Abstract
The three members of the T1R class of taste-specific G protein-coupled receptors have been hypothesized to function in combination as heterodimeric sweet taste receptors. Here we show that human T1R2/T1R3 recognizes diverse natural and synthetic sweeteners. In contrast, human T1R1/T1R3 responds to the umami taste stimulus l-glutamate, and this response is enhanced by 5′-ribonucleotides, a hallmark of umami taste. The ligand specificities of rat T1R2/T1R3 and T1R1/T1R3 correspond to those of their human counterparts. These findings implicate the T1Rs in umami taste and suggest that sweet and umami taste receptors share a common subunit.
Large-scale sequencing of a subtracted cDNA library derived from rat taste tissue identified a new C-family G protein-coupled receptor, T1R1, that is expressed selectively in taste receptor cells; T1R1-based degenerate PCR led to the identification of a related taste-specific receptor, T1R2 (1). Recently, a third and possibly final member of the T1R family, T1R3, was identified in the human DNA databank (2–7). Tellingly, mouse T1R3 maps to a genomic interval containing Sac, a locus that influences sweet taste in the mouse (8, 9). Recent high-resolution genetic mapping and complementation studies have strengthened the connection between mouse T1R3 and Sac (2–7). Although T1R1 and T1R2 appear to be expressed in predominantly nonoverlapping regions of the tongue, they each are coexpressed with T1R3 (1, 3, 4, 6). These overlapping expression patterns and precedent from the structurally related heterodimeric γ-aminobutyric acid type B receptor (10–13) suggested that T1R1 and T1R2 may combine with T1R3 to form heterodimeric sweet taste receptors. Indeed, rat T1R2 has been shown recently to function in combination with T1R3 to recognize a subset of sweet taste stimuli, a finding that has been proposed to reflect the involvement of additional combinations of T1Rs in sweet taste (6). In this study we cloned and functionally expressed human and rat T1Rs. Human and rat T1R2/T1R3 recognized all sweet taste stimuli tested, and human and rat T1R1/T1R3 recognized umami taste stimuli. These findings suggest that different combinations of T1Rs function as heterodimeric sweet and umami taste receptors.
Material and Methods
T1R Cloning.
Intronless human T1R expression constructs were generated in a pEAK10-derived vector (Edge Biosystems, Gaithersburg, MD) by a combination of cDNA-based and genomic DNA-based methods. To generate the full-length T1R1 expression construct, two 5′ coding exons identified in a cloned T1R1 interval (GenBank accession no. AL159177) were combined by PCR overlap and then joined to a 5′-truncated testis cDNA clone. The T1R2 expression construct was generated from a partially sequenced T1R2 genomic interval. Two missing T1R2 5′ introns were identified by screening shotgun libraries of the cloned genomic interval using probes derived from the corresponding rat coding sequence. Coding exons then were combined by PCR overlap to produce the full-length expression construct. The T1R3 expression construct was generated by PCR overlap from a sequenced T1R3 genomic interval (GenBank accession no. AL139287). Rat T1R3 was isolated from a taste-tissue-derived cDNA library by using a rat T1R3 exon fragment generated by human T1R3-based degenerate PCR.
Gα15 chimeras were generated in a pEAK10-derived vector by PCR with mutagenic primers. The five-residue C-terminal tail of Gα15, EINLL, was replaced with EYNLV (Gαq and Gα11), EFNLV (Gα14), QYELL (Gαs and Gαolf), DCGLF (Gαi1, Gαi2, Gαt1, Gαt2, and Gαgust), ECGLY (Gαi3), GCGLY (Gαo1 and Gαo2), YIGLC (Gαz), DIMLQ (Gα12), or QLMLQ (Gα13).
Taste Detection Thresholds.
Detection threshold values were determined following the methods of Schiffman et al. (14) and averaged over three trials for four subjects.
T1R Functional Expression.
HEK-293T and a HEK-293 derivative that stably expresses Gα15 (Aurora Biosciences, San Diego; ref. 15) were grown and maintained at 37°C in DMEM supplemented with 10% FBS and MEM nonessential amino acids (GIBCO/BRL); the medium for Gα15 cells also contained 3 μg⋅ml−1 blasticidin (GIBCO/BRL). For calcium-imaging experiments, cells first were seeded onto 24-well tissue-culture plates (≈100,000 cells per well) and transfected by using Mirus TransIt-293 (Panvera, Madison, WI). Transfection efficiencies, which were estimated by cotransfection with a red fluorescent protein expression vector, were typically ≈70%. To minimize glutamate-induced and glucose-induced desensitization, supplemented DMEM was replaced with low-glucose DMEM supplemented with GlutaMAX and 10% dialyzed FBS (GIBCO/BRL) ≈24 h after transfection. After an additional 24 h, cells were loaded with the calcium dye fluo-4 acetoxymethyl ester (Molecular Probes), 3 μM in Dulbecco's PBS buffer (DPBS, GIBCO/BRL), for 1.5 h at room temperature. After replacement with 250 μl of DPBS, stimulation was performed at room temperature by the addition of 200 μl of DPBS supplemented with taste stimuli. Calcium mobilization was monitored on an Axiovert S100 microscope equipped with an inverted ×10/0.5 long working distance plano fluor objective (Zeiss) and a cooled charge-coupled device camera (Princeton Instruments, Trenton, NJ). Fluorescence images were acquired at 480-nm excitation and 535-nm emission and analyzed with IMAGING WORKBENCH 4.0 software (Axon Instruments, Foster City, CA). T1R receptor activity was quantitated by counting the number of responding cells 30 sec after stimulus addition. Stimuli were tested at concentrations that do not elicit calcium responses from mock-transfected Gα15 cells. Compounds used (and typical concentrations) were: acesulfame K (2.5 mM), l-AP4 (10 mM), aspartame (2.5 mM), d-aspartate (50 mM), l-aspartate (50 mM), CMP (5 mM), cyclamate (5 mM), denatonium benzoate (5 mM), dulcin (0.1 mM), fructose (300 mM), galactose (300 mM), glucose (300 mM), d-glutamate (25 mM), l-glutamate (25 mM), l-glutamine (10 mM), glycine (250 mM), GMP (1 mM), l-histidine (10 mM), IMP (1 mM), lactose (250 mM), l-leucine (10 mM), l-lysine (10 mM), maltose (300 mM), monellin (0.01%), neotame (0.1 mM), perillartine (15 μM), l-proline (10 mM), quinine hydrochloride (0.25 mM), saccharin (1 mM), l-serine (10 mM), sodium chloride (100 mM), sucralose (1 mM), sucrose (300 mM), thaumatin (0.01%), d-tryptophan (10 mM), l-tryptophan (10 mM), and l-tyrosine (5 mM). DPBS adjusted to pH 4.5 with acetic acid was used as a sour taste stimulus.
Human T1R2/T1R3 stable cell lines were generated by transfecting linearized pEAK10-derived T1R2 and pCDNA3.1/ZEO-derived (Invitrogen) T1R3 vectors into Gα15 cells. Cells were selected in 0.5 μg⋅ml−1 puromycin (Calbiochem) and 100 μg⋅ml−1 zeocin (Invitrogen) at 37°C in low-glucose DMEM supplemented with GlutaMAX/10% dialyzed FBS/3 μg⋅ml−1 blasticidin. Resistant colonies were expanded, and their responses to sweet taste stimuli were evaluated by fluorescence microscopy. For automated fluorometric imaging on VIPR-II instrumentation (Aurora Biosciences), T1R2/T1R3 stable cells first were seeded onto 96-well plates (≈15,000 cells per well). After 24 h, cells were loaded with the calcium dye fluo-3 acetoxymethyl ester (Molecular Probes), 5 μM in PBS, for 1 h at room temperature. After replacement with 70 μl of PBS, stimulation was performed at room temperature by the addition of 70 μl of PBS supplemented with taste stimuli. Fluorescence (480-nm excitation and 535-nm emission) responses from 20 to 30 sec after compound addition were averaged, corrected for background fluorescence measured before compound addition, and normalized to the response to the calcium ionophore ionomycin (1 μM, Calbiochem).
Results and Discussion
We cloned human and rat T1Rs for functional expression experiments (Fig. 1). The G protein or G proteins that couple to the T1Rs in vivo are not known. Consequently, we transiently transfected the human T1Rs into a HEK-293-derived cell line that stably expresses Gα15, a promiscuous phospholipase C-linked G protein (15, 16). Sucrose elicited transient intracellular calcium increases in Gα15 cells cotransfected with human T1R2 and T1R3 but not in cells transfected with T1R2 or T1R3 alone. T1R2/T1R3 activity was inhibited by the sweet taste inhibitor lactisole (17); this inhibition likely reflects antagonism at the T1R2/T1R3 receptor, because lactisole did not inhibit the Gα15-dependent activity of endogenous β2-adrenergic receptor (Fig. 2A). In addition to sucrose, T1R2/T1R3 responded to all other sweet taste stimuli tested: the sugars fructose, galactose, glucose, lactose, and maltose; the amino acids glycine and d-tryptophan (but not its bitter enantiomer); the sweet proteins monellin and thaumatin; and the synthetic sweeteners acesulfame K, aspartame, cyclamate, dulcin, neotame, saccharin, and sucralose (Fig. 2B).
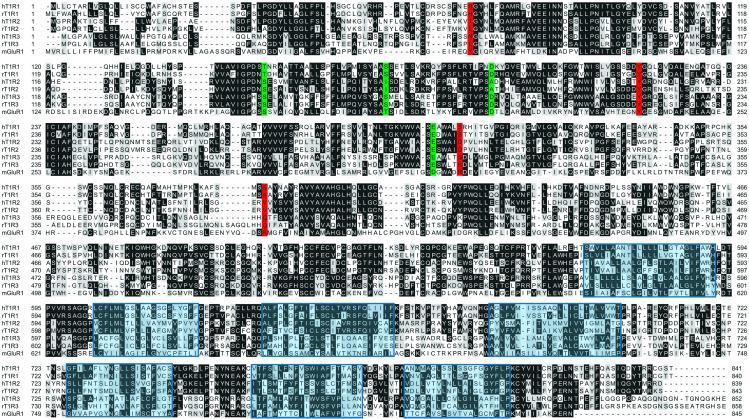
Figure 1.
Sequence alignment of human and rat T1Rs. T1R sequences determined in this study, human T1R1 (GenBank accession no. BK000153), T1R2 (accession no. BK000151), T1R3 (accession no. BK000152), and rat T1R3 (accession no. AF456324), are aligned here with previously described rat T1Rs (accession nos. AAD18069 and AAD18070) and the rat mGluR1 metabotropic glutamate receptor (accession no. P23385). The mGluR1 C terminus is not shown. Potential transmembrane segments are boxed in blue. mGluR1 ligand-binding residues are highlighted following the color scheme used in Fig. 3A.
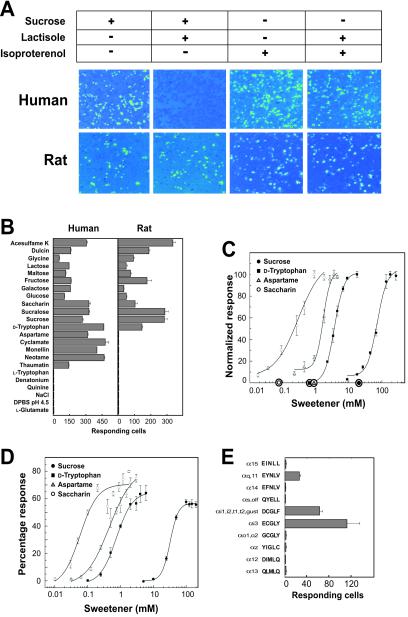
Figure 2.
Human and rat T1R2/T1R3 recognize sweet taste stimuli. (A) Gα15 cells transiently transfected with human T1R2 and T1R3 and HEK-293T cells transiently transfected with rat T1R2, T1R3, and Gα15/i1 were assayed for intracellular calcium increases in response to 300 mM sucrose in the presence (+) and absence (−) of 1.25 mM lactisole and to 10 nM isoproterenol in the presence and absence of 1.25 mM lactisole. Each imaged field shown contains ≈1,000 confluent cells. (B) The responses of human T1R2/T1R3 and rat T1R2/T1R3 were determined for sweet taste stimuli at the concentrations listed in Materials and Methods. (C) Human T1R2/T1R3 dose responses were determined for sucrose, d-tryptophan, aspartame, and saccharin. Dose responses were normalized to the maximal percentage of responding cells, which ranged from 10 to 40% for different sweeteners. Values represent the mean ± SE of four independent responses. The x axis circles represent average psychophysical detection threshold values for these four sweeteners. (D) The dose responses of Gα15 cells stably transfected with human T1R2 and T1R3 were determined for sucrose, d-tryptophan, aspartame, and saccharin. Responses are shown as the percentage of fluorescence values relative to fluorescence increases elicited by 1 μM ionomycin. Values represent the mean ± SE of four independent responses. (E) HEK-293T cells were transiently transfected with rat T1R2, rat T1R3, and each Gα15 chimera, and assayed for intracellular calcium increases in response to 75 mM sucrose. The five C-terminal residues of each Gα15 chimera are shown. The activities in B and E represent the mean ± SE number of responding cells for four imaged fields of ≈1,000 confluent cells.
The weak responses of T1R2/T1R3-transfected cells precluded quantitating receptor activity by monitoring summated fluorescence changes over fields of calcium-dye-loaded cells as, for example, in Chandrashekar et al. (15). However, increased receptor activity in Gα15-based assays is reflected not only in increased magnitudes of the calcium responses of individual cells but also in increased numbers of responding cells. Therefore, we quantitated T1R receptor activity by counting the number of responding cells after stimulus addition. To validate this method, EC50 values were determined for several test receptors including the mGluR4 metabotropic glutamate receptor and mT2R5 cycloheximide receptor and found to correspond to published values (data not shown). The dose responses of T1R2/T1R3 were determined in this manner for several sweeteners, which activated T1R2/T1R3 at physiologically relevant concentrations with a rank order similar to human taste (Fig. 2C). Moreover, stable expression in Gα15 cells markedly increased T1R2/T1R3 activity and allowed us to quantitate receptor activity by monitoring summated calcium signals with automated fluorometric imaging. The increased activity of the T1R2/T1R3 stable cell line resulted in 2–5-fold left-shifted dose-response curves relative to cells transiently transfected with T1R2/T1R3 (Fig. 2D). This close correspondence further validates quantification of receptor activity by counting cells.
The response of human T1R2/T1R3 to all sweet taste stimuli tested contrasted with the limited response of rat T1R2/T1R3 determined previously by functional expression in heterologous cells and coupling to Gα16/z44 (a Gα16 variant containing the C-terminal 44 residues of Gαz; refs. 6 and 18). To address this discrepancy, we transfected rat T1R2 and T1R3 into Gα15 cells and found that only high concentrations of a small number of sweet taste stimuli elicited detectable responses. These weak responses prompted us to investigate the possibility that rat T1R2/T1R3 responses to additional sweet taste stimuli might be obscured by deficiencies in the Gα15- and Gα16/z44-based assays such as poor expression of the rat T1Rs or inefficient G protein coupling. Several Gαi-linked receptors have been shown to couple more efficiently to a Gαq variant containing the C-terminal five residues of Gαi2 than to Gαq (19) and more efficiently to Gα16/z44 than to Gα16 (18). Accordingly, we reasoned that replacing the Gα15 C terminus with that of the G protein that couples to T1R2/T1R3 in vivo would improve coupling efficiency. Because the identity of this G protein is not known, we generated a panel of Gα15 chimeras in which the five-residue C-terminal tail of Gα15 was replaced by the tails of all other G proteins. The response of rat T1R2/T1R3-transfected HEK-293T cells to sucrose was enhanced in the presence of chimeras containing the C termini of Gαi-related G proteins or a chimera containing the C terminus of Gαq (Fig. 2E). Transient cotransfection of a Gα15 chimera containing the C-terminal tail of Gαi1, Gα15/i1, with rat T1R2 and T1R3 into HEK-293T cells revealed responses to all sweet taste stimuli tested (except for compounds such as aspartame and monellin that taste sweet to humans but are not palatable to rodents; Fig. 2B; refs. 20 and 21). Lactisole, which does not inhibit sweet taste in rats (17), did not inhibit rat T1R2/T1R3 activity (Fig. 2A).
The inclusive ligand specificity of human and rat T1R2/T1R3 suggested that T1R1/T1R3 mediates a different taste modality. We speculated that T1R1/T1R3 might function as an umami taste receptor because key ligand-binding residues of the mGluR1 metabotropic glutamate receptor are conserved in T1R1 (Figs. 1 and 3A; ref. 22). Indeed, l-glutamate elicited transient intracellular calcium increases in Gα15 cells cotransfected with human T1R1 and T1R3 but not in cells transfected with T1R1 or T1R3 alone (Fig. 3 B and C).
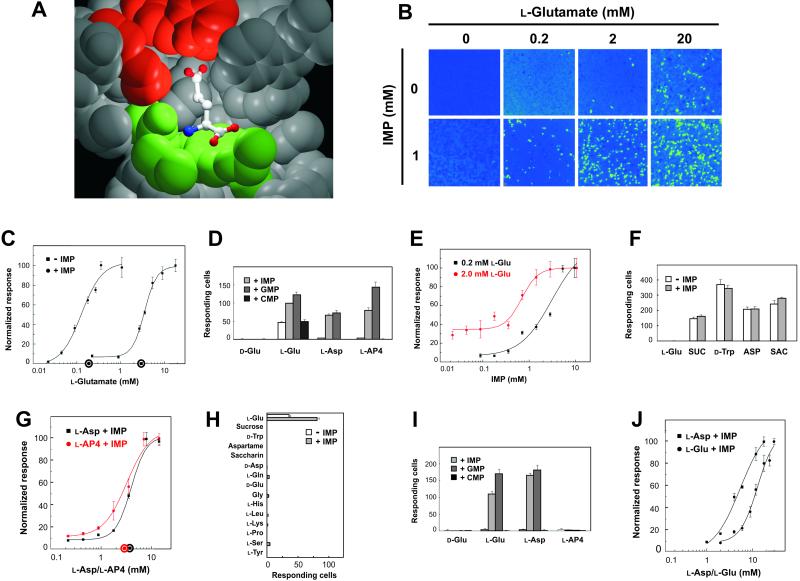
Figure 3.
Human and rat T1R1/T1R3 recognize umami taste stimuli. (A) mGluR1 residues (PDB entry no. 1EWK) that contact the l-glutamate side chain carboxylate are shown in red, and residues that contact the l-glutamate α-amino acid moiety are shown in green. (B) Gα15 cells transiently transfected with human T1R1 and T1R3 were assayed for intracellular calcium increases in response to increasing concentrations of l-glutamate in the presence or absence of 1 mM IMP. Each imaged field shown contains ≈1,000 confluent cells. (C) Human T1R1/T1R3 dose responses were determined for l-glutamate in the presence and absence of 0.2 mM IMP. Dose responses were normalized to the maximal percentage of responding cells, which was ≈5% for l-glutamate and ≈10% for l-glutamate plus IMP. Values represent the mean ± SE of four independent responses. The x axis circles represent average psychophysical detection threshold values for l-glutamate in the presence and absence of 0.2 mM IMP. (D) Human T1R1/T1R3 responses to 25 mM d-glutamate, 25 mM l-glutamate, 25 mM l-aspartate, 25 mM l-AP4, and binary mixtures of these compounds with 2.5 mM IMP, 2.5 mM GMP, or 2.5 mM CMP were determined. (E) Human T1R1/T1R3 dose responses were determined for IMP in the presence of 0.2 and 2 mM l-glutamate and normalized to the maximal percentages of responding cells, which were ≈10%. Values represent the mean ± SE of four independent responses. (F) Human T1R2/T1R3 responses to 25 mM l-glutamate, 100 mM sucrose (SUC), 2.5 mM d-tryptophan, 1.5 mM aspartame (ASP), and 0.4 mM saccharin (SAC) were determined in the presence and absence of 2.5 mM IMP. (G) Human T1R1/T1R3 dose responses were determined for l-aspartate and l-AP4 in the presence of 0.2 mM IMP. Dose responses were normalized to the maximal percentage of responding cells, which was ≈5% for l-aspartate plus IMP and ≈10% for l-AP4 plus IMP. Values represent the mean ± SE of four independent responses. The x axis circles represent average psychophysical detection threshold values for l-aspartate plus 0.2 mM IMP and l-AP4 plus 0.2 mM IMP. (H) Human T1R1/T1R3 responses to sucrose (100 mM), d-tryptophan (20 mM), aspartame (2 mM), saccharin (1 mM), l-tyrosine (5 mM), and d-glutamate, l-glutamine, d-aspartate, glycine, l-histidine, l-leucine, l-lysine, l-proline, and l-serine (each at 10 mM) were determined in the presence and absence of 1 mM IMP. (I) HEK-293T cells were transiently transfected with rat T1R1, rat T1R3, and Gα15/i1 and assayed for increases in intracellular calcium in response to 25 mM d-glutamate, 25 mM l-glutamate, 25 mM l-aspartate, 25 mM l-AP4, and binary mixtures with 2.5 mM IMP, 2.5 mM GMP, or 2.5 mM CMP. (J) Rat T1R1/T1R3 dose responses were determined for l-aspartate and l-glutamate in the presence of 0.2 mM IMP. Dose responses were normalized to the maximal percentage of responding cells, which was ≈10% for l-aspartate plus IMP and ≈15% for l-glutamate plus IMP. Values represent the mean ± SE of four independent responses. The activities in D, F, H, and I represent the mean ± SE number of responding cells for four imaged fields of ≈1,000 confluent cells.
Robust synergism between l-glutamate and the 5′ ribonucleotides IMP and GMP is a hallmark of umami taste (23). IMP or GMP alone did not activate human T1R1/T1R3, but these 5′ ribonucleotides potentiated the T1R1/T1R3 response to l-glutamate; CMP, which does not enhance umami taste, had no effect (Fig. 3 B and D). In the presence of 0.2 mM IMP, the EC50 of T1R1/T1R3 for l-glutamate was shifted 30-fold; this dramatic effect on T1R1/T1R3 activity was similar to the effect of these compounds on umami taste (Fig. 3C). The effect of IMP on T1R1/T1R3 was saturable (Fig. 3E) and selective; human T1R2/T1R3 was not activated by l-glutamate in the presence of IMP, and IMP did not enhance the response of human T1R2/T1R3 to sweet taste stimuli (Fig. 3F). In contrast to l-glutamate, T1R1/T1R3-transfected cells did not respond to the weak umami taste stimuli l-aspartate and l-AP4 (23). In the presence of IMP and GMP, however, these compounds activated T1R1/T1R3 at physiologically relevant concentrations (Fig. 3 D and G). The response of T1R1/T1R3 was selective for umami taste stimuli; T1R1/T1R3 did not respond to sweet taste stimuli, amino acids such as d-glutamate and d-aspartate, or binary mixtures of these compounds with IMP (Fig. 3H).
As in human taste, IMP enhances l-glutamate taste in rodents, and rodent taste tests suggest that l-aspartate and l-AP4 mimic l-glutamate (24–26). Cotransfection of Gα15/i1 with rat T1R1 and T1R3 into HEK-293T cells revealed responses to binary mixtures of IMP with l-glutamate and l-aspartate but not l-AP4; responses to l-glutamate and l-aspartate were observed also in the presence of GMP but not CMP (Fig. 3I). l-Glutamate and l-aspartate in the presence of 2.5 mM IMP activated rat T1R1/T1R3 with similar potency (Fig. 3J). Rat T1R1/T1R3 did not respond to sweet taste stimuli such as sucrose and d-tryptophan, d-glutamate, d-aspartate, or binary mixtures of these compounds with IMP (data not shown).
In summary, our findings suggest that different combinations of T1Rs function as sweet and umami taste receptors. Intriguingly, human and rat T1R2/T1R3 were activated by all sweet taste stimuli tested. T1R2/T1R3 therefore may be the only sweet taste receptor. Analogously, the finding that all umami taste stimuli tested acted in combination with 5′ ribonucleotides at human T1R1/T1R3 raises the possibility that T1R1/T1R3 is the only umami taste receptor. Discrepancies between T1R1/T1R3 activity and umami taste (such as the lack of response of rat T1R1/T1R3-transfected cells to l-AP4) may reflect deficiencies in the Gα15-based assay or the involvement in umami taste of other receptors such as two previously identified candidate umami taste receptors (27, 28). Evaluation of these hypotheses awaits further experimentation such as characterization of T1R-null mice.
Preliminary analysis of T1R surface expression with epitope-tagged receptors revealed that the T1Rs are predominantly localized intracellularly (L.S., unpublished data). Such negligible amounts of surface receptors, although evidently sufficient for activity, confound coimmunoprecipitation experiments with whole-cell lysates and preclude biochemical analysis of T1R interactions on the surface of heterologous cells. However, the hypothesis that mammalian T1Rs function as heterodimeric sweet and umami taste receptors is supported by functional codependence, coincident expression in taste receptor cells (3, 4, 6), and precedent from the structurally related heterodimeric γ-aminobutyric acid type B receptor (10–13). The presence of three T1R1-like and two T1R3-like genes in the pufferfish genome suggests that the emergence of heterodimeric C-family chemosensory receptors preceded the emergence of terrestrial animals.
Acknowledgments
We thank S. Zozulya, S. O'Connell, R. Campbell, F. Echeverri, K. Wang, A. Watkins, A. Chang, M. Qi, and F. Wong for their scientific contributions and R. Tsien, L. Stryer, B. Moyer, N. Callamaras, and G. Servant for comments on the manuscript. The partial human T1R1 cDNA, partial human T1R2 genomic sequence, rat T1R1 and T1R2 cDNAs, and rat taste-tissue-derived cDNA library were provided by C. Zuker.
Footnotes
References
- 1.Hoon M A, Adler E, Lindemeier J, Battey J F, Ryba N J, Zuker C S. Cell. 1999;96:541–551. doi: 10.1016/s0092-8674(00)80658-3. [DOI] [PubMed] [Google Scholar]
- 2.Kitagawa M, Kusakabe Y, Miura H, Ninomiya Y, Hino A. Biochem Biophys Res Commun. 2001;283:236–242. doi: 10.1006/bbrc.2001.4760. [DOI] [PubMed] [Google Scholar]
- 3.Max M, Shanker Y G, Huang L, Rong M, Liu Z, Campagne F, Weinstein H, Damak S, Margolskee R F. Nat Genet. 2001;28:58–63. doi: 10.1038/ng0501-58. [DOI] [PubMed] [Google Scholar]
- 4.Montmayeur J P, Liberles S D, Matsunami H, Buck L B. Nat Neurosci. 2001;4:492–498. doi: 10.1038/87440. [DOI] [PubMed] [Google Scholar]
- 5.Sainz E, Korley J N, Battey J F, Sullivan S L. J Neurochem. 2001;77:896–903. doi: 10.1046/j.1471-4159.2001.00292.x. [DOI] [PubMed] [Google Scholar]
- 6.Nelson G, Hoon M A, Chandrashekar J, Zhang Y, Ryba N J P, Zuker C S. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- 7.Bachmanov A A, Li X, Reed D R, Ohmen J D, Li S, Chen Z, Tordoff M G, de Jong P J, Wu C, West D B, et al. Chem Senses. 2001;26:925–933. doi: 10.1093/chemse/26.7.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuller J L. J Hered. 1974;65:33–36. doi: 10.1093/oxfordjournals.jhered.a108452. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Inoue M, Reed D R, Huque T, Puchalski R B, Tordoff M G, Ninomiya Y, Beauchamp G K, Bachmanov A A. Mamm Genome. 2001;12:13–16. doi: 10.1007/s003350010236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones K A, Borowsky B, Tamm J A, Craig D A, Durkin M M, Dai M, Yao W J, Johnson M, Gunwaldsen C, Huang L Y, et al. Nature (London) 1998;396:674–679. doi: 10.1038/25348. [DOI] [PubMed] [Google Scholar]
- 11.Kaupmann K, Malitschek B, Schuler V, Heid J, Froestl W, Beck P, Mosbacher J, Bischoff S, Kulik A, Shigemoto R, Karschin A, Bettler B. Nature (London) 1998;396:683–687. doi: 10.1038/25360. [DOI] [PubMed] [Google Scholar]
- 12.White J H, Wise A, Main M J, Green A, Fraser N J, Disney G H, Barnes A A, Emson P, Foord S M, Marshall F H. Nature (London) 1998;396:679–682. doi: 10.1038/25354. [DOI] [PubMed] [Google Scholar]
- 13.Kuner R, Kohr G, Grunewald S, Eisenhardt G, Bach A, Kornau H C. Science. 1999;283:74–77. doi: 10.1126/science.283.5398.74. [DOI] [PubMed] [Google Scholar]
- 14.Schiffman S S, Frey A E, Luboski J A, Foster M A, Erickson R P. Physiol Behav. 1991;49:843–854. doi: 10.1016/0031-9384(91)90193-r. [DOI] [PubMed] [Google Scholar]
- 15.Chandrashekar J, Mueller K L, Hoon M A, Adler E, Feng L, Guo W, Zuker C S, Ryba N J. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 16.Offermanns S, Simon M I. J Biol Chem. 1995;270:15175–15180. doi: 10.1074/jbc.270.25.15175. [DOI] [PubMed] [Google Scholar]
- 17.Sclafani A, Perez C. Physiol Behav. 1997;61:25–29. doi: 10.1016/s0031-9384(96)00316-2. [DOI] [PubMed] [Google Scholar]
- 18.Mody S M, Ho M K, Joshi S A, Wong Y H. Mol Pharmacol. 2000;57:13–23. [PubMed] [Google Scholar]
- 19.Conklin B R, Farfel Z, Lustig K D, Julius D, Bourne H R. Nature (London) 1993;363:274–276. doi: 10.1038/363274a0. [DOI] [PubMed] [Google Scholar]
- 20.Brouwer J N, Hellekant G, Kasahara Y, van der Wel H, Zotterman Y. Acta Physiol Scand. 1973;89:550–557. doi: 10.1111/j.1748-1716.1973.tb05549.x. [DOI] [PubMed] [Google Scholar]
- 21.Sclafani A, Abrams M. Physiol Behav. 1986;37:253–256. doi: 10.1016/0031-9384(86)90228-3. [DOI] [PubMed] [Google Scholar]
- 22.Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K. Nature (London) 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- 23.Yamaguchi S. Physiol Behav. 1991;49:833–841. doi: 10.1016/0031-9384(91)90192-q. [DOI] [PubMed] [Google Scholar]
- 24.Delay E R, Beaver A J, Wagner K A, Stapleton J R, Harbaugh J O, Catron K D, Roper S D. Chem Senses. 2000;25:507–515. doi: 10.1093/chemse/25.5.507. [DOI] [PubMed] [Google Scholar]
- 25.Stapleton J R, Roper S D, Delay E R. Chem Senses. 1999;24:449–457. doi: 10.1093/chemse/24.4.449. [DOI] [PubMed] [Google Scholar]
- 26.Chaudhari N, Yang H, Lamp C, Delay E, Cartford C, Than T, Roper S. J Neurosci. 1996;16:3817–3826. doi: 10.1523/JNEUROSCI.16-12-03817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teeter J H, Kumazawa T, Brand J G, Kalinoski D L, Honda E, Smutzer G. In: Sensory Transduction. Corey D P, Roper S, editors. New York: Rockefeller Univ. Press; 1992. pp. 291–306. [Google Scholar]
- 28.Chaudhari N, Landin A M, Roper S D. Nat Neurosci. 2000;3:113–119. doi: 10.1038/72053. [DOI] [PubMed] [Google Scholar]