by creating an SPN mutant (R27A) that can bind to TMG caps and Xpo1 (Figure 2), but not to importin-β (Figure 6A). Interestingly, this protein relocalizes from the cytoplasm to the nucleus upon LMB treatment (Figure 6C), suggesting that SPN(R27A) is imported together with snRNPs via the cap-independent, Sm-core import pathway. Consistent with this interpretation, in vitro transport assays showed that import of the R27A mutant depended upon addition of the SMN complex and importin-β (Figure 7). Thus, SPN(R27A) is able to bind to the TMG cap and “piggyback” into the nucleus via the cap-independent snRNP import pathway.
Cargo Binding and the Directionality of snRNP Transport
Transport adaptors such as SPN or importin-α shuttle continuously between the nucleus and cytoplasm. In importin-α, the high nuclear concentration of RanGTP is critical for release of importin-β from the nuclear side of the NPC during import, whereas import of SPN-bound complexes can be achieved in the absence of Ran (Huber et al., 2002). Our data reveal that cargo binding is not a requirement for SPN import (Figures 3 and 4) and that internal deletion/substitution mutations disrupting TMG-cap binding also inhibited Xpo1 binding (Figure 2). Thus, we conclude that SPN nucleocytoplasmic shuttling is relatively insensitive to the presence of cargo and that the mutually exclusive nature of TMG versus Xpo1 binding (Paraskeva et al., 1999 and Table 1) provides a mechanism by which newly imported snRNPs are prevented from being reexported to the cytoplasm.
On arrival in the nucleus, newly assembled snRNPs are thought to target to Cajal bodies before proceeding on to their final nucleoplasmic destinations (reviewed in Kiss, 2004). Whether SPN accompanies all U snRNP import complexes to Cajal bodies or not is unclear, however, we found that the protein accumulated in these structures when 12S U2 snRNPs were used as import cargoes or when export was blocked with LMB. Curiously, similar experiments with U1 snRNPs revealed that neither U1 nor SPN targeted to Cajal bodies after nuclear import in vitro. The mechanistic underpinnings of this difference will be a subject of future investigation.
The molecular mechanism that triggers cargo release from SPN in the nucleoplasm is not well understood. Huber et al. (2002) showed that RanGTP is not required for SPN translocation across the nuclear pore. However, Ran could still play a role in release of cargo or in dissociation of the import complex. In this context, it is important to note that Ran(Q69L)GTP destabilizes complexes between importin-β and either wild-type (Paraskeva et al., 1999) or mutant SPN constructs (Supplemental Figure S3). It is possible that cargo dissociation might even be facilitated by factors present in Cajal bodies, such as Xpo1. Under steady-state conditions, GFP-SPN(25-27A) localized in both the cytoplasm and Cajal bodies (Figure 6C). Thus, SPN can bind to TMG-capped RNAs while in the nucleus. Given that SPN(25-27A) is slightly defective in binding to Xpo1, perhaps perturbation of SPN recycling results in its accumulation in these structures. Whether the interaction with importin-β plays a role in modulating SPN′s affinity for TMG cargo is also unknown. Future experiments will be required to address these issues.
While this manuscript was under revision, Strasser et al. (2005) reported the crystal structure of the TMG-binding domain of human SPN and showed that two tryptophan residues (W107 and W276) make important contacts with the TMG cap binding pocket. Strasser et al. (2005) show that the structure of the TMG domain is primarily composed of two nearly coplanar β sheets, and the TMG pocket is located between them. We identified these same residues by phylogenetic analysis and showed that they were required for TMG-binding (Figure 2A and Table 1). Furthermore, mutation of these two residues to alanine also had a significant effect on binding to Xpo1 in vitro (Figure 2B), perhaps due to misfolding of the β strands. Unfortunately, Strasser et al. (2005) were unable to recover crystals of full-length SPN or of the TMG binding domain in the absence of bound TMG cap dinucleotide. Future efforts to characterize potential interdomain interactions within SPN (in the presence and absence of bound cargo) should be informative in this regard.
SPN Autoregulation via an Intramolecular Interaction?
Access to the IBB domain of importin-α is thought to be regulated by sequences within the C-terminal NLS-binding domain (Kobe 1999). Disruption of this so-called “autoinhibitory” interaction was shown to have functional consequences in yeast (Harreman et al., 2003a,b). Our discovery that the SPN N and C termini interact suggests that SPN may function in a similar manner. Thus the current perceived modular character of the SPN IBB domain must be reevaluated. We favor a snRNP import model wherein folding of the C terminus regulates the availability of the N-terminal IBB domain. Consistent with this interpretation, we found that mutation or removal of the C-terminal domain increased the binding of SPN to importin-β (Figure 8A and Supplemental Figure S1) and that interactions between isolated N- and C-terminal fragments of SPN were disrupted by mutations within either of these subdomains (Figure 8B). Thus, we conclude that SPN forms an intramolecular interaction and that cross-talk between subdomains may modulate the efficiency of nuclear import.
To facilitate snRNP import, SPN must form a complex with both snRNPs and importin-β. The order of complex formation is unknown. After export and release from Xpo1 in the cytoplasm, SPN is presumably free to bind to the receptor, the cargo or to itself via an intramolecular interaction. Because an intramolecular interaction would be kinetically favorable, we propose that sequestering of the SPN IBB might help prevent cargo-less SPN molecules from binding to importin-β in the cytoplasm, thus reducing the number of futile import cycles.
A recent structure-function study of Exportin1 also has also to an autoinhibitory hypothesis regarding the Ran binding loop of this transport protein (Petosa et al., 2004). Similarly-detailed structural studies, comparing the TMG bound and unbound states, will be required to demonstrate the existence of an intramolecular interaction within SPN. However, our finding that the SPN N and C termini can interact reveals a common theme among two different transport adaptors for importin-β. In the future, it will be interesting to see whether other transport factors use similar mechanisms.
Supplementary Material
Acknowledgments
We thank U. Narayanan, K. Shpargel, T. K. Rajendra, and M. Walker for scientific discussions. We are indebted to J. Yong, G. Dreyfuss, I. Lemm, R. Lührmann, M. Fornerod, K. Weis, and T. Nilsen for reagents. This work was supported by National Institutes of Health Grants R01-GM53034 and R01-NS41617 (to A.G.M.). J.K.O. was supported in part by National Institutes of Health predoctoral traineeship T32-GM08613. E.D. was supported by the Polish Ministry of Science grant KBN2 P04A 00628.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-04-0316) on July 29, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Adam, E. J., and Adam, S. A. (1994). Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J. Cell Biol. 125, 547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam, S. A., and Gerace, L. (1991). Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell 66, 837-847. [DOI] [PubMed] [Google Scholar]
- Adam, S. A., Marr, R. S., and Gerace, L. (1990). Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111, 807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade, M. A., Petosa, C., O'Donoghue, S. I., Muller, C. W., and Bork, P. (2001). Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309, 1-18. [DOI] [PubMed] [Google Scholar]
- Askjaer, P., Jensen, T. H., Nilsson, J., Englmeier, L., and Kjems, J. (1998). The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J. Biol. Chem. 273, 33414-33422. [DOI] [PubMed] [Google Scholar]
- Bednenko, J., Cingolani, G., and Gerace, L. (2003). Importin-β contains a COOH-terminal nucleoporin binding region important for nuclear transport. J. Cell Biol. 162, 391-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon, S., Verheggen, C., Jady, B. E., Girard, C., Pescia, C., Paul, C., Ospina, J. K., Kiss, T., Matera, A. G., Bordonne, R., and Bertrand, E. (2004). PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol. Cell 16, 777-787. [DOI] [PubMed] [Google Scholar]
- Cingolani, G., Petosa, C., Weis, K., and Muller, C. W. (1999). Structure of importin-β bound to the IBB domain of importin-α. Nature 399, 221-229. [DOI] [PubMed] [Google Scholar]
- Conti, E., Uy, M., Leighton, L., Blobel, G., and Kuriyan, J. (1998). Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 94, 193-204. [DOI] [PubMed] [Google Scholar]
- Dreyfuss, G., Kim, V. N., and Kataoka, N. (2002). Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3, 195-205. [DOI] [PubMed] [Google Scholar]
- Fechter, P., and Brownlee, G. G. (2005). Recognition of mRNA cap structures by viral and cellular proteins. J. Gen. Virol. 86, 1239-1249. [DOI] [PubMed] [Google Scholar]
- Fischer, U., Sumpter, V., Sekine, M., Satoh, T., and Lührmann, R. (1993). Nucleo-cytoplasmic transport of U snRNPs: definition of a nuclear location signal in the Sm core domain that binds a transport receptor independently of the m3G cap. EMBO J. 12, 573-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod, M., Ohno, M., Yoshida, M., and Mattaj, I. W. (1997). CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90, 1051-1060. [DOI] [PubMed] [Google Scholar]
- Frey, M. R., and Matera, A. G. (1995). Coiled Bodies Contain U7 Small Nuclear RNA and Associate with Specific DNA Sequences in Interphase Cells. Proc. Natl. Acad. Sci. USA 92, 5915-5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried, H., and Kutay, U. (2003). Nucleocytoplasmic transport: taking an inventory. Cell Mol. Life Sci. 60, 1659-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich, D., Henklein, P., Laskey, R. A., and Hartmann, E. (1996). A 41 amino acid motif in importin-α confers binding to importin-beta and hence transit into the nucleus. EMBO J. 15, 1810-1817. [PMC free article] [PubMed] [Google Scholar]
- Görlich, D., and Mattaj, I. (1996). Nucleocytoplasmic transport. Science 271, 1513-1518. [DOI] [PubMed] [Google Scholar]
- Harel, A., and Forbes, D. J. (2004). Importin β: conducting a much larger cellular symphony. Mol. Cell 16, 319-330. [DOI] [PubMed] [Google Scholar]
- Harreman, M. T., Cohen, P. E., Hodel, M. R., Truscott, G. J., Corbett, A. H., and Hodel, A. E. (2003a). Characterization of the auto-inhibitory sequence within the N-terminal domain of importin alpha. J. Biol. Chem. 278, 21361-21369. [DOI] [PubMed] [Google Scholar]
- Harreman, M. T., Hodel, M. R., Fanara, P., Hodel, A. E., and Corbett, A. H. (2003b). The auto-inhibitory function of importin alpha is essential in vivo. J. Biol. Chem. 278, 5854-5863. [DOI] [PubMed] [Google Scholar]
- Hieda, M., Tachibana, T., Yokoya, F., Kose, S., Imamoto, N., and Yoneda, Y. (1999). A monoclonal antibody to the COOH-terminal acidic portion of Ran inhibits both the recycling of Ran and nuclear protein import in living cells. J. Cell Biol. 144, 645-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, J., Cronshagen, U., Kadokura, M., Marshallsay, C., Wada, T., Sekine, M., and Lührmann, R. (1998). Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 17, 4114-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, J., Dickmanns, A., and Lührmann, R. (2002). The importin-β binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro. J. Cell Biol. 156, 467-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde, E., Kutay, U., von Kobbe, C., Mattaj, I. W., and Gorlich, D. (1997). The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 16, 6535-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski, A., Boelens, W. C., Izaurralde, E., and Mattaj, I. W. (1994). Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol. 124, 627-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss, T. (2004). Biogenesis of small nuclear RNPs. J. Cell Sci. 117, 5949-5951. [DOI] [PubMed] [Google Scholar]
- Klebe, C., Nishimoto, T., and Wittinghofer, F. (1993). Functional expression in Escherichia coli of the mitotic regulator proteins p24ran and p45rcc1 and fluorescence measurements of their interaction. Biochemistry 32, 11923-11928. [DOI] [PubMed] [Google Scholar]
- Kobe, B. (1999). Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin α. Nat. Struct. Biol. 6, 388-397. [DOI] [PubMed] [Google Scholar]
- Kudo, N., Wolff, B., Sekimoto, T., Schreiner, E. P., Yoneda, Y., Yanagida, M., Horinouchi, S., and Yoshida, M. (1998). Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242, 540-547. [DOI] [PubMed] [Google Scholar]
- Marshallsay, C., and Lührmann, R. (1994). In vitro nuclear import of snRNPs: cytosolic factors mediate m3G-cap dependence of U1 and U2 snRNP transport. EMBO J. 13, 222-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama, K., Taniguchi, I., Kataoka, N., and Ohno, M. (2004). RNA length defines RNA export pathway. Genes Dev. 18, 2074-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister, G., Eggert, C., and Fischer, U. (2002). SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol. 12, 472-478. [DOI] [PubMed] [Google Scholar]
- Moroianu, J., Blobel, G., and Radu, A. (1995). Previously identified protein of uncertain function is karyopherin alpha and together with karyopherin beta docks import substrate at nuclear pore complexes. Proc. Natl. Acad. Sci. USA 92, 2008-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast, N., and Pemberton, L. F. (2004). Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14, 547-556. [DOI] [PubMed] [Google Scholar]
- Mouaikel, J., Bujnicki, J. M., Tazi, J., and Bordonne, R. (2003). Sequence-structure-function relationships of Tgs1, the yeast snRNA/snoRNA cap hypermethylase. Nucleic Acids Res. 31, 4899-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouaikel, J., Verheggen, C., Bertrand, E., Tazi, J., and Bordonne, R. (2002). Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell 9, 891-901. [DOI] [PubMed] [Google Scholar]
- Narayanan, U., Achsel, T., Lührmann, R., and Matera, A. G. (2004). Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein. Mol. Cell 16, 223-234. [DOI] [PubMed] [Google Scholar]
- Narayanan, U., Ospina, J. K., Frey, M. R., Hebert, M. D., and Matera, A. G. (2002). SMN, the spinal muscular atrophy protein, forms a pre-import snRNP complex with snurportin1 and importin β. Hum. Mol. Genet. 11, 1785-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesic, D., Tanackovic, G., and Krämer, A. (2004). A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J. Cell Sci. 117, 4423-4433. [DOI] [PubMed] [Google Scholar]
- Nilsson, J., Askjaer, P., and Kjems, J. (2001). A role for the basic patch and the C terminus of RanGTP in regulating the dynamic interactions with importin β, CRM1 and RanBP1. J. Mol. Biol. 305, 231-243. [DOI] [PubMed] [Google Scholar]
- Ohno, M., Segref, A., Bachi, A., Wilm, M., and Mattaj, I. W. (2000). PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101, 187-198. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari, B., Bachelerie, F., and Dargemont, C. (1997). Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 278, 141-144. [DOI] [PubMed] [Google Scholar]
- Palacios, I., Hetzer, M., Adam, S. A., and Mattaj, I. W. (1997). Nuclear import of U snRNPs requires importin β. EMBO J. 16, 6783-6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pante, N. (2004). Nuclear pore complex structure: unplugged and dynamic pores. Dev. Cell 7, 780-781. [DOI] [PubMed] [Google Scholar]
- Paraskeva, E., Izaurralde, E., Bischoff, F. R., Huber, J., Kutay, U., Hartmann, E., Lührmann, R., and Gorlich, D. (1999). CRM1-mediated recycling of snurportin 1 to the cytoplasm. J. Cell Biol. 145, 255-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni, L., Yong, J., and Dreyfuss, G. (2002). Essential role for the SMN complex in the specificity of snRNP assembly. Science 298, 1775-1779. [DOI] [PubMed] [Google Scholar]
- Petosa, C., Schoehn, G., Askjaer, P., Bauer, U., Moulin, M., Steuerwald, U., Soler-Lopez, M., Baudin, F., Mattaj, I. W., and Muller, C. W. (2004). Architecture of CRM1/Exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex. Mol. Cell 16, 761-775. [DOI] [PubMed] [Google Scholar]
- Quiocho, F. A., Hu, G., and Gershon, P. D. (2000). Structural basis of mRNA cap recognition by proteins. Curr. Opin. Struct. Biol. 10, 78-86. [DOI] [PubMed] [Google Scholar]
- Rout, M. P., and Aitchison, J. D. (2001). The nuclear pore complex as a transport machine. J. Biol. Chem. 276, 16593-16596. [DOI] [PubMed] [Google Scholar]
- Segault, V., Will, C. L., Sproat, B. S., and Lührmann, R. (1995). In vitro reconstitution of mammalian U2 and U5 snRNPs active in splicing: Sm proteins are functionally interchangeable and are essential for the formation of functional U2 and U5 snRNPs. EMBO J. 14, 4010-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman, J. E., and Lamond, A. I. (1999). Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol. 9, 1065-1074. [DOI] [PubMed] [Google Scholar]
- Strasser, A., Dickmanns, A., Lührmann, R., and Ficner, R. (2005). Structural basis for m(3)G-cap-mediated nuclear import of spliceosomal UsnRNPs by snurportin1. EMBO J. 24, 2235-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser, A., Dickmanns, A., Schmidt, U., Penka, E., Urlaub, H., Sekine, M., Lührmann, R., and Ficner, R. (2004). Purification, crystallization and preliminary crystallographic data of the m3G cap-binding domain of human snRNP import factor snurportin 1. Acta Crystallogr. D. Biol. Crystallogr. 60, 1628-1631. [DOI] [PubMed] [Google Scholar]
- Sumpter, V., Kahrs, A., Fischer, U., Kornstadt, U., and Lührmann, R. (1992). In vitro reconstitution of U1 and U2 snRNPs from isolated proteins and snRNA. Mol. Biol. Rep. 16, 229-240. [DOI] [PubMed] [Google Scholar]
- Suntharalingam, M., and Wente, S. R. (2003). Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell 4, 775-789. [DOI] [PubMed] [Google Scholar]
- Tanackovic, G., and Krämer, A. (2005). Human splicing factor SF3a, but not SF1, is essential for pre-mRNA splicing in vivo. Mol. Biol. Cell 16, 1366-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen, C., Lafontaine, D. L., Samarsky, D., Mouaikel, J., Blanchard, J. M., Bordonne, R., and Bertrand, E. (2002). Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 21, 2736-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis, K., Mattaj, I., and Lamond, A. (1995). Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science 268, 1049-1053. [DOI] [PubMed] [Google Scholar]
- Will, C. L., and Lührmann, R. (2001). Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13, 290-301. [DOI] [PubMed] [Google Scholar]
- Will, C. L., Urlaub, H., Achsel, T., Gentzel, M., Wilm, M., and Lührmann, R. (2002). Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 21, 4978-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong, J., Golembe, T. J., Battle, D. J., Pellizzoni, L., and Dreyfuss, G. (2004). snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol. Cell Biol. 24, 2747-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.