Abstract
Nuclear aggregates formed by proteins containing expanded poly-glutamine (poly-Q) tracts have been linked to the pathogenesis of poly-Q neurodegenerative diseases. Here, we show that a protein (GFP170*) lacking poly-Q tracts forms nuclear aggregates that share characteristics of poly-Q aggregates. GFP170* aggregates recruit cellular chaperones and proteasomes, and alter the organization of nuclear domains containing the promyelocytic leukemia (PML) protein. These results suggest that the formation of nuclear aggregates and their effects on nuclear architecture are not specific to poly-Q proteins. Using GFP170* as a model substrate, we explored the mechanistic details of nuclear aggregate formation. Fluorescence recovery after photobleaching and fluorescence loss in photobleaching analyses show that GFP170* molecules exchange rapidly between aggregates and a soluble pool of GFP170*, indicating that the aggregates are dynamic accumulations of GFP170*. The formation of cytoplasmic and nuclear GFP170* aggregates is microtubule-dependent. We show that within the nucleus, GFP170* initially deposits in small aggregates at or adjacent to PML bodies. Time-lapse imaging of live cells shows that small aggregates move toward each other and fuse to form larger aggregates. The coalescence of the aggregates is accompanied by spatial rearrangements of the PML bodies. Significantly, we find that the larger nuclear aggregates have complex internal substructures that reposition extensively during fusion of the aggregates. These studies suggest that nuclear aggregates may be viewed as dynamic multidomain inclusions that continuously remodel their components.
INTRODUCTION
Newly synthesized proteins must be properly folded and modified to function correctly. Eukaryotic cells have developed extensive folding machineries to ensure the fidelity of protein processing. Nevertheless, misfolding can occur due to mutations within a protein, outside stresses, or the overexpression of proteins. Misfolded proteins often expose their hydrophobic domains, which leads to nonproductive protein associations and results in aggregation. Aggregated proteins tend to coalesce and form large deposits termed inclusion bodies, Russell bodies, or aggresomes, depending on their composition and location. Formation of such inclusions underlies a number of aggresomal diseases, including Alzheimer's disease, Parkinson's disease, familial amyotrophic lateral sclerosis, and the poly-glutamine (poly-Q) neuropathologies (reviewed in Zoghbi and Orr, 2000; Garcia-Mata et al., 2002).
The biological processes leading to protein aggregation have been actively investigated (reviewed in Kopito, 2000; Garcia-Mata et al., 2002; Goldberg, 2003; Selkoe, 2003). Aggregation of proteins most likely occurs cotranslationally, while nascent peptide chains are synthesized on polyribosomes. If the nascent peptides cannot fold correctly, they will aggregate to form aggresomal particles. Small aggresomal particles form throughout the cell and are quickly transported toward the microtubule (MT)-organizing center, where they coalesce to form aggresomes (Johnston et al., 1998; Garcia-Mata et al., 1999). Aggresome formation is blocked by drugs that depolymerize microtubules, and by the expression of p50/dynamitin, suggesting that a dynein-based transport along microtubules is required for aggresome formation (Johnston et al., 1998; Garcia-Mata et al., 1999). Cytoplasmic aggresomes are enriched in molecular chaperones (including Hsc70, Hdj1 and Hdj2, and the chaperonin TCP) and in proteasomal subunits (Wojcik et al., 1996; Wigley et al., 1999). The active recruitment of refolding and degradative machineries suggests that the formation of aggresomes is a dynamic process that cells use to cope with misfolded proteins. The preferential localization of aggresomes to the peri-centriolar region in mammalian cells suggests that the cytoplasmic milieu contains regions specialized to sequester and clear misfolded proteins.
In addition to cytoplasmic aggresomes, nuclear inclusions are often found in patients with Huntington's disease (HD) or spinocerebellar ataxias (SCAs) (DiFiglia et al., 1997; Perez et al., 1998; Chai et al., 2001; Waelter et al., 2001; Yamada et al., 2001). HD and SCAs are neurodegenerative diseases caused by expanded poly-Q repeats in huntingtin and ataxins, respectively. The mutant proteins are aggregation prone and form both cytoplasmic and intranuclear inclusions. In vitro studies with purified disease-causing proteins show that aggregation is based on a nucleated polymerization reaction, suggesting that self-aggregation of poly-Q proteins may occur when the protein concentration reaches a critical level (Scherzinger et al., 1999; Chen et al., 2002). The formation of nuclear inclusions depends on the length of poly-Q repeats and on as yet unidentified factors in the host cells. Studies in cell culture systems and in transgenic mice show that the nuclear inclusions recruit molecular chaperones, ubiquitin, and proteasomal subunits (Cummings et al., 1998; Chai et al., 1999b; Kim et al., 2002). The association of the degradative machineries suggests that nuclear inclusions may be involved in the proteolytic clearing of poly-Q aggregated substrates. Such nuclear inclusions may be analogous to cytoplasmic aggresomes, suggesting that the nucleus may also contain specialized sites to compartmentalize and clear misfolded proteins. The link between poly-Q content and the ability to form nuclear aggregates has led to the suggestion that the formation of nuclear aggregates may involve poly-Q-dependent mechanisms.
Here, we show that a nonpoly-Q protein (GFP170*), which contains green fluorescent protein (GFP) fused to an internal segment (amino acids 566-1375) of the Golgi Complex Protein 170 (GCP170), is deposited in nuclear aggregates. The aggregates share characteristic features of aggregates formed by poly-Q proteins, because they recruit chaperones and proteasomes. Importantly, we show that GFP170* and the G3 domain of aggrecan, another unrelated nonpoly-Q protein shown previously to form nuclear inclusions (Chen et al., 2001), disrupt the organization of promyelocytic leukemia (PML) bodies. Thus, the formation of nuclear aggresomes may be a common response to misfolded proteins that gain access to the nucleus. Our studies suggest that the mammalian nucleus is compartmentalized with respect to handling misfolded proteins. Using fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP), we show that GFP170* aggregated within nuclear inclusions undergoes rapid exchange with a pool of soluble GFP170* molecules. This observation suggests that nuclear aggregates may be dynamic localizations of proteins, rather than precipitated stores. Using GFP170* as a model substrate, we explored the mechanistic details of nuclear aggregate formation. We show that GFP170* deposition is initiated at or adjacent to PML bodies. Time-lapse imaging of live cells shows extensive movements and fusion of small GFP170* aggregates to form large structures within the nucleus. The coalescence of the foci is accompanied by spatial rearrangements of the PML bodies. The nuclear GFP170* aggregates exhibit a complex internal architecture that is extremely dynamic and undergoes extensive remodeling during the fusion of aggregates. Our studies indicate that nuclear components may be entrapped in the GFP170* aggregates and that the internal organization of the aggregates may be facilitated by phase partitioning between their components.
MATERIALS AND METHODS
Antibodies and Reagents
Anti-giantin antibody was a gift from Dr. Hans P. Hauri (University of Basel, Basel, Switzerland). Anti-Hdj2 polyclonal antibody was a gift from Dr. Douglas Cyr (University of North Carolina, Chapel Hill, NC). Anti-GFP polyclonal antibody (catalog no. Ab290) was purchased from Abcam (Cambridge, United Kingdom). Anti-FLAG monoclonal antibody was purchased from Eastman Kodak (Rochester, NY). Anti-β-tubulin, anti-coilin, and anti-SC35 monoclonal antibodies were purchased from Sigma-Aldrich (St. Louis, MO). Anti-lamin A (catalog no. sc-6215) and anti-PML (PG-M3) (catalog no. sc-966) monoclonal antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Hsc70 (catalog no. SPA-815) and anti-Hsp70 (catalog no. SPA-810) monoclonal antibodies were purchased from StressGen Biotechnologies (San Diego, CA). Anti-20S proteasome (α-subunit) polyclonal antibody was purchased from EMD Biosciences (San Diego, CA). Anti-human CFTR (C-terminus specific) antibody was purchased from R&D Systems (Minneapolis, MN). Rabbit polyclonal antibody to ubiquitin-protein conjugates was from Affiniti Research Products (Exeter, United Kingdom). Oregon green-labeled goat anti-rabbit IgG antibody, Texas red-labeled goat anti-mouse IgG antibody, and Texas red-labeled goat anti-rabbit IgG antibody were from Molecular Probes (Eugene, OR). Horseradish peroxidase (HRP)-labeled sheep anti-rabbit IgG antibody and HRP-labeled goat anti-rabbit IgG antibody were from GE Healthcare (Piscataway, NJ). Nocodazole was purchased from Sigma-Aldrich and used at the indicated concentration. Super-Signal West Pico chemiluminescence substrate was from Pierce Chemical (Rockford, IL). Restriction enzymes and molecular reagents were from Promega (Madison, WI), New England BioLabs (Beverly, MA), or QIAGEN (Valencia, CA). All other chemicals were from Sigma-Aldrich or Fisher Scientific (Pittsburgh, PA).
DNA Constructs
To make a chimera of enhanced green fluorescent protein (EGFP) and GCP170, a primer containing the XhoI restriction enzyme site was designed in front of the start codon of GCP170. The 770-base pair PCR fragment containing sequences from the start codon of GCP170 to the EcoRI site of FQSY1024 (Misumi et al., 1997) was cloned into the XhoI and EcoRI sites of pEGFP-C2 plasmid (BD Biosciences Clontech, Palo Alto, CA). A 5558-base pair EcoRI fragment from FQSY1024 was then cloned into the EcoRI sites of the plasmid mentioned above to generate an EGFP-tagged full-length GCP170. GFP170* construct was then generated by removing the BglII fragment and SacII fragment from the N-terminal and C-terminal end of EGFP-GCP170, respectively. The resultant construct expresses an EGFP-tagged GCP170 fragment from amino acid 566-1375. The G3-FLAG construct has been described previously (Chen et al., 2001). It encodes the putative aggrecan signal sequence (the first 23 N-terminal amino acids of aggrecan); a GAG5 consensus sequence, G3; and C-terminally attached His and FLAG epitopes. GFP-250 and ΛF508-CFTR constructs were described previously (Garcia-Mata et al., 1999). The plasmid expressing Q82-GFP was a gift from Dr. Richard Morimoto (Northwestern University, Evanston, IL) and was described previously (Kim et al., 2002).
Cell Culture, Transfections, and Immunofluorescence Microscopy
COS-7 and COS-1 cells were grown in DMEM with glucose and glutamine (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). Cells were transfected with the FuGENE transfection reagent (Roche Diagnostics, Indianapolis, IN), with TransIT polyamine transfection reagents (Mirus, Madison, WI), or with Lipofectin (Invitrogen), according to manufacturer protocols. At 18-48 h after transfection, cells were fixed with 3% paraformaldehyde, or in some cases with cold methanol and processed for immunofluorescence microscopy as described previously (Alvarez et al., 1999; Chen et al., 2001).
Electron Microscopy and Immunogold Labeling
COS-7 cells were transfected with the GFP170* construct. At 48 h after transfection, cells were washed with phosphate-buffered saline (PBS), detached from the plate by trypsinization, and collected by centrifugation at 300 × g for 5 min at 4°C. Cells were washed twice with PBS and then fixed for 90 min with 1.5% glutaraldehyde in 0.1 M sodium cacodylate, pH 7.4. Cells were washed three times with sodium cacodylate and postfixed with 1% OsO4 in 0.1 M sodium cacodylate, pH 7.4, for 60 min on ice. After washing three times with 0.1 M sodium cacodylate, pH 7.4, cells were dehydrated by incubation with a series of ethanol solutions (30, 50, 70, 90, 95, and 3 × 100%) followed by 2-h incubation in 1:1 Spurr's resin/propylene oxide. After two changes of fresh 100% resin, the cell pellets were transferred to gelatin molds and polymerized in fresh resin overnight at 60°C. Gold epoxy sections (100 nm thick) were generated with a Reichert Ultracut ultramicrotome and collected on 200 mesh copper grids. Grid specimens were stained for 20 min with saturated aqueous uranyl acetate (3.5%) diluted 1:1 with ethanol just before use, followed by staining with lead citrate for 10 min. Stained samples were examined on a JEOL 100CX electron microscope.
For immunogold labeling, COS-7 cells expressing GFP170* were harvested by trypsinization 24 h after transfection. Cells were washed three times with PBS and prefixed with 3% formaldehyde and 0.2% glutaraldehyde for 40 min, followed by dehydration with series of graded ethanol at room temperature. Cells were infiltrated and embedded with LR White. After polymerization, sections were cut with an ultramicrotome and collected onto nickel grids. The grids were incubated with anti-GFP primary antibody overnight at 4°C and goat anti-rabbit IgG conjugated to 12-nm gold particles for 1 h at room temperature (Jackson ImmunoResearch Laboratories, West Grove, PA), followed by postfixation with 2% glutaraldehyde for 5 min at room temperature, and counterstained with 2% uranyl acetate for 5 min at room temperature.
Analysis of Soluble and Insoluble GFP170*
COS-7 cells were transfected with GFP170*. At 48 h after transfection, cells were washed and harvested in ice-cold PBS. Equal amounts of cells were lysed for 1 h on ice with 100 μl of either 1% Triton X-100 in PBS or RIPA buffer (50 mM Tris-HCl, pH 8.0, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, and 150 mM NaCl) supplemented with protease inhibitor cocktail and 1.0 mM phenylmethylsulfonyl fluoride (PMSF). Lysates were sonicated for 5 s with a microtip sonicator followed by 15-min centrifugation at 15,000 × g. Pellets were resuspended in 100 μl of 1% SDS in PBS. Equal volumes of each pellet and supernatant were boiled in SDS-PAGE sample buffer and resolved on 8% SDS-PAGE. The gel was transferred to nitrocellulose membrane and processed for Western blotting as described previously (Gao and Sztul, 2001).
Analysis of Degradation Rate of GFP170* and GFP-250
COS-7 cells were transfected with either GFP170* or GFP-250 construct in a six-well plate for 24 h. Cells were then washed in PBS and incubated in methionine-free DMEM for 1 h. Cells were labeled with 200 μCi/ml [35S]methionine (PerkinElmer Life and Analytical Sciences, Boston, MA) for 60 min. Incorporation was terminated by washing the cells with PBS and chasing with DMEM medium supplemented with 0.2 mM methionine for indicated times. At each time point, cells were lysed with RIPA buffer supplemented with protease inhibitor cocktail and 1.0 mM PMSF. Equal amounts of lysate from each time point were resolved by 10% SDS-PAGE followed by autoradiography using a PhosphoImage screen. The relative radioactive intensity of GFP170* and GFP-250 bands was quantified and compared using ImageQuant software.
Time-Lapse Imaging Study
The movement of GFP170* was analyzed by time-lapse imaging as described previously (Garcia-Mata et al., 1999). Briefly, COS-7 cells grown on glass coverslips were transfected with GFP170* construct. At 20 h after transfection, coverslips were placed onto sealed silicon rubber chambers containing culture medium buffered with 25 mM HEPES, pH 7.5. Images were acquired with an Olympus IX70 inverted microscope equipped with a 40×/1.35 numerical aperture objective lens and a cooled charge-coupled device camera. IpLab Spectrum software (Signal Analytics; Scanalytics, Fairfax, VA) was used to control image acquisition and manipulation.
FRAP and FLIP Analysis
FRAP and FLIP analysis of the GFP170* and Q82-GFP was performed as described previously (Kim et al., 2002), using a Leica TCS SP2 confocal microscope with a 63× objective lens. COS-7 cells expressing GFP170* were subject to analysis 24-48 h after transfection. COS-7 cells expressing Q82-GFP were analyzed 72 h after transfection. During the experiment, COS-7 cells were kept at 37°C in a glass-bottom dish containing DMEM medium buffered with 25 mM HEPES, pH 7.5. For FRAP analysis, fluorescent regions outlined in figures were photobleached at full laser power with zoom, and images were taken at lower laser power (∼20%) before bleaching and every 10-30 s after photobleaching. Fluorescent recovery was calculated by comparing the intensity ratio in regions of bleached area before the bleach and after recovery. The postbleach intensities were normalized upward to correct for total loss of fluorescence due to the photobleach by comparing the fluorescent intensities outside the bleached area using IpLab Spectrum software (Signal Analytics). For FLIP analysis, cells were repeatedly bleached in the same defined region and imaged at 1-min intervals. At each time point after photobleaching, fluorescence intensities in the cytosolic and nuclear regions were measured separately using IpLab software and normalized to those values before bleaching.
RESULTS
GFP170* Deposits within Cytoplasmic and Nuclear Aggregates
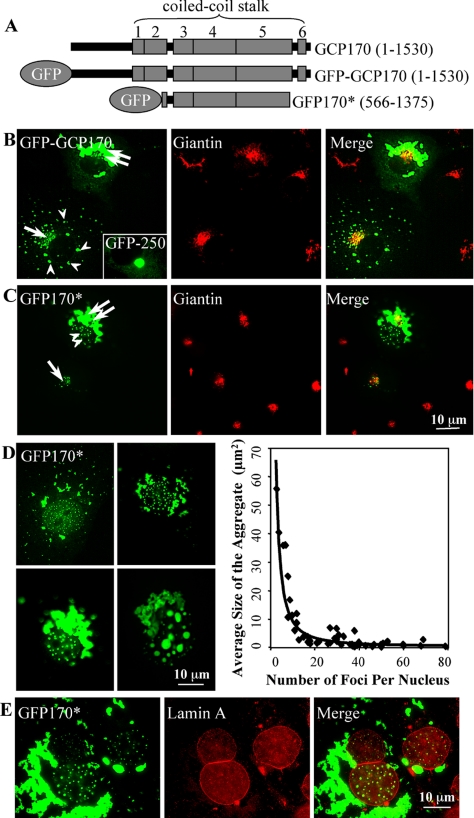
GCP170, also known as golgin-160, was identified as a human auto-antigen in patients with Sjögren syndrome (Fritzler et al., 1993). Patient sera reacted with an antigen localized in the Golgi, and subsequent studies led to the cloning of GCP170 (Misumi et al., 1997). GCP170 contains 1530 amino acids, arranged into an N-terminal head domain followed by a long coiled-coil stalk and a short C-terminal tail (Figure 1A). The stalk region is divided into six coiled-coil segments. Coiled-coil domains have been shown to mediate protein-protein interactions, and GCP170 may form a parallel homodimer through the intertwining of its coiled-coil segments (Hicks and Machamer, 2002). GCP170 is a soluble protein that localizes to the cytoplasmic face of Golgi membranes (Misumi et al., 1997; Hicks and Machamer, 2002). We have generated various GFP-tagged constructs of GCP170 to study targeting of GCP170 to the Golgi in vivo. The description of the signals that target GCP170 to the Golgi will be presented elsewhere. Here, GFP-tagged wild-type GCP170 and a construct called GFP170* that encodes amino acids 566-1375 of GCP170 and contains coiled-coils 3, 4, and 5 are described (Figure 1A).
Figure 1.
GFP170* deposits within the cytoplasm and in the nucleus. (A) Schematic diagram of full-length GCP170(1-1530), GFP-GCP170, and GFP170*(566-1375). The full-length GCP170 contains an amino-terminal head domain followed by a coiled-coil stalk of six-coiled coils (shaded boxes). GFP170* contains GFP fused to an internal segment (amino acid 566-1375 of GCP170). (B) COS-7 cells were transfected with a GFP-tagged full-length GCP170 or GFP-250 construct. After 48 h, cells were processed for indirect immunofluorescence using antibody against the Golgi marker protein giantin. In a cell expressing low levels of GFP-GCP170 (bottom left), most of the molecules are targeted to the Golgi region and colocalize with giantin (arrow). In addition, small peripheral aggregates are visible (arrowheads). In a cell expressing high levels of GFP-GCP170 (top right), the GFP-GCP170 forms large aggregates that surround the Golgi (double arrow). The GFP170* aggregates seem “ribbon-like” and are distinct from the spherical aggregates formed by GFP-250 (inset). (C) COS-7 cells were transfected with GFP170*. After 48 h, cells were processed for indirect immunofluorescence using antibody against giantin. In a cell expressing low levels of GFP170* (bottom left), most of the molecules are targeted to the Golgi region, as demonstrated by colocalization with giantin (arrow). In a cell expressing high levels of GFP170* (top right), GFP170* forms large aggregates that surround the Golgi (double arrow). In addition, GFP170* is detected in numerous nuclear foci (double arrowhead). (D) COS-7 cells were transfected with GFP170*. After 24-48 h, cells were processed for epifluorescence. Images of COS-7 cells containing different numbers of nuclear aggregates were selected. The average size of GFP170* nuclear foci was determined using IPLab software and plotted as a function of number of foci per nucleus. (E) COS-7 cells were transfected with GFP170*. After 24 h, cells were processed for immunofluorescence using anti-lamin A antibody. Spherical GFP170* foci are enclosed within the lamin A-defined space.
We analyzed the cellular localization of GFP-tagged GCP170 and GFP170*. GFP-tagged GCP170 is targeted to the Golgi in COS-7 cells expressing moderate levels of the recombinant protein (Figure 1B, cell at bottom left, arrow). The Golgi localization is shown by extensive overlap of the GFP signal with the Golgi marker giantin. In addition, dispersed cytoplasmic aggregates are also evident (arrowheads). In an adjacent cell (top right), perhaps expressing tagged GFP-tagged GCP170 for a longer period or at a higher level, the protein accumulates in large aggregates surrounding the Golgi complex (double arrow). The formation of cytoplasmic aggregates is consistent with previous biochemical findings that GCP170 is aggregation prone (Misumi et al., 1997).
GFP170* is also targeted to the Golgi when expressed at low levels (Figure 1C, cell at bottom left, arrow). In addition, in an adjacent cell, where GFP170* is present at high levels, most of the protein localizes to large cytoplasmic aggregates in the juxtanuclear region (double arrow). Examination of cells at different times after transfection with GFP170* suggests that the cytoplasmic aggregates grow by coalescence (Figure 1D). Initially, numerous small (<0.5 μm in diameter) particles are distributed throughout the cell. Subsequently, the peripheral aggregates relocate to the peri-centriolar zone, coalesce, and ultimately form a compact ribbon-like structure adjacent to the Golgi. The GFP170* cytoplasmic aggregates are morphologically similar to those formed by GFP-tagged wild-type GCP170. The ribbon-like morphology is distinct from the spherical aggresomes formed by overexpressing CFTR (Johnston et al., 1998), GFP-250 (Garcia-Mata et al., 1999), or poly-Q expanded huntingtin (Waelter et al., 2001). In those cases, a compact spherical structure is formed around the microtubule-organizing center (Figure 1B, inset). The ribbon-like morphology is observed when GFP170* is expressed in a number of different cell types (e.g., simian COS-7, human HeLa, and mouse embryonic fibroblast), suggesting that the structure of aggregates is defined by the nature of the aggregating protein, rather than by the cell type.
Unexpectedly, a portion of GFP170* localizes to discrete punctate foci within the nucleus (Figure 1C, cell at top right, double arrowhead). When COS-7 cells were transfected with GFP170*, at least 95% of transfected cells contained GFP170* nuclear aggregates of varying sizes (our unpublished data). A quantitative analysis of 50 randomly chosen transfected cells shows a relationship between the size and the number of the nuclear aggregates (Figure 1D). In nuclei containing >20 foci, the average size of individual foci is <5 μm2, whereas nuclei containing <10 foci have aggregates >10 μm2 (Figure 1D, graph). These results suggest that the larger structures form by coalescence of the small foci. This conclusion is also supported by the shift in the ratios of small, medium and large foci during GFP170* expression. At 24 h after GFP170* transfection, ∼42% of cells contain small (0.5 μm in diameter) aggregates, ∼54% contain medium (1-1.5 μm in diameter) aggregates, and ∼4% contain large (>2 μm in diameter) aggregates. These ratios change 48 h after GFP170* transfection, at which time ∼30% of cells contain small aggregates, ∼51% contain medium aggregates, and ∼22% contain large aggregates.
The nuclear GFP170* aggregates are contained in regions of the nucleoplasm enclosed within the nuclear membrane (Figure 1E). Lamin A forms a mesh-like matrix on the inner face of the nuclear membrane that delineates the nuclear space (Hozak et al., 1995). A focal plane through the nucleus shows GFP170* aggregates inside the lamin A-enclosed space. Aggregates are not found in association with the nuclear rim.
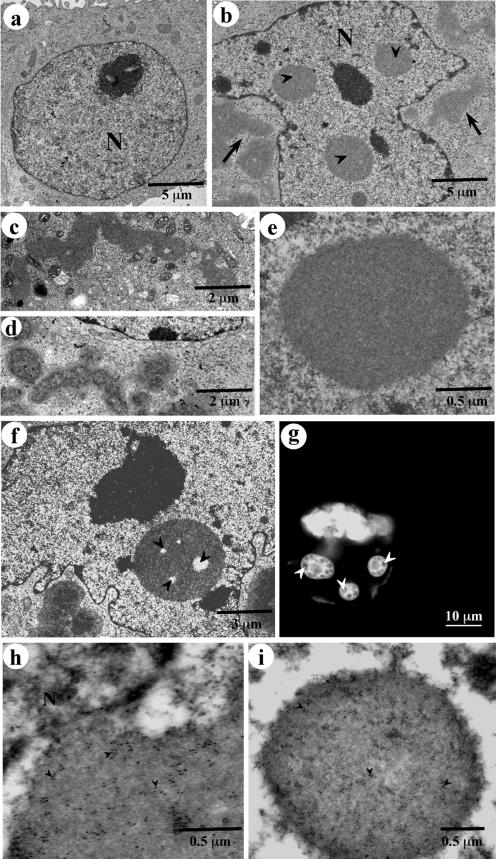
A direct confirmation of nuclear localization and morphological characterization of the cytoplasmic and nuclear GFP170* aggregates are provided by transmission electron microscopy and immunogold labeling (Figure 2). COS-7 cells transfected with GFP170* contain irregular cytosolic aggregates (Figure 2B, arrows) and varying numbers of nuclear aggregates (Figure 2B, arrowheads). Nontransfected control cells never contain such structures (Figure 2A). The cytosolic aggregates look ribbon-like and can extend to >15 μm in length. They display an uneven distribution of components and seem to have an internal architecture (Figure 2, C and D). The cytoplasmic aggregates are often surrounded by mitochondria (Figure 2C), similar to the association of mitochondria with aggresomes formed by CFTR (Johnston et al., 1998) or GFP-250 (Garcia-Mata et al., 1999). The nuclear aggregates are spherical or ovoid and range in diameter from 0.5 to 3 μm. Sometimes they look like homogenous accumulations of granular material without apparent sub-domain structure (Figure 2E). Often, however, they contain internal electron lucent spaces (Figure 2F, arrowheads). The presence of internal substructures within the GFP170* nuclear aggregates is also observed by fluorescence microscopy (Figure 2G, arrowheads). The deposition of GFP170* within the morphologically defined cytoplasmic and nuclear aggregates was confirmed by immunogold labeling with anti-GFP antibodies. Gold particles label both cytoplasmic and nuclear aggregates (Figure 2, H and I).
Figure 2.
Ultrastructure of GFP170* aggregates. Nontransfected COS-7 cells (a) or COS-7 cells transfected with GFP170* (b-i) were processed for transmission electron microscopy (b-f), fluorescence (g), or immunogold labeling (h and i). (a) Nontransfected COS-7 cell lacks aggregates. (b) Transfected COS-7 cell contains cytoplasmic aggregates (arrows) and nuclear aggregates (arrowheads). (c and d) Cytoplasmic aggregates are ribbon-like and are surrounded by mitochondria. (e) Nuclear aggregates are either spherical or ovoid. (f) Nuclear aggregates contain internal electron-lucent spaces (arrowheads). (g) Internal GFP170* substructure within the nuclear aggregates is also visible by fluorescence (arrowheads). (h and i) The content of the cytoplasmic and nuclear aggregates was confirmed by immunogold labeling with anti-GFP antibodies followed by secondary antibodies conjugated to 12-nm gold particles (arrowheads). N, nucleus.
GFP170* Aggregates Are Cytoplasmic and Nuclear Aggresomes
Cytoplasmic aggregates (formed by either poly-Q proteins or nonpoly-Q proteins) and nuclear aggregates (formed by poly-Q proteins) have been described as aggresomes, based on a number of defining characteristics. However, an in-depth and dynamic analysis of nuclear aggregates formed by a nonpoly-Q protein has not been reported previously. GFP170* provides a unique tool to characterize the cytoplasmic and nuclear aggregates within the same cell.
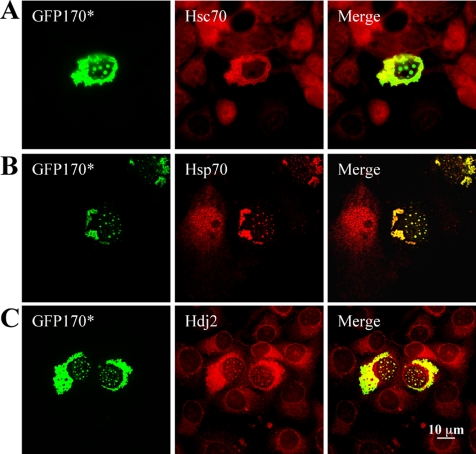
One of the defining characteristics of aggresomes is the recruitment of molecular chaperones. Chaperones have been detected in association with cytoplasmic aggregates of poly-Q expanded huntingtin (Waelter et al., 2001) and with aggregates of the nonpoly-Q proteins CFTR (Johnston et al., 1998) and GFP-250 (Garcia-Mata et al., 1999). Chaperones are also recruited to nuclear aggregates of poly-Q expanded ataxin-3 and huntingtin (Chai et al., 1999a; Waelter et al., 2001). The cytoplasmic and nuclear aggregates containing GFP170* seem to be aggresomes, based on their recruitment of Hsc70, Hsp70, and Hdj2 (representatives of the Hsp70 and the Hsp40 families of chaperones, respectively) in transfected cells (Figure 3). Hsc70 is recruited to the periphery of cytosolic aggregates, but it is excluded from the nucleus (Figure 3A). In contrast, the stress-responsive Hsp70 localizes to both the nuclear and cytoplasmic aggregates (Figure 3B). Hdj2 also colocalizes with both cytosolic and nuclear aggregates (Figure 3C). Our observations suggest distinct roles for these chaperones in the formation of cytoplasmic versus nuclear aggregates.
Figure 3.
Recruitment of molecular chaperones to GFP170* aggregates. COS-7 cells were transfected with GFP170*. After 48 h, cells were processed for indirect immunofluorescence using antibodies against Hsc70 (A), Hsp70 (B), or Hdj2 (C). Chaperones are recruited to GFP170* aggregates.
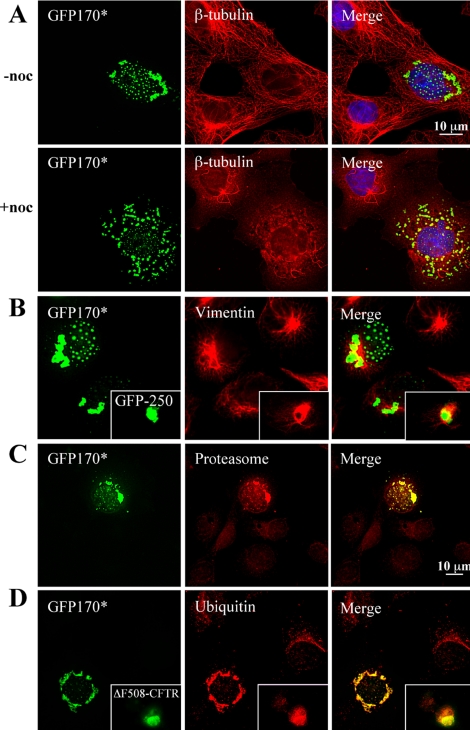
The motor-dependent movement of aggregated particles on microtubules is a hallmark of cytoplasmic aggresome formation (Johnston et al., 1998; Garcia-Mata et al., 1999). However, the importance of MT-mediated transport to nuclear aggresome formation has not been explored. We therefore examined the role of MTs in the formation of cytoplasmic and nuclear GFP170* aggregates. In cells treated with nocodazole during GFP170* expression, cytosolic aggregates are smaller and dispersed throughout the cell (Figure 4A). In such cells, peripheral aggregates do not coalesce into a compact perinuclear structure. A similar dispersed phenotype has been reported for GFP-250 aggregates in cells defective in MT- and dynein-mediated transport (Garcia-Mata et al., 1999). Interestingly, the nuclear aggregates are also significantly smaller in nocodazole-treated cells (Figure 4A). Large nuclear aggregates are not detected at times when larger aggregates are present in untreated cells. This observation suggests that nuclear aggregates form less efficiently in the absence of MT-dependent traffic. MTs have not been detected within the nucleus, but the delivery of various components to the nucleus has been shown to be MT dependent (Sodeik et al., 1997). In the absence of MTs, these molecules may become limited within the nucleus and influence the coalescence of GFP170* foci.
Figure 4.
Involvement of microtubules in the formation of GFP170* aggregates and the recruitment of vimentin and proteasomal components to GFP170* aggregates. (A) COS-7 cells were transfected with GFP170*. After 8 h, cells were left untreated (-noc) or supplemented with 1.5 μM nocodazole (+noc), and cultured for additional 25 h. Cells were processed for indirect immunofluorescence using anti β-tubulin antibody. The nuclei were stained with Hoechst 33258. Nocodazole treatment leads to disruption of the microtubule cytoskeleton. In cells treated with nocodazole, cytosolic and nuclear aggregates of GFP170* are smaller and more dispersed. (B-D) COS-7 cells were transfected with GFP170*, GFP-250, or ΔF508-CFTR. After 48 h, cells were processed for indirect immunofluorescence using antibodies against vimentin (B), α-subunit of proteasome (C), or ubiquitin (D). Vimentin envelopes the cytosolic GFP170* and GFP-250 aggregates (B and inset). Proteasomal subunits are recruited to cytoplasmic and nuclear GFP170* aggregates (C). Ubiquitin is recruited to GFP170* and ΔF508-CFTR aggregates (D and inset).
The formation of cytoplasmic aggresomes has been correlated with the collapse of the vimentin intermediate filament cytoskeleton (Johnston et al., 1998; Garcia-Mata et al., 1999). Specifically, vimentin has been shown to relocate to form a “cage” around the cytosolic aggresomes of GFP-250 (Figure 4B, inset; Garcia-Mata et al., 1999) or CFTR (Johnston et al., 1998). We found that vimentin also surrounds the ribbon-like GFP170* cytoplasmic aggregates (Figure 4B).
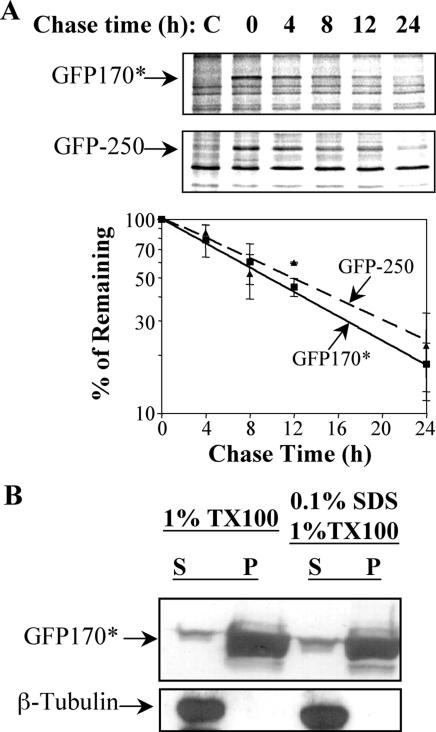
Another characteristic feature of cytoplasmic and nuclear aggresomes is the recruitment of proteasomes (Johnston et al., 1998; Garcia-Mata et al., 1999; Waelter et al., 2001). Proteasomes are composed of a 20S proteolytic core and two 19S regulatory caps responsible for recognizing and unfolding the substrates. The 20S core is composed of two antechambers, each lined with nonproteolytic α-subunits, and a central cavity lined with proteolytic β-subunits (Voges et al., 1999). Antibodies against the α-subunits label the cytoplasmic and nuclear GFP170* aggregates (Figure 4C). Proteasomes seem to be recruited to the GFP170* aggregates. Cytoplasmic and nuclear aggregates have been shown to be positive for ubiquitin (Johnston et al., 1998; Waelter et al., 2001). In agreement with a previous report, anti-ubiquitin antibodies label cytoplasmic aggregates of mutant CFTR (ΔF508-CFTR) (Figure 4D, inset). The same antibodies also label GFP170* aggregates (Figure 4D). The association of chaperones, proteasomes and ubiquitin with GFP170* aggresomes is likely to increase the local concentration of molecules necessary for unfolding and degradation of the aggregated protein and may contribute to its clearance. The degradation rate of GFP170* was therefore examined. Pulse-chase analyses defined a half-life of ∼8 h for GFP170* (Figure 5A). This degradation rate is comparable with that of previously described aggresomal proteins such as GFP-250 (Figure 5A; Garcia-Mata et al., 1999).
Figure 5.
Degradation and solubility of GFP170*. COS-7 cells were either mock transfected with PBS (C, control) or transfected with GFP170* or GFP-250. (A) 24 h after transfection, cells were pulse labeled with [35S]methionine for 1 h and chased for indicated times. Cells were lysed and the lysates analyzed by SDS-PAGE and autoradiography. A representative autoradiogram is shown. The density of GFP170* and GFP250 bands was quantified with ImageQuant software. Results from three independent experiments are presented in the graph. (B) Forty-eight hours after transfection, cells were lysed using indicated buffers. The supernatant (S) and the pellet (P) fractions from each lysis were analyzed by SDS-PAGE, followed by immunoblotting with antibody against either GFP or β-tubulin. Most of GFP170* is present in the insoluble fraction.
Proteins deposited within aggresomes are largely detergent insoluble (Ward et al., 1995; Scherzinger et al., 1997; Garcia-Mata et al., 1999; Kopito, 2000). We examined the solubility of GFP170* in either Triton X-100 or SDS-containing buffers. Both extraction conditions are sufficient to solubilize 100% of cellular β-tubulin. In contrast, the majority of GFP170* is present in the insoluble fraction, with <5% in the extractable fraction (Figure 5B). Together, our findings indicate that the cytoplasmic and nuclear aggregates of GFP170* share key characteristics of aggresomes.
Cytoplasmic and Nuclear GFP170* Aggresomes Are Dynamic
Previous studies of nuclear and cytoplasmic aggregates of poly-Q proteins suggest that some poly-Q expanded proteins (e.g., ataxin-1) are dynamic and exchange their components (Stenoien et al., 2002), whereas others (e.g., ataxin-3 and Q82-GFP) are immobile (Kim et al., 2002). We therefore used FRAP and FLIP to explore the dynamics of nuclear and cytoplasmic GFP170* aggregates.
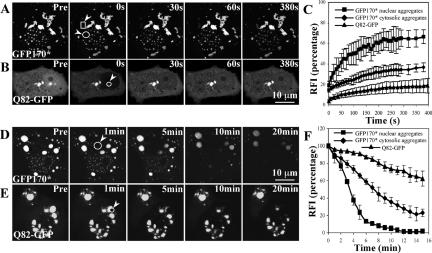
The fluorescence within nuclear aggregates of GFP170* recovers to ∼65%, 5 min after photobleaching (Figure 6A, circle, and C). This result indicates that a large portion of GFP170* molecules deposited within the aggregates is mobile and exchangeable with soluble molecules in the nucleoplasm. The t1/2 of fluorescence recovery to steady state levels is ∼50 s. The recovery of GFP170* is slower than that of a poly-Q expanded GFP-ataxin-1-84Q (t1/2 of <2 s) deposited in nuclear inclusions of similar size (Stenoien et al., 2002). However, GFP170* recovers significantly faster than the almost immobile poly-Q expanded ataxin-3 (Chai et al., 2002) or Q82-GFP (Kim et al., 2002). To provide a direct comparison between dynamics of GFP170* and an immobile aggregate, we analyzed FRAP of Q82-GFP. The fluorescence recovery of Q82-GFP in nuclear aggregates is limited to only ∼12% (Figure 6B, circle, and C). This result is consistent with the observation reported previously (Kim et al., 2002) and validates our experimental parameters. The different FRAP dynamics of GFP170* relative to the more mobile GFP-ataxin-1-84Q or the largely immobile Q82-GFP may reflect differences in biophysical characteristics, stabilization by interacting with distinct nuclear components, or other factors.
Figure 6.
FRAP and FLIP analysis of GFP170* aggregates. COS-7 cells were transfected with GFP170* (A and D) or Q82-GFP (B and E). (A-C) FRAP analysis of GFP170* and Q82-GFP aggregates. (A) A defined cytosolic (rectangle, arrowhead) or nuclear (circle, arrowhead) GFP170* aggregate was photobleached and the recovery of fluorescence into that region was monitored. (B) A nuclear Q82-GFP aggregate (circle, arrowhead) was photobleached and the recovery of fluorescence into that region was monitored. (C) The relative fluorescence intensity (RFI) was determined for each time after bleaching and is represented as the average analysis of three to five cells. Error bars represent standard deviations of FRAP results from different cells. (D-F) FLIP analysis of GFP170* and Q82-GFP aggregates. (D) A defined cytosolic region (circle, arrowhead) of a cell containing GFP170* aggregates was repeatedly bleached and the loss of fluorescence from nuclear and cytosolic GFP170* aggregates was monitored. (E) A defined region (circle, arrowhead) of a cell containing Q82-GFP aggregates was repeatedly bleached and the loss of fluorescence from the Q82-GFP aggregates was monitored. (F) RFI was determined for each time point and is represented as the average analysis of three to five cells. Error bars represent standard deviations of FLIP results from different cells.
The dynamics of GFP170* within cytoplasmic aggregates were also analyzed by FRAP. The fluorescence recovers to ∼35%, 5 min after photobleaching (Figure 6A, rectangle, and C). Measuring the initial slope of the recovery indicates a t1/2 of ∼2 min. This result indicates that a portion of GFP170* molecules within the cytoplasmic aggregates is mobile and constantly exchanges with a soluble pool of GFP170*. The different kinetics of FRAP between the nuclear and cytosolic GFP170* aggregates may reflect the different structural properties of aggregates in the cytosol and the nucleus (Figure 2).
We also used FLIP to compare the dynamics of GFP170* molecules within the cytosolic and nuclear aggregates. A cytosolic region of a cell containing cytoplasmic and nuclear aggregates was repeatedly photobleached and the loss of fluorescence from the cytosolic and nuclear aggregates was monitored over time. The fluorescence within the nuclear aggregates decreases to ∼25% after 5 min of bleaching and is undetectable after 15 min (Figure 6D, circle, and F). The t1/2 of FLIP to background level is ∼3.5 min. This result suggests that GFP170* molecules are efficiently exported from the nucleus to the cytosol. In contrast, the fluorescence of the cytosolic aggregates only decreases to ∼65% after 5 min of bleaching, and is still ∼20% of the initial fluorescence after 15 min of photobleaching (Figure 6, D and F). In this case, the rate of fluorescence loss is slower than from the nuclear aggregates, with a t1/2 of ∼8 min. This result is consistent with the FRAP data, and both data sets indicate that GFP170* molecules within cytosolic aggregates are less mobile than those within nuclear aggregates. As expected, only ∼10% of fluorescence is lost from Q82-GFP aggregates after 5 min photobleaching, and ∼70% of initial fluorescence still remains after 15 min of photobleaching (Figure 6E, circle, and F). This finding is in agreement with our FRAP results and with previously reported findings (Kim et al., 2002).
Nuclear GFP170* Aggresomes Show a Relationship to PML Bodies
The nucleus is organized into defined territories specialized to perform distinct functions (Spector, 2001). Disruption of nuclear structures, as exemplified by the (t15;17:q22;q21) translocation that results in a fusion of the PML protein and the retinoic acid receptor α, leads to acute promyelocytic leukemia (Weis et al., 1994). PML bodies are also called nuclear domain 10 (ND10) bodies or PML oncogenic domains. The mammalian nucleus contains 10-30 PML bodies, which vary in size from 0.2 to 1 μm. They are thought to function in transcriptional regulation, cell cycle progression, and apoptosis, based on their content of proteins such as Sp100, PML, Daxx, pRB, CBP, and p53, which are involved in these processes (Yasuda et al., 1999; Maul et al., 2000; Zhong et al., 2000). PML bodies have been shown to associate with nuclear aggregates of poly-Q expanded proteins (Dovey et al., 2004). Whether this association is related to the poly-Q track or represents a general response to aggregated protein within the nucleus has not been explored. Therefore, we examined the relationship between PML bodies and GFP170* aggregates.
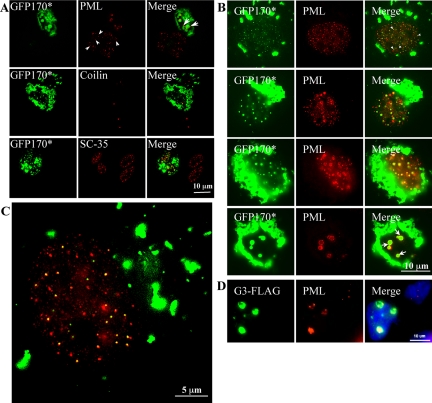
The nuclear aggregates of GFP170* show a close relationship with PML bodies (Figure 7). In nontransfected cells, anti-PML antibodies label numerous small nuclear foci (Figure 7A, arrowheads). In a transfected cell containing large GFP170* nuclear foci, the number and the distribution of PML bodies in the nucleus are altered. PML-labeled structures seem to be associated with the GFP170* aggregates (Figure 7A, arrows). In contrast, we did not observe significant changes in nuclear Cajal bodies and nuclear speckles. The number and distribution of Cajal bodies marked by the protein coilin and the number and distribution of nuclear speckles marked by the SC35 splicing factor are not altered by GFP170* expression (Figure 7A), suggesting that GFP170* aggregates do not indiscriminately disrupt nuclear architecture.
Figure 7.
Association of nuclear GFP170* and G3 aggregates with PML bodies. COS cells were transfected with GFP170* (A-C) or G3-FLAG (D). After 48 h, cells were processed for immunofluorescence using anti-PML antibody (A-D), anti-Sc35 antibody (A), anti-coilin antibody (A), or anti-FLAG antibody (D). (A) A nontransfected COS-7 cell (bottom left) contains numerous PML bodies in the nucleus (arrowheads). In a cell with nuclear GFP170* aggregates, PML is concentrated on the surface of GFP170* aggregates (arrows). In contrast, the distribution of nuclear Cajal bodies marked by coilin and nuclear speckles marked by Sc35 is not changed in the presence of GFP170* aggregates. (B) A panel of COS-7 cells containing GFP170* nuclear aggregates of different sizes. Small GFP170* aggregates usually colocalize with, or are adjacent to individual PML bodies (first row, arrowheads). Larger GFP170* aggregates contain PML bodies on their surface (arrows). (C) An enlarged image shows the preferential association of nuclear GFP170* aggregates with PML bodies. (D) G3 nuclear inclusions also associate with PML bodies. Nuclei are visualized with 4,6-diamidino-2-phenylindole staining.
The association of PMLs with GFP170* aggregates led us to further explore this process. Examination of cells at different times after transfection suggests that the initial deposition of GFP170* often occurs in or adjacent to PML bodies (Figure 7B, first row, arrowheads). Colocalization is first observed when foci are very small, before PML distribution is altered (Figure 7C). In this representative cell, ∼82% of GFP170* aggregates are associated with PML bodies. Quantitation of colocalization in multiple transfected cells indicates that 80-90% of GFP170* puncta are adjacent to or overlap with PML bodies in all examined cells. As the small GFP170* aggregates merge to form larger structures, PMLs are repositioned on the surface of the larger aggregates (Figure 7B, second and third rows). In cells containing only a few large aggregates, PML bodies form beaded necklace-like structures on the surface of the aggregates (fourth row, arrows). Thus, deposition of GFP170* aggregates induces dramatic changes in the nuclear architecture of PML bodies. Analogous rearrangements are observed in other cell lines (e.g., human HeLa cells and mouse embryonic fibroblasts), suggesting a commonality of the response. The observed phenotype is distinct from “clumping” of PML bodies during mitotic prophase and metaphase (Everett et al., 1999). It is also morphologically distinct from the increased number of small PML bodies observed during heat shock (Everett et al., 1999). The PML body rearrangements observed in cells expressing GFP170* are likely to be induced by the aggregates.
The association of PML bodies with nuclear aggregates of GFP170*, in addition to the reported association with nuclear aggregates of poly-Q proteins, suggested that it may be a common nuclear response to nuclear aggregates. We therefore explored the relationship between PMLs and nuclear inclusions formed by another nonpoly-Q protein, the G3 domain fragment of aggrecan. Aggrecan is an extracellular matrix proteoglycan abundant in cartilage, but an engineered G3 domain fragment has been shown to mistarget to the nucleus and form nuclear foci in transfected cells (Chen et al., 2001). G3 aggregates are also associated with PML bodies (Figure 7D). The association of PMLs with G3 as well as with GFP170*, in addition to poly-Q proteins, suggests that the association is independent of the primary sequence of the misfolded protein.
Cytosolic and Nuclear GFP170* Aggresomes Form by Fusion
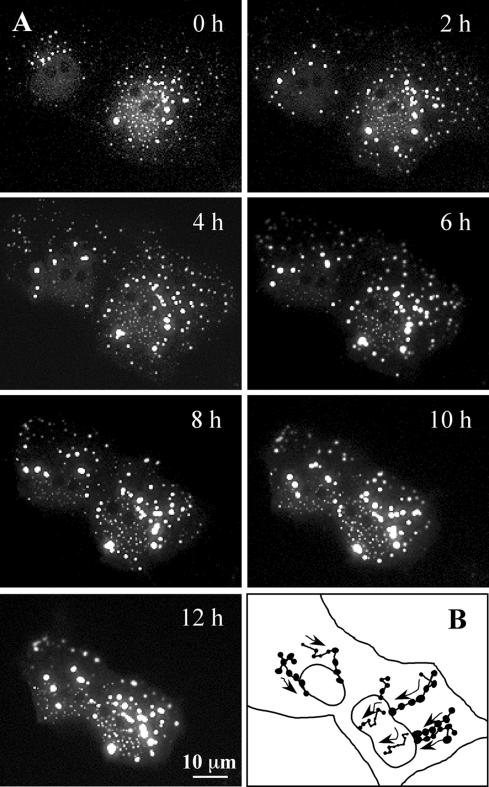
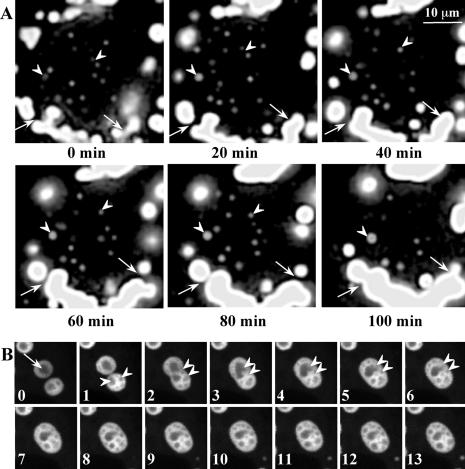
The formation of cytoplasmic aggresomes by the poly-Q expanded androgen receptor occurs by joining of smaller cytoplasmic aggregates (Stenoien et al., 1999). The size variations of GFP170* aggregates in transfected cells suggested that fusion events may underlie formation of large aggresomes. We used time-lapse imaging of live cells to explore aggresome formation. Two cells with small cytoplasmic and nuclear GFP170* aggregates were selected and imaged for 12 h (Figure 8A and QuikTime Movie 1). Tracking the behavior of individual peripheral particles in the cytoplasm indicates that they move inward and coalesce to form larger peri-nuclear aggregates (tracings in Figure 8B). The movements seem quasi-linear, as would be expected for MT-based transport. The centripetal movement of the particles is not constant over time, and the particles move and then hover in place before moving again. Particle fusions are common and occur preferentially within the peri-nuclear zone. A higher magnification series of time-lapse images clearly shows the fusion of cytoplasmic aggregates (Figure 9A, arrows, and QuickTime Movie 2). Note that spherical aggregates move toward the larger ribbon-like aggregate and fuse with it.
Figure 8.
Dynamic movements of GFP170* aggregates. COS-7 cells were transfected with GFP170*. After 20 h, two cells expressing low levels of GFP170* were imaged every 5 min for additional 12 h. (A) A panel of images at the time points indicated. A QuickTime movie file is attached as supplemental material. (B) Tracing of the movement of selected GFP170* aggregates. Arrows indicate the directions of the movement, and dots indicate fusion events between aggregates.
Figure 9.
Fusions and rearrangements of nuclear GFP170* aggregates. COS-7 cells were transfected with GFP170*. After 48 h, cells were subject to time-lapse imaging, with images acquired every 20 s for 2 h. Two QuickTime movie files are attached as supplemental materials. (A) A panel of images at time points indicated. Arrows indicate fusion between cytosolic aggregates. Arrowheads indicate fusion between nuclear aggregates. (B) A panel of images at 1-min intervals. Fusion of two nuclear aggregates is accompanied by extensive rearrangements of internal structures. An arrow shows an inclusion containing one fluorescent-lucent internal structure. Arrowheads indicate multiple internal structures within an inclusion. Double arrowheads mark two aggregates that undergo fusion and rapid internal reorganization.
Events leading to the formation of nuclear aggregates (either by poly-Q or nonpoly-Q proteins) have not been previously described by live imaging. Time-lapse analysis of the nuclear aggregates of GFP170* illustrates three novel phenomena. First, a gradual increase in the fluorescence intensity of nuclear foci (Figure 9A and QuickTime Movie 2) suggests that GFP170* molecules are constantly deposited into preexisting aggregates. The continuous incorporation may utilize the pool of soluble GFP170* within the nucleus. Second, small nuclear aggregates move and merge to form larger structures (Figure 9A, arrowheads, and QuickTime Movie 2). Tracking of the nuclear movements indicates that some particles move directionally, which is inconsistent with unrestricted Brownian movement. In addition, smaller aggregates seem to preferentially fuse with medium to large nuclear aggregates, rather than with other small foci. Third, the nuclear aggregates enclose internal substructures that are extremely dynamic and undergo rapid rearrangements (Figure 9B and QuickTime Movie 3). In some aggregates, one single substructure is surrounded by GFP170* protein (Figure 9B, arrow). In other aggregates, multiple substructures of different sizes are evident (Figure 9B, arrowhead). The substructures are dynamic and undergo continuous movements within the aggregates. Fusion of two aggregates is paralleled by the reorganization and fusion of the substructures (Figure 9B, double arrowheads). This observation indicates that the components of nuclear aggregates may have preferential affinity for each other and separate by phase partitioning.
DISCUSSION
Extensive literature documents the formation of nuclear inclusions by proteins with expanded poly-Q tracks (reviewed in Zoghbi and Orr, 2000; Ross, 2002). These reports have led to the suggestion that nuclear aggregation may be regulated by mechanisms specific to poly-Q expansions. Here, we show that GFP170*, a nonpoly-Q protein, also forms nuclear aggresomes. The GFP170* aggregates seem similar to those formed by poly-Q proteins by a number of criteria: their morphology, the recruitment of chaperones, and the association with proteosomes. Our findings suggest that the formation of nuclear aggresomes is not a poly-Q restricted process and may represent a common response to the accumulation of misfolded proteins within the nucleus. We investigated the process of nuclear aggresome formation by time-lapse imaging of GFP170*. Our studies indicate that the formation of nuclear GFP170* aggresomes is a dynamic process. It is initiated by the deposition of GFP170* at or adjacent to PML bodies followed by the movement of small aggregates toward larger structures and their coalescence to form still larger aggregates. We also show that G3, an unrelated nonpoly-Q protein reported previously to deposit in nuclear inclusions, forms similar associations with PML bodies. Unexpectedly, we observed internal substructures that exclude GFP170* within the nuclear aggregates. Internal substructures were also observed within G3 nuclear inclusions (Chen et al., 2001). Time-lapse analysis of GFP170* shows that the internal substructures undergo extensive rearrangements during fusion of the aggregates. Our results suggest that phase partitioning may underlie the formation of GFP170* aggregates.
Deposition of Nuclear Aggregates by Nonpoly-Q GFP170*
The GFP170* protein, consisting of the GFP and an internal fragment of the Golgi matrix protein GCP170, forms aggregates within the cytoplasm and within the nucleus. Overexpression of GFP170* represents an artificial experimental system that is similar to the cellular or transgenic overexpression of various poly-Q expanded proteins (Steffan et al., 2000; Nucifora et al., 2001; Lieberman et al., 2002) and to analyses in which proteolytic inhibitors are used to define cellular responses to aggregated proteins (Johnston et al., 1998; Bence et al., 2001). Such experimental manipulations, although artificial, provide insight into cellular processes that deal with misfolded proteins.
The nucleation process for aggregation of GFP170* may begin at the site of translation in the cytoplasm, because high local concentrations of GFP170* may be produced during the translation of a single mRNA. GFP170* has 1075 amino acids and a 3225-base pair coding region that would be represented in its mRNA. Ribosomes attach every ∼80 base pairs on an mRNA, suggesting that ∼40 ribosomes could be simultaneously translating a single GFP170* message. As a result, ∼40 GFP170* molecules may be produced in a localized area and could serve to seed an aggregation particle. It is unknown whether GFP170* is transported across the nuclear membrane in a nonaggregated form, similar to the import pathway of most transcription factors, or as an oligomeric particulate, similar to the import pathway used by viral particles (Swindle et al., 1999; Tang et al., 2000).
A nuclear localization sequence (NLS) has been described in the amino terminus of GCP170 between residues 232 and 239 (Hicks and Machamer, 2002). However, GFP170* lacks this region (GFP170* initiates at residue 566 of GCP170), and consequently uses another mechanism for entry into the nucleus. It is possible that nuclear import is mediated by a redundant NLS contained within the 566-1375 residues of GFP170*. Alternatively, nuclear localization may be mediated by GFP, because GFP localizes to the nucleus when expressed in mammalian cells (Garcia-Mata et al., 2000). It is also possible that GFP170* enters the nucleus passively, by associating with a protein that normally shuttles between the cytoplasm and the nucleus. The exact pathway and cellular components involved in GFP170* import into the nucleus remain to be identified. The enlargement of GFP170* nuclear aggregates is MT-dependent. It is currently unclear whether the microtubule cytoskeleton is required for the nuclear import of GFP170* itself. Trafficking of GFP170* toward the peri-nuclear region is clearly MT dependent. It is likely that the proximity to nuclear pores may be the major determinant of GFP170* entry into the nucleus. In addition, it is possible that MT assist in the trafficking of nuclear components, which facilitate the formation of the nuclear aggregates.
Dynamic Nature of GFP170* Aggregates
FRAP and FLIP analyses of GFP170* within cytoplasmic and nuclear aggregates show that GFP170* is largely mobile and rapidly exchanges in and out of the aggregates. Similar dynamic mobility is exhibited by aggregates of poly-Q expanded ataxin-1 molecules (Stenoien et al., 2002). Both contrast with the behavior of aggregates of poly-Q expanded ataxin-3 that are largely immobile and do not exchange their components (Chai et al., 2002). These results indicate that structures defined morphologically as visible inclusions can be true aggregates (as those formed by ataxin-3) or concentrated depositions of rapidly exchanging components (as those formed by ataxin-1 and GFP170*). The dynamic behavior of GFP170* observed in vivo is in contrast with its physical state after cells are disrupted. Although the majority of GFP170* seems mobile in live cells, the majority of GFP170* is recovered in stable aggregates after cell extraction (compare Figures 5B and 6A). A possible explanation for this apparent contradiction is suggested by the recruitment of chaperones to the GFP170* aggregates. We suggest that in vivo, the continuous action of ATP-dependent molecular chaperones mediates the mobility of GFP170* molecules. In disrupted cells, falling ATP levels may be insufficient to support chaperone function and instead result in insoluble GFP170* aggregates. This hypothesis is supported by the finding that the association of Hsp70 with htt-82Q aggregates has the same diffusion coefficient as thermally unfolded substrates, suggesting that Hsp70 associates with the aggregates as part of its functional folding cycle (Kim et al., 2002).
Relationship between Nuclear Aggregates and PML Bodies
A relationship between PML bodies and poly-Q aggregates has been documented previously. PML rearrangements have been described in Purkinje cells of transgenic mice and in transfected COS-1 cells expressing poly-Q expanded ataxin-1 (Skinner et al., 1997). PML alterations also occur in cells expressing poly-Q expanded ataxins responsible for the Machado-Joseph disease (Yamada et al., 2002) and SCA7 (Takahashi et al., 2002). These reports suggest that PML bodies may represent sites specialized to recognize and sequester foreign proteins within the nucleus. This hypothesis is supported by the alterations in PML bodies described during DNA and RNA virus infections (reviewed in Doucas and Evans, 1996). In cells infected with Hepatitis delta virus (Bell et al., 2000), herpes simplex virus-1, human cytomegalovirus (Ishov et al., 2002), or human papilloma virus (Becker et al., 2004), the virus localizes to enlarged PMLs, and in some cases subsequently disrupts PML bodies (Everett and Maul, 1994). It should be stressed that viruses are highly ordered protein aggregates. Here, we show that two unrelated proteins, GFP170* and G3, which are distinct from poly-Q expanded proteins or viral components, also associate with and alter PML bodies. The preferential deposition of GFP170* at such sites may occur by association with nuclear components present in those sites, or by aggregation-promoting environments in those sites. Our results support the hypothesis that PML bodies represent dedicated domains specialized to handle foreign particulates within the nucleus. The concentration of aggregated material by spatial sequestration, and the concurrent recruitment of unfolding and degradative machineries to such structures may improve the efficiency of clearance.
A proteolytic function for PML bodies has been suggested by the increased number of PML bodies in cells treated with a proteasomal inhibitor (Burkham et al., 2001). The association of the protein unfolding and degradative machineries with the nuclear GFP170* aggregates is consistent with the participation of PML bodies in proteasomal degradation. Protein clearance within the cytoplasm seems to be consigned to a specialized peri-centriolar proteolytic region (Wojcik and DeMartino, 2003). The association of nuclear GFP170* and G3 aggregates with PML bodies suggests that like the cytoplasm, the nucleus may contain spatially defined degradative regions. We suggest that the overexpression of GFP170* amplifies these nuclear regions.
In addition to recruiting PML components, poly-Q nuclear inclusions also alter the distribution and activity of specific transcription factors. The cAMP response element-binding protein binding protein (CBP), Sp1, and p53 associate with poly-Q huntingtin fragments (Steffan et al., 2000; Nucifora et al., 2001). The association seems to repress CBP- and p53-mediated transcription (Steffan et al., 2000; Nucifora et al., 2001). In addition to transcriptional repression, expression of poly-Q proteins such as mutant huntingtin and atrophin-1 in cultured cells, or in animal disease models, leads to cellular toxicity (Nucifora et al., 2001). These responses seem not to be restricted to poly-Q-containing aggregates because we find that GFP170* expression represses p53 transcriptional activity and is cytotoxic (Fu et al., 2005). The similarity in cellular responses elicited by poly-Q proteins and the nonpoly-Q GFP170* suggests that the responses are common to the presence of any misfolded proteins in the nucleus.
Organization of Nuclear Aggresomes
Time-lapse imaging of nuclear aggresomes in live cells documents three novel phenomena. First, GFP170* is continuously deposited at preexisting nuclear regions as demonstrated by the increased fluorescence intensity of nuclear GFP170* foci. The growth of GFP170* aggregates may be a combined result of direct targeting and selective stabilization of GFP170* molecules at the nuclear foci. The concentration of GFP170* at defined foci suggests that, like the deposition of cytoplasmic aggresomes to the peri-centriolar region, formation of aggregates is spatially restricted within the nucleus.
Second, small nuclear aggregates of GFP170* undergo extensive movements and fusions to form larger aggregates. The movement of small GFP170* aggregates seems similar to that reported for endogenous nuclear structures. Cajal bodies move through the nucleus with similar kinetics, and can fuse and split to modify their size (Platani et al., 2000). PML bodies also can move; although most PML bodies show only localized oscillations (∼63%) or are immobile (∼25%), a small percentage (∼12%) undergo rapid and more extended movements (Muratani et al., 2002). Nuclear movements are most consistent with passive diffusion within a volume that is limited by a constraint (Platani et al., 2002). A model that is compatible with the available data suggests that nuclear structures move within submicron channels formed by the exclusion of nuclear components, rather than by vectorial transport on molecular tracks (Carmo-Fonseca et al., 2002). Actin-dependent myosins may control the diffusible volume because the movement of PML bodies can be blocked by an inhibitor of actin-dependent myosins (Muratani et al., 2002). The driving force that regulates nuclear movements of the various nuclear structures and of GFP170* aggregates remains to be elucidated.
Third, our imaging of nuclear GFP170* aggregates uncovered a novel characteristic of nuclear aggregates: the presence of complex internal structures. These internal subdomains are themselves dynamic and undergo constant movements, fusions and fissions. These observations suggest that the aggregating protein encapsulates nuclear components, and that such encapsulation is not static but undergoes extensive remodeling. Thus, we should no longer view nuclear aggregates as static precipitates, but rather, as rapidly reforming entities subject to the coordinated restructuring of their constituents.
Supplementary Material
Acknowledgments
We thank Drs. Richard Morimoto, Douglas Cyr, and Hans-Peter Hauri for plasmids and antibodies. We thank Drs. James Collawn, Anne Theibert, and Guillermo Marques for helpful discussions. We thank Leigh Millican for technical support during the electron microscopy study. This work was supported by National Institutes of Health postdoctoral fellowship T32 HL07553 (to L. F.), National Institutes of Health Grant GM-062696 (to E. S.), and a research grant from the Arthritis Foundation (to B.M.V.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-01-0019) on July 29, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Alvarez, C., Fujita, H., Hubbard, A., and Sztul, E. (1999). ER to Golgi transport: requirement for p115 at a pre-Golgi VTC stage. J. Cell Biol. 147, 1205-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, K. A., Florin, L., Sapp, C., Maul, G. G., and Sapp, M. (2004). Nuclear localization but not PML protein is required for incorporation of the papillomavirus minor capsid protein L2 into virus-like particles. J. Virol. 78, 1121-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, P., Brazas, R., Ganem, D., and Maul, G. G. (2000). Hepatitis delta virus replication generates complexes of large hepatitis delta antigen and antigenomic RNA that affiliate with and alter nuclear domain 10. J. Virol. 74, 5329-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bence, N. F., Sampat, R. M., and Kopito, R. R. (2001). Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292, 1552-1555. [DOI] [PubMed] [Google Scholar]
- Burkham, J., Coen, D. M., Hwang, C. B., and Weller, S. K. (2001). Interactions of herpes simplex virus type 1 with ND10 and recruitment of PML to replication compartments. J. Virol. 75, 2353-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca, M., Platani, M., and Swedlow, J. R. (2002). Macromolecular mobility inside the cell nucleus. Trends Cell Biol. 12, 491-495. [DOI] [PubMed] [Google Scholar]
- Chai, Y., Koppenhafer, S. L., Bonini, N. M., and Paulson, H. L. (1999a). Analysis of the role of heat shock protein (Hsp) molecular chaperones in polyglutamine disease. J. Neurosci. 19, 10338-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, Y., Koppenhafer, S. L., Shoesmith, S. J., Perez, M. K., and Paulson, H. L. (1999b). Evidence for proteasome involvement in polyglutamine disease: localization to nuclear inclusions in SCA3/MJD and suppression of polyglutamine aggregation in vitro. Hum. Mol. Genet. 8, 673-682. [DOI] [PubMed] [Google Scholar]
- Chai, Y., Shao, J., Miller, V. M., Williams, A., and Paulson, H. L. (2002). Live-cell imaging reveals divergent intracellular dynamics of polyglutamine disease proteins and supports a sequestration model of pathogenesis. Proc. Natl. Acad. Sci. USA 99, 9310-9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, Y., Wu, L., Griffin, J. D., and Paulson, H. L. (2001). The role of protein composition in specifying nuclear inclusion formation in polyglutamine disease. J. Biol. Chem. 276, 44889-44897. [DOI] [PubMed] [Google Scholar]
- Chen, S., Ferrone, F. A., and Wetzel, R. (2002). Huntington's disease age-of-onset linked to polyglutamine aggregation nucleation. Proc. Natl. Acad. Sci. USA 99, 11884-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T. L., Wang, P. Y., Luo, W., Gwon, S. S., Flay, N. W., Zheng, J., Guo, C., Tanzer, M. L., and Vertel, B. M. (2001). Aggrecan domains expected to traffic through the exocytic pathway are misdirected to the nucleus. Exp. Cell Res. 263, 224-235. [DOI] [PubMed] [Google Scholar]
- Cummings, C. J., Mancini, M. A., Antalffy, B., DeFranco, D. B., Orr, H. T., and Zoghbi, H. Y. (1998). Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat. Genet. 19, 148-154. [DOI] [PubMed] [Google Scholar]
- DiFiglia, M., Sapp, E., Chase, K. O., Davies, S. W., Bates, G. P., Vonsattel, J. P., and Aronin, N. (1997). Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277, 1990-1993. [DOI] [PubMed] [Google Scholar]
- Doucas, V., and Evans, R. M. (1996). The PML nuclear compartment and cancer. Biochim. Biophys. Acta 1288, M25-.M29. [DOI] [PubMed] [Google Scholar]
- Dovey, C. L., Varadaraj, A., Wyllie, A. H., and Rich, T. (2004). Stress responses of PML nuclear domains are ablated by ataxin-1 and other nucleoprotein inclusions. J. Pathol. 203, 877-883. [DOI] [PubMed] [Google Scholar]
- Everett, R. D., Lomonte, P., Sternsdorf, T., van Driel, R., and Orr, A. (1999). Cell cycle regulation of PML modification and ND10 composition. J. Cell Sci. 112, 4581-4588. [DOI] [PubMed] [Google Scholar]
- Everett, R. D., and Maul, G. G. (1994). HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13, 5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzler, M. J., Hamel, J. C., Ochs, R. L., and Chan, E. K. (1993). Molecular characterization of two human autoantigens: unique cDNAs encoding 95- and 160-kD proteins of a putative family in the Golgi complex. J. Exp. Med. 178, 49-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, L., Gao, Y., and Sztul, E. (2005). Transcriptional repression and cell death induced by nuclear aggregates of non-polyglutamine protein. Neurobiol. Dis. (in press). [DOI] [PMC free article] [PubMed]
- Gao, Y., and Sztul, E. (2001). A novel interaction of the Golgi complex with the vimentin intermediate filament cytoskeleton. J. Cell Biol. 152, 877-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata, R., Bebok, Z., Sorscher, E. J., and Sztul, E. S. (1999). Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 146, 1239-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata, R., Gao, Y., Alvarez, C., and Sztul, E. S. (2000). The membrane transport factor p115 recycles only between homologous compartments in intact heterokaryons. Eur. J. Cell Biol. 79, 229-239. [DOI] [PubMed] [Google Scholar]
- Garcia-Mata, R., Gao, Y. S., and Sztul, E. (2002). Hassles with taking out the garbage: aggravating aggresomes. Traffic 3, 388-396. [DOI] [PubMed] [Google Scholar]
- Goldberg, A. L. (2003). Protein degradation and protection against misfolded or damaged proteins. Nature 426, 895-899. [DOI] [PubMed] [Google Scholar]
- Hicks, S. W., and Machamer, C. E. (2002). The NH2-terminal domain of Golgin-160 contains both Golgi and nuclear targeting information. J. Biol. Chem. 277, 35833-35839. [DOI] [PubMed] [Google Scholar]
- Hozak, P., Sasseville, A. M., Raymond, Y., and Cook, P. R. (1995). Lamin proteins form an internal nucleoskeleton as well as a peripheral lamina in human cells. J. Cell Sci. 108, 635-644. [DOI] [PubMed] [Google Scholar]
- Ishov, A. M., Vladimirova, O. V., and Maul, G. G. (2002). Daxx-mediated accumulation of human cytomegalovirus tegument protein pp71 at ND10 facilitates initiation of viral infection at these nuclear domains. J. Virol. 76, 7705-7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, J. A., Ward, C. L., and Kopito, R. R. (1998). Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143, 1883-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., Nollen, E. A., Kitagawa, K., Bindokas, V. P., and Morimoto, R. I. (2002). Polyglutamine protein aggregates are dynamic. Nat. Cell Biol. 4, 826-831. [DOI] [PubMed] [Google Scholar]
- Kopito, R. R. (2000). Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524-530. [DOI] [PubMed] [Google Scholar]
- Lieberman, A. P., Harmison, G., Strand, A. D., Olson, J. M., and Fischbeck, K. H. (2002). Altered transcriptional regulation in cells expressing the expanded polyglutamine androgen receptor. Hum. Mol. Genet. 11, 1967-1976. [DOI] [PubMed] [Google Scholar]
- Maul, G. G., Negorev, D., Bell, P., and Ishov, A. M. (2000). Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol. 129, 278-287. [DOI] [PubMed] [Google Scholar]
- Misumi, Y., Sohda, M., Yano, A., Fujiwara, T., and Ikehara, Y. (1997). Molecular characterization of GCP170, a 170-kDa protein associated with the cytoplasmic face of the Golgi membrane. J. Biol. Chem. 272, 23851-23858. [DOI] [PubMed] [Google Scholar]
- Muratani, M., Gerlich, D., Janicki, S. M., Gebhard, M., Eils, R., and Spector, D. L. (2002). Metabolic-energy-dependent movement of PML bodies within the mammalian cell nucleus. Nat. Cell Biol. 4, 106-110. [DOI] [PubMed] [Google Scholar]
- Nucifora, F. C., Jr., et al. (2001). Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science 291, 2423-2428. [DOI] [PubMed] [Google Scholar]
- Perez, M. K., Paulson, H. L., Pendse, S. J., Saionz, S. J., Bonini, N. M., and Pittman, R. N. (1998). Recruitment and the role of nuclear localization in polyglutamine-mediated aggregation. J. Cell Biol. 143, 1457-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platani, M., Goldberg, I., Lamond, A. I., and Swedlow, J. R. (2002). Cajal body dynamics and association with chromatin are ATP-dependent. Nat. Cell Biol. 4, 502-508. [DOI] [PubMed] [Google Scholar]
- Platani, M., Goldberg, I., Swedlow, J. R., and Lamond, A. I. (2000). In vivo analysis of Cajal body movement, separation, and joining in live human cells. J. Cell Biol. 151, 1561-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, C. A. (2002). Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington's disease and related disorders. Neuron 35, 819-822. [DOI] [PubMed] [Google Scholar]
- Scherzinger, E., Lurz, R., Turmaine, M., Mangiarini, L., Hollenbach, B., Hasenbank, R., Bates, G. P., Davies, S. W., Lehrach, H., and Wanker, E. E. (1997). Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90, 549-558. [DOI] [PubMed] [Google Scholar]
- Scherzinger, E., Sittler, A., Schweiger, K., Heiser, V., Lurz, R., Hasenbank, R., Bates, G. P., Lehrach, H., and Wanker, E. E. (1999). Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: implications for Huntington's disease pathology. Proc. Natl. Acad. Sci. USA 96, 4604-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe, D. J. (2003). Folding proteins in fatal ways. Nature 426, 900-904. [DOI] [PubMed] [Google Scholar]
- Skinner, P. J., Koshy, B. T., Cummings, C. J., Klement, I. A., Helin, K., Servadio, A., Zoghbi, H. Y., and Orr, H. T. (1997). Ataxin-1 with an expanded glutamine tract alters nuclear matrix-associated structures. Nature 389, 971-974. [DOI] [PubMed] [Google Scholar]
- Sodeik, B., Ebersold, M. W., and Helenius, A. (1997). Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136, 1007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector, D. L. (2001). Nuclear domains. J. Cell Sci. 114, 2891-2893. [DOI] [PubMed] [Google Scholar]
- Steffan, J. S., Kazantsev, A., Spasic-Boskovic, O., Greenwald, M., Zhu, Y. Z., Gohler, H., Wanker, E. E., Bates, G. P., Housman, D. E., and Thompson, L. M. (2000). The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl. Acad. Sci. USA 97, 6763-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien, D. L., Cummings, C. J., Adams, H. P., Mancini, M. G., Patel, K., DeMartino, G. N., Marcelli, M., Weigel, N. L., and Mancini, M. A. (1999). Polyglutamine-expanded androgen receptors form aggregates that sequester heat shock proteins, proteasome components and SR C-1, and are suppressed by the HDJ-2 chaperone. Hum. Mol. Genet. 8, 731-741. [DOI] [PubMed] [Google Scholar]
- Stenoien, D. L., Mielke, M., and Mancini, M. A. (2002). Intranuclear ataxin1 inclusions contain both fast- and slow-exchanging components. Nat. Cell Biol. 4, 806-810. [DOI] [PubMed] [Google Scholar]
- Swindle, C. S., Zou, N., Van Tine, B. A., Shaw, G. M., Engler, J. A., and Chow, L. T. (1999). Human papillomavirus DNA replication compartments in a transient DNA replication system. J. Virol. 73, 1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, J., et al. (2002). Two populations of neuronal intranuclear inclusions in SCA7 differ in size and promyelocytic leukaemia protein content. Brain 125, 1534-1543. [DOI] [PubMed] [Google Scholar]
- Tang, Q., Bell, P., Tegtmeyer, P., and Maul, G. G. (2000). Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J. Virol. 74, 9694-9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges, D., Zwickl, P., and Baumeister, W. (1999). The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68, 1015-1068. [DOI] [PubMed] [Google Scholar]
- Waelter, S., Boeddrich, A., Lurz, R., Scherzinger, E., Lueder, G., Lehrach, H., and Wanker, E. E. (2001). Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol. Biol. Cell 12, 1393-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, C. L., Omura, S., and Kopito, R. R. (1995). Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83, 121-127. [DOI] [PubMed] [Google Scholar]
- Weis, K., Rambaud, S., Lavau, C., Jansen, J., Carvalho, T., Carmo-Fonseca, M., Lamond, A., and Dejean, A. (1994). Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell 76, 345-356. [DOI] [PubMed] [Google Scholar]
- Wigley, W. C., Fabunmi, R. P., Lee, M. G., Marino, C. R., Muallem, S., DeMartino, G. N., and Thomas, P. J. (1999). Dynamic association of proteasomal machinery with the centrosome. J. Cell Biol. 145, 481-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik, C., and DeMartino, G. N. (2003). Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol. 35, 579-589. [DOI] [PubMed] [Google Scholar]
- Wojcik, C., Schroeter, D., Wilk, S., Lamprecht, J., and Paweletz, N. (1996). Ubiquitin-mediated proteolysis centers in HeLa cells: indication from studies of an inhibitor of the chymotrypsin-like activity of the proteasome. Eur. J. Cell Biol. 71, 311-318. [PubMed] [Google Scholar]
- Yamada, M., Sato, T., Shimohata, T., Hayashi, S., Igarashi, S., Tsuji, S., and Takahashi, H. (2001). Interaction between neuronal intranuclear inclusions and promyelocytic leukemia protein nuclear and coiled bodies in CAG repeat diseases. Am. J. Pathol. 159, 1785-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, M., Tsuji, S., and Takahashi, H. (2002). Involvement of lysosomes in the pathogenesis of CAG repeat diseases. Ann. Neurol. 52, 498-503. [DOI] [PubMed] [Google Scholar]
- Yasuda, S., et al. (1999). Triggering of neuronal cell death by accumulation of activated SEK1 on nuclear polyglutamine aggregations in PML bodies. Genes Cells 4, 743-756. [DOI] [PubMed] [Google Scholar]
- Zhong, S., Salomoni, P., and Pandolfi, P. P. (2000). The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2, E85-E90. [DOI] [PubMed] [Google Scholar]
- Zoghbi, H. Y., and Orr, H. T. (2000). Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 23, 217-247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.