Abstract
Yeast cells respond to mitochondrial dysfunction by altering the expression of a subset of nuclear genes, a process known as retrograde signaling (RS). RS terminates with two transcription factors, Rtg1p and Rtg3p. One positive regulator, Rtg2p, and four negative regulators, Lst8p, Mks1p, and the redundant 14-3-3 proteins, Bmh1p and Bmh2p, control RS upstream of Rtg1/3p. Mks1p is negatively regulated by binding to Rtg2p and positively regulated when bound to Bmh1/2p. Here we report that Grr1p, a component of the SCFGrr1 E3 ubiquitin ligase, modulates RS by affecting Mks1p levels. Grr1p polyubiquitinates Mks1p not bound to either Rtg2p or to Bmh1/2p, targeting it for degradation. An acidic domain region of Mks1p constitutes the portable Mks1p degron sequence. We have isolated dominant mutations in Grr1p leading to increased Mks1p degradation. These mutations result in a gain of positive charge on the concave surface of the leucine rich repeat (LRR) domain of Grr1p, the proposed substrate binding site. We propose that Mks1p is a central player of RS and is acted upon by multiple regulators of the pathway.
INTRODUCTION
The retrograde response is a mitochondria-to-nucleus signaling pathway that monitors and transmits changes in mitochondrial function to specific changes in nuclear gene expression (Butow and Avadhani, 2004). This response is, for the most part, adaptive, affecting diverse cellular activities that include metabolic, nutrient sensing, transport and stress pathways (Epstein et al., 2001; Biswas et al., 2003). These activities are adjusted to accommodate cells to the alterations in the mitochondrial state, for instance, to the loss of respiratory activity. In animal cells, mitochondrial dysfunctions often initiate changes in intracellular Ca2+ dynamics that lead to changes in nuclear gene expression via the activation of transcription factors, e.g., NFκB and NFAT (Luo et al., 1997; Biswas et al., 1999, 2003). In the budding yeast, Saccharomyces cerevisiae, the loss of respiratory activity results in increased expression of a defined set of nuclear genes encoding proteins that participate in activities that include anaplerotic pathways, peroxisome biogenesis, small molecule transport systems and pleiotropic drug resistance (Hallstrom and Moye-Rowley, 2000; Epstein et al., 2001).
Expression of some retrograde responsive genes in yeast, such as CIT2, encoding a glyoxylate cycle isoform of citrate synthase, requires three regulatory factors, Rtg1p, Rtg2p, and Rtg3p (Liao and Butow, 1993; Jia et al., 1997). This requirement places these genes in the RTG pathway, in contrast to other retrograde responsive genes whose elevated expression in cells with dysfunctional mitochondria is essentially independent of the RTG genes (Epstein et al., 2001). Rtg1p and Rtg3p are bHLH/Zip transcription factors that bind to the promoter region of RTG target genes (Jia et al., 1997). When the RTG pathway is activated, a hyperphosphorylated form of Rtg3p sequestered in the cytoplasm with Rtg1p becomes partially dephosphorylated, and both transcription factors translocate to the nucleus (Sekito et al., 2000). These processes require Rtg2p, a novel cytoplasmic protein with an N-terminal ATP binding domain whose integrity is required for Rtg2p function (Liao and Butow, 1993; Liu et al., 2003). The RTG pathway can also be activated by inhibition of TOR (target of rapamycin) kinase activity (Komeili et al., 2000). The intersection of these pathways underscores the intimate relation between mitochondrial function and nutrient sensing, one of the important activities of TOR signaling.
A key feature of the RTG pathway is its role in reconfiguring metabolism to meet the special demands of respiratory deficient cells. For instance, transcriptional regulation of the Krebs cycle genes, CIT1, ACO1, IDH1, and IDH2 switches from control by the HAP transcriptional complex to the Rtg1/3p complex in respiratory-deficient cells (Liu and Butow, 1999). The products of these genes produce α-ketoglutarate, the direct precursor of glutamate. Consequently, mutations in any one of the RTG genes lead to glutamate auxotrophy in cells that are respiratory deficient (Liu and Butow, 1999). Glutamate is also a potent repressor of the RTG pathway (Liu and Butow, 1999; Sekito et al., 2002), underscoring the importance of the pathway in glutamate homeostasis. These findings highlight how the RTG pathway functions to regulate the changing metabolic needs of cells with altered mitochondrial function.
Additional regulatory factors have been identified that act between Rtg2p and Rtg1/3p. These include Mks1p, Lst8p, and the 14-3-3 proteins Bmh1p and Bmh2p, all of which negatively regulate the RTG pathway (Liu et al., 2001; Dilova et al., 2002; Sekito et al., 2002; Tate et al., 2002; Liu et al., 2003). In cells that either lack or have mutant forms of these proteins, RTG-dependent gene expression is constitutive and is not dependent on Rtg2p. Recent studies have provided additional insights into the regulation of the RTG pathway involving a dynamic interplay among Mks1p, Rtg2p, and Bmh1/2p (Liu et al., 2003). Although the specific mechanism of how Mks1p, a phosphoprotein, acts as a negative regulator has not yet been established, what is clear is that when the RTG pathway is activated, Mks1p is largely dephosphorylated and is sequestered by Rtg2p, effectively relieving the negative regulation of the RTG pathway. Conversely, when the RTG pathway is repressed, for example, in respiratory-competent cells, or by the addition of glutamate to the medium, Mks1p is hyperphosphorylated and is in a complex with Bmh1/2p. Mks1p in that complex has been suggested to be the active, negative regulator of the RTG pathway (Liu et al., 2003).
The main objective of this study was identification and characterization of a new regulatory factor of the RTG pathway. Here we show that Mks1p levels are regulated by Grr1p, the F-box component of the SCFGrr1 (Skp1-Cullin-F box) E3 ubiquitin ligase (Deshaies, 1999), and that modulation of Mks1p levels can affect the regulation of the RTG pathway. We have isolated dominant GRR1 mutants with single amino acid changes within the putative substrate binding site localized to the concave surface of the Grr1p leucine-rich repeat (LRR) domain. These changes result in a net increase in positive charge and lead to instability of Mks1p and activation of the RTG pathway. We found that the Bmh1/2p act as negative regulators of the RTG pathway by protecting Mks1p from degradation by Grr1p. We have identified a novel, portable Mks1p degron sequence, which is an acidic domain in the central region of the protein. Finally, we provide evidence that the form of Mks1p targeted for degradation by Grr1p is not bound to either Rtg2p or to Bmh1/2p.
MATERIALS AND METHODS
Strains and Plasmids
Strains and plasmids used in this study are available at Molecular Biology of the Cell Online.
Growth Media
Yeast strains were grown at 30°C in YPD medium (1% yeast extract, 2% bactopeptone, and 2% dextrose), YNBcasD (0.67% yeast nitrogen base, 1% casamino acids, and 2% dextrose), SCRaffGal (0.67% yeast nitrogen base, 0.065% CSM-uracil-leucine dropout mix [Bio101, Carlsbad, CA], 2% raffinose, and 2% galactose), or minimal YNBD medium (0.67% yeast nitrogen base, 2% dextrose) with or without glutamate (concentrations indicated in the text and figures). MG132 (Sigma-Aldrich, St. Louis, MO) was dissolved in dimethyl sulfoxide and applied at a final concentration of 50 μM.
EMS Mutagenesis and Cloning of GRR1 Dominant Mutants
EMS mutagenesis was conducted as described in Liu et al. (2001). Cloning of dominant rtg2Δ bypass mutations was described in Liu et al. (2003). Briefly, a yeast genomic DNA library was constructed from strain G15 and transformed into rtg2Δ cells (PSY142-rtg2) to recover the mutant gene conferring the rtg2Δ bypass phenotype. This new rtg2Δ bypass mutant was due to a mutation in GRR1. GRR1 was then cloned by PCR from a collection of 18 other dominant rtg2 bypass mutants to determine whether these mutants harbor GRR1 mutations. Altogether, eight GRR1 mutations were uncovered, with five due to change of glutamic acid at position 522 to lysine (E522K), one E548K mutation, one D657N mutation, and one S444R mutation.
Total mRNA Isolation and Northern Blot Analysis
Total yeast RNA was isolated from 50-ml logarithmic-phase cultures, fractionated on 1·2% agarose gels, transferred to Nylon membrane and hybridized at 65°C with probes specific for transcripts of the CIT2 and ACT1 genes. Hybridization signals were detected with a Molecular Dynamics Phosphor-Imager (Sunnyvale, CA).
β-galactosidase Activity Assays for CIT2-lacZ Reporter Gene Expression
Liquid cultures were inoculated with a pool of several independent transformants in YNBcasD medium and grown to OD600 0.6-0.8. The preparation of cell extracts and β-galactosidase assays were carried out as described by Rose et al. (1990).
Cell Extracts, Immunoblotting, Immunoprecipitations, and Ubiquitin Assay
Whole cell extracts were prepared and coimmunoprecipitation experiments were performed as described by Sekito et al. (2000) except for the following changes: for ubiquitination assay of Mks1p, buffer A (20 mM Tris-HCl, pH 7.6, 0.5% Triton X-100, 0.7% deoxycholate, 0.1% SDS, and protease inhibitors) was used; for interaction between Mks1p and Bmh1p, Rtg2p, or Grr1p, buffer B was used (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 0.5% Triton X-100, and protease inhibitors). Samples were resolved by SDS-PAGE. Detection of HA- or myc-tagged proteins on Western blots was conducted as described (Liu et al., 2003). Anti-ubiquitin antibody (P4D1, Santa Cruz Biotechnology, Santa Cruz, CA) was used to detect ubiquitinated species of Mks1p.
Cycloheximide Chase Assay
Cells expressing indicated HA-tagged proteins were grown in liquid YNBcasD medium to OD600 ∼1. Cycloheximide (50 μg/ml) was added to start the chase. Every 15 or 20 min, a 1-ml aliquot of the cell culture was withdrawn, and the cell pellets were subject to TCA precipitation as described (Sekito et al., 2000).
In Vitro Translation of Mks1p and GST Pulldown Assay with GST or GST-Grr1p
MKS1 and MKS1 (ΔAD) cloned in pET24a (Novagen, Madison, WI) were in vitro translated and labeled with 35S-methionine using the TnT T7 Quick coupled transcription/translation labeling kit (Promega, Madison, WI). GST and GST-Grr1p was expressed in bacteria BL21(DE3) cells (Novagen) and purified using Sepharose 4B beads (Amersham Pharmacia, Piscataway, NJ). For GST pulldown assay, GST fusion proteins bound to beads were equilibrated with GST buffer (25 mM HEPES, pH 7.5, 100 mM NaCl, 0.05% Nonidet NP-40, 5 mM dithiothreitol, 10% glycerol, 50 μg/ml bovine serum albumin), and the binding reactions were performed in the same buffer. After incubation of GST or GST-Grr1p with 35S-labeled Mks1p or Mks1p (ΔAD) for 2 h at 4°C, the beads were washed five times with 1 ml GST buffer. Bound proteins were eluted by boiling in protein loading buffer, separated by SDS-PAGE, and dried gels were analyzed by autoradiography.
RESULTS
Dominant GRR1 Mutants Bypass the rtg2Δ Mutation
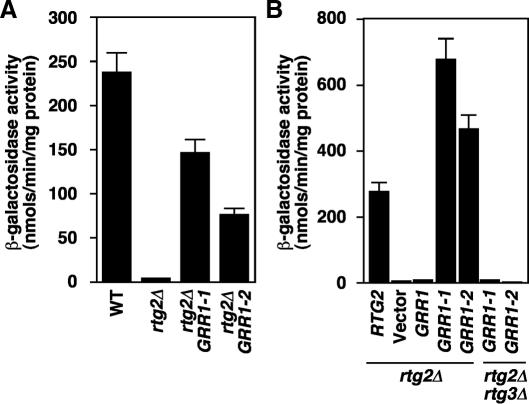
To search for new regulatory components of the RTG pathway, we used a genetic screen for rtg2Δ bypass mutants that was successfully used to identify the RTG regulators, Lst8p, Mks1p, and Bmh1/2p (Liu et al., 2001; Sekito et al., 2002; Liu et al., 2003). The current screen revealed eight dominant GRR1 mutants representing four different alleles, GRR1-(1-4). GRR1 encodes a subunit of the SCFGrr1 E3 ubiquitin ligase complex. As is shown for two of the alleles, GRR1-1 and GRR1-2, these mutants confer an rtg2Δ bypass phenotype to the expression of a CIT2-lacZ reporter gene (Figure 1A). Expression of these mutant alleles from centromeric plasmids restored the block in CIT2 reporter gene expression in rtg2Δ GRR1 cells, indicating that they are dominant mutations (Figure 1B). Neither mutant was able to rescue CIT2 reporter gene expression in rtg2Δ rtg3Δ cells, showing that they affect the RTG pathway downstream of RTG2 but upstream of RTG1/3. Similar results were obtained for the GRR1-3 and GRR1-4 mutants (unpublished data). Finally, none of the mutants affect the steady state level of Grr1p, and overexpression of wild-type GRR1 from a 2 μm plasmid failed to bypass the rtg2Δ mutation (unpublished data).
Figure 1.
GRR1 mutations activate Rtg2p-independent CIT2-lacZ reporter gene expression. (A) Two dominant GRR1 mutations, GRR1-1 and GRR1-2, restore CIT2-lacZ reporter gene expression in rtg2Δ cells. Wild-type (WT) and mutant haploid strains as indicated were examined for CIT2-lacZ expression as described in Materials and Methods. (B) The GRR1-1 and GRR1-2 mutant alleles are dominant and their rtg2Δ bypass phenotype is dependent on RTG3. rtg2Δ and rtg2Δ rtg3Δ mutant cells transformed with centromeric plasmids containing RTG2, GRR1, GRR1-1, GRR1-2 or no insert (vector) were examined for CIT2-lacZ expression.
Mks1p Is Unstable in rtg2Δ GRR1-1 and rtg2Δ GRR1-2 Mutant Cells
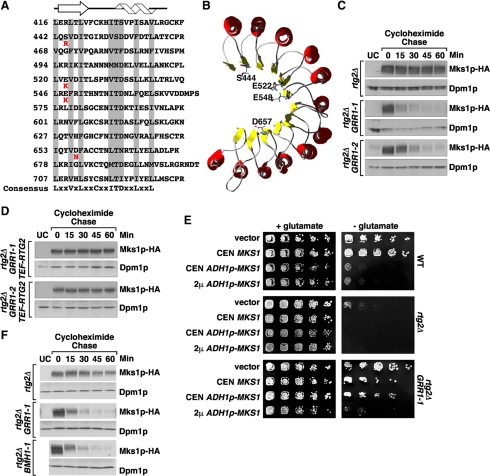
Sequence analysis of the GRR1 mutant alleles revealed that each contains a single amino acid change in the LRR domain of Grr1p, resulting in a net gain of one or two positive charges (E522K in GRR1-1, E548K in GRR1-2, S444R in GRR1-3, and D657N in GRR1-4; Figure 2A). LRR domains are generally involved in protein-protein interactions. The structure of the LRR domain from porcine liver ribonuclease inhibitor reveals a horseshoe-shaped solenoid with a concave surface of parallel β-sheets and a convex surface of α-helices (Kobe and Deisenhofer, 1995). The mutations in the Grr1p LRR domain map to the predicted concave surface of the protein (Figure 2B), which has been proposed to be the binding site for Cln2p (Hsiung et al., 2001).
Figure 2.
GRR1 dominant mutations destabilize Mks1p in rtg2Δ cells. (A) Sequence alignment of the Grr1p leucine-rich repeats (LRR). The conserved residues are highlighted in gray and the consensus sequence shown at the bottom indicates amino acid residues present in at least five of the repeats. The altered residue of each of the four dominant mutations of GRR1-(1-4) is indicated in red underneath the wild-type residue. The predicted secondary structure (helix for α-helix and arrow for β-sheet) is indicated at the top. (B) The dominant GRR1 mutations localize to the concave surface of the LRR of Grr1p. The modeled 3D structure is adapted from Hsiung et al. (2001). The residues mutated in the dominant GRR1 mutants are indicated by showing their side chains. (C) Mks1p expressed from the ADH1 promoter is unstable in rtg2Δ GRR1-1 and rtg2Δ GRR1-2 cells. Mks1p stability was examined in indicated cells using cycloheximide chase assay as described in Materials and Methods.. Mks1p-HA levels were determined by probing Western blots with anti-HA antibody. Dpm1p, dolichol phosphate mannose synthase, was used as loading control. UC, untagged control. (D) Rtg2p protects Mks1p from degradation in GRR1-1 and GRR1-2 mutants. Mks1p stability was examined following a cycloheximide chase in rtg2Δ GRR1-1 and rtg2Δ GRR1-2 cells, each containing a copy of RTG2 integrated at the LEU2 locus under the control of the TEF1 promoter. (E) Overexpression of MKS1 reverses the rtg2Δ bypass phenotype of the GRR1-1 mutation. Serial dilutions of culture of indicated cells were prepared and cells were spotted onto solid YNBD medium with or without 0.02% glutamate. Cells were grown for three days at 30°C. (F) Stability assay of Mks1p-HA expressed from its own promoter. Mks1p stability was examined using cycloheximide chase assay.
Because substrates for E3 ubiquitin ligases are generally phosphoproteins (Deshaies, 1999), a candidate substrate for Grr1p is the RTG negative regulator, Mks1p. Mks1p is hyperphosphorylated when the retrograde pathway is shut down, for example, in rtg2Δ cells (Sekito et al., 2002). We therefore asked whether the GRR1 dominant mutants might bypass the rtg2Δ mutation because of increased degradation of Mks1p. Accordingly, we examined the levels of a functional, C-terminal HA-tagged derivative of Mks1p expressed from a centromeric plasmid under the control of the ADH1 promoter in rtg2Δ cells at various times after the addition of cycloheximide. In otherwise wild-type rtg2Δ cells, Mks1p-HA was relatively stable over the 60-min cycloheximide chase period (Figure 2C and Supplementary Figure S1). In contrast, in both rtg2Δ GRR1-1 and rtg2Δ GRR1-2 mutant cells, Mks1p-HA was unstable, as most of it disappeared by 30 min after cycloheximide addition. These findings are consistent with the notion that the rtg2Δ bypass phenotype in GRR1-1/2 mutant cells is due to Mks1p degradation. Increased degradation of Mks1p-HA in rtg2Δ GRR1-1 cells was also observed in a Gal shut-off assay in which Mks1p-HA expression was placed under the control of the galactose-inducible GAL1 promoter (Supplementary Figure S2). This indicates that the Mks1p-HA instability observed in Figure 2C is not an artifact due to the growth inhibitory effects of cycloheximide.
The Grr1p-dependent degradation of Cln2 has been well studied (Lanker et al., 1996; Hsiung et al., 2001). To determine whether the GRR1 dominant mutants also affect Cln2 stability, we carried out a Gal shut-off assay in which Cln2-HA expression was placed under the control of the GAL1 promoter. Neither the GRR1-1 or GRR1-2 mutation has an affect on Cln2-HA stability (Supplementary Figure S3), indicating the specificity of the GRR1-1/2 mutations toward Mks1p.
In wild-type cells with robust RTG pathway activity, Mks1p is hypophosphorylated and in a complex with Rtg2p (Liu et al., 2003). To determine the stability of that form of Mks1p in GRR1-1 and GRR1-2 cells, Mks1p-HA was expressed as above, and in the same cells, Rtg2p was overexpressed from a wild-type RTG2 allele integrated at the LEU2 locus under the control of the strong TEF1 promoter so as to drive most, if not all, of the Mks1p-HA into a complex with Rtg2p. The validity of this strategy is indicated by the fact that the glutamate auxotrophy resulting from Mks1p-HA overexpression (due to down-regulation of the RTG pathway) can be completely reversed by overexpression of Rtg2p (unpublished data). As shown in Figure 2D, Mks1p-HA expressed in GRR1-1 and GRR1-2 cells is now stable in transformants overexpressing Rtg2p. As shown previously (Sekito et al., 2002; Liu et al., 2003), Mks1p is hypophosphorylated in cells coexpressing Rtg2p. The protection effect of overexpression of Rtg2p on Mks1p stability is unlikely to be indirect, for example, by inhibiting SCFGrr1, because overexpression does not affect Cln2 stability (Supplementary Figure S3).
To show directly that the rtg2Δ bypass phenotype of GRR1-1/2 mutants is specifically the result of increased degradation of Mks1p, we increased the expression of Mks1p in GRR1-1 cells, reasoning that the rtg2Δ bypass phenotype might be suppressed by saturation of the Mks1p degradation system. GRR1-1 cells were transformed with a centromeric plasmid expressing Mks1p from its own promoter (CEN MKS1), from the stronger ADH1 promoter (ADH1p-MKS1), or from a multicopy 2μ plasmid also under the control of the ADH1 promoter (2μ ADH1p-MKS1). Down-regulation of the RTG pathway in those cells was then scored by the appearance of glutamate auxotrophy. In wild-type cells, expression of Mks1p from ADH1p-MKS1 or 2μ ADH1p-MKS1 is sufficient to repress the RTG pathway (Figure 2E). In rtg2Δ cells, which show a slightly leaky glutamate auxotrophy, full repression of the RTG pathway can be achieved when Mks1p was expressed from CEN MKS1. In contrast, significant suppression of the rtg2Δ bypass phenotype in GRR1-1 mutant cells requires that Mks1p be expressed from 2μ ADH1p-MKS1. These results show that there is an Mks1p dosage-dependent reversal of the GRR1-1 mutant phenotype, suggesting that the rtg2Δ bypass phenotype of the GRR1-1/2 dominant mutants is specifically because of an increase in the degradation of Mks1p.
The level of Mks1p expressed from the ADH1 promoter is about fivefold higher than that expressed from its own promoter (unpublished data). To exclude the possibility that instability of Mks1p in Figure 2C is due to Mks1p overexpression, we examined the stability of Mks1p expressed from its own promoter and found it is unstable in rtg2Δ GRR1-1 cells (Figure 2F and Supplementary Figure S4). Moreover, the steady state level of Mks1p expressed from its own promoter was reduced by >10-fold in rtg2Δ GRR1-1 cells (Supplementary Figure S5), consistent with the notion that rtg2Δ bypass phenotype of the GRR1-1 mutation is due to accelerated Mks1p degradation.
Bmh1/2p protect Mks1p from Grr1p-dependent Degradation in Wild-type Cells
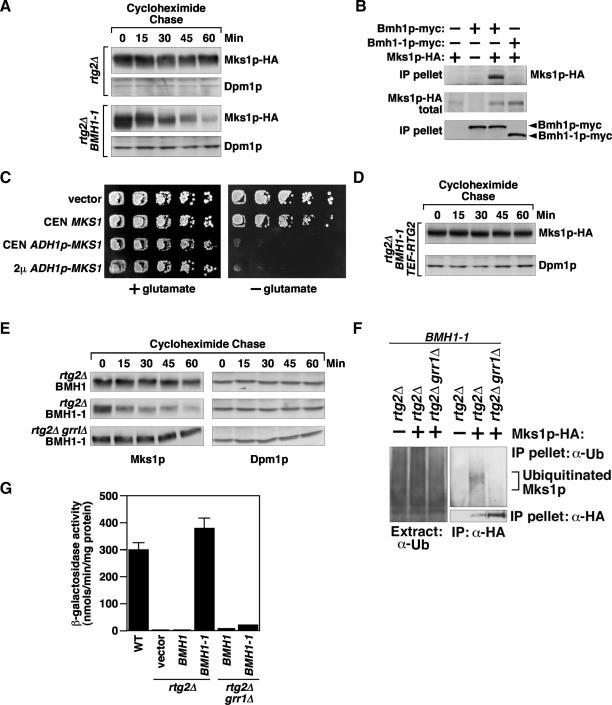
We previously suggested that the form of Mks1p that inhibits the RTG pathway is a hyperphosphorylated species complexed with the 14-3-3 proteins, Bmh1p and Bmh2p (Liu et al., 2003). That conclusion was based in part on the analysis of a dominant-negative BMH1 mutant, BMH1-1, which shows a high level of CIT2 expression and loss of glutamate auxotrophy in rtg2Δ cells due to depletion of a functional Bmh2p pool. Bmh1p is the major isoform (Gelperin et al., 1995), and Bmh1p and Bmh2p can function as homo- or heterodimers (Chaudhri et al., 2003). Moreover, we also observed that the steady state level of Mks1p is substantially lower in bmh1/2 mutant cells than in wild-type cells. These findings raise the possibility that Mks1p is not bound to the 14-3-3 proteins in BMH1-1 cells and is rapidly degraded, thereby activating the RTG pathway. To test this, we first compared the stability of Mks1p expressed under the control of the ADH1 promoter in rtg2Δ cells expressing either wild-type BMH1 or the BMH1-1 dominant mutant allele. As shown in Figure 3A, Mks1p-HA is unstable in rtg2Δ BMH1-1 cells but is relatively stable in the rtg2Δ BMH1 wild-type control. Similar data were obtained using Mks1p-HA expressed from its own promoter (Figure 2F and Supplementary Figure S4). Consistent with increased Mks1p-HA degradation in rtg2Δ BMH1-1 cells, the steady-state level of Mks1p expressed from its own promoter was reduced by >10-fold in rtg2Δ BMH1-1 cells (Supplementary Figure S5).
Figure 3.
GRR1-dependent Mks1p instability in rtg2Δ BMH1-1 cells. (A) Mks1p is unstable in rtg2Δ BMH1-1 cells. Mks1p stability was examined in rtg2Δ or rtg2Δ BMH1-1 cells expressing Mks1p under the control of the ADH1 promoter using cycloheximide chase assay. (B) Mks1p-HA interacts with wild-type Bmh1p, but not mutant Bmh1-1p. Cell extracts of bmh1Δ bmh2Δ mks1Δ cells expressing the tagged proteins as indicated were obtained for immunoprecipitation. Anti-myc antibody was used to precipitate Bmh1p-myc and Bmh1-1p-myc. (C) Overexpression of MKS1 reverses the rtg2Δ bypass phenotype of the BMH1-1 mutation. rtg2Δ BMH1-1 cells expressing different levels of MKS1 were assayed for glutamate auxotrophy phenotype as described for Figure 2E. (D) Rtg2p protects Mks1p from degradation in BMH1-1 cells. Mks1p stability was examined in rtg2Δ BMH1-1 cells transformed with an integrated copy of RTG2 under the control of TEF1 promoter. (E) A grr1Δ mutation leads to Mks1p stability in rtg2Δ cells expressing BMH1-1. Mks1p stability was examined in rtg2Δ or rtg2Δ grr1Δ cells expressing wild-type BMH1 or BMH1-1 from a centromeric plasmid. (F) A grr1Δ mutation abolishes ubiquitination of Mks1p expressed from the ADH1 promoter in rtg2Δ cells expressing the mutant Bmh1-1p. Total cell extracts were prepared from cells as indicated expressing Mks1p-HA under the control of the ADH1 promoter. Mks1p-HA was immunoprecipitated using anti-HA antibody conjugated agarose beads. Anti-ubiquitin antibody was used to detect Mks1p ubiquitination. (G) A grr1Δ mutation reverses BMH1-1-induced CIT2-lacZ expression in rtg2Δ cells.
Next, we determined whether the increased degradation rate of Mks1p-HA is due to its failure to interact with Bmh1-1p. We performed coimmunoprecipitation experiments from extracts of cells in which Mks1p-HA was coexpressed with either wild-type Bmh1p-myc or the Bmh1-1p-myc dominant mutant. Immunoprecipitation of these proteins with anti-myc antibody shows that mutant Bmh1-1p has lost the ability to interact with Mks1p (Figure 3B). We can exclude the possibility that failure of Mks1p to interact with Bmh1-1p is due to misfolding of Bmh1-1p, such that it cannot form a heterodimer with Bmh2p, because coimmunoprecipitation experiments indicate that Bmh1-1p interacts as well with Bmh2p as does wild-type Bmh1p (Supplementary Figure S6). Further evidence that Mks1p depletion accounts for the rtg2Δ bypass phenotype in BMH1-1 cells is the findings that overexpression of Mks1p in those cells from either a centromeric or 2μ plasmid under the control of the ADH1 promoter restores glutamate auxotrophy, indicating that the RTG pathway has been down-regulated (Figure 3C).
To determine the stability of Mks1p when bound to Rtg2p in BMH1-1 cells, Rtg2p was overexpressed from a TEF1 promoter. Under these conditions, Mks1p was now stable (Figure 3, compare panels A and D). These data are similar to the preceding observations that the Rtg2p-bound form of Mks1p was also stable in GRR1 dominant mutant cells (Figure 2D). To show that wild-type Grr1p targets Mks1p for degradation in BMH1-1 cells, we inactivated GRR1 in rtg2Δ cells expressing the dominant BMH1-1 allele from a centromeric plasmid. Mks1p was now stable in those cells (Figure 3E).
To determine whether GRR1 dependent instability of Mks1p in rtg2Δ BMH1-1 cells is due to ubiquitination, Mks1p-HA expressed from the ADH1 promoter was immunoprecipitated from a total cell extract using anti-HA antibody and Mks1p ubiquitination in the immunoprecipitate was probed with anti-ubiquitin antibody. Figure 3F shows that ubiquitination of Mks1p could be easily detected in rtg2Δ BMH1-1 cells expressing Mks1p-HA and that ubiquitination was dependent on GRR1. To provide direct evidence that the Rtg2p- or Bmh1p-bound form of Mks1p is not a substrate for Grr1p-dependent ubiquitination, Bmh1p-myc was immunoprecipitated with anti-myc antibody and a 12xHis tagged Rtg2p was precipitated with Ni2+ beads from extracts of cells coexpressing Mks1p-HA. After a second precipitation with anti-HA antibody, the recovered Mks1p-HA was then examined for its ubiquitination state using anti-ubiquitin antibody. The Mks1p-HA recovered under these conditions was not ubiquitinated (Supplementary Figure S7), consistent with the notion that Rtg2p- or Bmh1p-bound form of Mks1p is not a substrate for Grr1p-dependent ubiquitination.
That GRR1-dependent degradation of Mks1p in rtg2Δ cells expressing the dominant BMH1-1 mutant allele under-lies the rtg2Δ bypass phenotype is evident from the findings that expression of the mutant BMH1-1 allele, but not wild-type BMH1, in rtg2Δ cells results in increased CIT2-lacZ expression that is dependent on GRR1 (Figure 3G). Together, these data suggest that Mks1p not bound to either Bmh1/2p or to Rtg2p is targeted by Grr1p for degradation. As expected, mutations in two other SCFGrr1 components, Cdc53p and Skp1p (Skowyra et al., 1997; Patton et al., 1998), also block BMH1-1 induced degradation of Mks1p in rtg2Δ cells (Supplementary Figure S8).
Functional Proteasomes Are Required for the rtg2Δ Bypass Phenotype in BMH1-1 and GRR1-1 Cells
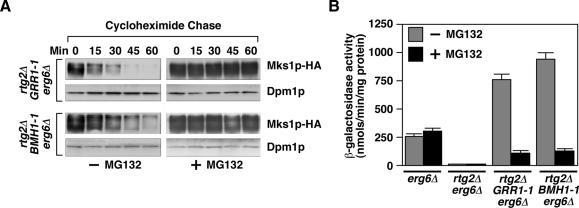
To demonstrate that the instability of Mks1p in GRR1-1 and BMH1-1 mutant cells is proteasome dependent, we treated cells with the proteasome inhibitor, MG132 (Lee and Goldberg, 1996). To facilitate uptake of MG132 into cells, an erg6Δ mutation was also introduced into rtg2Δ GRR1-1 and rtg2Δ BMH1-1 mutant strains. The resultant rtg2Δ GRR1-1 erg6Δ and rtg2Δ BMH1-1 erg6Δ cells were treated with or without 50 μM MG132 for 90 min before the cycloheximide chase. The instability of Mks1p expressed from the ADH1 promoter in rtg2Δ GRR1-1 erg6Δ and rtg2Δ BMH1-1 erg6Δ cells was blocked in cells treated with MG132 (Figure 4A), indicating that Mks1p instability is dependent on functional proteasomes. To determine whether proteasomes are required for rtg2Δ bypass phenotype in GRR1-1 and BMH1-1 cells, CIT2-lacZ expression was examined in cells treated with or without MG132. The addition of MG132 had little effect on basal level of CIT2-lacZ expression in erg6Δ and rtg2Δ erg6Δ cells (Figure 4B). However, the high level of CIT2-lacZ expression observed in rtg2Δ GRR1-1 erg6Δ and rtg2Δ BMH1-1 erg6Δ cells was largely reversed by MG132. Thus the rtg2Δ bypass phenotypes of BMH1-1 and GRR1-1 mutant cells can be accounted for by proteasome-dependent degradation of Mks1p.
Figure 4.
Functional proteasome is required for rtg2Δ bypass phenotype in GRR1-1 and BMH1-1 cells. (A) Mks1p is stabilized by MG132. Mks1p stability was examined in cells as indicated treated with or without MG132 (50 μM). Cells were grown in YNBcasD medium. (B) MG132 inhibits CIT2-lacZ expression in GRR1-1 and BMH1-1 cells. Indicated strains were grown in YNBcasD medium with or without MG132.
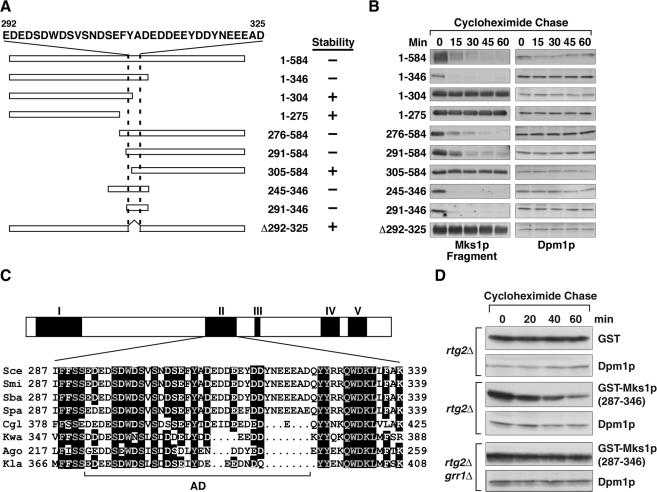
An Acidic Domain Region of Mks1p Contains the Degron Sequence
We next wanted to identify the domain of Mks1p conferring its Grr1p-dependent instability. Taking advantage of the robust degradation of Mks1p in rtg2Δ GRR1-1 cells, we examined the stability of a series of Mks1p fragments (Figure 5A) with a 3xHA tag at their C-terminus expressed from centromeric plasmids under the control of the ADH1 promoter. Figure 5B shows that an N-terminal Mks1p fragment (1-346) is unstable, whereas the shorter N-terminal fragments,1-304 and 1-275, are stable. The C-terminal fragment 276-584 and a shorter C-terminal fragment, 291-584, are also unstable in rtg2Δ GRR1-1 cells, but further truncation to 305-584 results in a stable fragment. These results, summarized in Figure 5A, suggest that an acidic amino acid-rich region of Mks1p located between residues 291 and 346 is required to confer Mks1p instability in rtg2Δ GRR1-1 cells. That fragment alone is unstable, and deleting a stretch of residues rich in acidic amino acids from 292 to 325 (Figure 4A) from full-length Mks1p to yield Mks1p (Δ292-325) confers stability to the protein. This acidic domain region of Mks1p is also required for wild-type GRR1-dependent Mks1p degradation in rtg2Δ BMH1-1 cells (unpublished data). Sequence alignment of Mks1p from eight fungal species reveals five blocks of sequence conservation, shown diagrammatically in Figure 5C, and in complete form in Supplementary Figure S9. Block II largely overlaps with the Mks1p acidic domain degron region (residues 291-346), underlying the importance of acidic domain region for Mks1p function.
Figure 5.
An acidic domain region of Mks1p contains the portable degron sequence. (A) Diagrammatic representation of constructs expressing various fragments of Mks1p. Numbers indicate the Mks1p residues expressed. Two dashed lines indicate a stretch of acidic amino acid-rich residues of Mks1p. (B) Residues 291-346 of Mks1p are required to confer Mks1p instability. The stability of various Mks1p constructs in rtg2Δ GRR1-1 cells was determined using the cycloheximide chase assay. (C) A diagrammatic representation of sequence conservation of Mks1p from eight fungal species. Five conserved sequence blocks, I-V, are indicated, with Block II shown in detail. AD indicates a conserved acidic amino acid-rich subdomain. Sce, S. cerevisiae; Spa, S. paradoxus; Smi, S. mikatae; Sba, S. bayanus; Cgl, Candida glabrata; Kla, Kluyveromyces lactis; Kwa, Kluyveromyces waltii; Ago; Ashbya gossypii. (D) Mks1p residues 287-346 confer Grr1p-dependent instability to GST. GST or GST-Mks1p (287-346) fusion protein expressed in cells as indicated were examined for stability.
To show that the acidic domain region of Mks1p is indeed a Grr1p degradation signal, we constructed a fusion protein containing residues 287-346 of Mks1p fused to the C-terminus of Schistosoma japonicum glutathione S-transferase (GST) and expressed this construct in rtg2Δ and rtg2Δ grr1Δ cells. In cells expressing wild-type Grr1p, the GST-Mks1p (287-346) fusion protein, by contrast to GST alone, is unstable over a 60-min cycloheximide chase, and this instability is dependent on wild-type Grr1p (Figure 5D). These data indicate that the acidic domain region of Mks1p contains the Mks1p degron sequence and that this degron is portable.
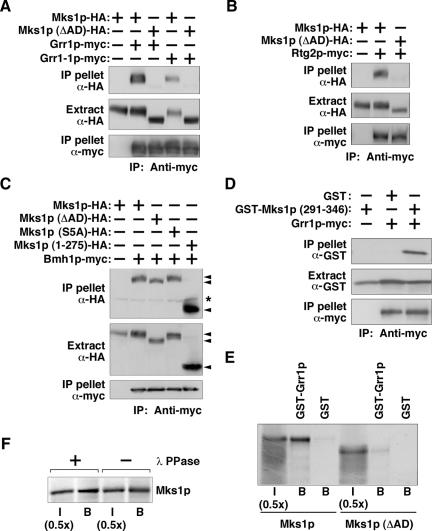
The Acidic Domain Region of Mks1p Is Required for Interaction with Rtg2p and Grr1p, but not with Bmh1p
Previous studies have shown that an Mks1p C-terminal fragment, 276-584, which includes the acidic domain, binds to Rtg2p (Liu et al., 2003). The finding that Mks1p is stable when bound to Rtg2p, even in GRR1 dominant mutant cells, raises the possibility that the acidic domain is not only a recognition site for Grr1p, but is also important for the binding of Mks1p to Rtg2p. If so, the acidic domain could be inaccessible to Grr1p in an Mks1p-Rtg2p complex. Conversely, the acidic domain may be less important for interaction of Mks1p with Bmh1/2p. These considerations would explain why Mks1p is insensitive to degradation when bound to Rtg2p but not to Bmh1/2p in cells expressing the dominant GRR1 alleles.
To test these various possibilities, we asked whether the acidic domain region is important for binding to Grr1p, to the dominant mutant form of the protein, Grr1-1p, as well as to Rtg2p and Bmh1p. First, coimmunoprecipitation experiments were carried out with extracts of cells expressing HA-tagged Mks1p or its acidic domain deletion mutant, Mks1p (ΔAD) (Δ292-325), both under the control of GAL1 promoter. Those same cells were also expressing either myc-tagged wild-type Grr1p, or mutant Grr1-1p, also under the control of GAL1 promoter. Extracts were immunoprecipitated with anti-myc antibody and then probed by Western blotting for the various HA-tagged Mks1p derivatives. These experiments show that, in contrast to wild-type Mks1p-HA, the acidic domain deletion mutant, Mks1p (ΔAD)-HA, fails to interact with either Grr1p-myc or Grr1-1p-myc (Figure 6A).
Figure 6.
The Mks1p acidic domain (AD) is required for interaction with Grr1p and Rtg2p, but not Bmh1p. (A) The acidic domain of Mks1p is required for interaction with both wild-type Grr1p and with mutant Grr1-1p. rtg2Δ erg6Δ cells expressing HA- or myc-tagged proteins as indicated were grown in SCRaffGal medium. Coimmunoprecipitation experiments were carried out. (B) The acidic domain of Mks1p is required for Mks1p interaction with Rtg2p. Total cell extracts from mks1Δ cells expressing tagged proteins as indicated were subject to a coimmunoprecipitation assay. (C) Interaction between Bmh1p-myc and HA-tagged Mks1p. Total cell extracts from bmh1Δ bmh2Δ mks1Δ cells expressing tagged proteins as indicated were subject to a coimmunoprecipitation assay. “*” indicates the heavy chain of anti-myc antibody used for immunoprecipitation. (D) Mks1p acidic domain region is sufficient for interaction with Grr1p. Total cell extracts from rtg2Δ erg6Δ cells expressing tagged proteins as indicated were subject to a coimmunoprecipitation assay. GST also contains 48 amino acids at its C-terminus translated from an HA3 coding sequence and the polylinker, so its mobility is the same as the GST-Mks1p (291-346) fusion protein. Ub, ubiquitin. (E) In vitro-translated Mks1p interacts with Grr1p. Mks1p and Mks1p (ΔAD) were translated in vitro and labeled with 35S-methionine as described in Materials and Methods.. The GST pulldown assay is described in Materials and Methods.. I, input fraction; B, glutathione sepharose beads bound fraction. (F) Interaction between GST-Grr1p and protein phosphatase treated Mks1p. In vitro-translated, 35S-labeled Mks1p was treated with or without lambda protein phosphates (λ PPase) as indicated and subjected to a GST pulldown assay as described in E.
Similar coimmunoprecipitation experiments were carried out to assess the role of the Mks1p acidic domain region in binding to Rtg2p (Figure 6B) and Bmh1p (Figure 6C). Figure 6B shows that wild-type Mks1p interacts with Rtg2p, whereas Mks1p (ΔAD) binds poorly to Rtg2p. These results suggest that Rtg2p protects Mks1p from degradation by blocking the binding site for Grr1p and Grr1-1p. In contrast, Bmh1p interacts with Mks1p (ΔAD) as well it does with wild-type Mks1p (Figure 6C). Previously, we showed that Bmh1p interacts with an N-terminal fragment of Mks1p, (1-346), but not with an overlapping C-terminal fragment (276-584; Liu et al., 2003). 14-3-3 proteins usually bind to RxxpS/pT and RxxxpS/pT motifs (Yaffe et al., 1997), and five such sites are located within residues 1-346 of Mks1p. However, mutation of the S/T residues in all five of those sites to alanines in the full-length protein does not affect the interaction of Mks1p with Bmh1p (Figure 6C) nor does it affect Mks1p functionality (unpublished data). Importantly, the Mks1p fragment 1-275 interacts well with Bmh1p. These data suggest that Bmh1p binds to Mks1p through the N-terminal 275 residues of Mks1p via novel binding sites.
To show that the acidic domain region of Mks1p is sufficient for interaction with Grr1p, we fused Mks1p residues 291-346 to the amino terminus of GST, which was expressed together with myc-tagged wild-type Grr1p in rtg2Δ cells. Coimmunopreciptation experiments show that the Mks1p acidic domain-GST fusion protein interacts with wild-type myc-tagged Grr1p, whereas no interaction was observed with GST alone (Figure 6D). The GST-Mks1p (291-346) fusion protein migrates as a single band on SDS-PAGE gels, and its mobility is unaffected by phosphatase treatment (unpublished data), suggesting that it is not phosphorylated. To provide additional support for the notion that the acidic domain region rather than phosphorylation is an important determinant for the interaction of Mks1p with Grr1p, we translated Mks1p in vitro and examined its interaction with a GST-Grr1p fusion protein expressed and purified from bacteria. Figure 6E shows that Grr1p tagged with GST, but not GST alone, interacts with in vitro-translated Mks1p, whereas the binding of the Mks1p (ΔAD) mutant protein is reduced by 10-fold (Figure 6E). Moreover, treatment of the in vitro-translated Mks1p with lambda phosphatase did not affect its mobility on SDS-PAGE or its ability to interact with Grr1p (Figure 6F). Taken together, these data suggest that the acidic domain region of Mks1p is both necessary and sufficient for interaction with Grr1p and that Mks1p phosphorylation does not play an essential role in recognition by Grr1p.
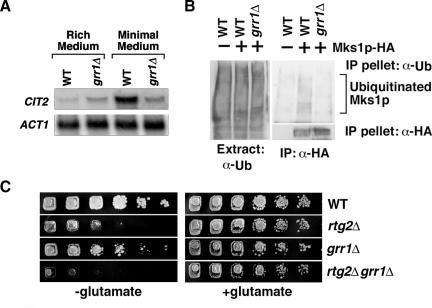
GRR1 Contributes to the Regulation of the RTG Pathway
To demonstrate that wild-type Grr1p plays a role in the regulation of the RTG pathway, we inactivated the wild-type GRR1 allele and determined the effect on both basal and induced expression of CIT2. These different levels of CIT2 expression are readily accomplished by growing cells in rich medium, in which there is a basal level of CIT2 expression, or in minimal medium in which CIT2 expression is highly induced (Liu and Butow 1999). Inactivation of GRR1, in contrast, has no effect on CIT2 basal expression, but it markedly inhibited the induction of CIT2 expression by growth of cells in minimal medium (Figure 7A). To provide additional support for Grr1p regulation of the RTG pathway via the regulation of Mks1p stability, we assayed for Mks1p ubiquitination in wild-type cells similar to the experiments described in Figure 3F. Figure 7B shows that Mks1p-HA expressed under the control of its own promoter is ubiquitinated in-wild type GRR1 cells, but not in grr1Δ cells. Together with the previous observations that Mks1p exerts its highest inhibitory effect on the RTG pathway in the absence of Rtg2p (Dilova et al., 2002; Sekito et al., 2002), it is unlikely that Mks1p degradation in cells carrying an rtg2Δ mutation shown in Figures 2C and 3A is due to mis-folding of the protein. Furthermore, the constitutive high level expression of CIT2 observed in mks1Δ cells (Sekito et al., 2002) is not reduced by the introduction of a grr1Δ mutation (unpublished data), indicating that MKS1 is epistatic to GRR1.
Figure 7.
(A) A grr1Δ mutation blocks full induction of CIT2 expression. Wild-type and grr1Δ cells were grown in rich YPD or minimal YNBD medium to midlog phase. Total mRNA was prepared and CIT2 mRNA levels were determined by Northern blot analysis. ACT1 was used as loading control. (B) A grr1Δ mutation blocks ubiquitination of Mks1p expressed from its own promoter. Ubiquitination of Mks1p was determined as described in Figure 3F. (C) The grr1Δ and rtg2Δ mutations additively reduce cell growth on medium lacking glutamate. Serial dilutions (5×) of cultures were prepared and cells were spotted onto solid YNBD medium with or without 0.02% glutamate. Cells were grown for 3-4 d at 30°C.
We have routinely observed that the glutamate auxotrophy phenotype of rtg2Δ cells is somewhat leaky (Figure 2E). If Grr1p contributes to the positive regulation of the RTG pathway, as suggested from the data above, then inactivation of GRR1 should exacerbate the glutamate auxotrophy phenotype of rtg2Δ cells. This is indeed the case because grr1Δ and rtg2Δ mutations have additive effects on reducing cell growth on minimal medium without glutamate (Figure 7C). Taken together, we conclude that Grr1p is a positive regulator of the RTG pathway.
DISCUSSION
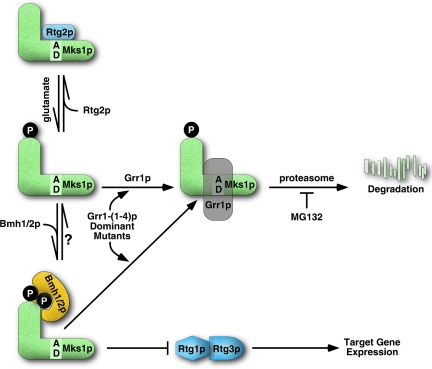
Regulation of the RTG pathway is achieved in part by factors that interact with Mks1p to modulate its activity. When bound to the redundant 14-3-3 proteins, Bmh1p and Bmh2p, Mks1p negatively regulates the RTG pathway; when bound to Rtg2p, Mks1p is inactive, resulting in high levels of RTG target gene expression. The present study reveals a third mode of regulation of Mks1p, whereby its abundance is controlled by SCFGrr1 E3 ubiquitin ligase-dependent polyubiquitination and degradation. This feature of Mks1p regulation appears to be important for the induction of CIT2, rather than for its basal level of expression. Our studies further suggest that the form of Mks1p targeted for degradation by wild-type Grr1p is not bound to either Rtg2p or Bmh1/2p. Why do cells need Grr1p as an additional modulator of Mks1p activity? The interaction of Mks1p with Rtg2p and Bmh1/2p is dynamic, switching the RTG pathway on and off depending on whether Mks1p is bound to Rtg2p or to Bmh1/2p (Liu et al., 2003). We suggest that the efficiency of this switch would be enhanced, as summarized in the model depicted in Figure 8, by maintaining a low level of unbound Mks1p. In accordance with previous results (Liu et al., 2003), we propose that a critical feature of Mks1p regulation is the two state interaction of Mks1p with either Bmh1/2p, which down-regulates the pathway by inhibiting the Rtg1/3p complex, or with Rtg2p, which prevents Mks1p from interacting with Bmh1/2p, thus activating the pathway. Mks1p not bound to either Rtg2p or to Bmh1/2p is proposed to be the preferred substrate for wild-type Grr1p, targeting Mks1p for degradation via the proteasome by polyubiquitination. This step would tighten control of the Mks1p on-off switch by keeping the unbound Mks1p pool low. We suggest that the Grr1p dominant mutants, by gaining one or two positive charges in the substrate binding surface, have a higher affinity for the acidic domain of Mks1p, allowing the mutant proteins access to Mks1p while still bound to Bmh1/2p. However, the acidic domain is required for binding of Mks1p to Rtg2p, but not to Bmh1/2p, and thus may not be accessible to the dominant Grr1p mutants in the Mks1p-Rtg2p complex.
Figure 8.
A model for regulation of the RTG pathway. Regulation relies on the dynamic interactions of Mks1p with Rtg2p and Bmh1/2p. When the RTG pathway is ON, Rtg2p binds to and inactivates Mks1p, which is hypophosphorylated. When the RTG pathway is OFF, Mks1p is released from Rtg2p, becomes hyperphosphorylated, and interacts with Bmh1/2p. The free Mks1p, unbound to either Rtg2p or Bmh1/2p, is degraded through SCFGrr1 dependent ubiquitination and degradation, which would keep free Mks1p level low to tighten control of Mks1p on-off switch and allow full activation of the RTG pathway. The acidic domain region in the central portion of Mks1p is Grr1p recognition motif. Dominant mutations on the concave surface of the Grr1p leucine-rich repeat domain allow Grr1p access to the acidic domain of Mks1p in the presence of Bmh1/2p, resulting Mks1p degradation.
The dominant GRR1 mutations, which lead to Rtg2p-independent activation of the RTG pathway, represent, to our knowledge, the first gain-of-function mutations described for a SCF E3 ubiquitin ligase. All of the independent, dominant GRR1 mutants we identified have a net gain of one or two positive charges in the concave surface of the Grr1p LRR domain. The SCF E3 ubiquitin ligase family usually targets phosphorylated substrates, consistent with the modeled structure of the LRR domain in which there is a high density of positively charged residues on the concave, substrate-binding surface (Hsiung et al., 2001). It has been shown that mutations of these basic residues to neutral or acidic amino acids results in an inability of the mutant protein to interact with one Grr1p substrate, Cln2p, resulting in its stabilization (Hsiung et al., 2001). Our results lend further support to the view that the concave surface of LRR is the binding site for its substrates, likely through electrostatic interactions.
The acidic domain region of Mks1p is both necessary and sufficient for interaction with Grr1p and is a novel feature of the recognition of a substrate by an E3 ubiquitin ligase. Although SCF E3 ubiquitin ligases are generally believed to recognize multiply phosphorylated substrates, for example, Sic1p, the target of Cdc4p (Verma et al., 1997), and Cln2p, a target of Grr1p (Lanker et al., 1996), we observed an acidic domain-dependent interaction of Grr1p with Mks1p in which Mks1p was synthesized in an unphosphorylated form by in vitro translation. However, we cannot exclude the possibility that Mks1p phosphorylation may play some role in modulating the efficiency by which Mks1p is recognized by Grr1p. A plausible explanation for the GRR1 dominant mutant phenotype is that the gain of positive charges in the LRR domain increases the affinity of the interaction between Grr1p and the acidic domain of Mks1p, thereby allowing Mks1p to be targeted for degradation when bound to Bmh1/2p.
The finding that the acidic domain of Mks1p is also required for its interaction with Rtg2p provides an explanation for the resistance of the Rtg2p-bound form of Mks1p to degradation in the GRR1 wild-type or dominant mutant background: in the Mks1p-Rtg2p complex, the acidic domain would likely be occluded from interaction with Grr1p. By contrast, the acidic domain is not required for Mks1p binding to Bmh1/2p, and thus when bound to Bmh1/2p it would still be available for interaction with a Grr1p mutant with a presumed higher affinity for the acidic domain. The Bmh1/2p-bound form of Mks1p may nevertheless result in some physical hindrance to the accessibility of the Mks1p acidic domain, because Bmh1/2p protects Mks1p from being targeted for degradation by wild-type Grr1p. Thus one important function of Bmh1/2p because negative regulators of the RTG pathway would be to protect Mks1p from being targeted for degradation by wild-type Grr1p. Precisely how retrograde signals are translated to switch Mks1p binding between Rtg2p to Bmh1/2p is presently unclear. However, we have observed that the dissociation of Mks1p from Rtg2p requires ATP hydrolysis (our unpublished data), suggesting the possibility that signaling may also involve some monitoring of ATP/ADP levels.
Inhibition of TOR kinase activity by rapamycin treatment activates the RTG pathway (Komeili et al., 2000; Shamji et al., 2000). There exist two TOR kinase complexes, TORC1 and TORC2, both of which contain Lst8p (Loewith et al., 2002; Wedaman et al., 2003). Isolation and characterization of lst8 mutations which bypass the rtg2Δ mutation suggest that TOR kinases play a role in regulation of the RTG pathway (Chen and Kaiser, 2003; Liu et al., 2003). The findings presented here raise questions whether activation of the RTG pathway by rapamycin treatment is due to increased Mks1p instability. However, we found that rapamycin fails to increase Mks1p turnover in either wild-type cells or in rtg2Δ mutant cells (unpublished data), indicating that Mks1p degradation represents a TOR-independent process in the regulation of the RTG pathway. Because activation of the RTG pathway by rapamycin treatment requires Rtg2p (Komeili et al., 2000 and our unpublished observations), we believe that increased Mks1p binding to Rtg2p is likely to be the mechanism of rapamycin-induced activation of the RTG pathway.
14-3-3 proteins are involved in multiple physiological pathways and usually bind their substrates through phospho-serine or threonine at Rxx(x)pS/pT motifs (Yaffe et al., 1997). Similar to the protection effect of Bmh1/2p on Mks1p, a 14-3-3 protein protects the tumor suppressor p53 from degradation by a E3 ubiquitin ligase, Mdm2 (Yang et al., 2003). Mks1p contains five consensus RxxpS/pT sites within an N-terminal region (1-275), which we have shown here binds to Bmh1/2p. We have mutated all five of those sites, but have not observed any effect on Bmh1p binding (Figure 6C). This suggests that Bmh1/2p may recognize novel sites in the N-terminal domain of Mks1p.
Grr1p is known to regulate the stability of the G1 cyclins, Cln1 and Cln2, and the putative Cdc42p regulator, Gic2p (Deshaies, 1999). Grr1p is also required for glucose induction of several hexose transporters, including Hxt1p, by increasing degradation of a negative regulator Mth1p (Li and Johnston, 1997; Flick et al., 2003). Grr1p has also been reported to be required for amino acid induced expression of several amino acid permeases, including Agp1p, and a peptide transporter, Ptr2p (Bernard and Andre, 2001). We have examined all four GRR1 dominant mutant alleles for their potential effects on expression of AGP1, HXT1, and PTR2 and no effects have been found thus far (our unpublished data). We have also failed to see an effect of the GRR1-1/2 mutations on Cln2 stability (Supplementary Figure S3). These results suggest that Grr1p utilizes different sites to interact with regulators in different pathways and are consistent with the finding that neutralization of positively charged residues in the concave surface of LRR of Grr1p results in defects in Cln2 degradation but has no effect on transcription of HXT1 and AGP1 (Hsiung et al., 2001).
Supplementary Material
Acknowledgments
We thank Gerald Fink, Mark Johnston, and Per Ljungdahl for yeast strains, Curt Wittenberg for CLN2 plasmid and the modeled structure of Grr1p LRR, José Ribamar Ferreira Júnior for technical support, and A. Tizenor for graphics. This work was supported by Grants GM22525 from the National Institutes of Health and I-0642 from The Robert A. Welch Foundation.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-06-0516) on August 10, 2005.
Abbreviations used: RS, retrograde signaling.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Bernard, F., and Andre, B. (2001). Ubiquitin and the SCF(Grr1) ubiquitin ligase complex are involved in the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. FEBS Lett. 496, 81-85. [DOI] [PubMed] [Google Scholar]
- Biswas, G., Adebanjo, O. A., Freedman, B. D., Anandatheerthavarada, H. K., Vijayasarathy, C., Zaidi, M., Kotlikoff, M., and Avadhani, N. G. (1999). Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 18, 522-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, G., Anandatheerthavarada, H. K., Zaidi, M., and Avadhani, N. G. (2003). Mitochondria to nucleus stress signaling: a distinctive mechanism of NFkappaB/Rel activation through calcineurin-mediated inactivation of IkappaBbeta. J. Cell Biol. 161, 507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butow, R. A., and Avadhani, N. G. (2004). Mitochondrial signaling: the retrograde response. Mol. Cell 14, 1-15. [DOI] [PubMed] [Google Scholar]
- Chaudhri, M., Scarabel, M., and Aitken, A. (2003). Mammalian and yeast 14-3-3 isoforms form distinct patterns of dimers in vivo. Biochem. Biophys. Res. Commun. 300, 679-685. [DOI] [PubMed] [Google Scholar]
- Chen, E. J., and Kaiser, C. A. (2003). LST8 negatively regulates amino acid biosynthesis as a component of the TOR pathway. J. Cell Biol. 161, 333-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies, R. J. (1999). SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435-467. [DOI] [PubMed] [Google Scholar]
- Dilova, I., Chen, C.-Y., and Powers, T. (2002). Mks1 in concert with TOR signaling negatively regulates RTG target gene expression in S. cerevisiae. Curr. Biol. 12, 389-395. [DOI] [PubMed] [Google Scholar]
- Epstein, C. B., Waddle, J. A., Hale, I. V., W., Davé, V., Thornton, J., Macatee, T. L., Garner, H. R., and Butow, R. A. (2001). Genome-wide responses to mitochondrial dysfunctions. Mol. Biol. Cell 12, 297-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick, K. M., Spielewoy, N., Kalashnikova, T. I., Guaderrama, M., Zhu, Q., Chang, H. C., and Wittenberg, C. (2003). Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol. Biol. Cell 14, 3230-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin, D., Weigle, J., Nelson, K., Roseboom, P., Irie, K., Matsumoto, K., and Lemmon, S. (1995). 14-3-3 proteins: potential roles in vesicular transport and Ras signaling in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 92, 11539-11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallstrom, T. C., and Moye-Rowley, S. (2000). Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275, 34347-37356. [DOI] [PubMed] [Google Scholar]
- Hsiung, Y. G., Chang, H. C., Pellequer, J. L., La Valle, R., Lanker, S., and Wittenberg, C. (2001). F-box protein Grr1 interacts with phosphorylated targets via the cationic surface of its leucine-rich repeat. Mol. Cell. Biol. 21, 2506-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y., Rothermel, B., Thornton, J., and Butow, R. A. (1997). A basic helix-loop-helix zipper transcription complex functions in a signaling pathway from mitochondria to the nucleus. Mol. Cell. Biol. 17, 1110-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe, B., and Deisenhofer, J. (1995). A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature 374, 183-186. [DOI] [PubMed] [Google Scholar]
- Komeili, A., Wedaman, K. P., O'Shea, E. K., and Powers, T. (2000). Mechanism of metabolic control: target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 151, 863-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanker, S., Valdivieso, M. H., and Wittenberg, C. (1996). Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science 271, 1597-1601. [DOI] [PubMed] [Google Scholar]
- Lee, D. H., and Goldberg, A. L. (1996). Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem. 271, 27280-27284. [DOI] [PubMed] [Google Scholar]
- Li, F. N., and Johnston, M. (1997). Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1, coupling glucose sensing to gene expression and the cell cycle. EMBO J. 16, 5629-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, X., and Butow, R. A. (1993). RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell 72, 61-71. [DOI] [PubMed] [Google Scholar]
- Liu, Z., and Butow, R. A. (1999). A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol. Cell. Biol. 19, 6720-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Sekito, T., Epstein, C. B., and Butow, R. A. (2001). RTG-dependent mitochondria to nucleus signaling is negatively regulated by the seven WD-repeat protein Lst8p. EMBO J. 20, 7209-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Sekito, T., Spirek, M., Thornton, J., and Butow, R. A. (2003). Retrograde signaling is regulated by the dynamic interaction between Rtg2p and Mks1p. Mol. Cell 12, 401-411. [DOI] [PubMed] [Google Scholar]
- Loewith, R., Jacinto, E., Wullschleger, S., Lorberg, A., Crespo, J. L., Bonenfant, D., Oppliger, W., Jenoe, P., and Hall, M. N. (2002). Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. Biol. 10, 457-468. [DOI] [PubMed] [Google Scholar]
- Luo, Y., Bond, J. D., and Ingram, V. M. (1997). Compromised mitochondrial function leads to increased cytosolic calcium and to activation of MAP kinases. Proc. Natl. Acad. Sci. USA 94, 9705-9710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, E. E., Willems, A. R., Sa, D., Kuras, L., Thomas, D., Craig, K. L., and Tyers, M. (1998). Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 12, 692-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M. D., Winston, F., and Heiter, P. (1990). Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Sekito, T., Liu, Z., Thornton, J., and Butow, R. A. (2002). RTG-dependent mitochondria-to-nucleus signaling is regulated by MKS1 and is linked to the formation of the yeast prion [URE3]. Mol. Biol. Cell 13, 795-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekito, T., Thornton, J., and Butow, R. A. (2000). Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol. Biol. Cell 11, 2103-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamji, A. F., Kuruvilla, F. G., and Schreiber, S. L. (2000). Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr. Biol. 10, 1574-1581. [DOI] [PubMed] [Google Scholar]
- Skowyra, D., Craig, K. L., Tyers, M., Elledge, S. J., and Harper, J. W. (1997). F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91, 209-219. [DOI] [PubMed] [Google Scholar]
- Tate, J. J., Cox, K. H., Rai, R., and Cooper, T. G. (2002). Mks1p is required for negative regulation of retrograde gene expression in Saccharomyces cerevisiae but does not affect nitrogen catabolite repression-sensitive gene expression. J. Biol. Chem. 277, 20477-20482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, R., Feldman, R. M., and Deshaies, R. J. (1997). SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol. Biol. Cell 8, 1427-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedaman, K. P., Reinke, A., Anderson, S., Yates, J., 3rd, McCaffery, J. M., and Powers, T. (2003). Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 1204-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, M. B., Rittinger, K., Volinia, S., Caron, P. R., Aitken, A., Leffers, H., Gamblin, S. J., Smerdon, S. J., and Cantley, L. C. (1997). The structural basis for 14-3-3, phosphopeptide binding specificity. Cell 91, 961-971. [DOI] [PubMed] [Google Scholar]
- Yang, H.-Y., Wen, Y.-Y., Chen, C.-H., Lozano, G., and Lee, M.-H. (2003). 14-3-3σ positively regulates p53 and suppresses tumor growth. Mol. Cell. Biol. 23, 7096-7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.