Abstract
The pathogenic fungus Histoplasma capsulatum escapes innate immune defenses and colonizes host macrophages during infection. After the onset of adaptive immunity, the production of the antimicrobial effector nitric oxide (.NO) restricts H. capsulatum replication. However, H. capsulatum can establish persistent infections, indicating that it survives in the host despite exposure to reactive nitrogen species (RNS). To understand how H. capsulatum responds to RNS, we determined the transcriptional profile of H. capsulatum to .NO-generating compounds using a shotgun genomic microarray. We identified 695 microarray clones that were induced ≥4-fold upon nitrosative stress. Because our microarray clones were generated from random fragments of genomic DNA, they did not necessarily correspond to H. capsulatum open reading frames. To identify induced genes, we used high-density oligonucleotide tiling arrays to determine the genomic boundaries and coding strand of 153 RNS-induced transcripts. Homologues of these genes in other organisms are involved in iron acquisition, energy production, stress response, protein folding/degradation, DNA repair, and .NO detoxification. Ectopic expression of one of these genes, a P450 nitric oxide reductase homologue, was sufficient to increase resistance of H. capsulatum to RNS in culture. We propose that H. capsulatum uses the pathways identified here to cope with RNS-induced damage during pathogenesis.
INTRODUCTION
Histoplasma capsulatum, the etiologic agent of histoplasmosis, is a systemic dimorphic fungal pathogen. H. capsulatum exists in two morphological forms: a mycelial (or filamentous) form in soil and a yeast form in the host. During infection, mycelial fragments and associated spores are inhaled by the host. Once inside the host, conversion of these cells to the budding yeast form is triggered within hours. Yeast cells evade killing and multiply within macrophages (Bullock, 1993; Eissenberg and Goldman, 1994). Subsequently, yeast cells use host phagocytic cells as vehicles to spread to multiple organs of the reticuloendothelial system such as the spleen, liver, lymph nodes, and bone marrow. In patients with disseminated disease, a variety of additional organs can be colonized (Eissenberg and Goldman, 1991).
As described above, naïve macrophages are susceptible to colonization by H. capsulatum. However, upon induction of a cell-mediated immune response, macrophages become activated and gain the ability to restrict H. capsulatum replication (Newman, 1999). The anti-Histoplasma activity of activated murine macrophages is dependent on production of .NO (Lane et al., 1994; Nakamura et al., 1994; Newman, 1999).
A number of studies indicate that .NO and related reactive species are important antimicrobial effectors produced by macrophages (for reviews on RNS and pathogenesis, see Fang, 1999, 2004; Nathan and Shiloh, 2000; Shiloh and Nathan, 2000; Missall et al., 2004). To produce .NO, macrophages induce the transcription of inducible nitric oxide synthase (NOS2, also known as iNOS). Whereas NOS2 mRNA levels rise moderately during initial infection, a much more significant induction occurs upon interferon (IFN)-γ stimulation of macrophages by T-cells (Shiloh and Nathan, 2000). Nos2p converts arginine into l-citrulline and the free radical .NO. .NO readily combines with thiols, metals, and reactive oxygen species (such as superoxide, O2-·) to form multiple reactive species, collectively referred to as reactive nitrogen species (RNS) (Nathan and Shiloh, 2000; Fang, 2004). RNS react with many cellular components, including transition metals, lipids, thiol moieties, and DNA bases. These perturbations result in DNA and membrane damage, inhibition of replication and respiration, and inactivation of other cellular enzymes.
Whereas cell culture experiments suggest that .NO is required to restrict H. capsulatum replication in activated macrophages (Lane et al., 1994; Nakamura et al., 1994; Newman, 1999), the inhibitory effect of .NO on H. capsulatum is fungi-static rather than fungicidal (Nakamura et al., 1994). This result suggests that H. capsulatum is able to resist killing by .NO in host cells. Additionally, the immune system is not able to completely eliminate H. capsulatum; instead, yeast cells remain latent in the host for many years (Eissenberg and Goldman, 1991, 1994; Bullock, 1993). Because the ability of H. capsulatum to withstand nitrosative stress is likely to contribute to pathogenesis, the goal of this work was to uncover candidate genes that might contribute to the response of this organism to RNS. Because molecular genetic tools in H. capsulatum are still limited, we took a functional genomics approach to identify H. capsulatum genes that are induced in response to treatment with .NO donors. We identified the first set of candidate genes and genetic pathways that H. capsulatum may use to cope with nitrosative stress.
MATERIALS AND METHODS
Strains and Culture Growth
H. capsulatum strain G217B (ATCC 26032; a kind gift of William Goldman, Washington University, St. Louis, MO) was grown in Histoplasma Macrophage Medium (HMM) broth (Worsham and Goldman, 1988) or on HMM agar plates supplemented with 5 mg/ml bovine serum albumin (BSA). H. capsulatum strain G217B ura5- (a kind gift of William Goldman; Woods and Goldman, 1992) was grown in HMM broth supplemented with 0.2 mg/ml uracil (Sigma-Aldrich, St. Louis, MO). After transformation with URA5-containing plasmids, G217B ura5- was grown in HMM broth or on HMM agarose plates with no uracil supplementation. H. capsulatum cultures were passaged every 2-3 d at 1:25 dilution. Liquid cultures were grown at 37°C under 5% CO2 on an orbital shaker. Plates were grown at 37°C under 5% CO2 in a humidified incubator.
Cell Collection for Microarray Analysis
For each microarray sample, 100 ml of cells were harvested by vacuum filtration, frozen in liquid nitrogen, and stored at -80°C until RNA preparation.
Dipropylenetriamine NONOate (DPTA NONOate) Dose-Response Experiments
DPTA NONOate (Cayman Chemical, Ann Arbor, MI) was resuspended in 4°C 10 mM NaOH to a final concentration of 1 M. The exact concentration of the stock solution was determined by measuring absorption at 252 nm. Because DPTA NONOate is very stable in alkaline solutions, nitric oxide release was not initiated until addition of the stock to HMM, pH 7.5.
The half-life of DPTA NONOate under our experimental conditions was determined using a protocol modified from Hrabie et al. (1993). A 1 M stock of DPTA NONOate/10 mM NaOH was diluted to a final concentration of 50 mM in HMM and placed in a 37°C incubator with 5% CO2. Spectrophotometric measurements at 252 nm were taken every 30 min for the first 2 h and then every 30-90 min for the next 6 h. A final A252 reading taken at 50 h was used as the A∞ measurement. ln(A - A∞) versus time was plotted to determine the rate constant for the reaction. According to the equation t1/2 = 0.6931 (1/k), where k equals the rate constant, the half-life under our experimental conditions was calculated to be 90 min.
Two-day-old H. capsulatum yeast cultures were diluted to 4 × 107 cells/ml and grown for 24 h until they reached mid-logarithmic phase. Two aliquots of cells representing untreated, zero-minute time points were harvested. The remaining cells were split into eight cultures. DPTA NONOate (1 M) resuspended in NaOH was added to six of the cultures to result in final concentrations of 0, 0.1, 0.5, 0.8 1, or 2 mM DPTA NONOate. Because the DPTA NONOate stock contains NaOH, we controlled for NaOH addition by supplementing the first five of these cultures with 10 mM NaOH to give a final concentration of 20 μM NaOH, which matches the NaOH concentration in the 2 mM DPTA NONOate sample. An equivalent volume of water was added to the seventh culture. To the last culture, 1 M DPTA NONOate was added to a final concentration of 5 mM DPTA NONOate/50 μM NaOH. Samples were harvested 2 h after treatment for subsequent RNA analysis. Spectrophotometric readings at 600 nm were taken in triplicate at 0, 2, 5, and 25 h after treatment for 0, 0.5, 1, and 2 mM DPTA NONOate. Spectrophotometric readings at 600 nm were taken for the remaining cultures at 0, 2, and 25 h after treatment. Serial dilutions of cells were prepared in triplicate for each culture at 0, 2, and 25 h after treatment. Dilutions were plated in duplicate on HMM agar plates containing bovine serum albumin (BSA). Colony forming units (CFUs) were assessed 12 d later.
DPTA NONOate Kinetic Time Course
DPTA NONOate (1 M) stock and cell cultures were prepared as described above. An aliquot of cells was harvested before the beginning of the time course. DPTA NONOate was added to a final concentration of 1.2 mM DPTA NONOate/12 μM NaOH. Samples were harvested 0.17, 0.5, 1, 2, 5, and 25 h after treatment. A mock treatment time course was performed after addition of 12 μM NaOH.
S-Nitrosoglutathione (GSNO) Dose-Response Experiment
Two-day-old H. capsulatum yeast cultures were diluted to 3 × 107 cells/ml and grown for 24 h until they reached mid-logarithmic phase. Two aliquots of cells representing untreated, zero-minute time points were harvested. The remaining cells were split into five cultures. GSNO (Axxora Life Sciences, San Diego, CA) resuspended in HMM was added to four of the cultures to result in final concentrations of 0.2, 0.4, 1, or 5 mM GSNO. As a control, an equivalent volume of HMM was added to the fifth culture. Spectrophotometric readings at 600 nm were taken in triplicate at 0, 2, 5, 10, and 25 h after GSNO addition. Serial dilutions of cells were prepared in triplicate for each culture at 0, 2, and 25 h after treatment. Dilutions were plated in duplicate on HMM agar plates containing BSA. CFUs were assessed 12 d later.
Environmental Stress Experiments
For each environmental stress experiment H. capsulatum was diluted to 1.8 × 107 cells/ml and grown for 48 h. An aliquot of cells was harvested before treatment, and then each time course was initiated by the addition of diamide, menadione, dithiothreitol (DTT), or sorbitol. For the diamide time course, 1 M diamide (Sigma-Aldrich) was added to the culture to result in a final concentration of 1.5 mM. Samples were removed at 10, 20, 30, 40, 60, and 90 min after treatment. For the menadione time course, 1 M menadione (Sigma-Aldrich) was added to the culture to result in a final concentration of 1 mM. Samples were removed at 10, 20, 30, 50, 80, and 340 min after treatment. For the DTT time course, 1 M DTT (Roche Diagnostics, Indianapolis, IN) was added to the culture to result in a final concentration of 2.5 mM. Samples were removed at 30, 60, 120, 240, and 480 min after treatment. For the sorbitol time course, an equal volume of 2 M sorbitol in HMM (Sigma-Aldrich) was added to the culture, resulting in a final concentration of 1 M sorbitol. Samples were removed at 5, 15, 30, and 60 min after treatment.
Log Phase Time Course
H. capsulatum was diluted to 1.8 × 107 cells/ml and grown for 48 h. Samples were harvested at 0, 15, 30, 60, 180, 240, 480 min, and 24 h after treatment.
Cell Lysis and RNA Isolation
Total RNA was isolated using a guanidine thiocyanate lysis protocol (Hwang et al., 2003). Poly-adenylated (poly-A) RNA was isolated from total RNA using an Oligotex mRNA kit (QIAGEN, Valencia, CA) or an oligonucleotide-dT cellulose (Ambion, Austin, TX) column. The 2 mM DPTA NONOate total RNA sample from dose-response experiment 2 was lost during manipulations.
Probe Preparation, Microarray Hybridization, and Data Acquisition for Shotgun Genomic Microarray
Fluorescently labeled cDNA was made by incorporating amino-allyl dUTP during reverse transcription of poly-A RNA with oligonucleotide-dT and random hexamers. Cy5 or Cy3 dyes (GE Healthcare, Piscataway, NJ) were coupled to the amino-allyl group as described previously (DeRisi et al., 1997). For each sample, the cDNA was coupled to Cy5. All poly-A RNAs from a given time course or dose-response experiment were combined to prepare a reference cDNA pool, which was coupled to Cy3. A Cy5-labeled cDNA probe was mixed with its corresponding Cy3-labeled cDNA reference and competitively hybridized on a shotgun genomic microarray. Arrays were scanned using either a GenePix 4000A or GenePix 4000B scanner (Axon Instruments/Molecular Devices, Union City, CA).
Shotgun Genomic Microarray Data Analysis
Arrays were analyzed using GenePix PRO version 3.0 (Axon Instruments/Molecular Devices), NOMAD (http://derisilab5.ucsf.edu/NOMAD), CLUSTER (Eisen et al., 1998), and Java Treeview 1.0.8 (available at http://sourceforge.net/project/showfiles.php?group_id=84593). Each spot on the microarray corresponds to a random genomic fragment that will be referred to hereafter as a microarray clone. To eliminate clones with low signal, we did not analyze clones for which the sum of the medians for the 635- and 532-nm channels was ≤500 intensity units. Because all samples were hybridized against reference pools, each ratio measurement was normalized relative to its respective untreated zero-minute time point by dividing the expression ratio for each clone in a sample by the expression ratio in its respective untreated zero-minute time point. For each clone in each dose-response experiment, the mean of the expression ratios from the duplicate untreated, zero-minute samples was calculated and used for the normalization described here.
Cluster analysis was performed on the two DPTA NONOate experiments and one GSNO dose-response experiment. We identified 714 induced clones and 964 repressed clones as ≥4-fold changed in at least one sample (compared with the zero-minute sample) and present in 80% of the samples. Further analysis was performed on clones that were induced in nitrosative stress. 678/718 clones were induced ≥2-fold in at least two dose-response experiments compared with their zero-minute sample. Another 17 clones were induced ≥4-fold in at least one dose-response sample versus the zero-minute sample but had data for <80% of the samples. The resultant set of 695 RNS-induced clones is shown in Figure 2.
Figure 2.
Cluster analysis of gene expression profiles of RNS-induced clones in a variety of stresses. Cells were subjected to a variety of stresses or control treatments as described in Materials and Methods. Gene expression profiling was performed on the resultant samples, and Cluster analysis was used to group genes with similar expression profiles. For each experiment, relative to its zero-minute time point, red boxes indicate up-regulation, green boxes indicate down-regulation, black boxes indicate no change, and gray boxes indicate missing data. YvM indicates a comparison of expression in yeast-phase cells (green) with mycelial-phase cells (red) as described in Hwang et al. (2003). Individual clusters A-E are described in the text and/or Figures 4, 5, 6, 7.
RNS-Induced Microarray Clone Annotation
Incyte Genomics (Wilmington, DE) sequenced one end of each clone using the M13-forward primer, and the Genome Sequencing Center (Washington University, St. Louis, MO) sequenced both ends of each clone twice using M13-forward and M13-reverse primers.
Sequence reads were aligned to the contigs of the H. capsulatum genome sequencing project (Genome Sequencing Center, http://genome.wustl.edu/projects/hcapsulatum) using BLASTN (Altschul et al., 1997). Alignments were required to have a maximum E-value of 1E-99 or high identity (>95%) over their length. Additional alignments were accepted upon manual curation. For clones with forward and reverse reads, the contig sequence between and including the sequence reads was designated the microarray clone sequence. Because the average size of the microarray clones was 1-2 kb (Hwang et al., 2003), the sequence of clones with only one end read was defined as the contig sequence beginning with the sequence read and continuing for 1 kb.
Some sequence reads aligned to multiple locations in the genome and were likely to represent repetitive sequence. All clone sequence reads were compared with a library of H. capsulatum repetitive sequences (provided by the Genome Sequencing Center) using BLASTN. Sequence reads with minimum alignment lengths of 100 base pairs and 80% identity were designated repetitive sequence. BLASTN was used to determine whether clones that could be located in the genome corresponded to transposon-related or rDNA sequences (Hwang et al., 2003). Of the 695 RNS-induced clones, 356 contained sequences identified as repetitive or transposon-related and three contained rDNA sequence.
In addition, we removed another 54 clones from further analysis. Twenty-one of these clones lacked sequence information, did not align to the genome sequence, or aligned to the genome sequence at more than one location but did not correspond to transposons/repeat sequences. The other 33 clones in this group were not pursued further because they clustered with the transposon/repeat clusters described in the text. These clusters showed induction in response to multiple stresses and therefore did not seem likely to represent a specific response to RNS.
Further annotation was performed on the remaining 282 array clones. Contigs with mapped array clones were subjected to BLASTX analysis against the nonredundant protein database (nr) maintained by the National Center for Biotechnology Information. As described in the Results, a total of 41 array clones could be unambiguously annotated. The remaining 241 clones were located in the genome sequence but could not be unambiguously annotated by BLASTX analysis for reasons described in the Results. The majority of these clones were pursued further as described below.
To determine how many of the 282 clones were specifically induced in RNS, the fold change of clones in each experiment was evaluated. Clones were designated as induced in RNS if they were up-regulated ≥4-fold in at least one RNS-treated sample compared with the zero-minute sample. Clones were designated as not induced in stress if they were <2-fold in all stress experiments (diamide, menadione, sorbitol, and DTT) and RNS controls (untreated dose-response 1 and 2, NaOH-treated dose-response 1 and 2, untreated GSNO dose response, and 12 μM NaOH time course). In addition, clones had to have data for 80% of the stress experiments to be considered in this analysis. Fifty-nine clones were up-regulated ≥4-fold in RNS (compared with the zero-minute sample) and <2-fold in stresses/controls. These were designated specifically induced by RNS.
Tiling Microarray Construction
Of the 241 unannotated clones described above, 198 were chosen for tiling analysis using high-density oligonucleotide tiling arrays (CombiMatrix, Mukilteo, WA). (All 241 could not be included because of space restrictions on the tiling arrays.) An additional 19 clones that had already been annotated were included as controls to verify that the tiling arrays identified RNS-induced transcripts. This brought the total number of tiled clones to 217.
Because some of the clones represented overlapping genomic regions, the 217 clones corresponded to 105 genomic regions. In addition, 19 genomic regions also were tiled for which we had additional data suggesting up-regulation in response to RNS, bringing the total number of tiled genomic regions to 124. The boundaries of each tiled region were chosen according to criteria described in the Results. The sequence of each tile was selected by CombiMatrix. The tiling oligonucleotides were between 20 and 50 base pairs in length. The oligonucleotide length was varied to minimize the range of melting temperatures; 99% of the tiles were separated by 9-10 base pairs relative to the contig sequence.
Tiling Microarray Probe Preparation, Hybridization, and Data Acquisition
Fluorescently labeled probes were generated as described previously with one modification; amino-modified C6 5′-labeled oligonucleotide-dT primer was used for reverse transcription. CombiMatrix microarrays were washed before use as described in the “shampoo” protocol (http://derisilab.ucsf.edu/core/resources/index.html). Cy5-coupled 0.8 mM or 0.5 mM DPTA NONOate (2-h time point) samples were competitively hybridized against Cy3-coupled NaOH (2-h time point) samples as described previously (DeRisi et al., 1997). Dose-response 1 samples were used for this experiment. Cy5-coupled 5 mM GSNO (2-h time point) samples were competitively hybridized against Cy3-coupled 0 mM GSNO (2-h time point) samples. Microarrays were hybridized 12-14 h at 55°C with constant orbital rotation on a Nutator (Clay Adams Brand, Fisher Scientific, Pittsburgh, PA).
Arrays were scanned using a GenePix 4000B scanner. To normalize the 635- and 532-nm signal, the 635- and 532-nm PMT levels were adjusted to give equivalent background signal from hybridization to the CombiMatrix quality control oligonucleotide QC-oligoB-AS-3, present 200 times on the array. Because QC-oligoB-AS-3 does not align to the H. capsulatum genome or mitochondrial sequence, binding by sample probes represents background and should be equivalent between the Cy3 and Cy5 probes. The data sets were analyzed using GenePix PRO version 3.0. The mean of the 635- and 532-nm median signal intensities for the 200 QC-oligoB-AS-3 spots was considered background and subtracted from the 635- and 532-nm median signal intensities, respectively.
Tiling Microarray Transcript Identification and Annotation
The 635- and 532-nm signal intensities were plotted as a function of tile position using the Generic Genome Browser (Gbrowse, http://www.gmod.org). Transcripts were chosen manually according to the following criteria: 1) at least five contiguous oligonucleotides with a minimum 635-nm intensity of 750 intensity units and 2) at least one tile with 635-nm intensity >1000 intensity units. Transcripts that seemed to have at least twofold higher 635-nm signal intensity verses 532-nm signal intensity were designated RNS-induced. We identified 135 RNS-induced transcripts from the 124 tiled genomic regions because some regions contained more than one RNS-induced gene. Transcripts that had <2-fold RNS induction were designated constitutive and not analyzed further. No transcripts were identified with higher 532- verses 635-nm signal intensity. One transcript was induced only in GSNO but not in DPTA NONOate, whereas three transcripts showed much stronger induction in DPTA NONOate than in GSNO. Some regions showed no induced transcripts, and microarray clones corresponding to these regions were annotated as “No Induction.” The GBrowse pages showing the tiling results, identified transcripts, and their Aspergillus nidulans, Saccharomyces cerevisiae, and Schizosaccharamyces pombe homologues can be found in the Supplemental Materials.
We defined background signal intensity as the median signal intensity of all tiles outside the constitutive and RNS-induced transcript areas. On each of the arrays, the background signal was <300 intensity units, indicating that our signal cutoff of 750 was sufficiently above background for transcript identification.
RNS-induced transcripts were aligned by BLASTX to nr and the following predicted gene sets: A. nidulans (Broad Institute, Oct. 2003), Magnaporthe grisea (Broad Institute, Oct. 2003), Fusarium graminearum (Broad Institute, Sep. 2003), Neurospora crassa (Munich Information Center for Protein Sequences, June 2004), Cryptococcus neoformans (National Center for Biotechnology Information GenBank, Feb. 2005), Candida albicans (Biotechnology Research Institute/National Research Council Canada, Dec. 2004), S. cerevisiae (Saccharomyces Genome Database/National Center for Biotechnology Information GenBank, Mar. 2004), S. pombe (Sanger Institute, Mar. 2005), Yarrowia lipolytica (Genolevures, May 2004), and Phanerochaete chrysosporium (DOE Joint Genome Institute, Mar. 2004). Only alignments that corresponded to the correct strand, extended across 75% of the query length, and had a maximum E-value of E-7 were designated homologues. Two exceptions to these criteria are noted in the Results.
Oligonucleotides
All oligonucleotides used for standard molecular biology manipulations are listed in Supplemental Table 4.
Northern Analysis
Total RNA (5 μg) from DPTA NONOate dose-response 2 samples was used for Northern analysis (Hwang et al., 2003). NOR1 and ACT1 probes were PCR amplified from genomic DNA using gene-specific primers.
NOR1 5′ Rapid Amplification of cDNA Ends (RACE), 3′ RACE, and cDNA Sequencing
Poly-A RNA from 0-h untreated and 2-h 0.1 mM DPTA NONOate-treated cells (dose-response 1) was used to map the NOR1 transcript ends and coding sequence. The 5′ and 3′ ends of cDNAs were determined using the First-Choice RNA-ligase mediated (RLM)-RACE kit (Ambion, Austin, TX) per manufacturer's instructions. cDNA reverse-transcribed from NONOate treated cells (dose-response 1) was used as a template to amplify the NOR1 coding sequence (cds). All amplified products were cloned using TOPO-TA (Stratagene, La Jolla, CA) and sequenced using a combination of M13-forward, M13-reverse, and gene-specific primers as necessary.
NOR1 Ectopic Expression Experiment
The copper responsive protein 1 (CRP1) promoter and catalase B (CATB) terminator were PCR amplified from G217B genomic DNA using primers containing restriction sites. The longer of the two NOR1 induced coding sequences was PCR amplified from cDNA, as described above, using primers containing restriction sites. All amplified products were cloned using TOPO-TA and sequenced as described above. After restriction digestion, the CRP1 promoter, CATB terminator, and NOR1 cds were gel purified and cloned into the Gateway entry vector, pENTR2B (Invitrogen, Carlsbad, CA), creating pDHM1. An analogous plasmid lacking the NOR1 cds also was created (pCC4).
To construct the Gateway destination vector, a polylinker was introduced into the BamHI site of H. capsulatum plasmid pWU55 (Woods et al., 1998) creating pDG29AS. To replace the Podospora anserina URA5 with H. capsulatum URA5, pDG29AS was first enzymatically digested with EcoRI, and the ends of the larger fragment were filled using Klenow. A plasmid containing H. capsulatum URA5 was created by PCR amplifying H. capsulatum URA5 from G217B genomic DNA, cloning the amplified product into TOPO-TA, and sequencing the product as described above. This plasmid was digested with the blunt cutting enzyme, HpaI, and the resulting URA5 fragment was ligated with the prepared pDG29AS blunt-ended backbone creating pDG31AS. Finally, the Gateway destination vector pDG33 was created by ligating the Gateway vector conversion cassette reading frame A into HpaI-digested pDG31AS.
Gateway recombination of pDHM1 with pDG33 resulted in pDHM5, an H. capsulatum plasmid containing CRP1 promoter-NOR1 cds-CATB terminator, H. capsulatum URA5, and telomeric repeats flanking a kanamycin resistance cassette. Gateway recombination of pCC4 with pDG33 yielded pDG45, the vector control, lacking the NOR1 cds. pDHM5 and pDG45 were digested with PacI to expose the telomeric ends and transformed into G217B ura5- after gel purification. Prototrophic colonies were selected on HMM agar plates.
For ectopic expression experiments, 2-d cultures of G217B ura5- transformed with pDHM5 or pDG45 were diluted to 1.6 and 1.7 × 107 cells/ml, respectively. The cultures were grown for 24 h until they were in mid-logarithmic phase. Copper sulfate (10 mM; Fisher Scientific) was added to each culture to a final concentration of 10 μM. After 1.5 h, each culture was split into three. DPTA NONOate (1 M), prepared as described previously, and 10 mM NaOH were added to each culture to achieve a final concentration of 0, 1, or 2 mM DPTA NONOate/20 μM NaOH. Spectrophotometric readings at 600 nm were taken in triplicate 0 and 24 h after DPTA NONOate treatment.
RESULTS
Identification of RNS-regulated H. capsulatum Genomic Regions Using a Shotgun Genomic Microarray
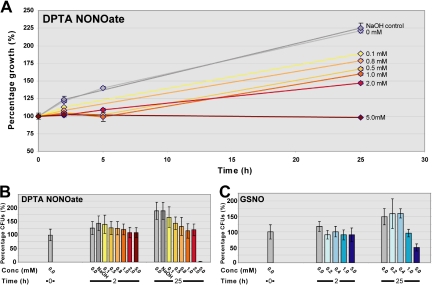
To examine the response of H. capsulatum to RNS-induced damage, cells were exposed in vitro to two different sources of nitrosative stress and then collected for further genomic analysis. In one set of experiments, yeast cultures were exposed to increasing doses of DPTA NONOate (0, 0.1, 0.5, 0.8, 1, 2, or 5 mM), which releases .NO at a slow, sustained rate at neutral pH. In a second set of experiments, cells were exposed to GSNO, a naturally occurring RNS that can release .NO or cause transnitrosylation of proteins. Cells were exposed to 0, 0.2, 0.4, 1, or 5 mM GSNO. The immediate effects of nitrosative stress on the growth rate of H. capsulatum cultures were measured spectrophotometrically (OD600) at 0, 2, 5, and 25 h after treatment (Figure 1A). We observed dose-dependent effects on growth rate, which became apparent as early as 2 h after DPTA NONOate treatment. In addition, the viability of H. capsulatum cultures was assessed by determination of CFUs from samples harvested 0, 2, and 25 h after treatment (Figure 1, B and C). CFU data did not show a large difference in viability at the 2-h time point, especially given the inherent variability in the efficiency of colony formation for H. capsulatum. However, by 25 h after treatment, CFU analysis revealed that 5 mM DPTA NONOate and 5 mM GSNO each had significant effects on viability. Specifically, treatment with 5 mM DPTA NONOate resulted in a 99% decrease in CFUs, and treatment with 5 mM GSNO resulted in a 50% decrease in CFUs compared with the initial time point. In all dose-response experiments, cells were harvested for transcriptional analysis immediately before and 2 h after RNS addition. We chose the 2-h time point for transcriptional analysis because the cells were experiencing sufficient RNS stress to slow growth rate without a significant reduction in viability.
Figure 1.
Nitrosative stress causes dose-dependent growth inhibition of H. capsulatum. H. capsulatum G217B cultures were grown to mid-log phase and treated with a range of concentrations of DPTA NONOate or GSNO. (A) Spectrophotometric readings at OD600 were taken at 0, 2, 5, and 25 h after the addition of DPTA NONOate. The starting OD was set at 100% growth. (B) Colony-forming units relative to the 0-h, 0-mM time point were determined at 0, 2, and 25 h after addition of DPTA NONOate. (C) Colony-forming units relative to the 0-h, 0-mM time point were determined at 0, 2, and 25 h after addition of GSNO. Data shown for A and B are from dose-response experiment 1. SD is indicated for each measurement in A, B, and C.
To examine the transcriptional profile of H. capsulatum in response to nitrosative stress, we used a shotgun genomic microarray. In brief, each of the 10,000 spots on the shotgun genomic microarray corresponds to a random fragment from the H. capsulatum genome. This array has been used to perform large-scale analysis of transcript abundance in the absence of a fully sequenced and annotated genome (Hwang et al., 2003). To determine the transcriptional response to nitrosative stress, poly-adenylated RNA isolated from each RNS-treated sample was subjected to microarray analysis and compared with controls as described in Materials and Methods.
Cluster analysis (Eisen et al., 1998), which groups array clones with common gene expression profiles, was used to compare the expression profiles observed in three RNS dose-response experiments (two DPTA NONOate experiments and one GSNO experiment; Figure 2). Overall, 714 clones were induced ≥4-fold in at least one dose-response sample compared with their relevant zero-minute sample, and 964 clones were repressed ≥4-fold in at least one dose-response sample compared with their relevant zero-minute sample. A comparison of the two DPTA NONOate dose-response experiments showed that a similar set of genes was induced in both experiments but not with the same dose dependence. Interestingly, no cluster was observed to be specific to a particular nitrosative stress, and in fact, the intensity of induction in GSNO was very similar to the second DPTA NONOate dose-response experiment. Subsequent analysis focused on array clones that were induced in at least two of the three RNS dose-response experiments compared with their zero-minute samples (695 clones). Because neither the shotgun microarray nor the H. capsulatum genome was annotated at the time of this work, it took considerable effort to determine the identity of transcripts that corresponded to each regulated clone from the microarray. Thus, we chose to focus on annotating only the induced (but not the repressed) clones, because these are the most likely to represent genes necessary for RNS response or resistance.
Annotation of Array Clones
The next step of our analysis was to determine which genes were represented by the 695 clones on the shotgun genomic microarray. Complete details of how the microarray clones were annotated can be found in Materials and Methods. To summarize, single-sequence reads that marked the boundaries of the genomic insert in each array clone were mapped onto contigs from the ongoing H. capsulatum genome sequencing project at the Washington University in St. Louis Genome Sequencing Center (http://genome.wustl.edu/projects/hcapsulatum). BLASTX analysis of the GSC contigs against the nr peptide sequence database maintained by the National Center for Biotechnology Information allowed us to locate homologues on the contig sequences. We determined the overlap between the location of microarray clones of interest and nr homologues relative to the genome sequence. Based on information from sequenced fungal genomes, we hypothesized that the minimum upstream regulatory sequence in H. capsulatum is 300 base pairs, and the minimum 3′-untranslated sequence is 200 base pairs. Therefore, if a microarray clone contained either >300 base pairs 5′ of a region of nr homology or >200 base pairs 3′ of a region of nr homology, it had the potential to overlap a second gene. Forty-one clones were unambiguously annotated because 1) the microarray clone and nr homologue completely overlapped or 2) the microarray clone and nr homologue did not completely overlap, but the remaining clone sequence was of insufficient size to contain a second gene. Two hundred seventy-four clones could not be annotated because 1) the microarray clone overlapped more than one nr homologue, 2) the microarray clone had the potential to overlap more than one nr homologue, or 3) the microarray clone did not overlap any nr homologues. In addition to the 41 clones mentioned above, 356 microarray clones were annotated as transposons/repeats because they aligned with either previously identified transposon-related sequences (Hwang et al., 2003) or H. capsulatum repeat families identified by the GSC. Another three microarray clones were annotated as rDNA because they aligned with previously identified rDNA sequence (Hwang et al., 2003). The remaining 21 clones could not be analyzed because they either lacked sequence information or could not be uniquely located in the genome sequence.
Comparison of Gene Expression Profiles of RNS-induced Clones in Other Stresses
Before proceeding with further annotation, we determined which clones were induced specifically in nitrosative stress. Work in S. cerevisiae and S. pombe demonstrated that a common set of genes changes in expression in response to all stresses. In contrast to this general environmental stress response, individual stresses also trigger a specific gene expression response (Gasch et al., 2000; Causton et al., 2001; Chen et al., 2003). To categorize the RNS-induced clones into those 1) induced in all stresses, 2) induced in a subset of stresses, and 3) induced specifically in nitrosative stress, we examined the transcriptional profiles of these clones in response to other stresses. To generate these data, H. capsulatum was exposed to a superoxide generator (menadione), a sulfhydryl oxidant (diamide), a disulfide reductant (DTT), and osmotic shock (sorbitol). Doses and time courses were chosen to match previous studies in S. cerevisiae (Gasch et al., 2000). To identify clones that change in expression during and after exponential growth, we determined the gene expression profile of log phase (this study) and stationary phase cultures (Hwang et al., 2003). In addition, a time course of 1 mM DPTA NONOate treatment (or mock treatment with 12 μM NaOH) consisting of 0-, 0.5-, 1-, 2-, 5-, and 25-h time points was performed to evaluate the kinetic effects of nitrosative stress.
Cluster analysis was used to compare RNS-induced array clones across all of the stresses (Figure 2 and Supplemental Table 1). The section of the cluster labeled “E” indicates 395 clones induced in all stresses as well as in response to RNS. We had previously annotated 356 induced clones as repeats/transposons, as described above. Three hundred and fifty of these clones are located in region E, constituting 89% (350/395) of the clones in this area. The induction of these clones in multiple stresses is interesting, but not surprising. Work in S. cerevisiae has shown that retrotransposons are induced when cells are stressed by nitrogen starvation or DNA-damaging agents (Bradshaw and McEntee, 1989; Morillon et al., 2000). The remainder of the clones in section E (33) were not annotated because these clones are not likely to represent pathways used by H. capsulatum to cope specifically with RNS-induced damage.
For our subsequent analysis, we focused on the remaining 282 clones that could be located in the genome sequence and were not repeats/transposons or rDNA. The majority of these clones are induced in RNS and minor subsets of the other stresses. Fifty-nine clones are induced only in RNS, but they do not cluster together into one group. The simplest interpretation of these data is that multiple transcriptional circuits are activated by nitrosative stress, although transcriptional profiling with a whole-genome array is necessary to definitively test this hypothesis.
Identification of RNS-induced H. capsulatum Transcripts Using Tiling Arrays
The BLASTX analysis described above allowed us to annotate only 41 of the 282 clones of interest. The remaining 241 clones defined genomic regions of interest that overlapped with an RNS-induced transcript of unknown identity. Because H. capsulatum has numerous small introns, the location of genes cannot be determined simply by finding open reading frames in the vicinity of microarray clones. Consequently, we used alternative methods to map RNS-induced transcripts.
We determined the precise location and coding strand of the remaining RNS-induced transcripts using high-density oligonucleotide tiling microarrays. Work in Arabidopsis thaliana, Escherichia coli, and Homo sapiens has demonstrated the utility of tiling arrays to map transcript locations (Shoemaker et al., 2001; Tjaden et al., 2002; Yamada et al., 2003; Schadt et al., 2004). In this type of array, both strands of a region of interest are represented on a microarray by a series of end-to-end oligonucleotides. Labeled cDNA probes representing the sequence of the 5′ untranslated region (UTR), 3′ UTR, and exons will bind to their complementary oligonucleotides on the microarray and generate an increase in intensity at these targets. In contrast, intron sequences and non-UTR intergenic sequences are not present in the cDNA probe, and thus oligonucleotides representing these areas will not bind the cDNA probes. Therefore, by hybridizing labeled cDNA probes to a tiling array, the boundaries of a transcript and its coding strand can be determined.
For each of the clones that were represented on tiling microarrays, tiling regions were determined in the following manner. For clones that overlapped two nr homologues, the tiled region extended 500 base pairs beyond the end of the first homologue to 500 base pairs beyond the end of the second homologue, thus allowing us to determine which of the two potential transcripts was induced by nitrosative stress (Figure 3A). For clones that had the potential to overlap 2 nr homologues as described above, the tiled region began 500 base pairs outside the nr homologue, continued through the clone sequence, and extended at least 1 kb outside the clone sequence (Figure 3C). For clones that did not overlap an nr homologue, the tiled region consisted of the clone sequence and at least 1 kb on either side (Figure 3E). In all cases, tiles were designed to be a maximum of 50 base pairs in length and no more than 10 base pairs apart relative to the contig sequence. The transcript for one of our control genes, H. capsulatum alternative oxidase 1 (AOX1), had previously been mapped (Johnson et al., 2003). For this region, we tiled 1 kb upstream and downstream of the defined coding sequence (Figure 3G). In total, 540.85 kb of genomic DNA were tiled, requiring 21,536 tiles. As described in Materials and Methods, the tiling arrays represented 217 microarray clones and 124 genomic regions.
Figure 3.
Examples of genomic regions that were represented on tiling microarrays and subjected to gene expression profiling. A, C, and E represent generalized schematic examples of genomic regions that were tiled for further analysis. The heavy black horizontal arrows represent genomic sequence. The gray rectangles represent the region of genomic sequence represented in the microarray clone. The green arrows represent the location of BLASTX hits for homologues in other organisms. The blue squares represent individual tiling oligonucleotides that together represent the entire genomic region of interest. A is an example of a microarray clone that overlaps two homologues, C is an example of a microarray clone that has partial overlap with one homologue but contains sufficient sequence to potentially overlap a second unknown transcript as described in Results, and E represents a microarray clone with no overlap to any BLASTX hits. B, D, and F show examples of actual gene expression data for three specific genomic regions that correspond to the generalized categories depicted in A, C, and E, respectively. The top black line represents the genomic sequence with hatch marks every 100 base pairs, and the gray rectangles represent microarray clone sequence. The regions of BLASTX homology are labeled and shown in green (A. nidulans), yellow (S. cerevisiae), or red (S. pombe). Arrowheads reflect the direction of the BLASTX hit. The intensity units for each tile are graphed for both the minus and plus strands in the lower part of each figure. Cy5 signal (red) reflects the transcript profile in RNS stress, whereas Cy3 signal (green) reflects the transcript profile in the absence of RNS. B shows the gene expression data for H.c. FAT1, which is present on the plus strand. D shows the gene expression data for H.c. ATF1, which is present on the minus strand. F shows the gene expression data for the novel transcript NIT72, which is present on the plus strand. The boundaries of each of these three transcripts are delineated by vertical black arrows. G shows the tiling data for the control transcript AOX1 as described in the text. The known H. capsulatum AOX1 coding sequence (Johnson et al., 2003) is shown in yellow and intron sequences are represented as gaps between the yellow exons. The 5′ and 3′ UTR sequences from AOX1 clone 1 sequence are represented as gray boxes. The regions of the genomic sequence that are represented in the transcript (5′ UTR, 3′ UTR, and exons) are shaded in gray to emphasize the correspondence between these regions and the tiles that give a high intensity signal on the plus strand graph.
Labeled cDNAs from existing RNS-treated samples were hybridized to the tiling microarrays. In each case, the RNS-treated sample was labeled with Cy5, and the corresponding uninduced sample was labeled with Cy3. The median intensity of the 635-nm channel (red) and 532-nm channel (green) was plotted as a function of oligonucleotide position for the plus and minus strands. To verify that the arrays were correctly and accurately identifying transcribed regions, the signal from the AOX1 control region was examined. The transcript boundaries predicted by the tiling data matched the published sequence of the longest AOX1 transcript remarkably well (Figure 3G). Figure 3, B, D, and F, show specific examples of data obtained for the categories of clones defined in Figure 3, A, C, and E, respectively. These data confirmed that H. capsulatum RNS-induced transcripts could be identified using the tiling microarrays. Intensity graphs for each of the defined transcripts are shown in Supplemental Materials.
Each of the tiled regions was examined for RNS-induced transcripts. We chose a minimum signal cutoff of 750 intensity units to qualify a tile as potentially representing part of a transcript because the mean intensity of tiles on the noncoding strand and intergenic regions was <300 intensity units in each of the arrays examined. Because we observed rare isolated oligonucleotides that gave a signal intensity >750 intensity units, we designated genomic regions as transcripts if they had 1) at least five contiguous oligonucleotides with a minimum 635-nm intensity of 750 and 2) at least one tile with intensity >1000. The first base pair of the 5′-induced tiling oligonucleotide and the last base pair of the 3′-induced tiling oligonucleotide were arbitrarily designated as the transcript boundaries, although the actual ends of the transcript are likely to fall within the relevant tiling oligonucleotide. In some cases, the signal intensity did not decline to baseline at the ends of the tiled region, indicating that the tiled region did not contain the entire transcript (Figure 3B). These transcripts are indicated as “partial” in Table 1 and Supplemental Tables 2 and 3. In each case, the strand of the induced transcript also was noted. Transcripts that were expressed at equivalent levels in the RNS-treated and untreated samples were not pursued. Only 14 tiled regions did not contain any transcripts by our criteria.
Table 1.
H. capsulatum RNS-induced transcripts with named homologues
| Transcript namea | Organism | Closest homologueb | E-Valuec | Accession no. of closest homologue | Organism | Characterized homologue | E-Valued | Accession no. of characterized homologue | Transcripte | Shotgunf | Tilingg |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Iron acquisition | |||||||||||
| NPS1 | Af | Nonribosomal peptide synthetase 6 (Nps6p) | 0E+00* | AAX11421.1 | Partial | x | x | ||||
| LOM1 | An | Ornithine monooxygenase (SidAp) | 1E-104 | AAP56238.1 | Complete | x | x | ||||
| NIT1 | An | AN6400.2 | 3E-93 | EAA58422.1 | Cn | Putative ferricchelate reductase | 9E-50 | AAW44420.1 | Partial | x | x |
| STR1 | An | Siderophore iron transporter (MirCp) | 0E+00 | Q870L3 | Sp | Siderophore iron transporter 1 (Str1p) | 5E-88 | O74395 | Unknown | x | |
| Respiration | |||||||||||
| COX12 | An | AN6255.2 | 9E-17**h | EAA58639.1 | Sc | Cytochrome c oxidase polypeptide Vib (Cox12p) | 2E-9h | NP_013139.1 | Complete | x | x |
| MCP2 | An | AN2977.2 | 1E-133 | EAA63548.1 | At | Mitochondrial phosphate transporter | 3E-71 | BAA31585.1 | Partial | x | x |
| SFC1 | Nc | Probable succinate-fumarate transporter | 1E-112 | CAE85498.1 | Sc | Succinate/fumarate mitochondrial transporter (Sfc1p) | 2E-75 | NP_012629.1 | Partial | x | x |
| NDH1 | Mg | MG04999.4 | 1E-102 | EAA52307.1 | Yl | Alternative NADH-dehydrogenase (Ndh2p) | 9E-56 | CAA07265.1 | Complete | x | x |
| CYC1 | Tl | Cytochrome c | 3E-35 | P00047 | Complete | x | x | ||||
| AOX1 | Hc | Alternative Oxidase 1 (Aox1p) | 0E+00 | AF133236 | Complete | x | x | ||||
| Lipid metabolism/oxidation | |||||||||||
| FAT1 | An | AN5192.2 | 1E-144** | EAA62373.1 | Sc | Fatty acid transporter and very long-chain fatty acyl-CoA synthetase (Fat1p) | 7E-66** | NP_009597.2 | Partial | x | x |
| FAA1 | An | AN6014.2 | 0E+00* | EAA57655.1 | Sc | Long-chain-fatty-acid–CoA ligase (Faa1p) | 1E-144 | NP_014962.1 | Complete | x | x |
| FOX2 | Fg | Peroxisomal hydratase-dehydrogenase-epimerase (FOX2_NEUCR) | 0E+00* | EAA76166.1 | Unknown | x | |||||
| SCS7 | An | AN0918.2 | 1E-138 | EAA65947.1 | Sc | Fatty acid hydroxylase (Scs7p) | 2E-98 | NP_013999.1 | Complete | x | x |
| Stress response | |||||||||||
| WIS1 | An | AN0931.2 | 0E+00 | EAA65960.1 | Sp | Protein kinase (Wis1p) | 1E-110 | NP_595457.1 | Partial | x | x |
| ATF1 | An | Transcription factor (AtfAp) | 1E-120 | AAN75015.1 | Sp | Transcription factor (Atf1p) | 5E-36 | NP_595652.1 | Complete | x | x |
| Protein folding/degradation | |||||||||||
| CDC37 | An | AN2051.2 | 1E-162 | EAA64883.1 | Sc | Hsp90p cochaperone (Cdc37p) | 5E-53 | NP_010452.1 | Complete | x | x |
| YDJ1 | An | AN2731.2 | 1E-159 | EAA63029.1 | Sc | Yeast dnaJ homologue (Ydj1p) | 3E-95 | NP_014335.1 | Partial | x | x |
| STI1 | An | AN9124.2 | 0E+00* | EAA61957.1 | Sc | Heat shock protein (Sti1p) | 1E-136 | NP_014670.1 | Partial | x | x |
| UFD1 | Nc | NCU05582.1 | 0E+00* | XP_325437.1 | At | UFD1-like protein | 7E-29 | AAM63245.1 | Partial | x | x |
| HSP70 | Hc | HSP70 | 0E+00 | AAC05418.1 | Sc | ATPase involved in protein folding and the response to stress (Ssa3p) | 1E-164** | NP_009478.1 | Unknown | x | |
| CUT8 | An | AN4411.2 | 1E-122 | EAA60328.1 | Sp | Cut8p | 2E-27 | NP_594593.1 | Partial | x | x |
| YME1 | Nc | Intermembrane space AAA protease (IAP-1p) | 0E+00* | AAG48698.1 | Sc | Intermembrane space AAA protease IAP-1 (Yme1p) | 1E-165 | NP_015349.1 | Partial | x | x |
| UBC1 | Fg | Ubiquitin-conjugating enzyme E2 (Ubc1p) | 4E-51 | EAA75159.1 | Complete | x | |||||
| UBI1 | Cg | CAGL0D05082g | 1E-155 | CAG58542.1 | Sc | Polyubiquitin (Ubi4p) | 1E-155 | NP_013061.1 | Complete | x | |
| DNA repair | |||||||||||
| HMP1 | Nc | Related to mismatched base pair and cruciform DNA recognition protein | 1E-15 | CAD37005.1 | Um | Mismatch base pair and cruciform DNA recognition (Hmp1) | 3E-07 | AAA86754.1 | Complete | x | x |
| NIT2 | Af | AfA5C5.060 | 1E-115 | CAF31982.1 | Sc | Wss1p | 1E-07 | NP_012002.1 | Partial | x | x |
| YNG2 | An | AN9126.2 | 1E-140 | EAA61959.1 | Sc | NuA4 histone acetyltransferase complex component (Yng2p) | 4E-20** | NP_011958.1 | Partial | x | |
| Nitric oxide detoxification/NOS2 inhibition | |||||||||||
| NOR1 | Ct | Cytochrome P450 nitric oxide reductase 2 (P450nor2) | 1E-100 | Q12599 | Fo | P450nor | 7E-96 | BAA03390.1 | Complete | x | x |
| CAR1 | An | Arginase (Arg1p) | 1E-134 | EAA63472.1 | Complete | x | x | ||||
| IDO1 | Nc | NCU05752.1 | 1E-113** | XP_325607.1 | Sc | Indoleamine-pyrrole 2,3-dioxygenase (Bna2p) | 1E-63 | NP_012612.1 | Complete | x | x |
| Oxidative response | |||||||||||
| CATA | Hc | Catalase A (CatAp) | 0E+00 | AAM53416.1 | Complete | x | x | ||||
| Signaling and transcriptional regulation | |||||||||||
| NIT3 | Ao | Putative transcriptional activator | 1E-130 | BAB47239.1 | Partial | x | x | ||||
| CRE1 | Te | Catabolite repressor (CreTp) | 1E-162 | AAL33631.4 | Complete | x | x | ||||
| PHO4 | An | PHO4-like protein, transcription factor (PalcAp) | 1E-114 | AAT02190.1 | Nc | Phosphorus acquisition controlling protein (Nuc1p) | 6E-71 | P20824 | Partial | x | x |
| NIT4 | An | AN2265.2 | 0E+00 | EAA63838.1 | Sp | Serine/threonineprotein kinase yeast ksp1-like | 7E-65 | NP_595795.1 | Partial | x | x |
| NIT5 | An | AN4935.2 | 1E-131 | EAA61013.1 | Nh | Ran1-like protein kinase | 5E-97 | AAA96531.1 | Complete | x | x |
| NIT6 | An | Putative transcription factor (RfeDp) | 1E-96 | AAO14632.1 | Partial | x | x | ||||
| RPC34 | An | AN9125.2 | 1E-109 | EAA61958.1 | Sp | DNA-directed RNA polymerase iii subunit (Rpc34p) | 5E-44 | NP_588398.1 | Complete | x | x |
| Transporters | |||||||||||
| ABC1 | Ao | ABC transporter (SidTp) | 0E+00* | BAC78652.1 | Partial | x | x | ||||
| ABC5 | Pb | Putative ATP-binding cassette transporter protein (Pfr1p) | 0E+00* | CAD90041.1 | Af | Multidrug resistance protein 2 (Mdr2p) | 0E+00 | AAB88660.1 | Partial | x | x |
| DNF1 | Sc | Potential phospholipid-transporting ATPase (Dnf1p) | 0E+00* | NP_011093.1 | Partial | x | |||||
| NIT7 | An | AN8791.2 | 1E-138 | EAA60584.1 | Sc | Putative metal transporter (Mmt2p) | 5E-52 | NP_015100.1 | Partial | x | x |
| SUL1 | An | AN4645.2 | 1E-147 | EAA60447.1 | Dp | Sulfate permease (SulPp) | 1E-41 | YP_064024.1 | Unknown | x | |
| ABC3 | An | ABC transporter protein (AtrFp) | 0E+00* | CAC42217.1 | Unknown | x | |||||
| NIT8 | Nc | NCU06341.1 | 1E-140 | XP_326196.1 | At | Transporter related | 2E-36 | NP_189965.2 | Unknown | x | |
| CRP1 | Ca | Copper-transporting P-type ATPase (Crp1p) | 0E+00* | AAF04593.1 | Unknown | x | |||||
| Intracellular trafficking | |||||||||||
| NIT9 | Sp | C2-domain, putative synaptic vesicle protein | 0E+00* | NP_587862.1 | Partial | x | |||||
| VPS13 | Sc | Vacuolar protein sorting-associated protein (Vps13p) | 0E+00* | NP_013060.1 | Partial | x | |||||
| NIT10 | Sp | Synaptobrevin-like V snare protein (Ykt6p) | 2E-17 | NP_596561.1 | Complete | x | |||||
| NIT11 | Nc | NCU05067.1 | 1E-121 | XP_324424.1 | Ca | Potential intra-Golgi transport complex subunit 5 (Cog5p) | 9E-15 | EAL04719.1 | Partial | x | x |
| NGT1 | Hc | N-glycosyl-transferase | 0E+00 | AAF90184.1 | Unknown | x | |||||
| Protein and DNA metabolism | |||||||||||
| NIT12 | An | AN5753.2 | 2E-30 | EAA62846.1 | Nc | Related to mitochondrial ribosomal protein | 5E-24 | 7nc651_010i | Complete | x | x |
| DPS1 | An | AN0314.2 | 0E+00* | EAA65720.1 | Sc | Aspartyl-tRNA synthetase (Dps1p) | 1E-122 | NP_013083.1 | Partial | x | |
| PPO1 | An | AN4780.2 | 6E-81 | EAA60350.1 | Sp | Pyridoxamine 5′-phosphate oxidase | 1E-17 | SPAC1952.08cj | Complete | x | x |
| CYD1 | Rb | Cytosine deaminase | 3E-09 | NP_868564.1 | Complete | x | x | ||||
| PNP1 | An | AN7550.2 | 0E+00* | EAA62130.1 | Zm | Phosphodiesterase-nucleotide pyrophosphatase | 3E-68 | YP_163339.1 | Partial | x | |
| MET1 | An | Cobalamin-independent methionine synthase (MetH/Dp) | 0E+00* | AAF82115.1 | Unknown | x | |||||
| GLN1 | An | AN4159.2 | 1E-156 | EAA59420.1 | An | Glutamine synthetase (GlnAp) | 1E-154 | AAK70354.1 | Unknown | x | |
| Other | |||||||||||
| CBR1 | An | AN6366.2 | 1E-129 | EAA58750.1 | Ma | NADH-cytochrome b5 reductase | 2E-72 | BAA85587.1 | Partial | x | x |
| FBP26 | Fg | FG04343.1 | 1E-166 | EAA70953.1 | Sc | Fructose-2,6-bisphosphate 2-phosphatase (Fbp26p) | 1E-129 | NP_012380.1 | Partial | x | x |
| RIB7 | An | AN6979.2 | 0E+00 | EAA61625.1 | Sc | HTP reductase (Rib7) | 7E-37 | NP_009711.1 | Partial | x | x |
| NIT13 | Lp | lpl2034 | 1E-106 | YP_127370.1 | Bf | Threonine dehydrogenase and related Zn-dependent dehydrogenases: COG1063 | 1E-105 | ZP_00284346.1 | Partial | x | x |
| NIT14 | An | AN2470.2 | 1E-137 | EAA63788.1 | Av | Threonine dehydrogenase and related Zn-dependent dehydrogenases: COG1063 | 4E-56 | ZP_00162515.1 | Complete | x | x |
| NIT15 | An | AN8053.2 | 0E+00* | EAA59675.1 | Sp | Probable inositol polyphosphate phosphatase | 1E-148 | T39233 | Partial | x | |
| NIT16 | An | AN9140.2 | 1E-152 | EAA61973.1 | Sc | Mitochondrial protein (Fmp42p) | 1E-104 | NP_013948.1 | Partial | x | |
| NIT17 | An | AN4539.2 | 2E-40 | EAA60882.1 | At | Yippee family protein | 2E-17 | NP_973645.1 | Partial | x | |
| NIT18 | An | AN1950.2 | 0E+00* | EAA65115.1 | Nc | Calcium-related spray protein | 0E+00 | XP_326529.1 | Partial | x | |
| NIT19 | Nc | Related to samB | 0E+00* | XP_326745.1 | Partial | x | x | ||||
| NIT20 | An | AN1875.2 | 1E-116 | EAA65040.1 | Sp | Zinc finger protein | 8E-61 | T41390 | Complete | x | x |
| NIT21 | Mg | MG00041.4 | 2E-44 | EAA48383.1 | Dh | Predicted O-methyltransferase: COG4122 | 2E-16 | ZP_00343262.1 | Complete | x | x |
| NIT22 | Fg | FG04688.1 | 7E-57 | EAA72605.1 | Nc | Related to estradiol 17β-dehydrogenase | 7E-58 | xnc107_200i | Complete | x | x |
| NIT23 | An | AN4779.2 | 1E-145 | EAA60349.1 | Dr | NIPSNAP1 protein | 5E-36 | CAB56701.1 | Partial | x | x |
| NIT24 | An | AN8817.2 | 1E-107 | EAA60610.1 | Nc | Clock-controlled protein 8 | 4E-24 | XP_330336.1 | Partial | x | x |
| NIT25 | Nc | NCU03781.1 | 1E-108 | XP_323082.1 | Pm | Putative GTPase, G3E family | 5E-28 | NP_875899.1 | Partial | x | x |
| NIT26 | Mg | MG07406.4 | 1E-25 | EAA53129.1 | Fs | Cytochrome P450 monooxygenase (Tri11p) | 6E-08 | AAD12755.1 | Partial | x | x |
| NIT27 | An | AN0446.2 | 0E+00 | EAA66545.1 | Nc | Related to suppressor protein PSP1 | 1E-170 | XP_328216.1 | Partial | x | |
| NIT28 | Af | Putative zinc finger protein | 1E-152 | CAD29608.1 | Complete | x | x | ||||
| DBP1 | An | AN5931.2 | 0E+00** | EAA57794.1 | Sc | RNA helicase (Dbp2p) | 1E-152** | NP_014287.1 | Partial | x | |
| RED1 | An | AN0895.2 | 1E-137 | EAA65924.1 | Ch | Reductase (Red1p) | 2E-96 | AAM88292.1 | Unknown | x | |
| NIT29 | An | AN8530.2 | 1E-140 | EAA66883.1 | Gf | Cytochrome P450 monooxygenase (P450Ip) | 5E-93** | CAA75565.1 | Unknown | x |
Af, A. fumigatus; An, A. nidulans; Ao, A. oryzae; At, A. thaliana; Av, A. variabilis; Bf, B. fungorum; Ca, C. albicans; Cg, C. glabrata; Ch, C. heterostrophus; Cn, C. neoformans; Ct, C. tonkinense; Dr, D. rerio; Dh, D. hafniense; Dp, D. psychrophila; Fo, F. oxysporum; Fs, F. sporotrichioides; Gf, G. fujikuroi; Fg, F. graminearum; Hc, H. capsulatum; Lp, L. pneumophila; Ma, M. alpina; Mg, M. grisea; Nc, N. crassa; Nh, N. haematococca; Pb, P. brasiliensis; Pm, P. marinus; Rb, R. baltica; Sc, S. cerevisiae; Sp, S. pombe; Te, T. emersonii; Tl, T. lanuginosus; Um, U. maydis; Yl, Y. lipolytica; and Zm, Z. mobilis.
Nitrosative stress-induced transcription
Closest homologue shown unless closest homologue also is characterized, in which case it is under `Characterized homologue'
E-value comparing H. capsulatum transcript and its closest homologue
E-value comparing H. capsulatum transcript and its characterized homologue
Designates if the defined transcript is likely to be complete, partial, or of unknown length
`x' denotes transcripts represented on the shotgun genomic microarray
`x' denotes transcripts represented on the tiling microarray
Homologue is <100 aa leading to lower E-value despite high homology
Munich Information Center for Protein Sequences (http://mips.gsf.de/proj/neurospora/)
S. pombe Gene Database (http://www.genedb.org/genedb/pombe/index.jsp)
More than one homologue exists with this E-value
More than one blast alignment to homologue. Lowest E-value alignment shown
In sum, a total of 153 RNS-induced transcripts were identified using the combination of shotgun genomic and tiling microarrays described above. We named these transcripts and submitted them to GenBank.
Annotation of RNS-induced Transcripts
For each of the identified transcripts, homologues were sought that would provide insight into gene function. BLASTX analysis was conducted on each identified transcript against the nr peptide database and predicted gene sets from a variety of fungal species (A. nidulans, M. grisea, F. graminearum, N. crassa, S. pombe, S. cerevisiae, C. albicans, C. neoformans, Y. lipolytica, and P. chrysosporium). A transcript was annotated as having a homologue if 1) the H. capsulatum transcript extended across 75% of the homologue's length, 2) the E-value was less than E-7 (with 2 exceptions for homologues with sequences <100 amino acids), and 3) the transcript was on the same strand as the homologue. Fifty-three percent of the identified transcripts had a named homologue in another organism, 25% were homologous to “hypothetical” proteins in another organism, and 22% had no identifiable homologues in any organism (Table 1 and Supplemental Tables 2 and 3). For the 149 previously unidentified H. capsulatum genes, we named them based on their putative homologue in another organism. In cases where this nomenclature scheme was not applicable, we named genes nitrosative stress-induced transcript (NIT). All annotations were applied to the cluster comparing RNS-induced clones to other environmental stresses (Figure 2)
Clusters of Induced Clones Suggest Particular Pathways Are Triggered by Exposure to RNS
The ultimate goal of this work is to identify genes and pathways that play an important role in the ability of H. capsulatum to respond to nitrosative stress. Genes that function in the same pathway are often transcriptionally coregulated and thus may be grouped together by Cluster analysis of transcriptional data over a series of conditions (Hughes et al., 2000). Because we used a shotgun genomic microarray that did not cover the entire genome, we did not have all genes represented on the microarray, and therefore, we could not generate all relevant clusters. Thus, some genes of interest clearly fall into clusters, whereas others do not. Here, we describe both RNS-induced pathways as defined by groups of genes with similar function that cluster together as well as individual genes that do not fall into obvious clusters at this time.
Iron Acquisition
Cluster A, shown in Figures 2 and 4, has homologues of several genes involved in iron acquisition in other fungi. This cluster contains a homologue of l-ornithine monooxygenase (H.c. LOM1; Hwang and Rine, personal communication), an enzyme that performs the first committed step in siderophore production. Siderophores are small molecules secreted by microbes to scavenge iron from the environment. In Ustilago maydis, the l-ornithine monooxygenase and nonribosomal peptide synthetase required for siderophore production are grouped together in the genome (Yuan et al., 2001). Our iron acquisition cluster also contains a nonribosomal peptide synthetase (H.c. NPS1), which, similar to U. maydis, is grouped together in the genome with H.c. LOM1. Two other genes of unknown function from this cluster are adjacent to NPS1 and LOM1 in the genome. The cluster analysis indicates these neighboring genes are coregulated with LOM1 and NPS1, suggesting that they may function in siderophore production as well. This iron acquisition cluster also includes a putative ferric chelate reductase (H.c. NIT1) that may function either to remove iron from siderophores after uptake of siderophore/Fe3+ complexes, or to reduce iron to Fe2+ outside of the cell before iron import (De Luca and Wood, 2000). H.c. STR1, a homologue of a siderophore iron transporter from S. pombe, also is induced upon RNS exposure but is present in a different cluster (Table 1).
Figure 4.
Iron acquisition cluster. Cluster A from Figure 2 is expanded to show the characteristics of the iron acquisition cluster. Iron acquisition genes discussed in the text are highlighted in red. The graph below the cluster diagram shows the average log2 fold change for all the genes in the cluster over each of the experimental conditions. In the DPTA NONOate and GSNO dose-response experiments, control treatments are represented by the gray bars.
Up-regulation of iron acquisition genes in response to nitrosative stress has been observed in a number of organisms (Crawford and Goldberg, 1998; Mukhopadhyay et al., 2004; Hromatka et al., 2005; Sarver and DeRisi, 2005). In E. coli, .NO inactivates the transcription factor Fur, which normally represses iron acquisition genes under high iron conditions. Thus, in nitrosative stress, Fur-regulated repression is relieved, and iron acquisition genes are induced (D'Autreaux et al., 2002; Mukhopadhyay et al., 2004). Induction of H. capsulatum iron acquisition genes may reflect .NO-induced modification of an unidentified iron-regulated transcriptional repressor.
Energy Production
Genes involved in energy production also are induced upon RNS exposure as seen in cluster B (Figures 2 and 5. Reactive nitrogen species are known to inhibit components of the electron transport chain [complexes I, II, III, and IV] [Brown, 1999]). Exposure to RNS may trigger up-regulation of genes that encode the components themselves or genes that allow bypass of standard complexes of the electron transport chain. We observed induction of homologues of cytochrome c oxidase subunit VIb (H.c. COX12) and cytochrome c (H.c. CYC1), which are standard components of the electron transport chain used to produce ATP for cellular processes. We also observed induction of a protein homologous to a mitochondrial succinate-fumarate transporter (H.c. SFC1), which, in other organisms, transports succinate into the mitochondrial matrix either for use by the tricarboxylic acid (TCA) cycle or to donate electrons to complex II (Palmieri et al., 1997; Joseph-Horne et al., 2001). A phosphate transporter homologue (H.c. MCP2) also was induced in response to RNS. In other organisms, this protein transports phosphate into the mitochondrial matrix for use in ATP production by the ATP synthase that is coupled to the electron transport chain (Laloi, 1999). Alternative components of the electron transport chain, such as alternative oxidase (H.c. AOX1), were also up-regulated. In other organisms, Aox1p is able to perform the terminal reduction of O2 to water and bypass complexes III and IV of the electron transport chain (Joseph-Horne et al., 2001). Unlike complex IV, Aox1p is refractory to inhibition by .NO; thus, its up-regulation may help to ensure that energy production can continue in the presence of .NO (Joseph-Horne et al., 2001). Alternatively, induction of Aox1p may relieve the oxidative stress associated with inhibition of complex IV (Joseph-Horne et al., 2001). Additionally, a homologue of alternative NADH dehydrogenase (H.c. NDH1) is also up-regulated upon RNS exposure but is found in a different cluster. In other organisms, this protein facilitates transfer of electrons from NADH to ubiquinone, the shuttle protein between complexes I and III (Joseph-Horne et al., 2001), thus allowing electrons from NADH to bypass complex I and enter the electron transport chain. In addition to maintaining electron flow, this process regenerates oxidized NAD+. As long as sufficient quantities of NAD+ exist, energy could continue to be produced via glycolysis in the absence of a functioning electron transport chain.
Figure 5.
Energy production cluster. Part of cluster B from Figure 2 is expanded to show the characteristics of the energy production cluster. Energy production genes discussed in the text are highlighted in blue. WIS1 of the stress response gene set is highlighted in green. The graph below the cluster diagram shows the average log2 fold change for all the genes shown over each of the experimental conditions. In the DPTA NONOate and GSNO dose-response experiments, control treatments are represented by the gray bars.
Other RNS-induced genes outside this cluster also may function in energy production. For example, several genes function in the metabolism and oxidation of lipids. Homologues of genes involved in fatty acid transport and activation (H.c. FAT1 and H.c. FAA1) as well as a gene involved in fatty acid β-oxidation (H.c. FOX2) are up-regulated (Table 1) (van Roermund et al., 2003). Fatty acid β-oxidation produces acetyl-CoA, which can be used in the TCA cycle directly or in the glyoxylate pathway to produce succinate. Succinate can be transported into the mitochondria, possibly via H.c. SFC1 (see above), where it can be used in the TCA cycle or the electron transport chain. Because the β-oxidation of fatty acids represents the highest energy-yielding pathway in the cell, up-regulation of these genes may reflect alternate pathways of energy production induced by the cell during nitrosative stress.
Stress Response
A small cluster of clones all corresponding to a homologue of the S. pombe transcription factor atf1 is shown in cluster C (Figures 2 and 6). S. pombe Atf1p is the primary transcription factor downstream of the Sty1 stress response pathway (Chen et al., 2003). This pathway operates through Wis1p, a mitogen-activated protein kinase kinase. We also observed that the H. capsulatum homologue of WIS1 is induced by RNS (Figure 5). In S. pombe, this pathway responds to a variety of stresses, including DNA damage, heat shock, oxidative stress, and high osmolarity (Hohmann, 2002). Atf1p induces transcription of a significant fraction of the core environmental stress response in S. pombe (Chen et al., 2003). We observed induction of H. capsulatum ATF1 in response to all stresses tested, including RNS exposure (Figure 6). This uniform pattern of induction suggests that the H. capsulatum Atf1p may function in a core environmental stress response.
Figure 6.
Stress response (ATF1) cluster. Cluster C from Figure 2 is expanded to show the characteristics of ATF1 induction. All of the microarray clones in this cluster correspond to H.c. ATF1. The graph below the cluster diagram shows the average log2 fold change for all of these clones over each of the experimental conditions. In the DPTA NONOate and GSNO dose-response experiments, control treatments are represented by the gray bars.
Protein Folding and Degradation
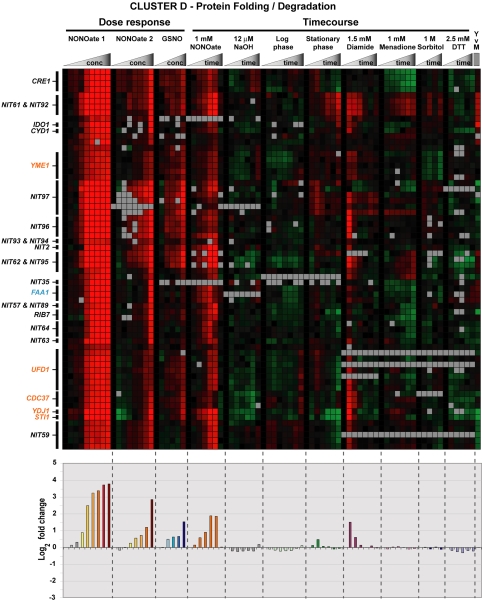
A gene cluster with many homologues of genes involved in protein folding and degradation is also induced in response to RNS (cluster D; Figures 2 and 7). We have identified homologues of S. cerevisiae CDC37, STI1, YDJ1, and YME1 as well as the Arabidopsis thaliana UFD1-like protein. S. cerevisiae Cdc37p, Sti1p, and Ydj1p are known to associate with heat-shock protein (Hsp)70p and Hsp90p to assist protein folding (Cyr et al., 1994; Bohen et al., 1995; Lee et al., 2004). We also observed induction of an Hsp70 family member (H.c. HSP70) in response to nitrosative stress, although this gene was located in a different cluster. S. cerevisiae Ydj1p also is involved in endoplasmic reticulum (ER)-associated degradation (ERAD), the process whereby misfolded proteins in the ER are removed to the cytoplasm for degradation by the proteasome (Huyer et al., 2004; Youker et al., 2004). Whereas the function of the A. thaliana UFD1-like protein has not been elucidated, S. cerevisiae Ufd1p also participates in ERAD by recognizing polyubiquitinated proteins and targeting them to the proteasome (Ye et al., 2001; Bays and Hampton, 2002). S. cerevisiae Yme1p functions in mitochondrial degradation of misfolded proteins (Nakai et al., 1995). Homologues of other RNS-induced genes outside this cluster also seem to be involved in proteasomal degradation (Table 1). For example, the homologue of H.c. CUT8 targets proteins to the proteasome for degradation in S. pombe (Tatebe and Yanagida, 2000). We also observed induction of ubiquitin (H.c. UBI1) and a ubiquitin-conjugating enzyme (H.c. UBC1) in response to RNS. Interestingly, the protein degradation cluster shows little induction in response to stresses other than RNS with the only exception being diamide, a stress known to induce protein unfolding (Figure 7). Thus, induction of these genes is relatively specific to RNS exposure in H. capsulatum, and likely reflects the cell's attempt to deal with unfolded proteins generated by nitrosative stress.
Figure 7.
Protein folding and degradation cluster. Cluster D from Figure 2 is expanded to show the characteristics of the protein folding and degradation cluster. Protein folding and degradation genes discussed in the text are highlighted in orange. The energy production gene FAA1 is highlighted in blue. The graph below the cluster diagram shows the average log2 fold change for all the genes in the cluster over each of the experimental conditions. In the DPTA NONOate and GSNO dose-response experiments, control treatments are represented by the gray bars.
Regulation of RNS-induced Genes in Other Stresses
Large-scale gene expression profiling over a variety of conditions allowed us to identify clusters of genes that were induced in nitrosative stress, as described above. In addition, we also were able to examine the response of these clusters under other stresses. Notably, two of the RNS-response clusters, iron acquisition and energy production, were down-regulated upon oxidative treatment (Figures 4 and 5). This transcriptional regulation is consistent with the known roles these proteins play in the production of oxidative species. Iron produces reactive oxygen species (ROS) by reaction with oxygen and hydrogen peroxide (De Luca and Wood, 2000), and respiration produces low concentrations of the ROS superoxide (Liu, 1997). Down-regulation of these two classes of genes under oxidative stress may help H. capsulatum avoid damage by limiting ROS production.
Homologues of Known RNS-Response or Stress-Response Genes
Some of the interesting RNS-response genes did not fall into specific clusters, yet still had intriguing connections to nitrosative stress response, as described below. Additionally, novel genes and genes with homologues of unknown function also were identified. Table 1 lists all of the identified RNS-induced transcripts that have named homologues. These transcripts are grouped according to the function of their homologous gene. RNS-induced transcripts homologous to “hypothetical” genes in other organisms are listed in Supplemental Table 2, and novel RNS-induced transcripts (with no homologues) are listed in Supplemental Table 3.
Some of the transcripts in Table 1 have homologues that have been observed to be necessary for or up-regulated during RNS response in other organisms. For example, the C. albicans transcriptional response to RNS stress includes up-regulation of a copper transporting P-type ATPase and an alternative oxidase (Hromatka et al., 2005), which we also observed. A copper transporting P-type ATPase also was induced in the E. coli response to RNS (Mukhopadhyay et al., 2004). In addition, the C. albicans as well as the S. cerevisiae RNS-induced transcriptional response includes up-regulation of a catalase (Hromatka et al.; 2005; Sarver and DeRisi, 2005). Screens in Mycobacterium tuberculosis for mutants unable to grow after RNS exposure identified proteins involved in proteasomal degradation (Darwin et al., 2003). Moreover, treatment of M. tuberculosis with inhibitors of the proteasome resulted in increased susceptibility to RNS, suggesting that protein degradation is necessary for the response of M. tuberculosis to nitrosative stress. Finally, the mammalian homologue of H.c. DBP2, p68, was strongly induced in human and murine keratinocytes upon .NO stimulation (Kahlina et al., 2004).
Homologues of other genes in Table 1 have intriguing connections to RNS stress. First, response to DNA damage is likely to be a key element of response to nitrosative stress. We observed that homologues of S. cerevisiae WSS1 (H.c. NIT2) and YNG2 (H.c. YNG2) were induced in response to RNS. In S. cerevisiae, disruption of these genes results in increased sensitivity to DNA damage, suggesting they have a role in DNA repair (Choy and Kron, 2002; O'Neill et al., 2004). A homologue of U. maydis Hmp1 (H.c. HMP1) also is induced. This protein exhibits cruciform binding activity thought to be necessary for the resolution of Holliday structures during homologous recombination (Dutta et al., 1997), which can be important for DNA repair. Second, we observed induction of a homologue of indoleamine-2,3-dioxygenase (H.c. IDO1), an enzyme in the kynurenine pathway. This protein modifies tryptophan during the first step in NAD+ synthesis (Panozzo et al., 2002). NAD+ is required for energy production and DNA repair, among other cellular processes. The up-regulation of IDO1 may allow H. capsulatum to increase levels of NAD+ during nitrosative stress. Alternatively, H. capsulatum could be using IDO1 to deplete tryptophan and thereby impair macrophage .NO production: in culture, depletion of tryptophan prevents NOS2 induction by IFN-γ in macrophages (Chiarugi et al., 2003). Additionally, a metabolite of indoleamine-2,3-dioxygenase has been shown to down-regulate NOS2 expression and inhibit Nos2p activity in a macrophage cell line (Sekkai et al., 1997; Oh et al., 2004). Finally, whereas homologues of H. capsulatum arginase (H.c. CAR1) are part of the glutamine biosynthesis pathway, other interesting possibilities exist for the role of arginase in RNS response. One of the products of the arginase reaction is l-ornithine (http://www.genome.jp/dbget-bin/www_bget?enzyme+3.5.3.1). l-ornithine is utilized by Lom1p during siderophore biosynthesis as discussed previously. By producing l-ornithine, arginase could facilitate siderophore biosynthesis and iron acquisition. Alternatively, because Nos2p uses arginine as a substrate to produce .NO, reduction of environmental levels of arginine by arginase could reduce macrophage Nos2p activity. Studies in Helicobacter pylori suggest that H. pylori arginase inhibits .NO production by macrophages by depleting extracellular arginine concentrations (Gobert et al., 2001). It is possible that H. capsulatum induces arginase in response to nitrosative stress to reduce further .NO production by host cells.
H. capsulatum P450 Nitric Oxide Reductase (P450nor) Is Sufficient to Provide Increased RNS Resistance In Vitro
In other organisms, genes involved in .NO detoxification have been observed to be up-regulated during nitrosative stress and to be required for RNS resistance. For example, flavohemoglobins, such as E. coli Hmp, are able to convert .NO into nitrate under aerobic conditions and into nitrous oxide under anaerobic conditions (reviewed in Poole and Hughes, 2000). Studies in a range of organisms including E. coli, S. typhimurium, S. cerevisiae, C. albicans, and C. neoformans have shown that deletion of flavohemoglobin results in .NO sensitivity (Crawford and Goldberg, 1998; Membrillo-Hernandez et al., 1999; Liu et al., 2000; de Jesus-Berrios et al., 2003; Ullmann et al., 2004; Hromatka et al., 2005; Sarver and DeRisi, 2005). Flavorubredoxin and NADH:(flavo)rubredoxin oxidoreductase, exemplified by E. coli NorVW, are capable of reducing .NO under anaerobic conditions (Gardner et al., 2002). Deletion of these enzymes in E. coli also results in RNS sensitivity (Gardner et al., 2002; Hutchings et al., 2002; Justino et al., 2005). BLASTX searches of the recently assembled H. capsulatum genome sequence failed to identify homologues to either of these classes of enzymes. H. capsulatum does have a homologue to GSNO reductase, an enzyme that contributes to the nitrosative stress response in other organisms by detoxifying GSNO (Liu et al., 2001; de Jesus-Berrios et al., 2003). Although the H. capsulatum GSNO reductase is transcriptionally induced upon GSNO addition (our unpublished data), the H. capsulatum gene has no apparent start codon, and thus its role in combating nitrosative stress is unclear. The only obvious candidate for a .NO detoxifying enzyme observed in these experiments was a P450nor (Figure 8A). P450nor was originally identified in F. oxysporum where it detoxifies .NO to nitrous oxide (N2O) during the process of denitrification (Shoun and Tanimoto, 1991; Nakahara et al., 1993).
Figure 8.
Nitric oxide reductase (NOR1) confers increased resistance to nitrosative stress. (A) The log2 fold change for NOR1 is shown for each of the experimental conditions from Figure 2. In the DPTA NONOate and GSNO dose-response experiments, control treatments are represented by the gray bars. (B) Northern blot analysis of NOR1 transcripts at 0 and 2 h after DPTA NONOate treatment. These samples are identical to those used for gene expression profiling in dose-response experiment 2. This blot was stripped and reprobed for the ACT1 transcript as shown (bottom). An RNA ladder is shown to the left of the gel. (C) An alignment between the predicted H.c. Nor1p and F. oxysporum P450nor protein (Fo P450nor). Identical amino acids are shaded in black. The first ATG after the transcription start site was designated as the starting methionine of each of the two mapped NOR1 transcripts. The red arrowhead marks the start of the smallest predicted Nor1 protein. (D) Ectopic expression of NOR1 enhances growth in the presence of RNS. H. capsulatum G217B ura5- cells transformed with either a vector control or NOR1 under the control of a copper-inducible promoter (PCRP1-NOR1) were grown in 10 μM copper sulfate for 1 h and then subjected to RNS stress by the addition of 0, 1, or 2 mM DPTA NONOate. Spectrophotometric readings at OD600 were taken at 0 and 25 h after treatment was initiated. The starting OD was set at 100% growth. SD is indicated for each measurement
We chose to use H. capsulatum P450nor (H.c. NOR1) to illustrate how future experiments will determine the role of RNS-induced genes in the nitrosative stress response. First, to verify the induction results seen by microarray, Northern analysis using a probe against NOR1 was performed on RNA extracted from DPTA NONOate treated H. capsulatum cultures (Figure 8B). Three NOR1 transcripts were observed; the largest transcript seemed to be constitutive, whereas two smaller transcripts were induced upon RNS stimulation with 0.1, 0.5, 0.8, and 1 mM DPTA NONOate. For unknown reasons, the cells treated with 5 mM DPTA NONOate did not show detectable transcript by Northern analysis. A combination of 5′ RACE, 3′RACE, and cDNA sequencing allowed mapping of two potential NOR1 transcripts that correspond in size to the two induced transcripts observed by Northern. The smallest of these transcripts initiates in the coding sequence. Figure 8C shows an alignment between F. oxysporum P450nor protein and the H. capsulatum Nor1p-predicted protein sequence.
To determine whether NOR1 plays a role in H. capsulatum RNS resistance, we attempted to disrupt the NOR1 gene. Unfortunately, we were unable to do so, likely because gene disruption technology in H. capsulatum still requires optimization. However, we were able to test whether ectopic expression of NOR1 could confer RNS resistance. NOR1 was expressed under the control of the CRP1 copper-inducible promoter (Gebhart et al., unpublished data), and copper-induced expression was verified by Northern blot analysis (our unpublished data). The growth of H. capsulatum strains bearing either a vector control or the ectopic expression construct (PCRP1-NOR1) was compared after DPTA NONOate treatment. Whereas both strains grew similarly in the absence of RNS, we reproducibly observed that the ectopic expression strain grew better than the vector control in the presence of DPTA NONOate. Specifically, spectrophotometric analysis showed that the NOR1 ectopic expression strain had a comparable increase in growth relative to the control culture in NaOH (75 vs. 72%) but had a significantly larger increase in growth relative to the control culture (55 vs. 23%) in 1 mM DPTA NONOate (Figure 8D). A corresponding increase in the long-term viability of cells, as judged by CFUs, also was observed (our unpublished data). Thus, expression of NOR1 is sufficient to increase RNS resistance in H. capsulatum.
DISCUSSION
Using a combination of shotgun genomic and tiling microarrays, we have identified 153 H. capsulatum transcripts that are induced in response to RNS. Even though our genomic microarray does not represent the entire genome, this analysis represents the first large-scale study of RNS response in a systemic dimorphic fungus. We identified RNS-induced genes that are implicated in many cellular processes, including iron acquisition, energy production, stress response, protein folding/degradation, DNA damage repair, and .NO detoxification (Figure 9). Any or all of these pathways may be required for resistance to RNS and fungal pathogenesis. We tested whether one identified gene, H. capsulatum NOR1, plays a role in RNS resistance. Ectopic expression of NOR1 is sufficient to provide increased RNS resistance in vitro, thus validating this approach for identifying genes involved in the RNS response. Future experiments will explore the role of genes and pathways identified in this work in host-microbe interactions and pathogenesis.
Figure 9.
A model for how gene induction in nitrosative stress causes enhanced survival in the host. A macrophage phagosome containing an H. capsulatum yeast cell is depicted. Pathways that were induced in nitrosative stress are shown. See text for details.
Several hypotheses might explain why specific transcripts are induced upon exposure to RNS. First, induced genes may represent repair or response pathways activated to cope with RNS-induced damage. RNS are known to cause damage to many cellular components, including DNA, proteins, and lipids. We observed up-regulation of proteins involved in DNA repair, which could facilitate repair of RNS-induced DNA lesions. Similarly, induction of protein folding and degradation pathways may help cells cope with proteins damaged by RNS. Additionally, RNS are known to inhibit respiration by modifying proteins of the electron transport chain (Brown, 1999), which results in increased ROS generation due to excess reduction of ubiquinone and reduced ATP synthesis due to decreased proton flux across the mitochondrial inner membrane. Induction of alternative components of the electron transport chain, such as AOX1, may allow H. capsulatum to generate limited quantities of ATP and reduce ROS production. Significantly, H. capsulatum AOX1 is induced in the presence of electron transport chain inhibitors, as well as in the presence of the ROS hydrogen peroxide (Johnson et al., 2003). Induction of catalase, which also reduces ROS levels, may provide additional protection from increased oxidative stress caused by inhibition of respiration. We also observed induction of genes involved in lipid oxidation and metabolism. This induction may reflect a need for alternate pathways for energy production or a need to replace damaged lipids.
Second, RNS-induced genes may be targets of RNS damage. We observed induction of homologues of glutamine synthetase (H.c. GLN1), catalase (H.c. CATA), and fatty acid-coA ligase (H.c. FAA1). Homologues of these genes are S-nitrosylated in M. tuberculosis upon RNS treatment (Rhee et al., 2005). Because S-nitrosylation can inhibit protein function (Rhee et al., 2005), some genes may be induced to up-regulate levels of proteins that are directly compromised by RNS-induced modifications. This hypothesis may explain why we observe the induction of standard components of the electron transport chain, such as cytochrome c and cytochrome c oxidase subunit VIb.
Third, because H. capsulatum is likely to encounter RNS during its sojourn in host macrophages, RNS might be interpreted as a host-specific signal, especially in conjunction with additional host signals, such as growth at 37°C. Thus, exposure to RNS in culture at 37°C may cause induction of genes normally required for survival in the host. An example of this type of regulation is seen in M. tuberculosis, where .NO exposure in culture triggers expression of genes involved in dormancy that are normally induced in the host (Ohno et al., 2003; Voskuil et al., 2003). In H. capsulatum, we observed induction of putative iron acquisition genes in response to RNS treatment. Because iron restriction by the host limits H. capsulatum replication (Lane et al., 1991; Newman et al., 1994), up-regulation of iron acquisition genes in response to RNS exposure may facilitate intracellular growth of H. capsulatum in host cells. Similarly, other RNS-induced genes may contribute to intracellular survival. To cope with the carbon-poor environment found in the host, some pathogens alter nutrient acquisition and metabolism pathways (Lorenz and Fink, 2002). On exposure to macrophages, C. albicans and M. tuberculosis up-regulate genes involved in the glyoxylate pathway and gluconeogenesis. If RNS is interpreted by H. capsulatum as a signal that it is in host cells, up-regulation of lipid metabolism and oxidation genes may reflect an attempt to shift to nutrient acquisition pathways that are appropriate for an intracellular lifestyle.
Finally, RNS may up-regulate genes required for detoxification of RNS. The clearest example of this type of gene is NOR1, which encodes a homologue of the F. oxysporum P450nor. F. oxysporum P450nor was originally identified as having a role in denitrification, a process used by soil microbes to generate energy under anaerobic conditions (Zumft, 1997). P450nor reduces .NO to the nontoxic nitrous oxide during fungal denitrification (Shoun and Tanimoto, 1991; Nakahara et al., 1993). Because H. capsulatum grows in the soil in the mycelial form, it also may perform NOR1-dependent denitrification. Interestingly, when H. capsulatum is grown in the yeast form, NOR1 is induced only in the presence of RNS; however, in the mycelial form, NOR1 is expressed in the absence of RNS (YvM; Figure 8A). It is tempting to hypothesize that H. capsulatum has co-opted an enzyme normally used during denitrification in the soil to overcome antimicrobial defenses in the host. To elucidate the role of NOR1 in virulence, we are continuing to try a number of different approaches to disrupt this gene. Once we have successfully inactivated NOR1, we will determine whether it contributes to H. capsulatum pathogenesis in both a macrophage model and a mouse model of disease. To our knowledge, H.c. NOR1 is the first P450nor homologue identified in a fungal pathogen of humans. Flavohemoglobins, which detoxify .NO, have been identified in the fungal pathogens C. neoformans and C. albicans; however, these organisms do not have homologues of P450nor. Because H. capsulatum has no obvious flavohemoglobin homologue in its genome, it may rely on a novel flavohemoglobin-independent mechanism for survival in host cells.
Importantly, several of the genes identified in this work have previously been implicated in pathogenesis in other organisms. H. pylori arginase mutants show decreased viability in murine macrophages (Gobert et al., 2001). Mutants in M. tuberculosis glutamine synthetase and C. neoformans alternative oxidase have been reported to have virulence defects in mouse models (Akhter et al., 2003; Tullius et al., 2003). Preliminary experiments with C. albicans fox2 mutants also suggest that FOX2 is required for pathogenesis (Lorenz, personal communication). Additionally, catalase mutants in M. tuberculosis, H. pylori, and C. albicans show decreased virulence in mice (Wysong et al., 1998; Harris et al., 2003; Ng et al., 2004). These findings further encourage us that the genes identified in this study will be relevant for H. capsulatum pathogenesis.
Many of the H. capsulatum RNS-induced transcripts either do not have homologues in other organisms (34 transcripts) or have homologues that are uncharacterized (38 transcripts). A number of transcripts in the latter category have homologues only in the closely related fungi A. nidulans, F. graminearum, M. grisea, and N. crassa. These data suggest that these transcripts and the novel transcripts with no homology represent a group of RNS-response genes that do not have counterparts in other well-studied fungi such as S. cerevisiae and C. albicans. Characterizing the functions of these genes will provide further insight into the unique response of H. capsulatum and closely related fungi to RNS.
While examining the regulation of H. capsulatum RNS-induced genes in other stresses, we did not see much evidence of a common environmental stress response. As discussed previously, global transcriptional profiling in S. cerevisiae and S. pombe revealed a core set of genes that changes in expression in all stresses tested (Gasch et al., 2000; Causton et al., 2001; Chen et al., 2003). We observed induction of the H. capsulatum homologue of atf1, a key transcription factor of the S. pombe environmental stress response (Chen et al., 2003), in all stresses examined. However, unlike S. cerevisiae and S. pombe, we do not observe changes in gene expression for a core set of genes (other than the repetitive sequence/transposon cluster), suggesting that the stress response in H. capsulatum may be fundamentally different from the response of previously characterized fungi.
Supplementary Material
Acknowledgments
We thank Sarah Elson for determining GSNO experimental conditions and Shirley Kovacs for performing GSNO microarray hybridizations. We thank Margareta Andersson and Adam Bahrami for help with the stress response experiments, microarray hybridizations, and microarray analyses. We thank Linda Eissenberg, William Goldman, and Jon Woods for the kind gift of strains, plasmids, and protocols. We thank Dana Gebhart and Christopher Carlton for plasmids used in these studies. We are grateful to Michael Marletta and Ferric Fang for expert technical advice on performing RNS experiments and for useful discussions about microbial responses to RNS. We thank Joseph DeRisi and Suzanne Noble for helpful suggestions at critical points in this work. We also thank Joseph DeRisi for help with microarray techniques and analysis. We thank Kael Fischer for discussions about CombiMatrix array design and hybridization protocols. We thank Tara Cinotte, Michael Tognotti, Ryan Orbus, and Andy Antoniewicz (CombiMatrix) for help with CombiMatrix array design and production. We are grateful to Bethann Hromatka, Alexander Johnson, Aaron Sarver, Joseph DeRisi, Lily Chao, Michael Marletta, Lena Hwang, and Jasper Rine for sharing data before publication. We thank Elaine Mardis, Michael Nhan, John Spieth, Li Ding, and Aniko Sabo (Genome Sequencing Center) for array clone sequencing and help with genome analysis. We thank Jeffery Cox, Jake Mayfield, and members of the Sil laboratory for helpful discussions. We are grateful to Diane Inglis, other members of the Sil Lab, Aaron Sarver, Alexander Johnson, Michael Shiloh, Lily Chao, Bethann Hromatka, Jake Mayfield, Limin Liu, and Sarah Green for helpful comments on this manuscript. This work was supported by a National Science Foundation Graduate Fellowship to M.P.N., grants from the Ellison Medical Foundation (ID-NS-0108-03), American Cancer Society (RSG-01-039-01-MBC), National Institutes of Health (PO1 AI063302) (to A. S.), the Sandler Program in Basic Sciences, and the Howard Hughes Medical Institute Biomedical Research Support Program Grant 5300246 to the University of California, San Francisco School of Medicine.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-05-0434) on July 19, 2005.
Abbreviations used: DPTA NONOate, dipropylenetriamine NONOate; GSNO, S-nitrosoglutathione; IFN, interferon; NIT, nitrosative stress-induced transcript; .NO, nitric oxide; NOS2, nitric oxide synthase 2; RNS, reactive nitrogen species; ROS, reactive oxygen species.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Akhter, S., McDade, H. C., Gorlach, J. M., Heinrich, G., Cox, G. M., and Perfect, J. R. (2003). Role of alternative oxidase gene in pathogenesis of Cryptococcus neoformans. Infect. Immun. 71, 5794-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays, N. W., and Hampton, R. Y. (2002). Cdc48-Ufd1-Npl 4, stuck in the middle with Ub. Curr. Biol. 12, R366-R371. [DOI] [PubMed] [Google Scholar]
- Bohen, S. P., Kralli, A., and Yamamoto, K. R. (1995). Hold 'em and fold 'em: chaperones and signal transduction. Science 268, 1303-1304. [DOI] [PubMed] [Google Scholar]
- Bradshaw, V. A., and McEntee, K. (1989). DNA damage activates transcription and transposition of yeast Ty retrotransposons. Mol. Gen. Genet. 218, 465-474. [DOI] [PubMed] [Google Scholar]
- Brown, G. C. (1999). Nitric oxide and mitochondrial respiration. Biochim. Biophys. Acta 1411, 351-369. [DOI] [PubMed] [Google Scholar]
- Bullock, W. E. (1993). Interactions between human phagocytic cells and Histoplasma capsulatum. Arch. Med. Res. 24, 219-223. [PubMed] [Google Scholar]
- Causton, H. C., Ren, B., Koh, S. S., Harbison, C. T., Kanin, E., Jennings, E. G., Lee, T. I., True, H. L., Lander, E. S., and Young, R. A. (2001). Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12, 323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., Toone, W. M., Mata, J., Lyne, R., Burns, G., Kivinen, K., Brazma, A., Jones, N., and Bahler, J. (2003). Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14, 214-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarugi, A., Rovida, E., Dello Sbarba, P., and Moroni, F. (2003). Tryptophan availability selectively limits NO-synthase induction in macrophages. J. Leukoc. Biol. 73, 172-177. [DOI] [PubMed] [Google Scholar]
- Choy, J. S., and Kron, S. J. (2002). NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol. Cell. Biol. 22, 8215-8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, M. J., and Goldberg, D. E. (1998). Role for the Salmonella flavohemoglobin in protection from nitric oxide. J. Biol. Chem. 273, 12543-12547. [DOI] [PubMed] [Google Scholar]
- Cyr, D. M., Langer, T., and Douglas, M. G. (1994). DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem. Sci. 19, 176-181. [DOI] [PubMed] [Google Scholar]
- D'Autreaux, B., Touati, D., Bersch, B., Latour, J. M., and Michaud-Soret, I. (2002). Direct inhibition by nitric oxide of the transcriptional ferric uptake regulation protein via nitrosylation of the iron. Proc. Natl. Acad. Sci. USA 99, 16619-16624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin, K. H., Ehrt, S., Gutierrez-Ramos, J. C., Weich, N., and Nathan, C. F. (2003). The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 302, 1963-1966. [DOI] [PubMed] [Google Scholar]
- de Jesus-Berrios, M., Liu, L., Nussbaum, J. C., Cox, G. M., Stamler, J. S., and Heitman, J. (2003). Enzymes that counteract nitrosative stress promote fungal virulence. Curr. Biol. 13, 1963-1968. [DOI] [PubMed] [Google Scholar]
- De Luca, N. G., and Wood, P. M. (2000). Iron uptake by fungi: contrasted mechanisms with internal or external reduction. Adv. Microb. Physiol. 43, 39-74. [DOI] [PubMed] [Google Scholar]
- DeRisi, J. L., Iyer, V. R., and Brown, P. O. (1997). Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278, 680-686. [DOI] [PubMed] [Google Scholar]
- Dutta, S., Gerhold, D. L., Rice, M., Germann, M., and Kmiec, E. B. (1997). The cloning and overexpression of a cruciform binding protein from Ustilago maydis. Biochim. Biophys. Acta 1352, 258-266. [DOI] [PubMed] [Google Scholar]
- Eisen, M. B., Spellman, P. T., Brown, P. O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg, L. G., and Goldman, W. E. (1991). Histoplasma variation and adaptive strategies for parasitism: new perspectives on histoplasmosis. Clin. Microbiol. Rev. 4, 411-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg, L. G., and Goldman, W. E. (1994). The Interplay between Histoplasma capsulatum and its host cells. Balliere's Clin. Infect. Dis. 1, 265-283. [Google Scholar]
- Fang, F. C. (1999). Nitric Oxide and Infection, New York: Kluwer Academic/Plenum Publishers.
- Fang, F. C. (2004). Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2, 820-832. [DOI] [PubMed] [Google Scholar]
- Gardner, A. M., Helmick, R. A., and Gardner, P. R. (2002). Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J. Biol. Chem. 277, 8172-8177. [DOI] [PubMed] [Google Scholar]
- Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D., and Brown, P. O. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert, A. P., McGee, D. J., Akhtar, M., Mendz, G. L., Newton, J. C., Cheng, Y., Mobley, H. L., and Wilson, K. T. (2001). Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc. Natl. Acad. Sci. USA 98, 13844-13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, A. G., Wilson, J. E., Danon, S. J., Dixon, M. F., Donegan, K., and Hazell, S. L. (2003). Catalase (KatA) and KatA-associated protein (KapA) are essential to persistent colonization in the Helicobacter pylori SS1 mouse model. Microbiology 149, 665-672. [DOI] [PubMed] [Google Scholar]
- Hohmann, S. (2002). Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66, 300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabie, J. A., Klose, J. R., Wink, D. A., and Keefer, L. K. (1993). New nitric oxide-releasing zwitterions derived from polyamines. J. Org. Chem. 58, 1472-1476. [Google Scholar]
- Hromatka, B. S., Noble, S. M., and Johnson, A. D. (2005). Transcriptional response of C. albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress and virulence. Mol. Biol. Cell 16, 4814-4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, T. R., et al. (2000). Functional discovery via a compendium of expression profiles. Cell 102, 109-126. [DOI] [PubMed] [Google Scholar]
- Hutchings, M. I., Mandhana, N., and Spiro, S. (2002). The NorR protein of Escherichia coli activates expression of the flavorubredoxin gene norV in response to reactive nitrogen species. J. Bacteriol. 184, 4640-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyer, G., Piluek, W. F., Fansler, Z., Kreft, S. G., Hochstrasser, M., Brodsky, J. L., and Michaelis, S. (2004). Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multi-spanning membrane protein and a soluble luminal protein. J. Biol. Chem. 279, 38369-38378. [DOI] [PubMed] [Google Scholar]
- Hwang, L., Hocking-Murray, D., Bahrami, A. K., Andersson, M., Rine, J., and Sil, A. (2003). Identifying phase-specific genes in the fungal pathogen Histoplasma capsulatum using a genomic shotgun microarray. Mol. Biol. Cell 14, 2314-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C. H., Prigge, J. T., Warren, A. D., and McEwen, J. E. (2003). Characterization of an alternative oxidase activity of Histoplasma capsulatum. Yeast 20, 381-388. [DOI] [PubMed] [Google Scholar]
- Joseph-Horne, T., Hollomon, D. W., and Wood, P. M. (2001). Fungal respiration: a fusion of standard and alternative components. Biochim. Biophys. Acta 1504, 179-195. [DOI] [PubMed] [Google Scholar]
- Justino, M. C., Vicente, J. B., Teixeira, M., and Saraiva, L. M. (2005). New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J. Biol. Chem. 280, 2636-2643. [DOI] [PubMed] [Google Scholar]
- Kahlina, K., Goren, I., Pfeilschifter, J., and Frank, S. (2004). p68 DEAD box RNA helicase expression in keratinocytes. Regulation, nucleolar localization, and functional connection to proliferation and vascular endothelial growth factor gene expression. J. Biol. Chem. 279, 44872-44882. [DOI] [PubMed] [Google Scholar]
- Laloi, M. (1999). Plant mitochondrial carriers: an overview. Cell Mol. Life Sci. 56, 918-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, T. E., Otero, G. C., Wu-Hsieh, B. A., and Howard, D. H. (1994). Expression of inducible nitric oxide synthase by stimulated macrophages correlates with their antihistoplasma activity. Infect Immun. 62, 1478-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, T. E., Wu-Hsieh, B. A., and Howard, D. H. (1991). Iron limitation and the gamma interferon-mediated antihistoplasma state of murine macrophages. Infect Immun. 59, 2274-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, P., Shabbir, A., Cardozo, C., and Caplan, A. J. (2004). Sti1 and Cdc37 can stabilize Hsp90 in chaperone complexes with a protein kinase. Mol. Biol. Cell 15, 1785-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., Hausladen, A., Zeng, M., Que, L., Heitman, J., and Stamler, J. S. (2001). A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410, 490-494. [DOI] [PubMed] [Google Scholar]
- Liu, L., Zeng, M., Hausladen, A., Heitman, J., and Stamler, J. S. (2000). Protection from nitrosative stress by yeast flavohemoglobin. Proc. Natl. Acad. Sci. USA 97, 4672-4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. S. (1997). Generating, partitioning, targeting and functioning of superoxide in mitochondria. Biosci. Rep. 17, 259-272. [DOI] [PubMed] [Google Scholar]
- Lorenz, M. C., and Fink, G. R. (2002). Life and death in a macrophage: role of the glyoxylate cycle in virulence. Eukaryot. Cell 1, 657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Membrillo-Hernandez, J., Coopamah, M. D., Anjum, M. F., Stevanin, T. M., Kelly, A., Hughes, M. N., and Poole, R. K. (1999). The flavohemoglobin of Escherichia coli confers resistance to a nitrosating agent, a “nitric oxide releaser,” and paraquat and is essential for transcriptional responses to oxidative stress. J. Biol. Chem. 274, 748-754. [DOI] [PubMed] [Google Scholar]
- Missall, T. A., Lodge, J. K., and McEwen, J. E. (2004). Mechanisms of resistance to oxidative and nitrosative stress: implications for fungal survival in mammalian hosts. Eukaryot. Cell 3, 835-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon, A., Springer, M., and Lesage, P. (2000). Activation of the Kss1 invasive-filamentous growth pathway induces Ty1 transcription and retrotransposition in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 5766-5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, P., Zheng, M., Bedzyk, L. A., LaRossa, R. A., and Storz, G. (2004). Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc. Natl. Acad. Sci. USA 101, 745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara, K., Tanimoto, T., Hatano, K., Usuda, K., and Shoun, H. (1993). Cytochrome P-450 55A1 (P-450dNIR) acts as nitric oxide reductase employing NADH as the direct electron donor. J. Biol. Chem. 268, 8350-8355. [PubMed] [Google Scholar]
- Nakai, T., Yasuhara, T., Fujiki, Y., and Ohashi, A. (1995). Multiple genes, including a member of the AAA family, are essential for degradation of unassembled subunit 2 of cytochrome c oxidase in yeast mitochondria. Mol. Cell. Biol. 15, 4441-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, L. T., Wu-Hsieh, B. A., and Howard, D. H. (1994). Recombinant murine gamma interferon stimulates macrophages of the RAW cell line to inhibit intracellular growth of Histoplasma capsulatum. Infect Immun. 62, 680-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan, C., and Shiloh, M. U. (2000). Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA 97, 8841-8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, S. L. (1999). Macrophages in host defense against Histoplasma capsulatum. Trends Microbiol. 7, 67-71. [DOI] [PubMed] [Google Scholar]
- Newman, S. L., Gootee, L., Brunner, G., and Deepe, G. S., Jr. (1994). Chloroquine induces human macrophage killing of Histoplasma capsulatum by limiting the availability of intracellular iron and is therapeutic in a murine model of histoplasmosis. J. Clin. Investig. 93, 1422-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, V. H., Cox, J. S., Sousa, A. O., MacMicking, J. D., and McKinney, J. D. (2004). Role of KatG catalase-peroxidase in mycobacterial pathogenesis: countering the phagocyte oxidative burst. Mol. Microbiol. 52, 1291-1302. [DOI] [PubMed] [Google Scholar]
- O'Neill, B. M., Hanway, D., Winzeler, E. A., and Romesberg, F. E. (2004). Coordinated functions of WSS1, PSY2 and TOF1 in the DNA damage response. Nucleic Acids Res. 32, 6519-6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, G. S., Pae, H. O., Choi, B. M., Chae, S. C., Lee, H. S., Ryu, D. G., and Chung, H. T. (2004). 3-Hydroxyanthranilic acid, one of metabolites of tryptophan via indoleamine 2,3-dioxygenase pathway, suppresses inducible nitric oxide synthase expression by enhancing heme oxygenase-1 expression. Biochem. Biophys. Res. Commun. 320, 1156-1162. [DOI] [PubMed] [Google Scholar]
- Ohno, H., Zhu, G., Mohan, V. P., Chu, D., Kohno, S., Jacobs, W. R., Jr., and Chan, J. (2003). The effects of reactive nitrogen intermediates on gene expression in Mycobacterium tuberculosis. Cell Microbiol. 5, 637-648. [DOI] [PubMed] [Google Scholar]
- Palmieri, L., Lasorsa, F. M., De Palma, A., Palmieri, F., Runswick, M. J., and Walker, J. E. (1997). Identification of the yeast ACR1 gene product as a succinate-fumarate transporter essential for growth on ethanol or acetate. FEBS Lett. 417, 114-118. [DOI] [PubMed] [Google Scholar]
- Panozzo, C., Nawara, M., Suski, C., Kucharczyka, R., Skoneczny, M., Becam, A. M., Rytka, J., and Herbert, C. J. (2002). Aerobic and anaerobic NAD+ metabolism in Saccharomyces cerevisiae. FEBS Lett. 517, 97-102. [DOI] [PubMed] [Google Scholar]
- Poole, R. K., and Hughes, M. N. (2000). New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol. Microbiol. 36, 775-783. [DOI] [PubMed] [Google Scholar]
- Rhee, K. Y., Erdjument-Bromage, H., Tempst, P., and Nathan, C. F. (2005). S-nitroso proteome of Mycobacterium tuberculosis: enzymes of intermediary metabolism and antioxidant defense. Proc. Natl. Acad. Sci. USA 102, 467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarver, A., and DeRisi, J. (2005). Fzflp regulates an inducible response to nitrosative stress in Saccharomyces cerevisiae. Mol. Biol. Cell 16, 4781—4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt, E. E., et al. (2004). A comprehensive transcript index of the human genome generated using microarrays and computational approaches. Genome Biol. 5, R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekkai, D., Guittet, O., Lemaire, G., Tenu, J. P., and Lepoivre, M. (1997). Inhibition of nitric oxide synthase expression and activity in macrophages by 3-hydroxyanthranilic acid, a tryptophan metabolite. Arch. Biochem. Biophys. 340, 117-123. [DOI] [PubMed] [Google Scholar]
- Shiloh, M. U., and Nathan, C. F. (2000). Reactive nitrogen intermediates and the pathogenesis of Salmonella and mycobacteria. Curr. Opin. Microbiol. 3, 35-42. [DOI] [PubMed] [Google Scholar]
- Shoemaker, D. D., et al. (2001). Experimental annotation of the human genome using microarray technology. Nature 409, 922-927. [DOI] [PubMed] [Google Scholar]
- Shoun, H., and Tanimoto, T. (1991). Denitrification by the fungus Fusarium oxysporum and involvement of cytochrome P-450 in the respiratory nitrite reduction. J. Biol. Chem. 266, 11078-11082. [PubMed] [Google Scholar]
- Tatebe, H., and Yanagida, M. (2000). Cut8, essential for anaphase, controls localization of 26S proteasome, facilitating destruction of cyclin and Cut2. Curr. Biol. 10, 1329-1338. [DOI] [PubMed] [Google Scholar]
- Tjaden, B., Saxena, R. M., Stolyar, S., Haynor, D. R., Kolker, E., and Rosenow, C. (2002). Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res. 30, 3732-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius, M. V., Harth, G., and Horwitz, M. A. (2003). Glutamine synthetase GlnA1 is essential for growth of Mycobacterium tuberculosis in human THP-1 macrophages and guinea pigs. Infect Immun. 71, 3927-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann, B. D., Myers, H., Chiranand, W., Lazzell, A. L., Zhao, Q., Vega, L. A., Lopez-Ribot, J. L., Gardner, P. R., and Gustin, M. C. (2004). Inducible defense mechanism against nitric oxide in Candida albicans. Eukaryot. Cell 3, 715-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roermund, C. W., Waterham, H. R., Ijlst, L., and Wanders, R. J. (2003). Fatty acid metabolism in Saccharomyces cerevisiae. Cell Mol. Life Sci. 60, 1838-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskuil, M. I., Schnappinger, D., Visconti, K. C., Harrell, M. I., Dolganov, G. M., Sherman, D. R., and Schoolnik, G. K. (2003). Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198, 705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods, J. P., and Goldman, W. E. (1992). In vivo generation of linear plasmids with addition of telomeric sequences by Histoplasma capsulatum. Mol. Microbiol. 6, 3603-3610. [DOI] [PubMed] [Google Scholar]
- Woods, J. P., Heinecke, E. L., and Goldman, W. E. (1998). Electrotransformation and expression of bacterial genes encoding hygromycin phosphotransferase and beta-galactosidase in the pathogenic fungus Histoplasma capsulatum. Infect Immun. 66, 1697-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsham, P. L., and Goldman, W. E. (1988). Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. J. Med. Vet. Mycol. 26, 137-143. [PubMed] [Google Scholar]
- Wysong, D. R., Christin, L., Sugar, A. M., Robbins, P. W., and Diamond, R. D. (1998). Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect Immun. 66, 1953-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K., et al. (2003). Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302, 842-846. [DOI] [PubMed] [Google Scholar]
- Ye, Y., Meyer, H. H., and Rapoport, T. A. (2001). The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414, 652-656. [DOI] [PubMed] [Google Scholar]
- Youker, R. T., Walsh, P., Beilharz, T., Lithgow, T., and Brodsky, J. L. (2004). Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol. Biol. Cell 15, 4787-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, W. M., Gentil, G. D., Budde, A. D., and Leong, S. A. (2001). Characterization of the Ustilago maydis sid2 gene, encoding a multidomain peptide synthetase in the ferrichrome biosynthetic gene cluster. J. Bacteriol. 183, 4040-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft, W. G. (1997). Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.