Abstract
A disintegrin and a metalloprotease (ADAM) family members have been implicated in many biological processes. Although it is recognized that recombinant ADAM disintegrin domains can interact with integrins, little is known about ADAM-integrin interactions in cellular context. Here, we tested whether ADAMs can selectively regulate integrin-mediated cell migration. ADAMs were expressed in Chinese hamster ovary cells that express defined integrins (α4β1, α5β1, or both), and cell migration on full-length fibronectin or on its α4β1 or α5β1 binding fragments was studied. We found that ADAMs inhibit integrin-mediated cell migration in patterns dictated by the integrin binding profiles of their isolated disintegrin domains. ADAM12 inhibited cell migration mediated by the α4β1 but not the α5β1 integrin. ADAM17 had the reciprocal effect; it inhibited α5β1- but not α4β1-mediated cell migration. ADAM19 and ADAM33 inhibited migration mediated by both α4β1 and α5β1 integrins. A point mutation in the ADAM12 disintegrin loop partially reduced the inhibitory effect of ADAM12 on cell migration on the α4β1 binding fragment of fibronectin, whereas mutations that block metalloprotease activity had no effect. Our results indicate that distinct ADAMs can modulate cell migration mediated by specific integrins in a pattern dictated, at least in part, by their disintegrin domains.
INTRODUCTION
Cell migration is essential for a variety of important events in both embryonic development and in the adult. Integrins, which interact with extracellular matrix (ECM) molecules, are key players in cell migration (Ridley et al., 2003). In particular, the fibronectin (FN) binding integrins α4β1 and α5β1 are well characterized for their roles in cell motility. The α5β1 integrin engages the central cell binding domain (CCBD) of FN and forms focal adhesions, which function both as anchors and as signaling centers. The integrin α4β1 engages the CS-1 region of FN and promotes lamellipodia protrusion in a focal adhesion-independent manner (Pinco et al., 2002; Goldfinger et al., 2003; Mostafavi-Pour et al., 2003). Integrins often work in cooperation with syndecans, transmembrane proteoglycans that foster cell adhesion, stress fiber formation, cell migration, and invasion (Woods and Couchman, 2000; Beauvais and Rapraeger, 2004).
Proteins containing a disintegrin and a metalloprotease (ADAM) domain are multidomain proteins that have been implicated in a spectrum of biological processes, including fertilization, myogenesis, axon extension, and axon guidance (Seals and Courtneidge, 2003; White, 2003; White et al., 2005). ADAMs are best known as ectodomain sheddases, a function of their metalloprotease domains. ADAMs shed a variety of important cell surface molecules, including cytokines (e.g., tumor necrosis factor-α), growth factors (e.g., epidermal growth factor family members), and adhesion molecules (e.g., L1, selectins, and vascular cell adhesion molecule) (Blobel, 2005). Downstream of their metalloprotease domains, ADAMs contain a disintegrin domain and a cysteine-rich domain. These domains have been implicated in interactions with integrins and syndecans, respectively. Many recombinant ADAM disintegrin domains have been shown to interact with a variety of β1 integrins, including α4β1 (Tomczuk et al., 2003; White, 2003; Bridges et al., 2004; White et al., 2005). However, the only disintegrin domains that have been reported to interact with α5β1 are those of human ADAM15, which is unique among ADAMs in having an RGD motif in its disintegrin loop (Zhang et al., 1998; Nath et al., 1999), and ADAM17 (Bax et al., 2004), which is a distal ADAM family member whose disintegrin domain differs in several respects from those of canonical ADAMs.
To date, only a few studies have addressed the possible role of ADAM-integrin interactions using full-length ADAM proteins in cell biological settings. It has been shown that cells expressing ADAM12 or ADAM15 can bind to cells expressing the α9β1 integrin (Eto et al., 2000) and that ADAM12 can influence β1 integrin mediated adipocyte differentiation (Kawaguchi et al., 2003). More recent work has shown that ADAM17 can associate with α5β1 integrin in HeLa cells (Bax et al., 2004) and that ADAM12 and α9β1 integrin function coordinately in myotube development (Lafuste et al., 2005). However, no studies have yet addressed the selectivity of ADAM-integrin interactions in a cell biological function such as cell migration. In this study we provide the first evidence that ADAMs can selectively modulate integrin-mediated cell migration.
MATERIALS AND METHODS
Antibodies and Plasmids
Mouse anti-α4 integrin functional blocking monoclonal antibody (mAb) HP2/1 and mouse anti-α5 integrin functional blocking antibody CBL497 were obtained from Chemicon International (Temecula, CA). Antibody against c-myc tag (9E10) was described previously (Smith et al., 2002). Human plasma fibronectin (FN) was obtained from Roche Diagnostics (Indianapolis, IN).
Constructs encoding GST-9.11, which contains the RGD and the synergy sites of FN, GST-V, which contains only the alternatively spliced CS-1 (or V) region of FN, and GST-12.15V0, which contains the binding site for syndecans, were provided by Douglas DeSimone (University of Virginia, Charlottesville, VA).
Mouse ADAM12 disintegrin domain in the vector pGEX-2T was provided by Anna Zolkiewski (Kansas State University, Manhattan, KS). To produce protein with higher purity, a 6Xhistidine (His) tag was added to the 3′ end of the disintegrin domain coding sequence, using a site-directed mutagenesis kit (Stratagene, La Jolla, CA). ADAM12 disintegrin loop mutants (R481A, D488A, and E491A), were generated using a site-directed mutagenesis kit (Stratagene), using the 6XHis-tagged ADAM12 disintegrin domain as the template.
Sequences encoding the disintegrin domains of mouse ADAM19 (residues G417-G504) or mouse ADAM 33 (residues P418-G505) in frame with a terminal 6Xhistidine tag were generated by PCR. Novel EcoRI and NotI restriction sites were incorporated during PCR at the 5′ and 3′ ends, respectively. PCR products were digested with EcoRI/NotI and ligated to EcoRI/NotI digested pGEX-4T-2 vector (GE Healthcare, Piscataway, NJ).
Sequence encoding full-length mouse ADAM12 (gift from Ulla Wewer, University of Copenhagen, Copenhagen, Denmark, and Jurgen Frey, University of Bielefeld, Bielefeld, Germany) was PCR amplified with cloning sites (BglII and ClaI) added to the 5′ and 3′ ends, respectively. Next, mADAM12 was digested and ligated into pCS2 vector, in frame with the 6Xmyc tag previously cloned into this vector. Site-directed mutagenesis was used to create a mutant ADAM12 by changing E349 to A as well as mutants encompassing the putative furin cleavage sites (199RARRHKR), where the italicized amino acids were changed into “SG” and “AA,” respectively.
The full coding sequence for mouse ADAM17 was generously provided by Roy Black (Amgen, Seattle, WA). Unique BamHI restriction sites were incorporated at the 5′ and 3′ ends of the ADAM17 sequence via PCR and the product was cloned into BamHI-digested pCS2 vector, in frame with 6Xmyc tags. Clones were screened for directionality and sequenced to ensure proper reading frame.
The entire coding sequence of mADAM19 was amplified by reverse transcription-PCR on mouse E8.5 oligo-dT primed cDNA with an XhoI site added to each end. The PCR product was inserted into pCRII-TOPO vector for amplification. The full-length clone was then cut out and ligated into pCS2 vector. To remove the stop codon and insert a ClaI site for cloning in frame with the 6Xmyc tag, another round of PCR amplification was performed on mADAM19 in pCS2. The PCR product was inserted into pCRII-TOPO, amplified, and cut out with a BclI/ClaI digestion before being introduced into pCS2 with the 6Xmyc tag.
A cDNA clone of ADAM33 without the first 300 base pairs was obtained by screening a mouse embryo day 11 cDNA library (BD Biosciences Clonetech, Palo Alto, CA). This clone was named D12 and inserted into pCS2 with an EcoRI partial digestion. The missing 5′ end was PCR amplified from OriGene Sure-RACE mouse embryo day 19 cDNA. After two rounds of amplification, the PCR product was cloned into pCRII using the TOPO-TA cloning kit (Invitrogen, Carlsbad, CA). 5′ end ADAM33 clone was isolated, digested with BglII and XhoI, and the resulting band was ligated into the pCS2 vector containing clone D12. To add a myc tag in frame with ADAM33, the full-length clone was PCR amplified with a forward primer that added a BclII site and a reverse primer that removed the stop codon and added a ClaI site. This PCR product was cloned into the pCRII-TOPO vector, amplified, cut out with BclII and ClaI, and then ligated into pCS2 in frame with 6Xmyc tags.
Preparation of Glutathione S-Transferase (GST)-Fusion Proteins
GST-chimeras encoding defined fibronectin fragments were purified exactly as described previously (Ramos and DeSimone, 1996). GST-tagged proteins containing the disintegrin domains of ADAM12 (wild type and mutant), ADAM19 and ADAM33 were produced exactly as described previously (Tomczuk et al., 2003).
Cell Culture and Transfection
CHOB2-α5 and CHOK1 cells (provided by A. Rick Horwitz, University of Virginia) were maintained in DMEM containing 10% fetal bovine serum (FBS) (Invitrogen), l-glutamine, nonessential amino acids, and antibiotics. CHOB2-α4 (provided by Cary Wu, University of Pittsburgh, Pittsburgh, PA) and CHO-α4/GFP cells (provided by Joy Yang, Johns Hopkins University, Baltimore, MD) were maintained in DMEM/F-12 (Invitrogen) containing 10% FBS, l-glutamine, antibiotics, and 0.8 mg/ml active G418 (Invitrogen). As described previously (Pinco et al., 2002), green fluorescent protein (GFP)-tagged α4 integrin is functionally normal. Molt 3 cells, obtained from American Type Culture Collection (Manassas, VA), were maintained in RPMI 1640 medium containing 10% FBS, l-glutamine, antibiotics, sodium pyruvate, and 15 mM HEPES buffer. Cells were transfected with different ADAM constructs using Lipofectamine 2000 reagent (Invitrogen) following manufacturer's instructions.
Cell Adhesion and Migration Assays
Cell adhesion assays were performed as described previously (Tomczuk et al., 2003).
Scratch wound migration assays were performed as described previously (Pinco et al., 2002). Briefly, transfected cells were plated onto 24-well tissue culture plates coated with 10 μg/ml human plasma FN (Roche Diagnostics) or GST-CCBD, or 2 μg/ml CS-1. After incubation for 2 h in a CO2 incubator at 37°C, confluent cell monolayer was scraped with a yellow Pipetman tip to generate scratch wounds. When migration assays were conducted on GST-CCBD, 2 μg/ml GST-12.15V0 (a region of FN that interacts with syndecan family heparan sulfate proteoglycans) was added to the culture media during incubation, to facilitate α5β1-mediated cell migration (Mostafavi-Pour et al., 2003). The cells were washed twice with phosphate-buffered saline (PBS) and then cultured in media containing 10% FBS in a tissue culture incubator. Plates were washed twice with PBS to remove unbound cells at designated time points after scratch wounding, and cells were viewed on a phase contrast microscope (Nikon, Melville, NY) and photographed with an attached camera (Diagnostic Instruments, Sterling Heights, MI). From these photographs, the wound area (area not occupied by cells) was measured using Photoshop7.0 software, and percentage of wound closure was calculated as an indication of rate of cell migration. To take sequential pictures and make movies, transfected cells were plated on 35-mm glass bottom culture dishes (MatTek, Ashland, MA) precoated with 2 μg/ml CS-1, scratch wounded, and placed on an open chamber with temperature controller (Harvard Apparatus, Holliston, MA). Pictures were then taken every 10 min for 6 h using Spot3.5 software.
Transwell migration assays were performed as described previously (Liu et al., 1999), with the following modifications. Transwell inserts were coated on the distal side with 10 μg/ml FN or CS-1 in serum-free F-12 medium for 2 h at 37°C. Cells were resuspended in serum free F-12 medium (5 × 105 cells/ml), added to the top chambers, and incubated for 4 h (for CHOK1 and CHO-α4/GFP cells) or 15 h (for CHOB2-α4 and CHOB2-α5 cells) at 37°C. After the incubation, cells in the top chambers were then removed with a cotton swab and membranes were fixed with 4% formaldehyde. Cells binding to the distal side of the membranes were stained with crystal violet, solubilized with SDS, and optical density at 595 nm was read.
Random migration assays were performed as described in Goldfinger et al., 2003. Briefly, 35-mm glass bottom culture dishes (MatTek) were precoated with 2 μg/ml CS-1 overnight at 4°C, and 5 × 105 mock or ADAM12-transfected cells were plated on a dish for 2 h. Cells were placed on an open chamber with temperature controller (Harvard Apparatus) and sequential pictures were taken every 10 min for 4 h, as described above. Migration tracks of individual cells were analyzed using Openlab 2.2.5 software.
Cell Surface Biotinylation and Immunoprecipitation
Twenty-four hours after transfection with ADAM constructs, cells in 6-cm dishes were washed twice with PBS (Mediatech, Herndon, VA), and then surface biotinylated by incubation with EZ-Link NHS-LC-biotin (Pierce Chemical, Rockford, IL) for 15 min at room temperature, followed by two more washes with PBS. Cells were lysed for 60 min at 4°C in ice-cold lysis buffer (1% Triton X-100, 10 mM 1,10-phenanthroline, 5 mM EDTA, and 2 mM phenylmethylsulfonyl fluoride in PBS), and cleared lysates were incubated with 20% avidin-agarose beads (Sigma-Aldrich, St. Louis, MO) in PBS at 4°C overnight. Beads were washed twice with 1% Triton X-100 in PBS, and proteins were eluted with SDS-sample buffer containing 50 mM dithiothreitol (Sigma-Aldrich). Eluted proteins were analyzed on Western blots for the presence of ADAM proteins by probing for their C-terminal myc epitope tag using mAb 9E10, followed by horseradish peroxidase-conjugated goat anti-mouse IgG (The Jackson Laboratory, Bar Harbor, ME). An experiment with cells transfected to express intracellular GFP showed that the biotinylation protocol did not label intracellular proteins (our unpublished data).
Flow Cytometry
Cells were detached from tissue culture dishes, resuspended at 1 × 106/ml in PBS containing 5% FBS (Invitrogen) and 1% bovine serum albumin (Calbiochem, San Diego, CA), and incubated for 30 min on ice. Primary antibody (mouse anti-human α4, HP2/1), diluted to 20 μg/ml in blocking buffer (PBS/fetal bovine serum/bovine serum albumin), was incubated with cells for 45 min at 4°C. Cells were then washed twice with PBS and resuspended in blocking buffer containing 20 μg/ml R-phycoerythrin-conjugated secondary antibody (Molecular Probes, Eugene, OR). After 45-min incubation on ice, cells were washed twice with PBS, resuspended in 0.3 ml of 2% paraformaldehyde in PBS, and analyzed on a FACSCalibur benchtop analyzer at the Flow Cytometry Core Facility at University of Virginia.
RESULTS
ADAM12 Inhibits α4β1- but Not α5β1 Integrin-mediated Cell Migration on Fibronectin
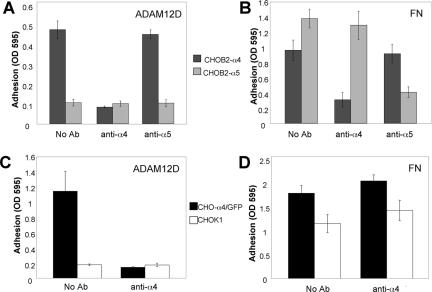
ADAM12 is phylogenetically related to Xenopus ADAM13, which has a documented role in cranial neural crest (CNC) cell migration (Alfandari et al., 2001). In addition, ADAM12 is expressed in mouse cranial neural crest (Tomczuk, 2004). Because of our interest in the potential role of ADAM-integrin interactions in cell migration, and because of the noted role of α4β1 integrin in cell migration events, including CNC migration (Kil et al., 1998), we first tested whether cells expressing α4β1 adhere to the mouse ADAM 12 disintegrin domain. We also tested whether cells expressing α5β1 integrin adhere because we are interested in the selectivity of ADAM-integrin interactions, and because very few ADAM disintegrin domains have been reported to interact with α5β1 integrin (White et al., 2005). As seen in Figure 1A, CHOB2 cells that express the human α4 integrin subunit (CHOB2-α4), bound to a recombinant ADAM12 disintegrin domain. In contrast, CHOB2-α5 cells, which express the human α5 integrin and bind to fibronectin (FN) (Figure 1B), did not adhere to the ADAM12 disintegrin domain (Figure 1A). Binding of CHOB2-α4 cells was inhibited by an antibody against the α4, but not one against the α5, integrin subunit (Figure 1A). CHOK1 cells that express an α4/GFP chimera as well as endogenous α5β1 integrin (CHO-α4/GFP), also bound to the ADAM12 disintegrin domain, and this interaction was disrupted by the anti-α4 antibody (Figure 1C). A previous study demonstrated that GFP-tagged α4 integrin behaves like untagged α4 integrin (Pinco et al., 2002). Parental CHOK1 cells, which express endogenous α5β1 integrin and which bind to FN (Figure 1D), did not adhere to the ADAM12 disintegrin domain (Figure 1C). These observations indicate that cells can adhere to a recombinant ADAM12 disintegrin domain using the α4β1 but not the α5β1 integrin.
Figure 1.
Cells expressing α4β1 but not α5β1 integrin bind to recombinant ADAM12 disintegrin domain. (A and B) CHOB2-α4 (black) and CHOB2-α5 (gray) cells were resuspended in Puck's saline A containing 0.5 mM Ca2+ and 0.5 mM Mg2+, pretreated with either no antibody, 5 μg/ml anti-α4 antibody (HP2/1), or 5 μg/ml anti-α5 antibody (CBL497) for 30 min, and then added to wells coated with 5 μg/ml ADAM12 disintegrin domain (A) or 5 μg/ml human plasma fibronectin (B). Cells were allowed to bind to coated wells for 1 h. After three washes, adherent cells were stained with crystal violet and solubilized with 1% SDS. Optical density of the wells at 595 nm was then read with a microplate reader. (C and D) CHO-α4/GFP (black) and CHOK1 cells (white) were pretreated with either no antibody or 5 μg/ml anti-α4 antibody (HP2/1) for 30 min before adding to wells coated with 5 μg/ml ADAM12 disintegrin domain (C) or human plasma FN (D). Adhesion assays were then conducted as described above. These experiments were repeated three times with similar results.
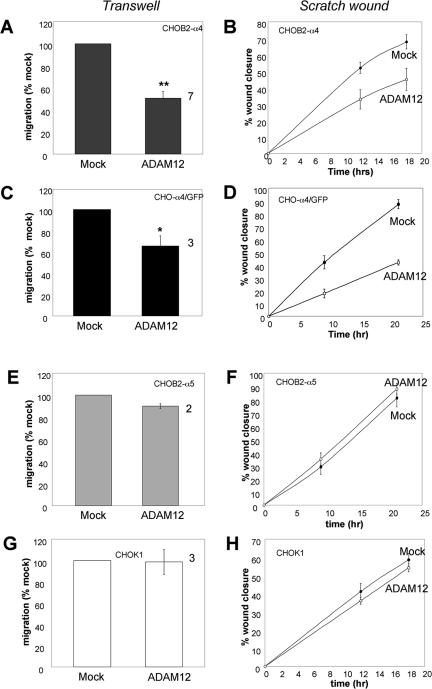
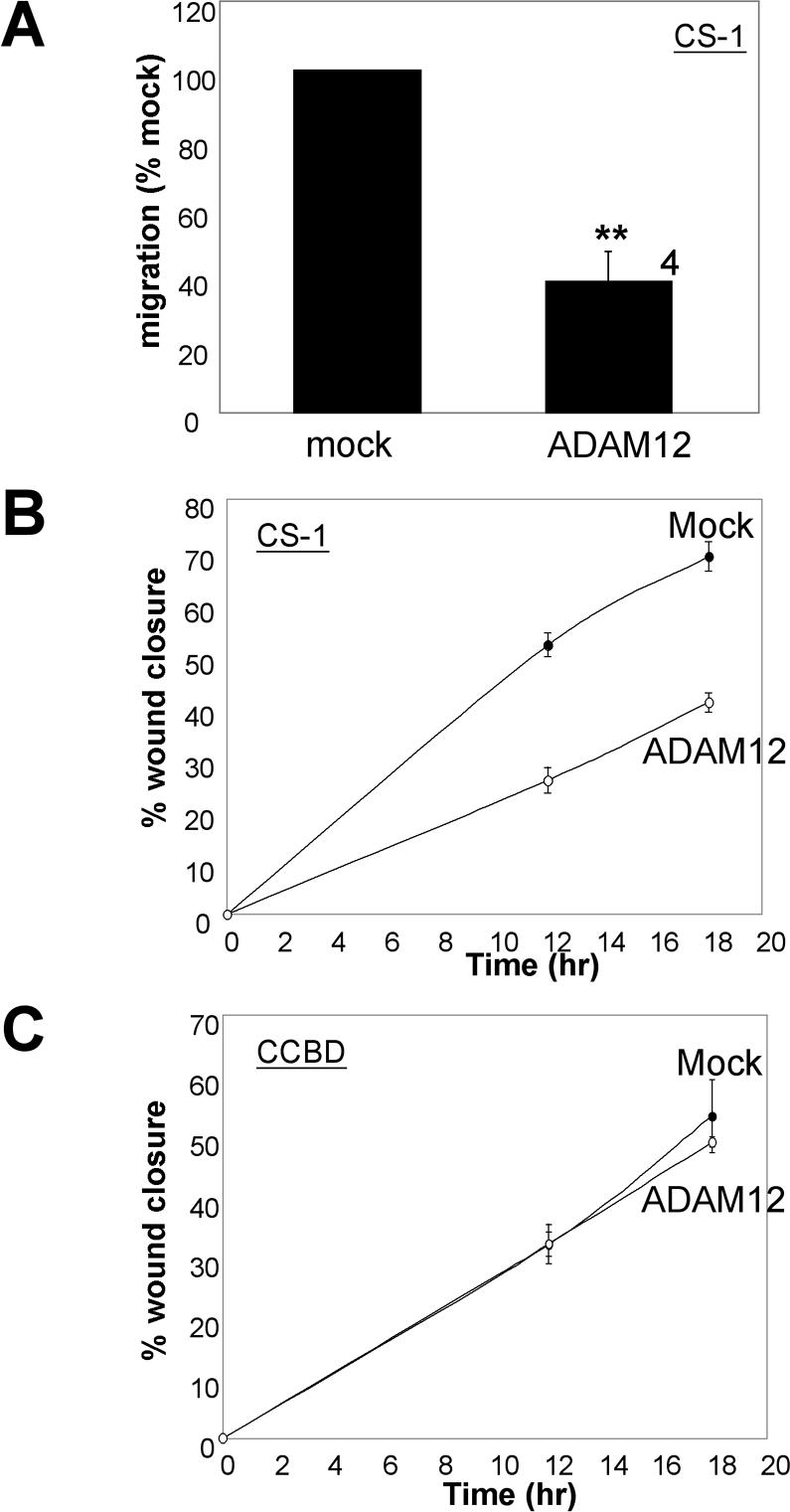
We next tested the effect of expressing full-length ADAM12 on directed cell migration on FN, using both transwell and scratch wound assays. Expression of ADAM12 reduced the motility of CHOB2-α4 cells in both transwell (Figure 2A) and scratch wound (Figure 2B) assays. Similarly, expression of ADAM12 in CHO-α4/GFP cells caused a reduction in cell motility in both transwell (Figure 2C) and scratch wound (Figure 2D) assays. The inhibition ranged from 35 to 50%.
Figure 2.
ADAM12 inhibits migration of α4β1- but not α5β1 integrin-expressing cells on fibronectin. Cells were either mock transfected or transfected to express full-length ADAM12, and transwell migration assays (A, C, E, and G) or scratch wound assays (B, D, F, and H) were conducted 24 h after transfection. For transwell migration assays, the distal membranes of transwell inserts were coated with 10 μg/ml FN, and number of cells that migrated through the pores were determined after an incubation of 15 h (for CHOB2-α4 and CHOB2-α5 cells) or 4 h (for CHO-α4/GFP and CHOK1 cells). All samples were analyzed in triplicate wells. For each experiment, the average migration rate of ADAM12-transfected cells was normalized to that of mock-transfected cells. The values plotted represent the average results from several independent experiments (numbers by the right of columns), plus or minus the SEM (error bar). Statistically significant differences between mock and ADAM12-transfected cells were found for CHOB2-α4 (A, ** indicates p < 0.02 in student t test) and CHO-α4/GFP cells (C, * indicates p < 0.05 in Student's t test), but not for CHOB2-α5 and CHOK1 cells (E and G). For scratch wound assays, 24-well dishes were precoated with 10 μg/ml FN, and migration assays were conducted as described in Materials and Methods. The average percentage of wound closure, plus or minus the SD (error bars) from triplicate wells, was plotted. The scratch wound assays were repeated twice with similar results.
We next assessed the effect of ADAM12 on α5β1-mediated cell migration on FN. Expression of ADAM12 had no significant effect on the migration of Chinese hamster ovary (CHO) cells expressing α5β1 integrin on FN, as assessed using transwell (Figure 2, E and G) and scratch wound (Figure 2, F and H) assays. Collectively, the results presented in Figure 2 (using two different cell migration assays) indicate that ADAM12 inhibits cell migration on full-length FN mediated by the α4β1 but not by the α5β1 integrin. Moreover, this pattern correlates with the selective adhesive activity of the ADAM12 disintegrin domain (Figure 1).
ADAM12 Inhibits Cell Migration on an α4β1- but Not on an α5β1-specific FN Fragment
To further investigate the selectivity of ADAM12 in integrin-mediated cell migration, we tested the effect of expressing full-length ADAM12 on cell migration on two fragments of FN: the CS-1 (or V) region, which is an adhesive substrate for α4β1 integrin, and the CCBD, which is used by the α5β1 integrin for cell adhesion. As seen in Figure 3, with cells expressing both the α4β1 and α5β1 integrins (CHO-α4/GFP), ADAM12 inhibited cell migration on the α4β1-specific substrate in both transwell (Figure 3A) and scratch wound (Figures 3B and 4A) assays. In contrast, expression of ADAM12 had no significant effect on the migration of the same cells (CHO-α4/GFP) on the α5β1 substrate (Figure 3C). These observations add support to our previous conclusion that ADAM12 inhibits cell migration mediated by the α4β1 but not by the α5β1 integrin.
Figure 3.
Full-length ADAM12 inhibits the migration of CHO-α4/GFP cells on the α4β1 binding CS-1 region but not on the α5β1 binding CCBD of fibronectin. (A) CHO-α4/GFP cells, which express both the α4β1 and α5β1 integrins, were either mock transfected or transfected to express full-length ADAM12. After 24 h they were analyzed in transwell migration assays as described in the legend to Figure 2, except that the transwell inserts were coated with 2 μg/ml CS-1. The values plotted represent the average results from four independent experiments (number by the right of column), plus or minus the SEM (error bar). ** indicates a statistically significant difference (p < 0.02 in Student's t test). (B) CHO-α4/GFP cells were transfected as indicated and then analyzed in scratch wound assays, as described in the legend to Figure 2, except that 24-well dishes were coated with 2 μg/ml CS-1. The experiment has been repeated six times with similar results. (C) CHO-α4/GFP cells were transfected as indicated and then analyzed in scratch wound assays, as described in the legend to Figure 2, except that 24-well dishes were coated with 10 μg/ml CCBD. The experiment has been repeated twice with similar results.
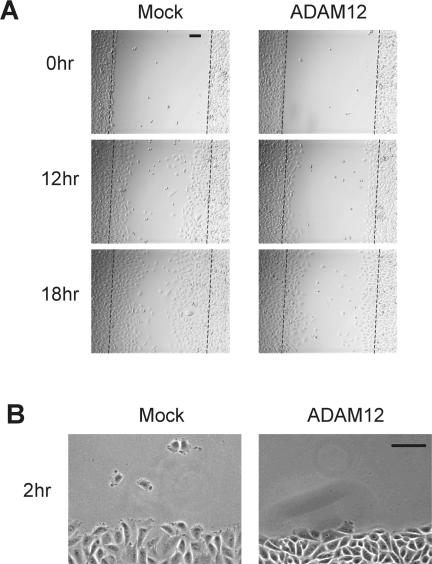
Figure 4.
Microscopy of mock- or ADAM12-transfected CHO-α4/GFP cells in scratch wound migration assays. (A) Images of cells from the 12-h time point of the experiment shown in Figure 3B. (B) Cells were plated on glass-bottom dishes precoated with 2 μg/ml CS-1. Confluent cell monolayers were then scratch wounded and observed by videomicroscopy, as described in the legend to Movie 1. Frames from the videos at 2-h postscratch wounding are shown for mock-(left) and ADAM12 (right)-transfected cells at the wound edges. Bars, 100 μm.
We next used time-lapse microscopy to analyze the effect of ADAM12 on the ability of CHO-α4/GFP cells to migrate on the CS-1 region of FN after scratch wounding. Mock-transfected cells at the wound edge protruded broad lamellipodia and displayed polarity toward the wound (Figure 4B, left, and Movie 1A). In contrast, lamellipodia protrusions and cell body translocation seemed less robust in ADAM12 transfected samples (Figure 4B, right, and Movie 1B).
Expression of ADAM12 inhibits α4β1-mediated cell migration (Figures 2, 3, 4). To test whether this effect is due to changes in expression of the α4 integrin subunit, we analyzed total and cell surface expression levels of the α4 subunit by flow cytometry. As seen in Table 1, expression of ADAM12 did not change either the total or the surface levels of the α4 integrin subunit.
Table 1.
ADAM12 does not alter expression of the integrin α4 subunit
| Surface α4 (antibody staining)
|
Total α4 (GFP)
|
|||
|---|---|---|---|---|
| DNA transfected | % Cells | MFU | % cells | MFU |
| Mock | 93.8 | 640 | 93.3 | 882 |
| ADAM12 | 92.2 | 643 | 92.2 | 887 |
CHO-α4/GFP cells were transfected with either pCS2 vector (mock) or pCS2 vector encoding ADAM12. Twenty-four hours after the transfection, cells were labeled with either a control antibody, or an antibody against the integrin α4 subunit (HP2/1), followed by phycoerythrin-conjugated goat anti-mouse IgG. Percentage of positive cells and mean fluorescence units (MFU) for both phycoerythrin and GFP were determined by flow cytometry.
A Disintegrin Loop Mutation That Impairs α4β1 Integrin Binding Reduces the Ability of ADAM12 to Inhibit α4β1 Integrin-mediated Cell Migration
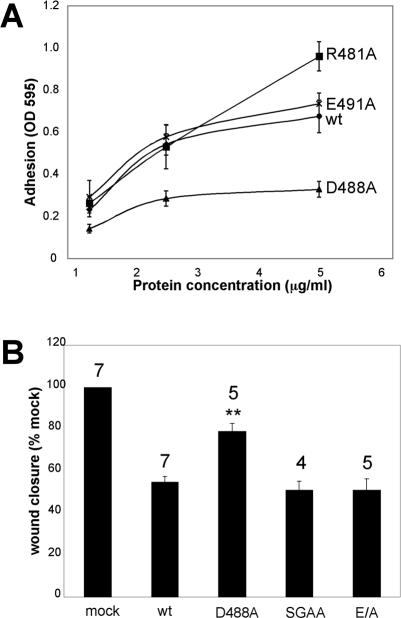
The disintegrin loop sequence of mouse ADAM12 is 480CRGSSNSCDLPEFC. Because previous reports have indicated that charged residues within the disintegrin loops of several ADAMs (including ADAM12) are important for integrin recognition (Eto et al., 2002), we mutated the three charged amino acids within the loop to alanines: R481A, D488A, and E491A. Recombinant mutant disintegrin domains were then tested for their ability to support integrin α4β1-dependent cell adhesion. Of these charge to alanine mutations, the mutant D488A was strongly impaired in its ability to support α4β1-mediated cell adhesion (Figure 5A). We therefore tested the effects of full-length ADAM12 containing the D488A mutation on α4β1-mediated cell migration. As seen in Figure 5B, the mutant D488A was compromised in its ability to inhibit α4β1-mediated cell migration compared with wild-type ADAM12.
Figure 5.
An ADAM12 disintegrin loop mutant has a lower apparent affinity to α4β1 integrin and a reduced ability to inhibit α4β1-mediated cell migration. (A) Microtiter wells were coated with the indicated concentrations of the indicated (wt and mutant) ADAM12 disintegrin domain: wild type (diamond), R481A (square), D488A (triangle), and E491A (cross). Adhesion assays were then conducted using Molt 3 human lymphoma cells as described in the legend to Figure 1, except that 1 mM Mn2+ was added to cell suspensions before the assays. The numbers plotted represent the average values from quadruplicate wells, plus or minus the SD (error bar). The experiment has been repeated three times with similar results. A similar observation with the D488A mutant was obtained using CHOB2-α4 cells in adhesion flow assays. (B) CHO-α4/GFP cells were transfected to express wild-type, D488A, E/A, or “poorly processed” (SGAA) ADAM12, and scratch wound migration assays were then conducted on the CS-1 region of FN, as described in the legend to Figure 2B. The extent of wound closure at 12 h postscratch wounding was analyzed. Wound closure obtained with mock-transfected cells was considered to be 100%. Average wound closure for cells transfected with different ADAM12 constructs was expressed as a percentage of that seen for mock-transfected cells. The average values (% of mock) from several independent experiments (numbers above the columns), plus or minus the SEM (error bars) are shown. The only mutant that showed significantly different behavior than wild type ADAM12 was the “D488A” mutant, as indicated by ** (p < 0.02 in Student's t test).
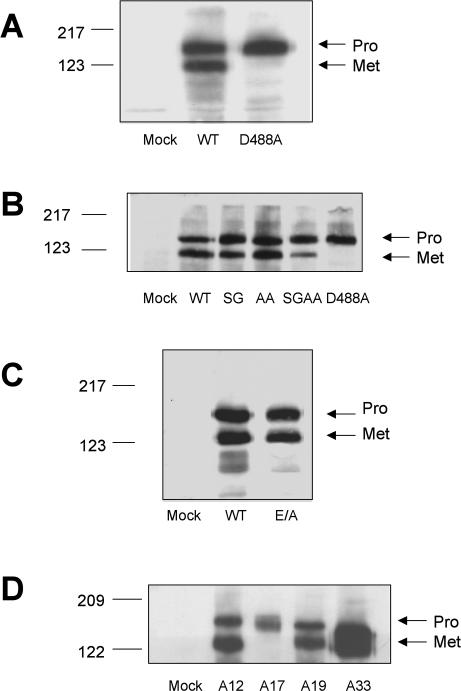
A biochemical analysis revealed, to our surprise, that although D488A ADAM12 is expressed and transported to the cell surface, it is not proteolytically processed (Figure 6A). To assess whether the D488A mutant is a less potent inhibitor of α4β1-mediated cell migration (Figure 5B) because it is not processed (Figure 6A), we set out to generate an “unprocessed” ADAM12. A previous report showed that the prodomain of human ADAM12 can be removed by a furin-like endopeptidase (Loechel et al., 1998). The sequence at the junction of the pro- and the catalytic domains in mouse ADAM12 is 199RARRHKR. Thus, it contains two potential furin recognition motifs (RX(K/R)R), which are199RARR and 202RHKR, respectively. We first mutated the two sites individually: 201RR was changed to SG (resulting construct designated “SG ADAM12”) and 204KR to AA (designated “AA ADAM12”). Both were processed equally well as wild-type ADAM12 (Figure 6B, lanes 3 and 4). When both mutations were introduced into the same construct (designated “SGAA ADAM12”), prodomain removal was significantly reduced, albeit not abolished (Figure 6B, lane 5), suggesting that other enzymes may be able to process ADAM12, as noted previously (Cao et al., 2002).
Figure 6.
Expression of wild-type and mutant full-length ADAMs on the surface of CHO-α4/GFP cells. CHO-α4/GFP cells were transfected with 4 μg of DNA (per 6-cm dish) encoding the indicated 6Xmyc-tagged constructs. Twenty-four hours after transfection, cells were labeled with the membrane-impermeable EZ-Link NHS-LC-biotin (Pierce Chemical), and avidin precipitation and immunoblotting were conducted as described in Materials and Methods. Wild-type and the disintegrin loop (D488A) ADAM12 (A); wild-type ADAM12 and ADAM12 with mutations in the putative furin cleavage sites (B); wild-type and E/A (protease inactive) ADAM12 (C); and wild-type ADAMs 12, 17, 19, and 33 (D). In these cells, transfected ADAM17 was predominantly expressed as the proform. The molecular weights of these ADAM constructs reflect the presence of the 6Xmyc tag appended at the C terminus of each construct. All gel analyses were repeated three times with similar results.
We also generated a mutant in which the catalytic glutamic acid in the protease domain was changed to alanine (“E/A ADAM12”). As seen in Figure 6C, E/A ADAM12 was transported to the cell surface and processed as well as wild-type ADAM12. We then tested the poorly processed (SGAA) and catalytic site (E/A) mutants for their effects on α4β1-mediated cell migration in scratch wound assays on the CS-1 region of FN. As seen in Figure 5B, both the poorly processed (SGAA) and the protease dead (E/A) mutants inhibited migration as much as wild-type ADAM12. Collectively, the results of Figures 5 and 6 show that the inhibitory effect of ADAM12 on α4β1-mediated cell migration does not require that it be proteolytically processed nor that it be proteolytically active. The D488A mutant therefore likely has a reduced effect on α4β1-mediated cell migration (Figure 5B) because its disintegrin domain has a reduced affinity for α4β1 integrin (Figure 5A).
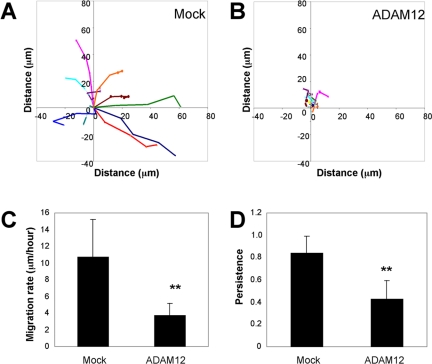
ADAM12 Inhibits Integrin α4β1-mediated Random Cell Motility
Our results so far indicate that ADAM12 inhibits α4β1 integrin-mediated cell migration in transwell and scratch wound assays. We also assessed the effect of expressing ADAM12 on random cell migration on the CS-1 region of FN. As seen in Figure 7, mock-transfected cells migrated persistently at a rate of ∼10 μm/h. In contrast, both the rate (Figure 7C) and persistence (Figure 7D) of migration of ADAM12-transfected cells were reduced and the cells showed little net translocation (Figure 7B). Analysis of movies showed that mock transfected cells displayed broad lamellipodia at the leading edge and the cells moved persistently in the direction of the protrusion (Movie 2A). In contrast, although some of the ADAM12-transfected cells extended membrane protrusions, these protrusions were less robust and did not lead to effective cell body translocation (Movie 2B). Thus, ADAM12 can also inhibit α4β1 mediated random motility of single cells.
Figure 7.
ADAM12 expression inhibits integrin α4β1-mediated random cell motility. Migration tracks for multiple (n = 10) control (A) and ADAM12 (B)-transfected cells were obtained using Openlab software, by monitoring the position of cell centroids over 4 h. (C) Average migration rates were calculated by dividing the total distance traveled by total time, based on the migration tracks for multiple cells (n = 10). (D) Average migration persistence was calculated by dividing the linear distance (the start point to the final position) by total distance traveled over 4 h, based on the migration tracks for multiple cells (n = 10). For both C and D, ** indicates statistically significant difference compared with mock-transfected cells (p < 0.02 in Student's t test). The experiment has been repeated three times with similar results.
Distinct ADAMs Selectively Inhibit Integrin-mediated Cell Migration
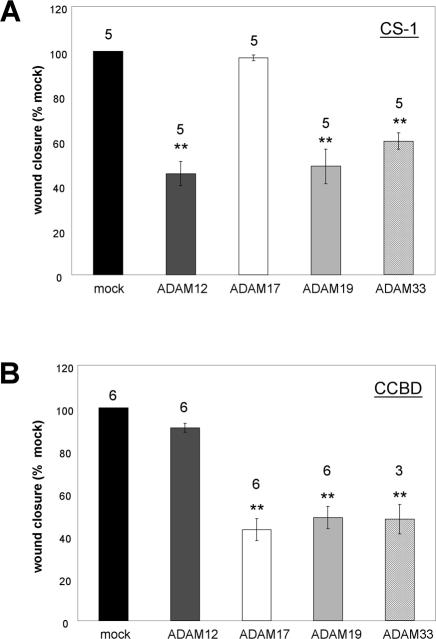
We next assessed whether other ADAM family members can selectively inhibit integrin-mediated cell migration. We first extended our analysis to ADAM17 (tumor necrosis factor-α converting enzyme), an important ADAM that was recently shown to interact with the α5β1 integrin (Bax et al., 2004). As seen in Figure 8, although ADAM17 had no effect on α4β1-mediated cell migration (Figure 8A), it significantly inhibited α5β1 integrin-mediated cell migration (Figure 8B). Thus, strikingly, ADAM12 and ADAM17 demonstrate reciprocal effects on α4β1 and α5β1 mediated cell migration (Figure 8, A and B).
Figure 8.
Selective modulation of α4- or α5-mediated cell migration by ADAMs. (A) Scratch wound assays were performed as described previously. CHO-α4GFP cells transiently expressing ADAM12, ADAM17, ADAM19, or ADAM33 were compared for their relative ability to inhibit wound closure after 12 h on the CS-1 region of FN. 4 μg (per 6-cm dish) of cDNA encoding ADAM12, ADAM17, and ADAM19 were used for transfection. Because ADAM33 was consistently expressed at the cell surface at higher levels than the other ADAMs (Figure 6D), we used a 1:1 mixture of vector and ADAM33 cDNA (2 μg each) for the ADAM33-transfected samples to achieve comparable expression levels of all ADAMs tested. (B) Scratch wound assays were repeated as in panel A, except that α5-mediated cell migration was scored on the CCBD region of FN. For both A and B, the average wound closure plus or minus the SEM (error bars) from several independent experiments (numbers above the columns) is shown. ** indicates statistically significant difference compared with mock-transfected cells (p < 0.02 in Student's t test). A higher expression level (4 μg per 6-cm dish) of ADAM33 inhibited α5β1-mediated cell migration, but its effect on α4β1-mediated migration was diminished (Huang, Bridges, and White, unpublished data).
We extended our analysis to ADAM19 and ADAM33, two other ADAMs that are phylogenetically related to Xenopus ADAM13 and that are found in mouse cranial neural crest cells (Tomczuk, 2004). Like ADAM12, ADAM19 and 33 are expressed at the surface of CHO cells in both pro- and metalloprotease active (Met) forms (Figure 6D). As seen in Figure 8A, expression of both ADAM19 and ADAM33 inhibited α4β1-mediated cell migration. As seen in Figure 8B, unlike ADAM12, expression of ADAM19 and 33 (individually) also inhibited α5β1-mediated cell migration.
DISCUSSION
ADAM (and ADAM-like) proteins have been implicated in several cell migration events (Nishiwaki et al., 2000; Alfandari et al., 2001; Martin et al., 2002; Huang et al., 2003; Seals and Courtneidge, 2003; White, 2003). However, the mechanisms by which ADAMs function in cell migration remain unclear, both for ADAMs with (e.g., Xenopus ADAM13) and for those without (e.g., Caenorhabditis elegans ADAM14; unc71) proteolytic activity. In addition to the role of ADAM proteolytic activity in cell migration (Alfandari et al., 2001), we are interested in whether the integrin binding properties of ADAM disintegrin domains can influence cell migration. To begin to explore this question we used tissue culture cells that express defined integrins (α4β1, α5β1, or both) and defined ADAMs (ADAMs 12, 17, 19, or 33), and we studied cell migration (in three different assays) on defined integrin binding substrates (FN or its α4β1 or α5β1 binding fragments). We show, for the first time, that ADAMs can selectively regulate integrin mediated cell migration.
ADAMs Selectively Modulate Integrin-mediated Cell Migration
In this report, we studied the effects of four ADAMs on cell migration mediated by either the α4β1or α5β1 integrins. We studied ADAMs 12, 19, and 33 because they are phylogenetically related to an ADAM (Xenopus ADAM13) that functions in Xenopus cranial neural crest migration (Alfandari et al., 2001), and because they are expressed in mouse cranial neural crest (Tomczuk, 2004). We also studied ADAM17, a key ectodomain sheddase (Blobel, 2005). We found that these four ADAMs display three distinct patterns of influence on integrin-mediated cell migration: 1) ADAM12 inhibits cell migration mediated by α4β1 but not α5β1 integrin. 2) ADAM17 has the opposite effects: it inhibits α5β1- but not α4β1-mediated cell migration. 3) ADAMs 19 and 33 inhibit migration mediated by both α4β1 and α5β1 integrins. The patterns of inhibition are consistent with the integrin binding profiles of the respective ADAM disintegrin domains on cells with activated β1 integrins (Bax et al., 2004) (Figure 1; our unpublished data). Moreover, a point mutation within the disintegrin loop of ADAM12 (D488A) that impairs its ability to support α4β1-mediated cell adhesion, is a less potent inhibitor of α4β1-mediated cell migration. Interestingly, a mutant allele of ADAM14 (UNC-71, a nonproteolytically active ADAM involved in two cell migration events in C. elegans) was found to have a substitution at the equivalent position (D504N) in its disintegrin loop. Further genetic experiments suggested that UNC-71 acts together with integrins for proper axon guidance (Huang et al., 2003). These observations suggest possible functional interactions between ADAM disintegrin domains and integrins during cell migration.
Possible Mechanisms by Which ADAMs Influence Integrin-mediated Cell Migration
ADAMs may participate in cell migration in many ways. For example, ADAM proteases may shed factors that stimulate or direct cell migration (Maheshwari et al., 2001; Mechtersheimer et al., 2001). Results presented in this study suggest that ADAMs may also influence cell migration in a manner independent of proteolytic activity. The ability of ADAM 12 to influence α4β1-mediated cell migration in this defined system was not altered by a point mutation that ablates its proteolytic activity, nor by mutations that prevent removal of its Prodomain, an obligate event for proteolytic activity. In contrast, as noted above, the disintegrin domain seems to be important for this activity.
How might ADAMs influence integrin-mediated cell migration independent of proteolytic activity? Our results suggest that ADAM12 does not inhibit α4β1-mediated cell migration by altering the expression level of α4β1 integrin (Table 1). Although ADAM12 expression can moderately increase cell adhesion to FN or FN fragments, this effect is not specific to α4β1 integrin (Huang and White, unpublished data). Although ADAMs and integrins may interact in neighboring cells (Eto et al., 2000), the fact that ADAM 12 inhibits α4β1-mediated migration of single cells (Figure 7) indicates that an ADAM can influence an integrin in the same cell. The ADAM may affect a partner integrin in the same cell by affecting its phosphorylation, association with adaptor proteins, intracellular trafficking, or localization to different regions of the cell surface (White, 2003), and it could do so through either direct or indirect mechanisms.
Implications of ADAM Modulation of Integrin-mediated Cell Migration
Our finding that different ADAMs can selectively affect cell migration mediated by different integrins suggests that ADAMs could influence integrin-mediated migration events in interesting and complex ways. For example, if cells in a population express multiple integrins that are capable of engaging multiple ECM substrates, the expression of an ADAM would not stop cell migration. Rather, by titering the functions of specific integrins, the ADAM may influence which integrin is used for cell migration. For example, if a cell expresses both α4β1 and α5β1 integrins, which bind to different regions of FN, expression of ADAM12 could promote use of α5β1 for migration. Conversely, the appearance of ADAM17 could promote use of α4β1. ADAMs could thus modify cell migration patterns, because α4β1 and α5β1 integrins promote the formation of different structures (lamellipodia, focal contacts, and focal adhesions) and hence promote cell migration by different mechanisms (Pinco et al., 2002; Goldfinger et al., 2003; Mostafavi-Pour et al., 2003; Na et al., 2003). If ADAMs selectively titer integrin-mediated cell migration, they may do so in complex ways. For example it seemed that if the expression level of ADAM33 is increased, it maintains its ability to inhibit α5β1 integrin-mediated cell migration, but loses it ability to inhibit α4β1-mediated cell migration (Huang, Brideges, and White, unpublished data). Interestingly, it has recently been shown that regulation of integrin function contributes to the complex behaviors of neural crest cells (Strachan and Condic, 2003, 2004).
In summary, our study provides the first evidence that ADAM family members can selectively regulate integrin-mediated cell migration. Further studies will investigate the mechanisms and consequences of this newly appreciated ADAM function.
Supplementary Material
Acknowledgments
We thank many colleagues for gifts of cells and plasmids as noted in Materials and Methods. We especially thank Katherine Smith for generating constructs expressing full-length ADAM12, ADAM19, and ADAM33 with myc tags. We also thank Donna Webb and Bette Dzamba for suggestions and critical comments on the manuscript. This work was supported by National Institutes of Health Grant GM-48739 (to J.M.W.). L.C.B. was supported by a Cancer Training Grant (University of Virginia).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-03-0258) on August 3, 2005.
Abbreviations used: ADAM, a disintegrin and metalloprotease; CCBD, central cell binding domain; CHO, Chinese hamster ovary; CNC, cranial neural crest; FBS, fetal bovine serum; GFP, green fluorescence protein; FN, fibronectin; PBS, phosphate buffered saline.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Alfandari, D., Cousin, H., Gaultier, A., Smith, K., White, J. M., Darribere, T., and DeSimone, D. W. (2001). Xenopus ADAM 13 is a metalloprotease required for cranial neural crest-cell migration. Curr. Biol. 11, 918-930. [DOI] [PubMed] [Google Scholar]
- Bax, D. V., Messent, A. J., Tart, J., van Hoang, M., Kott, J., Maciewicz, R. A., and Humphries, M. J. (2004). Integrin alpha5beta1 and ADAM-17 interact in vitro and co-localize in migrating HeLa cells. J. Biol. Chem. 279, 22377-22386. [DOI] [PubMed] [Google Scholar]
- Beauvais, D. M., and Rapraeger, A. C. (2004). Syndecans in tumor cell adhesion and signaling. Reprod. Biol. Endocrinol. 2, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel, C. P. (2005). ADAMs: key components in EGFR signalling and development. Nat. Rev. Mol. Cell. Biol. 6, 32-43. [DOI] [PubMed] [Google Scholar]
- Bridges, L. C., Sheppard, D., and Bowditch, R. D. (2004). ADAM disintegrin-like domain recognition by the lymphocyte integrins α4β1 and α4β7. Biochem J. 387, 101-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y., Kang, Q., Zhao, Z., and Zolkiewska, A. (2002). Intracellular processing of metalloprotease disintegrin ADAM12. J. Biol. Chem. 277, 26403-26411. [DOI] [PubMed] [Google Scholar]
- Eto, K., Huet, C., Tarui, T., Kupriyanov, S., Liu, H. Z., Puzon-McLaughlin, W., Zhang, X. P., Sheppard, D., Engvall, E., and Takada, Y. (2002). Functional classification of ADAMs based on a conserved motif for binding to integrin alpha 9beta 1, implications for sperm-egg binding and other cell interactions. J. Biol. Chem. 277, 17804-17810. [DOI] [PubMed] [Google Scholar]
- Eto, K., Puzon-McLaughlin, W., Sheppard, D., Sehara-Fujisawa, A., Zhang, X. P., and Takada, Y. (2000). RGD-independent binding of integrin alpha9beta1 to the ADAM-12 and -15 disintegrin domains mediates cell-cell interaction. J. Biol. Chem. 275, 34922-34930. [DOI] [PubMed] [Google Scholar]
- Goldfinger, L. E., Han, J., Kiosses, W. B., Howe, A. K., and Ginsberg, M. H. (2003). Spatial restriction of alpha4 integrin phosphorylation regulates lamellipodial stability and alpha4beta1-dependent cell migration. J. Cell Biol. 162, 731-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X., Huang, P., Robinson, M. K., Stern, M. J., and Jin, Y. (2003). UNC-71, a disintegrin and metalloprotease (ADAM) protein, regulates motor axon guidance and sex myoblast migration in C. elegans. Development 130, 3147-3161. [DOI] [PubMed] [Google Scholar]
- Kawaguchi, N., et al. (2003). ADAM12 induces actin cytoskeleton and extracellular matrix reorganization during early adipocyte differentiation by regulating beta1 integrin function. J. Cell Sci. 116, 3893-3904. [DOI] [PubMed] [Google Scholar]
- Kil, S. H., Krull, C. E., Cann, G., Clegg, D., and Bronner-Fraser, M. (1998). The alpha4 subunit of integrin is important for neural crest cell migration. Dev. Biol. 202, 29-42. [DOI] [PubMed] [Google Scholar]
- Lafuste, P., Sonnet, C., Chazaud, B., Dreyfus, P. A., Gherardi, R. K., Wewer, U. M., and Authier, F. J. (2005). ADAM12 and {alpha}9{beta}1 integrin are instrumental in human myogenic cell differentiation. Mol. Biol. Cell 16, 861-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S., Thomas, S. M., Woodside, D. G., Rose, D. M., Kiosses, W. B., Pfaff, M., and Ginsberg, M. H. (1999). Binding of paxillin to alpha4 integrins modifies integrin-dependent biological responses. Nature 402, 676-681. [DOI] [PubMed] [Google Scholar]
- Loechel, F., Gilpin, B. J., Engvall, E., Albrechtsen, R., and Wewer, U. M. (1998). Human ADAM 12 (meltrin α) is an active metalloprotease. J. Biol. Chem. 273, 16993-16997. [DOI] [PubMed] [Google Scholar]
- Maheshwari, G., Wiley, H. S., and Lauffenburger, D. A. (2001). Autocrine epidermal growth factor signaling stimulates directionally persistent mammary epithelial cell migration. J. Cell Biol. 155, 1123-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, J., Eynstone, L. V., Davies, M., Williams, J. D., and Steadman, R. (2002). The role of ADAM 15 in glomerular mesangial cell migration. J. Biol. Chem. 277, 33683-33689. [DOI] [PubMed] [Google Scholar]
- Mechtersheimer, S., et al. (2001). Ectodomain shedding of L1 adhesion molecule promotes cell migration by autocrine binding to integrins. J. Cell Biol. 155, 661-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi-Pour, Z., Askari, J. A., Parkinson, S. J., Parker, P. J., Ng, T. T., and Humphries, M. J. (2003). Integrin-specific signaling pathways controlling focal adhesion formation and cell migration. J. Cell Biol. 161, 155-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na, J., Marsden, M., and DeSimone, D. W. (2003). Differential regulation of cell adhesive functions by integrin alpha subunit cytoplasmic tails in vivo. J. Cell Sci. 116, 2333-2343. [DOI] [PubMed] [Google Scholar]
- Nath, D., Slocombe, P. M., Stephens, P. E., Warn, A., Hutchinson, G. R., Yamada, K. M., Docherty, A. J., and Murphy, G. (1999). Interaction of metargidin (ADAM-15) with alphavbeta3 and alpha5beta1 integrins on different haemopoietic cells. J. Cell Sci. 112, 579-587. [DOI] [PubMed] [Google Scholar]
- Nishiwaki, K., Hisamoto, N., and Matsumoto, K. (2000). A metalloprotease disintegrin that controls cell migration in Caenorhabditis elegans. Science 288, 2205-2208. [DOI] [PubMed] [Google Scholar]
- Pinco, K. A., He, W., and Yang, J. T. (2002). alpha4beta1 Integrin regulates lamellipodia protrusion via a focal complex/focal adhesion-independent mechanism. Mol. Biol. Cell 13, 3203-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, J. W., and DeSimone, D. W. (1996). Xenopus embryonic cell adhesion to fibronectin: position-specific activation of RGD/synergy site-dependent migratory behavior at gastrulation. J. Cell Biol. 134, 227-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley, A. J., Schwartz, M. A., Burridge, K., Firtel, R. A., Ginsberg, M. H., Borisy, G., Parsons, J. T., and Horwitz, A. R. (2003). Cell migration: integrating signals from front to back. Science 302, 1704-1709. [DOI] [PubMed] [Google Scholar]
- Seals, D. F., and Courtneidge, S. A. (2003). The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 17, 7-30. [DOI] [PubMed] [Google Scholar]
- Smith, K. M., Gaultier, A., Cousin, H., Alfandari, D., White, J. M., and DeSimone, D. W. (2002). The cysteine-rich domain regulates ADAM protease function in vivo. J. Cell Biol. 159, 893-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan, L. R., and Condic, M. L. (2003). Neural crest motility and integrin regulation are distinct in cranial and trunk populations. Dev. Biol. 259, 288-302. [DOI] [PubMed] [Google Scholar]
- Strachan, L. R., and Condic, M. L. (2004). Cranial neural crest recycle surface integrins in a substratum-dependent manner to promote rapid motility. J. Cell Biol. 167, 545-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomczuk, M. (2004). ADAMs in early mouse development, University of Virginia, Charlottesville.
- Tomczuk, M., et al. (2003). Role of multiple beta1 integrins in cell adhesion to the disintegrin domains of ADAMs 2 and 3. Exp. Cell Res. 290, 68-81. [DOI] [PubMed] [Google Scholar]
- White, J. M. (2003). ADAMs: modulators of cell-cell and cell-matrix interactions. Curr. Opin. Cell Biol. 15, 598-606. [DOI] [PubMed] [Google Scholar]
- White, J. M., Bridges, L. C., DeSimone, D. W., Tomczuk, M., and Wolfsberg, T. G. (2005). Introduction to the ADAM family. In: The ADAM Family of Proteases: Proteases in Biology and Disease, Vol. 4, ed. N. M. Hooper and V. Lendeckel, Dordrecht, The Netherlands: Springer, 1-28. [Google Scholar]
- Woods, A., and Couchman, J. R. (2000). Integrin modulation by lateral association. J. Biol. Chem. 275, 24233-24236. [DOI] [PubMed] [Google Scholar]
- Zhang, X. P., Kamata, T., Yokoyama, K., Puzon-McLaughlin, W., and Takada, Y. (1998). Specific interaction of the recombinant disintegrin-like domain of MDC-15 (metargidin, ADAM-15) with integrin alphavbeta3. J. Biol. Chem. 273, 7345-7350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.