Abstract
The Ras-Raf-MAPK cascade is a key growth-signaling pathway and its uncontrolled activation results in cell transformation. Although the general features of the signal transmission along the cascade are reasonably defined, the mechanisms underlying Raf activation remain incompletely understood. Here, we show that Raf-1 dephosphorylation, primarily at epidermal growth factor (EGF)-induced sites, abolishes Raf-1 kinase activity. Using mass spectrometry, we identified five novel in vivo Raf-1 phosphorylation sites, one of which, S471, is located in subdomain VIB of Raf-1 kinase domain. Mutational analyses demonstrated that Raf-1 S471 is critical for Raf-1 kinase activity and for its interaction with mitogen-activated protein kinase kinase (MEK). Similarly, mutation of the corresponding B-Raf site, S578, resulted in an inactive kinase, suggesting that the same Raf-1 and B-Raf phosphorylation is needed for Raf kinase activation. Importantly, the naturally occurring, cancer-associated B-Raf activating mutation V599E suppressed the S578A mutation, suggesting that introducing a charged residue at this region eliminates the need for an activating phosphorylation. Our results demonstrate an essential role of specific EGF-induced Raf-1 phosphorylation sites in Raf-1 activation, identify Raf-1 S471 as a novel phosphorylation site critical for Raf-1 and B-Raf kinase activities, and point to the possibility that the V599E mutation activates B-Raf by mimicking a phosphorylation at the S578 site.
INTRODUCTION
Raf-1 or c-Raf-1 is part of the Ras-Raf-MEK-MAPK pathway, which was among the first mammalian signaling cascades to be elucidated (Avruch et al., 1994). The Ras-Raf-MAPK pathway transmits signals from membrane-bound receptors to intracellular and nuclear targets, coordinating cellular response to a variety of environmental factors (Avruch et al., 2001; Chang and Karin, 2001). Aberrations at the receptor level and along the Ras-Raf-MAPK pathway are associated with a variety of diseases, especially cancer, with Ras mutations detected in >30% of all human cancers (Porter and Vaillancourt, 1998; Lyons et al., 2001; Herrera and Sebolt-Leopold, 2002). Recently, also B-Raf activating mutations were found in various cancers, most predominantly, melanomas (Davies et al., 2002; Mercer and Pritchard, 2003; Garnett and Marais, 2004). The elucidation of the Ras-Raf-MAPK pathway resulted from complimentary molecular and biochemical studies in mammalian cells and genetic studies in yeast, Drosophila, and Caenorhabditis elegans (Avruch et al., 2001). Although the general features of the pathway and its activation are known, the exact mechanisms at the molecular level remain incompletely understood. Initially, a simple linear activation process was proposed, i.e., Ras activates Raf, Raf activates mitogen-activated protein kinase kinase (MEK), and MEK activates extracellular signal-regulated kinase (ERK). However, it is clear now that this pathway is more complex than initially thought and that other processes and accessory proteins are involved in its regulation, with Raf being the most complexly and strictly regulated member along the pathway (Kerkhoff and Rapp, 2001; Dhillon and Kolch, 2002; Wellbrock et al., 2004a).
The mammalian Raf family of serine/threonine kinases consists of three highly conserved members, i.e., A-Raf, B-Raf, and Raf-1. Whereas Raf-1 is ubiquitously expressed, A- and B-Raf display a more tissue-specific expression. Raf-1, being the first Raf member to be identified is the most studied Raf isoform (Hagemann and Rapp, 1999). The first step in Raf-1 activation by receptor tyrosine kinases, such as the epidermal growth factor (EGF) receptor, involves Ras activation (i.e., Ras loading with GTP). The exchange of Ras from a GDP- to a GTP-bound form results in a conformational change allowing its high-affinity interaction with Raf-1. The association of Raf-1 with Ras is insufficient by itself for Raf-1 activation, and other modulations take place at the membrane, producing a stable active Raf-1. This complex process involves changes in Raf-1 localization, posttranslational modifications (e.g., phosphorylation), dimerization, and protein–protein interactions (Luo et al., 1996; Wellbrock et al., 2004a). The main unresolved question in the Ras-mediated Raf-1 activation relates to the events at the membrane resulting in stable Raf-1 activation.
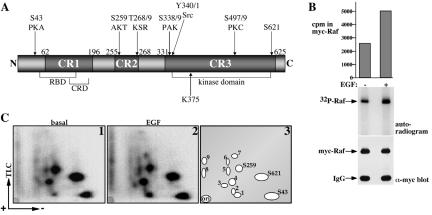
Phosphorylation is the major modification suggested to take place at the membrane. This idea is supported by studies demonstrating increased Raf-1 phosphorylation after mitogenic stimulation and studies showing that phosphatase treatment results in Raf-1 inactivation (Morrison et al., 1993; Dent et al., 1995; Marais et al., 1995). Several Raf-1 phosphorylation sites have been identified, including basal and mitogen-activated sites. The major basal in vivo Raf-1 phosphorylation sites are located at S43, S259, S621, and an as-of-yet-unidentified site (Figure 1A). The S43 site, a postulated inhibitory phosphorylation site, is targeted by protein kinase A (PKA); however, the functional significance of this site remains controversial. The kinases responsible for the constitutive phosphorylation of S259 and S621 have not been fully defined; however, several candidate kinases, including PKA, AKT, and AMPK have been proposed (Wellbrock et al., 2004a). S259 provides a binding point for the regulatory adapter protein 14-3-3 and serves as a negative regulatory site (Tzivion et al., 1998; Tzivion et al., 2001; Tzivion and Avruch, 2002). Phosphorylation of the site by AKT or PKA negatively regulates Raf-1, whereas its dephosphorylation by protein phosphatase 2A has been reported to be part of the Raf-1 activation mechanism. Conversely, Raf-1 phosphorylation at the S621 supports Raf-1 kinase activity by providing a second binding point for 14-3-3, which binding at this site is critical for sustaining Raf-1 kinase activity (Tzivion et al., 1998; Yip-Schneider et al., 2000). It is not known yet whether phosphorylation on S259 and S621 occur simultaneously or whether they represent two separate Raf-1 subgroups.
Figure 1.
Raf-1 phosphorylation sites. (A) Schematic diagram of Raf-1 showing known phosphorylation sites and potential kinases reported to phosphorylate these sites. The locations of the kinase domain, the Ras binding domain (RBD), the cysteine-rich domain (CRD), conserved regions 1–3 (CR 1–3), and the ATP binding site K375 are indicated. (B) Serum-deprived COS-7 cells expressing wild-type myc-Raf-1 were metabolically labeled with 32P for 2 h followed by stimulation with 100 ng/ml EGF or vehicle for 30 min. myc-Raf-1 proteins were immunoprecipitated and resolved using 7.5% SDS-PAGE. Ninety percent of the sample was analyzed by autoradiography (middle) and the remaining 10% were analyzed for myc-Raf-1 recovery (bottom). For quantifying 32P incorporation in myc-Raf-1, the corresponding Raf-1 bands were excised from the gel and 32P incorporation in Raf-1 was determined by Cherenkov counting (top). (C) myc-Raf-1 samples from B were subjected to phosphopeptide mapping as described in Materials and Methods. Representative autoradiograms of Raf-1 phosphopeptide maps (1 and 2) and a schematic representation of the phosphopeptide spots (3) are presented. The electrophoresis direction and the TLC chromatography orientation are indicated by arrows. “Ori” represents the spotting point. The location of phospho-S43, phospho-S621, and phospho-S259 peptides is indicated.
Beside the major sites, Raf-1 phosphorylation at several minor sites also has been reported. These include T268/269, S338/339, Y340/341, and S497/499. T268 is a proposed Raf-1 autophosphorylation site (Morrison et al., 1993), and T269 was reported to be phosphorylated by KSR (Zhang et al., 1997). Although the role of these phosphorylations in Raf-1 regulation remains unresolved, it seems that phosphorylation at these sites does not notably affect Raf-1 activation.
The Rac/CDC42-activated kinases PAK-2 and PAK-3 can phosphorylate Raf-1 on S338/339 and support Raf-1 activation by Ras and growth factors (King et al., 1998). However, phosphorylation at these sites does not activate Raf-1 per se, suggesting that other modifications may be required for stable Raf-1 activation (Wellbrock et al., 2004a). Similarly, Raf-1 phosphorylation at Y340/341, mediated by the tyrosine kinase Src, results in an only partial Raf-1 activation. S497 and S499 can be phosphorylated by protein kinase C; however, this phosphorylation does not result in Raf-1 activation, and phosphorylation at these sites is not required for Raf-1 activation by phorbol 12-myristate 13-acetate (PMA), growth factors, or Ras (Marais et al., 1998).
In addition to these established sites, several novel feedback in vivo Raf-1 phosphorylation sites targeted by ERK have been reported recently. These include S29, S289, S296, S301, and S642 (Dougherty et al., 2005; Hekman et al., 2005). The role of these phosphorylations in Raf-1 regulation remains, however, controversial, because both negative (Alessi et al., 1995; Dougherty et al., 2005; Hekman et al., 2005) and positive (Alessandrini et al., 1996; Zimmermann et al., 1997; Laird et al., 1999; Balan et al., 2005) MEK/ERK effects on Raf-1 have been observed.
The mechanism/s responsible for Raf-1 inactivation is much less known, and it is proposed that Raf-1 dephosphorylation should play a role in this process (Avruch et al., 2001).
Here, we demonstrate the necessity of EGF-induced Raf-1 phosphorylation in the EGF-induced Raf-1 activation and by using mass spectrometry, identify five novel in vivo Raf-1 phosphorylation sites. One of these sites, S471, located in subdomain VIB of the Raf-1 kinase domain, seems critical for Raf-1 kinase activity and for Raf-1 binding to MEK. Accordingly, mutation of the corresponding B-Raf site, S578, results in B-Raf inactivation, and, more importantly, is suppressed by the activating B-Raf V599E mutation, suggesting that introducing a charged residue at this region eliminates the need for an activating phosphorylation. Our results demonstrate a vital role of the EGF-induced Raf-1 phosphorylation in Raf-1 activation, identify Raf-1 S471 and B-Raf S578 as critical sites for Raf-1 and B-Raf kinase activities, and point to the possibility that the V599E mutation activates B-Raf by imitating phosphorylation at the S578 site.
MATERIALS AND METHODS
cDNA Constructs, Antibodies, and Reagents
pMT2 vector encoding wild-type myc-Raf, myc-Raf S259A, myc-Raf S259/621A, or HA-ERK-1, pGEX vector encoding GST-MEK-1 or GST-ERK-1, pEBG vector encoding GST-MEK-1, GST-14-3-3, GST-Raf-1, or GST-Bxb-Raf and pCDNA-3 vector encoding FLAG-Ras G12V were described previously (Tzivion et al., 1998; Shen et al., 2003b). pExchange 5A FLAG-B-Raf and FLAG-MEK-1 wild-type or DD were constructed by PCR amplification of B-Raf from pEBG-GST-B-Raf and MEK-1 DD from pCDNA 3.1 HA-MEK-1 wild type or DD (Alessandrini et al., 1996) and subcloning into pExchange 5A vector (Stratagene, La Jolla, CA) as Not I/Hind III and BamH I/Hind III, respectively. The QuikChange site-directed mutagenesis kit (Stratagene) was used to generate the following mutant proteins: myc-Raf S29A, S43A, S289/296/301A, S471A/C/D/E/T and T481A/C/D/E/S, GST-Bxb-Raf S471A (corresponding to Raf-1 S471A), and FLAG-B-Raf S578A, T588A, V599E, S578A/V599E, and T588A/V599E. Phospho-MEK, phospho-ERK, MEK, and ERK antibodies were purchased from Cell Signaling Technology (Beverly, MA); FLAG antibody from Sigma-Aldrich (St. Louis, MO); glutathione S-transferase (GST) antibody was from Upstate Biotechnology (Lake Placid, NY); HSC-70 and Raf-1 antibodies from Santa Cruz Biotechnology (Santa Cruz, CA); heat-shock protein (HSP)-70 antibody was from Affinity Bioreagents (Golden, CO); and hemagglutinin (HA) and myc epitope tag antibodies were produced from the 12CA5 and 9E10 hybridoma cell lines, respectively. EGF was purchased from Sigma-Aldrich; U0126 MEK inhibitor was from Promega (Madison, WI); and protein phosphatase 1-γ was from Calbiochem (San Diego, CA).
Cell Culture and Transfection
COS-7 and human embryonic kidney (HEK)-293 cells were maintained in DMEM supplemented with 10% fetal calf serum (FCS). For transient expression of proteins, cells were transfected using Lipofectamine (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. For mitogen stimulation, 24 h after transfection, cells were deprived of serum for 18 h before the addition of the agonist.
32P Metabolic Labeling
Serum-deprived COS-7 cells were washed once with medium lacking phosphate, followed by 30-min incubation in the same media for depleting intracellular phosphate. Cells were radiolabeled by incubation in the presence of 0.5 mCi/ml 32P for 2 h.
Cell Extraction and Protein Purification
Cells were lysed for 30 min using ice-cold extraction buffer containing 50 mM Tris-Cl, pH 7.5, 100 mM NaCl, 1% Triton X-100, 1 mM dithiothreitol (DTT), 1 mM EDTA, 1 mM EGTA, 2 mM Na3VO4, 50 mM β-glycerophosphate, and a protease inhibitor cocktail (GE Healthcare, Piscataway, NJ). For immunoprecipitation, cleared cell lysates were incubated at 4°C for 3 h with the appropriate antibody precoupled to protein A/G agarose-beads (Santa Cruz Biotechnology). The beads were washed twice with extraction buffer, twice with extraction buffer containing 0.5 M LiCl, and twice with kinase assay buffer (40 mM Tris-Cl, pH 7.5, 0.1 mM EDTA, 5 mM MgCl2, and 2 mM DTT).
Phosphopeptide Mapping
The two-dimensional phosphopeptide mapping was performed according to previously described protocols (Boyle et al., 1991; Luo et al., 1991). Briefly, the purified 32P-labeled myc-Raf proteins were resolved using 7.5% SDS-PAGE, transferred to a polyvinylidene difluoride (PVDF) membrane, excised, and the 32P incorporation in the myc-Raf was determined by Cherenkov counting. After incubation with 0.5% polyvinyl pyrrolidone in 100 mM acetic acid for 30 min at 37°C and extensive washes, the Raf protein was digested with 10 μg of sequencing grade modified trypsin (Promega) in 50 mM ammonium bicarbonate buffer for 2 h at 37°C and with additional 10 μg of trypsin for overnight (this method routinely allowed recovery of 90–95% of the initial radioactivity in myc-Raf). The eluted peptides were washed twice with 50 mM ammonium bicarbonate buffer and once with pH 1.9 thin layer chromatography (TLC)-electrophoresis buffer. Samples were spotted on cellulose TLC plates (Merck Research Labs, West Point, PA) and separated using Hunter thin layer electrophoresis system (CBS Scientific, Del Mar, CA) in pH 1.9 buffer for 25 min at 1000 V. The plates were dried overnight and subjected to second dimension chromatographic separation in a phospho-chromatography buffer. The plates were dried, and the phosphopeptide spots were visualized by autoradiography and phosphor imaging.
Raf Kinase Assay
Raf kinase activity was determined using a coupled MEK-ERK kinase assay using [γ-32P]ATP as described previously (Tzivion et al., 1998). Alternatively, Raf kinase activity was determined using a cold kinase assay followed by phospho-MEK or phospho-ERK immunoblotting. Briefly, after myc or GST pull-down, Raf containing beads were incubated in a kinase assay buffer (100-μl final volume) supplemented with 100 μM ATP and 0.4 μg of prokaryotic recombinant GST-MEK-1 for 20 min at 30°C. After adding 2 μg of prokaryotic recombinant kinase-inactive GST-ERK-1, the samples were incubated for additional 30 min at 30°C. Samples were separated on 10% SDS-PAGE and transferred to PVDF membrane. The phosphorylation of MEK-1 and ERK-1 were determined by phospho-MEK and phospho-ERK immunoblotting, respectively.
Mass Spectrometry Analysis
Raf protein for the mass spectrometry analysis was purified using myc-immunoprecipitation from myc-Raf expressing COS-7 cells treated as indicated in the figure legends. For the immunoprecipitation, 50 μg of myc antibody precoupled to 100 μl of protein A/G agarose-beads (Santa Cruz Biotechnology) was incubated with 30–40 mg of cell extract (precleared on 100 μl of protein A/G agarose-beads for 2 h at 4°C) for 18 h at 4°C, followed by two washes with 10 ml of lysis buffer, two washes with 10 ml of lysis buffer containing 1 M LiCl, and four washes with 10 ml of Raf kinase buffer. The immunoprecipitated proteins were eluted in SDS sample buffer and alkylated by incubation with 2% iodoacetamide for 30 min at room temperature. The protein samples were separated on 7.5% SDS-PAGE and visualized by Coomassie-Blue staining. Proteins of interest were excised from the gel and analyzed for identity using mass spectrometry. For Raf phosphorylation site identification, 4 to 5 μg of the myc-Raf protein was digested in the gel with trypsin and analyzed by mass spectrometry as described previously (Qin and Zhang, 2002).
Indirect In Vivo Raf Activity Assay
COS-7 cells were transfected with Raf expression vectors as indicated in the figure legends together with HA-ERK or FLAG-MEK as reporters. Thirty-six hours after transfection, cells were lysed, and ERK and MEK phosphorylation was determined in total cell extracts or in HA or FLAG immunoprecipitates using phospho-ERK or phospho-MEK immunoblotting, respectively.
Coimmunoprecipitation Assays
myc-Raf proteins were transfected together with either FLAG-Ras G12V, GST-MEK, GST-Raf, or GST-14-3-3 as described in the figure legends. FLAG-Ras was purified using FLAG immunoprecipitation and the GST-fusion proteins were purified using GSH-beads. After extensive washes, myc-Raf presence in the immunoprecipitates or GSH-pull-downs was determined using myc immunoblotting. Equal expression of myc-Raf in the various transfections was verified using myc-Raf immunoblotting in total cell extracts. For endogenous Raf-1-HSP-70 and HSC-70 association, 10 mg of cell extracts from exponentially growing or heat-shock treated HEK-293 cells were incubated with 20 μg of anti-HSP-70, anti-HSC-70, or control IgG precoupled to 50 μl of protein A agarose-beads (Pharmacia, Uppsala, Sweden) for 18 h at 4°C. The beads were washed twice with 10 ml of lysis buffer, twice with 10 ml of lysis buffer containing 0.5M LiCl, and twice with 10 ml of phosphate-buffered saline, and Raf-1 association was determined by Raf-1 immunoblotting.
RESULTS
EGF Induced Raf-1 Activation Relies on Raf-1 Phosphorylation at Specific EGF-induced Sites
We examined the role of phosphorylation in the EGF-induced Raf-1 activation by studying Raf-1 phosphorylation using in vivo 32P metabolic labeling (Figure 1). Purification of Raf-1 from 32P labeled cells shows that EGF stimulation results in an approximately twofold increase in overall Raf-1 phosphorylation (Figure 1B). To attain a stoichiometric comparison between Raf-1 phosphorylation sites under basal and EGF stimulation, equal counts of 32P-Raf-1 from each sample were used to generate two-dimensional tryptic phosphopeptide maps (Figure 1C). Raf-1 phosphopeptide maps derived from serum-deprived COS-7 cells show four major spots (Figure 1C, 1). Of these, three spots were correlated with pS43, pS621, and pS259 (Figure 1C, 3) by aligning our peptide maps with a previously published map (Morrison et al., 1993) and with a modeled phosphopeptide map generated using parameters published by Boyle et al. (1991). The identity of the fourth spot (Figure 1C, 3, spot 4) has not been determined yet. Beside the four major spots, there are several minor spots, marked as spots 1–3 and 5–9 (Figure 1C, 3), which identity also remains to be determined. EGF treatment results in an increase in the phosphorylation of some of these spots, notably spots 3, 6, and 7, and the appearance of several new spots (Figure 1C, compare 1 and 2). Thus, this comparison shows that the major Raf-1 phosphorylation sites are the same in EGF-stimulated and untreated cells and that EGF treatment causes Raf-1 phosphorylation on several minor sites. These results indicate that the overall increase in Raf-1 phosphorylation observed after EGF stimulation is mainly a consequence of increased Raf-1 phosphorylation on the basal sites and only partially because of phosphorylation on novel sites.
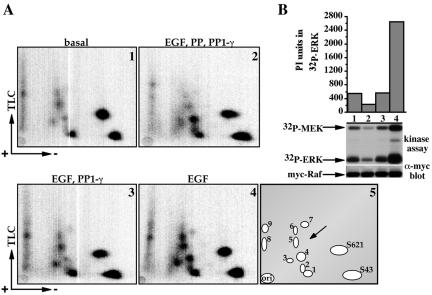
To examine the correlation between Raf-1 phosphorylation and Raf-1 kinase activity, we compared phosphopeptide maps of wild-type Raf-1 with that of a partially active Raf-1 mutant, Raf-1 S259A (Figure 2). These results show that Raf-1 S259A is already partially phosphorylated on the EGF-induced sites (notably spots 3 and 6) and that phosphorylation at these sites could be significantly increased after EGF stimulation (Figure 2A, compare 1 and 4), suggesting a role of these phosphorylation sites in Raf-1 activation. It is notable that the spot correlating to pS259 (Figure 2A, 5, marked with an arrow) is missing in the Raf-1 S259A mutant, supporting our alignments in Figure 1.
Figure 2.
Dephosphorylation of Raf-1, primarily at EGF-induced sites, results in Raf-1 kinase inactivation. (A and B) Serum-deprived COS-7 cells expressing myc-Raf-1 S259A mutant were metabolically labeled with 32P for 2 h, followed by stimulation with 100 ng/ml EGF (2–4) or vehicle (1) for 30 min. After myc immunoprecipitation, samples were incubated with vehicle (1, 2, and 4) or with 1 mM pS621 phosphopeptide corresponding to Raf-1 amino acids 613–627 to displace coassociated 14-3-3 (2) for 30 min at 4°C. After extensive washes, the samples were incubated with vehicle (1 and 4) or with protein phosphatase 1-γ (PP1-γ; 0.2 mU/ml, samples 2 and 3) for 30 min at 12°C. After extensive washes, 70% of the sample was analyzed by phosphopeptide mapping (A) and the remaining was assayed for Raf-1 kinase activity using an MEK-ERK-coupled kinase assay as described in Materials and Methods (B). Shown are representative autoradiograms of the phosphopeptide maps (A, 1–4) and a schematic representation of the phosphopeptide spots (A, 5; the location of the missing pS259 peptide spot is indicated with an arrow); and an autoradiogram of the Raf-1 kinase assay (B, middle), a myc-Raf-1 immunoblot (B, bottom), and a graphical representation of 32P incorporation in ERK quantified using phosphorimaging (top, PI units are arbitrary phosphorimager units).
To test whether phosphorylation at these sites is needed for Raf-1 kinase activity, we assayed the effect of Raf-1 dephosphorylation on Raf-1 kinase activity under specific dephosphorylation conditions that allow only partial Raf-1 dephosphorylation (Figure 2B). Comparing Raf-1 phosphopeptide maps before and after the phosphatase treatment (Figure 2A, compare 3 and 4) shows that under these specific dephosphorylation conditions, the main sites that are dephosphorylated are the EGF-induced sites, i.e., spots 3–7. Importantly, this dephosphorylation results in an almost complete loss of Raf-1 kinase activity (Figure 2B, compare lanes 3 and 4). To test whether 14-3-3 binding to Raf-1 has an effect on Raf-1 dephosphorylation pattern or Raf-1 inactivation, Raf-1 samples were treated in parallel with a 14-3-3 binding peptide to outcompete the Raf-bound 14-3-3 before treating with the phosphatase (Figure 2A, 2 and B, lane 2). This procedure resulted in a somewhat different dephosphorylation pattern: strong reduction in spots 3 and 5–7 but stabilization of spots 2 and 4 (Figure 2A, compare 2 and 3), suggesting that 14-3-3 binding has an effect on Raf-1 conformation and accessibility to the phosphatase. Raf-1 dephosphorylation after 14-3-3 removal resulted in a complete loss of Raf-1 kinase activity (Figure 2B, compare lane 2 with 3 and 4). It is important to note that in these experiments recombinant 14-3-3 was added after washing out the phosphatase to allow Raf-1 reactivation in the absence of a phosphatase treatment (Tzivion et al., 1998; Tzivion et al., 2000); thus, the lack of activity in this sample is a result of the dephosphorylation rather than the removal of 14-3-3. Collectively, this set of experiments pointed on a pivotal role of the EGF-induced Raf-1 phosphorylation sites in sustaining Raf-1 kinase activity.
Mass Spectrometry Analyses Identify HSP-70 and HSC-70 as Novel Raf-1-associated Proteins and Reveal a New EGF-induced Raf-1 Phosphorylation Site at a Tryptic Peptide Corresponding to Raf-1 471-483.
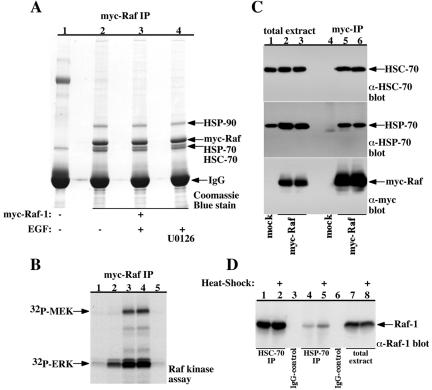
Because the EGF-induced Raf-1 phosphorylation sites are not well defined, we decided to identify basal and EGF-induced Raf-1 phosphorylation sites using electrospray mass spectrometry. In these experiments, Raf-1 samples immunopurified from serum-deprived or EGF-stimulated COS-7 cells were resolved on SDS-PAGE and analyzed for Raf-1 phosphorylation sites and coassociated proteins (Figure 3A). In parallel, aliquots of the Raf-1 samples were assayed for kinase activity (Figure 3B). As reported previously (Stancato et al., 1993), Raf-1 copurifies with a 90-kDa protein, confirmed to be HSP-90 by the mass spectrometry analysis (Figure 3A). Besides HSP-90, we observed that Raf-1 copurifies at a nearly 1:1 stoichiometric ratio with a doublet of ∼70-kDa proteins. Using mass spectrometry, these proteins were identified as HSP-70 (accession no. 462325) and HSC-70 (accession no. 123648). Notably, neither of these proteins was detected in the mass spectrometry analysis of a correspondingly migrating gel piece from a mock-immunoprecipitated lane. This association was confirmed by immunoblotting Raf-1 immunoprecipitates for HSP-70 (Figure 3C, middle) and HSC-70 (Figure 3C, top) and demonstrated also for the endogenous proteins by showing endogenous Raf-1 copurification with the endogenous HSP-70 and HSC-70 in HEK-293 cells (Figure 3D). Thus, these experiments show that Raf-1 is constitutively associated besides the previously reported HSP-90, 14-3-3 and p50 also with HSP-70 and HSC-70 and that heat-shock treatment increases to some extent Raf-1 association with HSP-70. The physiological significance of these associations remains to be determined.
Figure 3.
Identification of HSP-70 and HSC-70 as Raf-1 associated proteins. (A and B) Serum-deprived COS-7 cells expressing wild-type myc-Raf-1 (lanes 2–4) or control vector (lane 1) were incubated with vehicle (lanes 1–3) or with 20 μM MEK inhibitor (U0126, lane 4) for 1 h before stimulation with vehicle (lane 2) or with 100 ng/ml EGF (lanes 1, 3, and 4) for 30 min. Cell lysates were precleared using mouse IgG pre-coupled to protein A beads for 90 min at 4°C and then immunoprecipitated using 9E10 myc antibody for 12 h at 4°C. After extensive washes, 90% of the sample was resolved using 7% SDS-PAGE and stained with Coomassie-Blue (A), whereas the remaining 10% were assayed for Raf-1 kinase activity using an MEK-ERK-coupled kinase assay (B). Bands corresponding to HSP-90, myc-Raf-1, and the 70-kDa doublet band below myc-Raf-1 were excised from the gel and analyzed using electrospray mass spectrometry either for protein identity, or in the case of myc-Raf-1, for phosphorylation site identification. The 90-kDa protein was confirmed to be HSP-90, and the 70-kDa doublet was identified as HSP-70 and HSC-70. For attaining enough Raf-1 protein samples for the phosphorylation site identification (4 to 5 μg), twenty 15-cm dishes and 50 μg of myc antibody were used per each point. The migration points of HSP-90, myc-Raf-1, and IgG are indicated. (C) Serum-deprived COS-7 cells expressing wild-type myc-Raf-1 (lanes 2, 3, 5, and 6) or control vector (lanes 1 and 4) were stimulated with vehicle (lanes 2 and 5) or with 100 ng/ml EGF (lanes 1, 3, 4, and 6) for 30 min. Total cell extracts (lanes 1–3) and myc-immunoprecipitates (lanes 4–6) were resolved using 7.5% SDS-PAGE followed by immunoblot analysis using HSC-70 (top) HSP-70 (middle), or myc (bottom) antibodies. (D) Exponentially growing (lanes 1, 3, 4, 6, and 7) or heat-shock treated HEK-293 cells (30 min at 42°C, 6 h before cell extraction; lanes 2, 5, and 8) were lysed and cell extracts containing 10 mg of protein were used for immunoprecipitation s using HSC-70 (lanes 1 and 2), HSP-70 (lanes 4 and 5), or control IgG (lanes 3 and 6). The immunoprecipitates (lanes 1–6) and 50 μg of total cell extracts (lanes 7 and 8) were separated using 7.5% SDS-PAGE and immunoblotted for Raf-1.
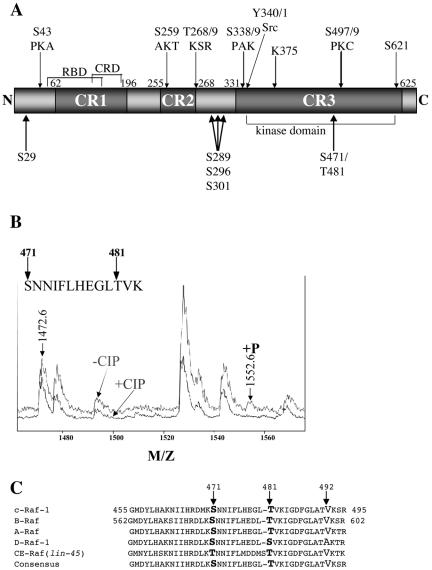
The mass spectrometry analysis of Raf-1 phosphorylation revealed some already known sites but also five new sites (Figure 4). Of these sites, four sites, S29, S289, S296, and S301, have been meanwhile reported (Dougherty et al., 2005; Hekman et al., 2005).
Figure 4.
Identification of Raf-1 S29, S289, S296, S301, and S471/T481 as novel Raf-1 phosphorylation sites. (A) Schematic diagram of Raf-1 showing known phosphorylation sites (top) and the newly identified phosphorylation sites (bottom). (B) myc-Raf-1 proteins were purified as described in Figure 3A and subjected to electrospray mass spectrometry to identify phosphorylation sites. The result pertaining to identification of phosphorylation at a peptide corresponding to Raf-1 471-483 is presented. Samples were analyzed after either treatment with calf intestinal phosphatase (+CIP) or with vehicle (-CIP). The mass picks corresponding to Raf-1 471-483 mass, 1472.6, and its phosphorylated form, 1552.6, are indicated. The sequence of Raf-1 471-483 peptide is provided with its two potential phosphorylation sites indicated. (C) Alignment of Raf-1 455-495 region containing subdomain VIB of the Raf-1 kinase domain with other Raf family members. The locations of Raf-1 S471 and T481 as well as V492, corresponding to the B-Raf activating mutation site V599, are indicated with arrows. D-Raf-1 and CE-Raf (lin-45) are the Drosophila and C. elegans Raf proteins, correspondingly.
The fifth site, is on a tryptic peptide corresponding to Raf-1 471-483 (Figure 4, A and B). This site is within subdomain VIB/VII of the Raf-1 kinase (the domain nomenclature is based on Hanks and Hunter (1995)), which is a highly conserved region among mammalian Raf family members and Raf from other species (Figure 4C). Phosphorylation at this peptide was detected only on Raf-1 from EGF-treated cells and was not affected by pretreatment of the cells with the MEK inhibitor U0126, indicating that phosphorylation at this site is not a result of a feedback phosphorylation by MEK downstream effectors.
Raf-1 S471 Site Is Critical for Raf-1 Kinase Activity
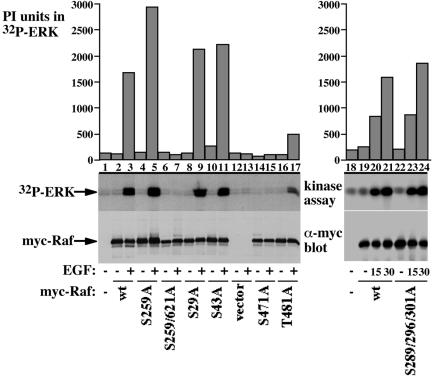
The Raf-1 471-483 tryptic peptide contains two potential phosphorylation sites: S471 and T481; however, the mass spectrometry analysis was unable to distinguish which of these sites is being phosphorylated. To determine the functional significance of the novel Raf-1 phosphorylation sites and to help in pinpointing which of the two potential phosphorylation sites within the Raf-1 471-483 tryptic peptide is important for Raf-1 kinase activity, we examined the effect of mutations at these sites on Raf-1 kinase activity. Initially, we generated alanine substituted Raf-1 mutants and assayed them for Raf-1 kinase activity in an in vitro-coupled kinase assay (Figure 5). As control, wild-type Raf-1 shows a low basal kinase activity in serum-deprived cells, which is substantially increased by EGF treatment (Figure 5, lanes 2 and 3). As an activating and inactivating Raf-1 mutation controls (Tzivion et al., 1998), S259A substitution results in a slight Raf-1 activation, and S259/621A substitution produces an inactive Raf-1 (Figure 5, lanes 4–7). S29A, and S43A substitutions do not seem to affect the basal Raf-1 kinase activity or the EGF-induced activity (Figure 5, lanes 8–11); importantly, however, the Raf-1 S471A substitution abolishes Raf-1 kinase activity (Figure 5, lanes 14 and 15). The T481A substitution also results in a diminished Raf-1 activity, although the extent of the inhibition is less profound than that observed with the S471A substitution (Figure 5, lanes 16 and 17). The S289/296/301A substitution did not seem to have an effect on Raf-1 kinase activity at the conditions tested here (Figure 5, compare lanes 19–21 with 22–24); however, in a subsequent study we show that these sites play a role in an ERK-mediated positive feedback Raf-1 regulation (Balan, V., Zhu, Balan, K., Leicht, Singh-Gupta, Qin, Ruan, Comb, and Tzivion, unpublished data).
Figure 5.
Kinase activity comparison of Raf-1 phosphorylation site mutants. The indicated Raf-1 phosphorylation site mutants were expressed in COS-7 cells, and their kinase activity was assayed using the coupled Raf kinase assay. Cells were stimulated with 100 ng/ml EGF or vehicle for 15 min (left) or for 15 or 30 min as indicated (right). Presented are an autoradiogram showing 32P incorporation in ERK (middle), a graphical representation of 32P incorporation in ERK quantified using phosphor imaging (top), and a myc immunoblot showing myc-Raf-1 recovery (bottom).
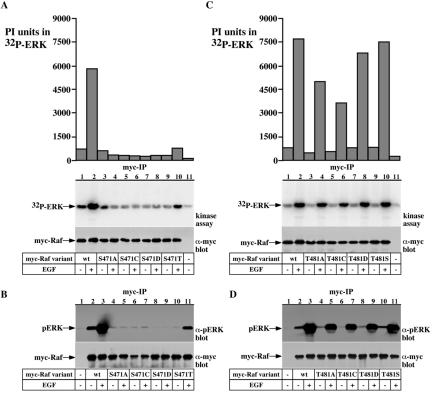
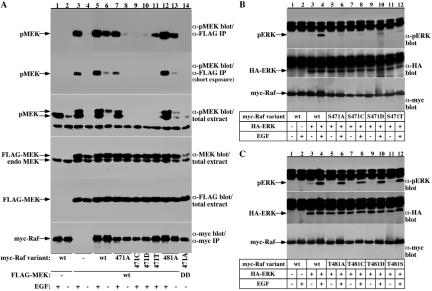
The striking effect of the S471A and T481A substitutions on Raf-1 kinase activity, their location within a conserved region in the Raf-1 kinase domain, and the fact that the peptide containing these sites was found phosphorylated only in EGF-treated cells, prompted us to explore more closely the role of these two sites in Raf-1 activation. Because, a serine/threonine substitution with an alanine could result in a conformational change, the effects of the S471A and T481A substitutions on Raf-1 activity do not necessarily indicate that phosphorylation at these sites is important for Raf-1 activity. To address this point, we generated Raf-1 mutants containing the substitutions S471C/D/T and T481C/D/S and assayed their effect on Raf-1 kinase activity in vitro and in vivo (Figures 6 and 7). The results show that any substitution at S471, besides to some extent Thr, results in a complete loss of Raf-1 kinase activity, assayed in vitro using a coupled Raf-1 kinase assay (Figure 6A, compare lanes 1 and 2 with lanes 3–10 and B, lanes 2 and 3 with 4–11), or in vivo using cotransfected FLAG-MEK or HA-ERK as reporters (Figure 7, A and B). Conversely, T481 substitution with cysteine, serine, or aspartic acid did not significantly affect Raf-1 kinase activity, assayed in vitro (Figure 6, C and D) or in vivo (Figure 7C). These results suggest that a phosphorylatable residue at the 481 site is not required for Raf-1 kinase activity but is obligatory at the 471 site. Thus, our results demonstrate a critical role of S471 for Raf-1 kinase activity and suggest that S471 is the phosphorylated residue on the Raf-1 471-483 peptide. This view is strengthened by the results with the Raf-1 S471T mutant, which repeatedly showed a residual kinase activity (Figure 6A, lane 10; B, lane 11; and 7A, lane 11). Considering that threonine is not more structurally related to serine than are cysteine or alanine, these results suggest that a phosphorylatable residue at the 471 site is needed for Raf-1 kinase activity. It is important to note that we could not detect activation of the S471 mutants also using PMA, nocodazole, or FCS (our unpublished data).
Figure 6.
Raf-1 S471 is a critical site for Raf-1 kinase activity. Serum-deprived COS-7 cells expressing wild-type myc-Raf-1 or the indicated myc-Raf-1 S471-substituted mutants (A and B), or the indicated myc-Raf-1 T481-substituted mutants (C and D) were stimulated with 100 ng/ml EGF or vehicle for 15 min. The myc-Raf-1 variants were immunoprecipitated and assayed for kinase activity in a coupled kinase assay, and ERK phosphorylation was determined either by 32P incorporation (A and C) or by immunoblotting with phospho-ERK specific antibodies (B and D). Presented are an autoradiogram showing 32P incorporation in ERK (A and C, middle), a graphical representation of 32P incorporation in ERK quantified using phosphor-imaging (A and C, top), a myc immunoblot showing myc-Raf-1 recovery (bottom) and an immunoblot showing ERK phosphorylation (B and D, top).
Figure 7.
Raf-1 S471 is critical for Raf-1 function in vivo. Serum-deprived COS-7 cells expressing myc-Raf-1 variants and FLAG-MEK (A) or HA-ERK (B and C), as indicated, were stimulated with 100 ng/ml EGF or vehicle for 15 min. Raf-1 activity in the cells was examined by determining phosphorylation at the activation sites of MEK (A) and ERK (B and C). MEK phosphorylation was determined both in FLAG-MEK immunoprecipitates (A, top two) and in total cell extracts (A, third from top). The endogenous MEK (endo), FLAG-MEK, and myc-Raf-1 variant expression in the cells was determined by anti-MEK (A, 4th panel from top), anti-FLAG (A, fifth from top) or anti-myc (A, bottom) immunoblotting correspondingly. ERK phosphorylation was determined in HA-ERK immunoprecipitates (B and C, top). HA-ERK recovery in the immunoprecipitates (B and C, middle) and myc-Raf-1 variant expression in the cells (B and C, bottom) were determined by anti-HA and anti-myc immunoblotting, respectively.
Raf-1 S471-corresponding Sites in Bxb-Raf and B-Raf Are Critical Sites for Kinase Activity
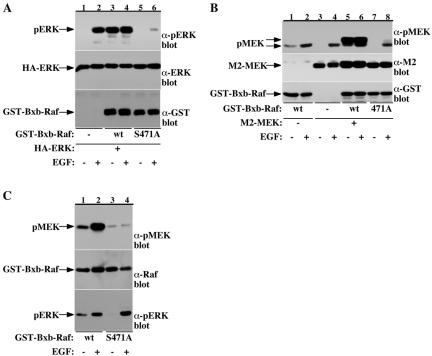
Several reports proposed that phosphorylation at S338/339 and Y340/341 may function via releasing an inhibitory effect of the Raf-1 N terminus on the Raf-1 C-terminal catalytic domain (Cutler et al., 1998; Chong and Guan, 2003). To examine whether the S471 site functions through a similar manner, we tested the effect of S471 substitutions on the kinase activity of a constitutively active form of Raf-1, Bxb-Raf (Bruder et al., 1992). Bxb-Raf has a deletion at the N-terminal regulatory domain (aa 26–303), resulting in an increased basal kinase activity, which remains, however, responsive to mitogenic stimulation. Mutation of S471 abolished both the basal kinase activity of Bxb-Raf and the EGF-induced activity (Figure 8). This abrogation is observed either when Raf kinase activity is assayed in vivo, using HA-ERK or FLAG-MEK as reporters (Figure 8, A and B), or in vitro, using a coupled kinase assay (Figure 8C). These experiments demonstrate that Raf-1 S471 is critical for the kinase activity of the Raf-1 catalytic kinase domain even when the inhibitory effect of the N terminus is released, and corroborate our findings with full-length Raf-1.
Figure 8.
Raf-1 S471 is required for the kinase activity of a constitutively active Raf form, Bxb-Raf, both in vivo and in vitro. (A and B) Serum-deprived COS-7 cells expressing HA-ERK (A) or FLAG-MEK (B) together with wild-type GST-Bxb-Raf or a mutant containing serine to alanine substitution at a site corresponding to Raf-1 S471 (S471A), as indicated, were stimulated with 100 ng/ml EGF or vehicle for 15 min. Raf activity in the cells was examined as in Figure 7 by determining phosphorylation of ERK in HA-ERK immunoprecipitates (A, top) and phosphorylation of MEK in total cell extracts (B, top). Shown are also HA-ERK recovery (A, middle) and FLAG-MEK (B, middle) and GST-Bxb-Raf (A and B, bottom) cellular expressions. (C) Serum-deprived COS-7 cells expressing wild-type GST-Bxb-Raf (lanes 1 and 2) or S471A GST-Bxb-Raf mutant (lanes 3 and 4) were stimulated with 100 ng/ml EGF or vehicle for 15 min. After GSH-pull-down, the kinase activity of the GST-Bxb-Raf forms was examined using the coupled Raf kinase assay. ERK and MEK phosphorylations were determined by phospho-ERK (bottom) and phospho-MEK (top) immunoblotting, respectively. GST-Bxb-Raf recovery was determined by anti-Raf-1 immunoblotting (middle).
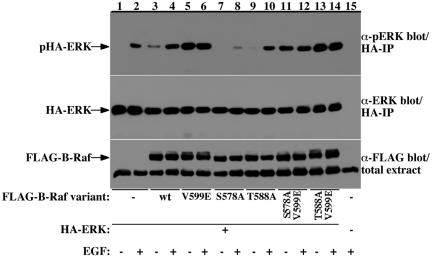
B-Raf V599 mutations occur in high frequencies in melanomas and several other cancers, producing a constitutively active kinase that has a transforming activity (Wellbrock et al., 2004b). Because the corresponding site in Raf-1, V492, is located in proximity to the S471 site, and the recent crystal structure resolution of the B-Raf kinase domain suggests that V599 substitution with charged residues might be mimicking a phosphorylation at this region that allows activation of the kinase (Wan et al., 2004), we hypothesized that phosphorylation of S471 in Raf-1, or the equivalent S578 in B-Raf, may be the activating phosphorylation site mimicked by the V599 substitution. To test this hypothesis, we generated serine 578 to alanine substitutions of wild-type and V599E B-Raf and tested their effect on B-Raf kinase activity in vivo (Figure 9). These experiments show that S578 is critical for B-Raf activation by growth factors similarly to its critical role in Raf-1 activation (Figure 9, compare lanes 3 and 4 and 7 and 8). More importantly, the V599E substitution was able represses the S578 mutation (Figure 9, compare lanes 7 and 8 and 11 and 12), indicating that a charged residue at this region eliminates the need to a phosphorylatable residue at the 578 site and that the S578A substitution does not result in an abrupt change in the kinase structure that prevents kinase activity. Substitution of the Raf-1 T481 corresponding site in B-Raf, T588, did not have much effect on B-Raf kinase activity, similarly to the results obtained with Raf-1 (Figure 9, compare lanes 3 and 4 and 9 and 10).
Figure 9.
B-Raf S578, corresponding to Raf-1 S471, is required for B-Raf kinase activation by EGF but not for the constitutive kinase activity induced by the V599E cancer associated mutation. Serum-deprived COS-7 cells expressing HA-ERK alone (lanes 1 and 2) or coexpressing wild-type FLAG-B-Raf (lanes 3 and 4), FLAG-B-Raf V599E (lanes 5 and 6), FLAG-B-Raf S578A (lanes 7 and 8), FLAG-B-Raf T588A (lanes 9 and 10), FLAG-B-Raf S578A/V599E (lanes 11 and 12) or FLAG-B-Raf T588A/V599E (lanes 13 and 14) were stimulated with 100 ng/ml EGF or vehicle for 15 min as indicated. B-Raf activity in the cells was assessed by HA immunoprecipitation and determining ERK phosphorylation at its activation sites (top). HA-ERK recovery and FLAG-B-Raf variant expression in the cells were determined by anti-ERK (middle) and anti-FLAG immunoblotting (bottom), respectively.
Raf-1 S471 Is Required for the Interaction with MEK
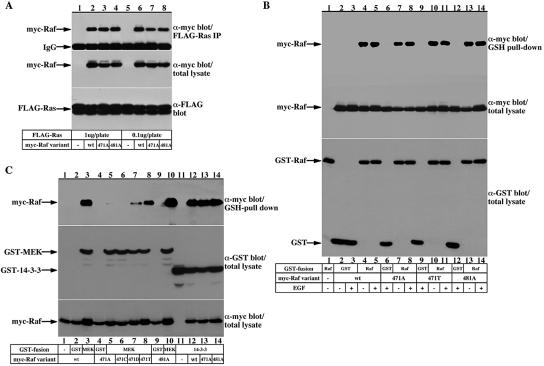
To identify functional determinants correlating with the abolished kinase activity of the Raf-1 S471 mutants and to gain further insight regarding their structural integrity, the interaction of the various Raf-1 mutants with known Raf-1 effectors was examined. Initially, we tested whether the inactivity of the Raf-1 S471 mutants is due to an altered ability to bind Ras (Figure 10A). In these experiments, Raf-1 S471A and T481A were coexpressed with constitutively active Ras, Ras G12V, and their binding to Ras was determined by immunoblotting Ras immunoprecipitates for coassociated Raf-1. This experiment shows that Raf-1 S471 and T481 substitutions do not affect Ras binding.
Figure 10.
Raf-1 S471 is required for Raf-1-MEK binding but not for Raf-1 dimerization or Raf-1 binding to Ras and 14-3-3. (A) COS-7 cells were transfected with FLAG-Ras G12V together with myc-Raf-1forms as indicated. myc-Raf-1 recovery in FLAG-Ras immunoprecipitates was determined using anti-myc immunoblotting (top). Also shown are myc-Raf expression in total cell lysates (middle) and FLAG-Ras recovery in the immunoprecipitates (bottom). (B) Serum-deprived COS-7 cells expressing a control GST vector or GST-Raf-1 fusion together with myc-Raf-1 variants as indicated were stimulated with 100 ng/ml EGF or vehicle for 15 min. myc-Raf-1 recovery in GST-pull-downs was determined using anti-myc immunoblotting (top). Also shown are myc-Raf-1 expression in total cell lysates (middle) and GST-fusion protein recovery in the GSH-pull-downs (bottom). (C) COS-7 cells were transfected with an empty vector (lane 1), GST control vector (lanes 2, 4, and 9), GST-MEK-1 (lanes 5–8 and 10), or GST-14-3-3 (lanes 11–14) together with the indicated myc-Raf-1 variants. myc-Raf-1 recovery in GST-pull-downs was determined using anti-myc immunoblotting (top). Also shown are GST-fusion protein recoveries in the GSH-pull-downs (middle) and myc-Raf-1 expression in total cell lysates (bottom).
It has been shown that Raf-1 forms dimers and that dimerization may have a significant role in Raf-1 activation (Luo et al., 1996). Recently, it has been demonstrated that Ras also dimerizes and that this dimerization is important for Raf activation (Inouye et al., 2000). To examine whether the impaired kinase activity of the Raf-1 S471 mutants is due to an impaired dimerization, we tested the ability of the Raf-1 mutants to form dimers (Figure 10B). In these experiments, myc epitope-tagged Raf-1 mutants were coexpressed with wild-type GST-Raf-1 and tested for dimerization by purification of GST-Raf-1 and immunoblotting for coassociated myc-Raf-1. These experiments showed normal dimerization capabilities of the S471 and T481 Raf-1 mutants.
The current Raf-1 activation model points that Raf-1 requires continuous association with 14-3-3. Because Raf-1 S471 site resembles a low-affinity 14-3-3 binding site, we tested the effect of S471 substitution on Raf-1/14-3-3 association by coexpressing myc-tagged Raf-1 mutants with GST-14-3-3 and examining Raf-1 recovery in GST-14-3-3 pull-downs (Figure 10C, lanes 11–14). These experiments showed that Raf-1 S471A- and T481A-substituted mutants bind 14-3-3 as efficiently as wild-type Raf-1.
Finally, the S471 and T481 substituted Raf-1 forms were tested for association with MEK (Figure 10C, lanes 1–10). In these experiments, myc-Raf-1 forms were coexpressed with GST-MEK, and Raf/MEK association was examined by purifying GST-MEK and immunoblotting for associated Raf. The results show that S471A substitution almost completely diminished Raf-1 association with MEK (Figure 10C, compare lane 3, wild-type Raf-1 with lane 5, Raf-1 S471A). Similarly, Raf-1 S471 substitutions with other amino acids result in a diminished MEK binding (Figure 10C, lanes 6–8). It is notable, however, that the threonine substituted Raf-1 shows a less impaired binding to MEK than the alanine, cysteine, or aspartic acid substituted Raf-1 (Figure 10A, compare lane 8 with lanes 5–7), correlating with the finding that this mutant has a residual kinase activity (Figures 6 and 7). The T481A Raf-1 mutant binds MEK as well as wild-type Raf-1 (Figure 10C, compare lanes 10 and 3).
These association experiments show that the S471 Raf-1 mutants behave similarly to wild-type Raf-1 in terms of their ability to bind Ras, dimerize, and bind 14-3-3, but they have impaired binding to MEK, suggesting that the S471 site may serve as the MEK binding site on Raf-1.
DISCUSSION
Raf is a key component of the Ras-Raf-MAPK pathway, and blocking Raf reverses transformation induced by oncogenic Ras or by abnormally active growth factor receptors. The search for specific Raf inhibitors calls for a detailed understanding of its activation mechanism; however, almost two decades after Raf discovery; our understanding of its regulation is far from complete. This shortcoming becomes even more stressing with the recent identification of B-Raf as a major oncogene in melanomas and other cancers (Wellbrock et al., 2004a). Elucidation of the Ras-component in Raf activation directed the initial drug discovery efforts aiming at blocking Ras–Raf interaction. However, with these endeavors resulting in limited success, much of the current efforts are focused on blocking Raf kinase activity, considered an essential feature for the Raf-mediated transformation (Kolch, 2002). Thus, understanding the mechanism regulating Raf kinase activity remains a focal point in the Raf study.
Phosphorylation has long been considered the main player in the Ras-mediated Raf activation; however, the mitogen-induced, Raf-activating phosphorylation site(s) has not been fully determined (Wellbrock et al., 2004a). Our result demonstrating that Raf-1 dephosphorylation primarily at EGF-induced sites results in Raf inactivation suggests that phosphorylation at these sites maintains the active Raf-1 conformation. Determining the relative significance of each of these sites requires, however, their identification. A main setback in identifying activating Raf-1 phosphorylation sites has been behind the fact that only a small fraction of cellular Raf-1 gets activated. Under certain conditions, Raf-1 phosphorylation at its basal sites can be increased by up to 10-fold in vivo (Shen et al., 2003a; Balan and Tzivion, unpublished observation), suggesting that a large pool of Raf-1 is in a nonphosphorylated form. Because Raf-1 phosphorylation on the S621 site is obligatory for Raf-1 activation, it seems that <10% of cellular Raf-1 can get activated. This, combined with previous published results, and the results shown in the present work showing that the EGF-induced phosphorylation sites have lower stoichiometry than the basal sites (Figures 1 and 2), indicate that a very small portion of cellular Raf-1 (>5%) gets activated. This low stoichiometry of Raf-1 phosphorylation on its activation sites has been a main obstacle in identifying Raf-1-activating phosphorylation sites. Thus, in our efforts to identify novel Raf-1 phosphorylation sites, we used large quantities of Raf-1 transfected COS-7 cells as the starting material, and nearly 4 to 5 μg of purified Raf-1 was used for each mass spectrometry analysis. Although this approach allowed the identification of five novel Raf-1 phosphorylation sites, it is important to note that this analysis is not complete. Based on the two-dimensional-phosphopeptide maps, Raf-1 has at least one more major unidentified basal phosphorylation site and several minor, basal and EGF-induced sites that the mass spectrometry analysis failed to pick up. Thus, completing the identification of all the sites will require further analysis and the use of complementary approaches because some phosphopeptides do not separate well using mass spectrometry. Of the five newly identified sites, the role of S289, S296, and S301 in mediating a positive feedback Raf-1 regulation by ERK was reported previously (Balan et al., 2005), and the role of S29 remains to be determined, although this site does not seem to be essential for Raf-1 kinase activation by EGF under our experimental conditions.
The fifth site, located at a tryptic peptide corresponding to Raf-1 471-483, turned to be the most significant finding of this analysis. This peptide is within the Raf-1 kinase domain (subdomain VIB-VII) in a region that is highly conserved among all Raf family members (Figure 4C) and considered to be nearby the kinase activation loop (Wellbrock et al., 2004a). Importantly, phosphorylation at this peptide was observed only in Raf-1 from EGF-treated cells but not in Raf-1 produced from serum-deprived cells. The 471-483 tryptic peptide contains two potential phosphorylation sites: S471 and T481. The mass spectrometry analysis failed to pinpoint which of these sites was actually phosphorylated; however, our functional mutational analysis showing a strict requirement for a phosphorylatable residue at the 471 site but not at the 481 site, suggests that the 471 site is the site that gets phosphorylated. Our analysis shows a strict requirement for a serine at the 471 site for Raf-1 kinase activity, which cannot be substituted by any other amino acid we tried besides to some extent threonine. The 481 site, on the other hand, showed much tolerance to substitutions, and even the nonphosphorylatable amino acid cysteine was able to support full Raf-1 kinase activity. Our analysis also demonstrated the requirement of serine at the 471 site for the activity of a constitutively active Raf kinase form, Bxb-Raf, suggesting that this site is needed for the actual catalytic activity of Raf. Most importantly, the corresponding site in B-Raf, S578, is required for wild-type B-Raf kinase activation by growth factors but not for the activity of the cancer-associated, naturally occurring B-Raf V599E mutant, suggesting that having a charged residue at the 599 site eliminates the need for S578 phosphorylation. Collectively, these findings point to a general role of the S471/S578 sites in the activation of various Raf family members and indicate that the S471/S578 mutations used in the study do not abruptly disrupt the Raf kinase structure but rather specifically prevent Raf activation by growth factors. This notion is further supported by the findings that Raf-1 S471-substituted mutants seem to maintain structural integrity as far as their ability to form dimers and interact with Ras and 14-3-3 is unchanged.
The exact site that mediates Raf-MEK binding is not well defined and a recent work pointed Raf T481 site as the MEK binding site (Pearson et al., 2000). Although this identification was in the context of a truncated Raf-1 mutant containing only the Raf-1 kinase domain, our work shows that T481 substitution does not affect MEK binding in the context of full-length Raf-1. Rather, our data show that S471 seems to be the critical site for MEK binding. Whether the S471 site is sufficient for the interaction or whether it is actually involved in the binding remains to be determined.
Our identification of Raf-1 S471 as a critical phosphorylation site is the first report of Raf-1 phosphorylation at a site near the kinase “activation loop” showing a significant effect on Raf kinase activity. A previous alanine scan analysis within the presumed Raf-1 “activation loop” failed to pinpoint a site important for Raf-1 kinase activity (Barnard et al., 1998). In an unrelated work, phosphorylation studies of B-Raf identified two sites in the presumed “activation loop,” which mutation impairs B-Raf kinase activity (Zhang and Guan, 2000). Their parallel sites in Raf-1, T491, and S494 seem to have an effect on the kinase activity; however, the evidence that these sites are indeed phosphorylated in Raf-1 is limited (Chong et al., 2001). In addition, mutation of these sites, as reported in the alanine scan study, does not seem to have much effect on Raf-1 kinase activation by Ras G12V, vSrc, or PMA (Barnard et al., 1998).
Acknowledgments
This work was supported by National Institute of Health Grant R01 GM-067134 (to G. T.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-02-0090) on August 10, 2005.
References
- Alessandrini, A., Greulich, H., Huang, W., and Erikson, R. L. (1996). Mek1 phosphorylation site mutants activate Raf-1 in NIH 3T3 cells. J. Biol. Chem. 271, 31612-31618. [DOI] [PubMed] [Google Scholar]
- Alessi, D. R., Cuenda, A., Cohen, P., Dudley, D. T., and Saltiel, A. R. (1995). PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J. Biol. Chem. 270, 27489-27494. [DOI] [PubMed] [Google Scholar]
- Avruch, J., Khokhlatchev, A., Kyriakis, J. M., Luo, Z., Tzivion, G., Vavvas, D., and Zhang, X. F. (2001). Ras activation of the Raf kinase: tyrosine kinase recruitment of the MAP kinase cascade. Recent Prog. Horm. Res. 56, 127-155. [DOI] [PubMed] [Google Scholar]
- Avruch, J., Zhang, X. F., and Kyriakis, J. M. (1994). Raf meets Ras: completing the framework of a signal transduction pathway. Trends Biochem. Sci. 19, 279-283. [DOI] [PubMed] [Google Scholar]
- Barnard, D., Diaz, B., Clawson, D., and Marshall, M. (1998). Oncogenes, growth factors and phorbol esters regulate Raf-1 through common mechanisms. Oncogene 17, 1539-1547. [DOI] [PubMed] [Google Scholar]
- Boyle, W. J., van der Geer, P., and Hunter, T. (1991). Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201, 110-149. [DOI] [PubMed] [Google Scholar]
- Bruder, J. T., Heidecker, G., and Rapp, U. R. (1992). Serum-, TPA-, and Ras-induced expression from Ap-1/Ets-driven promoters requires Raf-1 kinase. Genes Dev. 6, 545-556. [DOI] [PubMed] [Google Scholar]
- Chang, L., and Karin, M. (2001). Mammalian MAP kinase signalling cascades. Nature 410, 37-40. [DOI] [PubMed] [Google Scholar]
- Chong, H., and Guan, K. L. (2003). Regulation of Raf through phosphorylation and N terminus-C terminus interaction. J. Biol. Chem. 278, 36269-36276. [DOI] [PubMed] [Google Scholar]
- Chong, H., Lee, J., and Guan, K. L. (2001). Positive and negative regulation of Raf kinase activity and function by phosphorylation. EMBO J. 20, 3716-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler, R. E., Jr., Stephens, R. M., Saracino, M. R., and Morrison, D. K. (1998). Autoregulation of the Raf-1 serine/threonine kinase. Proc. Natl. Acad. Sci. USA 95, 9214-9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, H., et al. (2002). Mutations of the BRAF gene in human cancer. Nature 417, 949-954. [DOI] [PubMed] [Google Scholar]
- Dent, P., Jelinek, T., Morrison, D. K., Weber, M. J., and Sturgill, T. W. (1995). Reversal of Raf-1 activation by purified and membrane-associated protein phosphatases. Science 268, 1902-1906. [DOI] [PubMed] [Google Scholar]
- Dhillon, A. S., and Kolch, W. (2002). Untying the regulation of the Raf-1 kinase. Arch. Biochem. Biophys. 404, 3-9. [DOI] [PubMed] [Google Scholar]
- Dougherty, M. K., Muller, J., Ritt, D. A., Zhou, M., Zhou, X. Z., Copeland, T. D., Conrads, T. P., Veenstra, T. D., Lu, K. P., and Morrison, D. K. (2005). Regulation of Raf-1 by direct feedback phosphorylation. Mol. Cell 17, 215-224. [DOI] [PubMed] [Google Scholar]
- Garnett, M. J., and Marais, R. (2004). Guilty as charged: B-RAF is a human oncogene. Cancer Cell 6, 313-319. [DOI] [PubMed] [Google Scholar]
- Hagemann, C., and Rapp, U. R. (1999). Isotype-specific functions of Raf kinases. Exp. Cell Res. 253, 34-46. [DOI] [PubMed] [Google Scholar]
- Hanks, S. K., and Hunter, T. (1995). Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9, 576-596. [PubMed] [Google Scholar]
- Hekman, M., Fischer, A., Wennogle, L. P., Wang, Y. K., Campbell, S. L., and Rapp, U. R. (2005). Novel C-Raf phosphorylation sites: serine 296 and 301 participate in Raf regulation. FEBS Lett. 579, 464-468. [DOI] [PubMed] [Google Scholar]
- Herrera, R., and Sebolt-Leopold, J. S. (2002). Unraveling the complexities of the Raf/MAP kinase pathway for pharmacological intervention. Trends Mol. Med. 8, S27-S31. [DOI] [PubMed] [Google Scholar]
- Inouye, K., Mizutani, S., Koide, H., and Kaziro, Y. (2000). Formation of the Ras dimer is essential for Raf-1 activation. J. Biol. Chem. 275, 3737-3740. [DOI] [PubMed] [Google Scholar]
- Kerkhoff, E., and Rapp, U. R. (2001). The Ras-Raf relationship: an unfinished puzzle. Adv. Enzyme Regul. 41, 261-267. [DOI] [PubMed] [Google Scholar]
- King, A. J., Sun, H., Diaz, B., Barnard, D., Miao, W., Bagrodia, S., and Marshall, M. S. (1998). The protein kinase Pak3 positively regulates Raf-1 activity through phosphorylation of serine 338. Nature 396, 180-183. [DOI] [PubMed] [Google Scholar]
- Kolch, W. (2002). Ras/Raf signalling and emerging pharmacotherapeutic targets. Expert Opin. Pharmacother. 3, 709-718. [DOI] [PubMed] [Google Scholar]
- Laird, A. D., Morrison, D. K., and Shalloway, D. (1999). Characterization of Raf-1 activation in mitosis. J. Biol. Chem. 274, 4430-4439. [DOI] [PubMed] [Google Scholar]
- Luo, K. X., Hurley, T. R., and Sefton, B. M. (1991). Cyanogen bromide cleavage and proteolytic peptide mapping of proteins immobilized to membranes. Methods Enzymol. 201, 149-152. [DOI] [PubMed] [Google Scholar]
- Luo, Z., Tzivion, G., Belshaw, P. J., Vavvas, D., Marshall, M., and Avruch, J. (1996). Oligomerization activates c-Raf-1 through a Ras-dependent mechanism. Nature 383, 181-185. [DOI] [PubMed] [Google Scholar]
- Lyons, J. F., Wilhelm, S., Hibner, B., and Bollag, G. (2001). Discovery of a novel Raf kinase inhibitor. Endocr. Relat. Cancer 8, 219-225. [DOI] [PubMed] [Google Scholar]
- Marais, R., Light, Y., Mason, C., Paterson, H., Olson, M. F., and Marshall, C. J. (1998). Requirement of Ras-GTP-Raf complexes for activation of Raf-1 by PKC. Science 280, 109-112. [DOI] [PubMed] [Google Scholar]
- Marais, R., Light, Y., Paterson, H. F., and Marshall, C. J. (1995). Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. EMBO J. 14, 3136-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer, K. E., and Pritchard, C. A. (2003). Raf proteins and cancer: B-Raf is identified as a mutational target. Biochim. Biophys. Acta 1653, 25-40. [DOI] [PubMed] [Google Scholar]
- Morrison, D. K., Heidecker, G., Rapp, U. R., and Copeland, T. D. (1993). Identification of the major phosphorylation sites of the Raf-1 kinase. J. Biol. Chem. 268, 17309-17316. [PubMed] [Google Scholar]
- Pearson, G., Bumeister, R., Henry, D. O., Cobb, M. H., and White, M. A. (2000). Uncoupling Raf1 from MEK1/2 impairs only a subset of cellular responses to Raf activation. J. Biol. Chem. 275, 37303-37306. [DOI] [PubMed] [Google Scholar]
- Porter, A. C., and Vaillancourt, R. R. (1998). Tyrosine kinase receptor-activated signal transduction pathways which lead to oncogenesis. Oncogene 17, 1343-1352. [DOI] [PubMed] [Google Scholar]
- Qin, J., and Zhang, X. (2002). Identification of in vivo protein phosphorylation sites with mass spectrometry. Methods Mol. Biol. 194, 211-221. [DOI] [PubMed] [Google Scholar]
- Shen, Y. H., Godlewski, J., Bronisz, A., Zhu, J., Comb, M. J., Avruch, J., and Tzivion, G. (2003a). Significance of 14-3-3 self-dimerization for phosphorylation-dependent target binding. Mol. Biol. Cell 14, 4721-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y. H., Godlewski, J., Zhu, J., Sathyanarayana, P., Leaner, V., Birrer, M. J., Rana, A., and Tzivion, G. (2003b). Cross-talk between JNK/SAPK and ERK/MAPK pathways: sustained activation of JNK blocks ERK activation by mitogenic factors. J. Biol. Chem. 278, 26715-26721. [DOI] [PubMed] [Google Scholar]
- Stancato, L. F., Chow, Y. H., Hutchison, K. A., Perdew, G. H., Jove, R., and Pratt, W. B. (1993). Raf exists in a native heterocomplex with hsp90 and p50 that can be reconstituted in a cell-free system. J. Biol. Chem. 268, 21711-21716. [PubMed] [Google Scholar]
- Tzivion, G., and Avruch, J. (2002). 14-3-3 Proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J. Biol. Chem. 277, 3061-3064. [DOI] [PubMed] [Google Scholar]
- Tzivion, G., Luo, Z., and Avruch, J. (1998). A dimeric 14-3-3 protein is an essential cofactor for Raf kinase activity. Nature 394, 88-92. [DOI] [PubMed] [Google Scholar]
- Tzivion, G., Luo, Z. J., and Avruch, J. (2000). Calyculin A-induced vimentin phosphorylation sequesters 14-3-3 and displaces other 14-3-3 partners in vivo. J. Biol. Chem. 275, 29772-29778. [DOI] [PubMed] [Google Scholar]
- Tzivion, G., Shen, Y. H., and Zhu, J. (2001). 14-3-3 proteins; bringing new definitions to scaffolding. Oncogene 20, 6331-6338. [DOI] [PubMed] [Google Scholar]
- Wan, P. T., et al. (2004). Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 116, 855-867. [DOI] [PubMed] [Google Scholar]
- Wellbrock, C., Karasarides, M., and Marais, R. (2004a). The RAF proteins take centre stage. Nat. Rev. Mol. Cell. Biol. 5, 875-885. [DOI] [PubMed] [Google Scholar]
- Wellbrock, C., Ogilvie, L., Hedley, D., Karasarides, M., Martin, J., Niculescu-Duvaz, D., Springer, C. J., and Marais, R. (2004b). V599EB-RAF is an oncogene in melanocytes. Cancer Res 64, 2338-2342. [DOI] [PubMed] [Google Scholar]
- Yip-Schneider, M. T., Miao, W., Lin, A., Barnard, D. S., Tzivion, G., and Marshall, M. S. (2000). Regulation of the Raf-1 kinase domain by phosphorylation and 14-3-3 association. Biochem. J. 351, 151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. H., and Guan, K. L. (2000). Activation of B-Raf kinase requires phosphorylation of the conserved residues Thr598 and Ser601. EMBO J. 19, 5429-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Yao, B., Delikat, S., Bayoumy, S., Lin, X. H., Basu, S., McGinley, M., Chan-Hui, P. Y., Lichenstein, H., and Kolesnick, R. (1997). Kinase suppressor of Ras is ceramide-activated protein kinase. Cell 89, 63-72. [DOI] [PubMed] [Google Scholar]
- Zimmermann, S., Rommel, C., Ziogas, A., Lovric, J., Moelling, K., and Radzi-will, G. (1997). MEK1 mediates a positive feedback on Raf-1 activity independently of Ras and Src. Oncogene 15, 1503-1511. [DOI] [PubMed] [Google Scholar]