Abstract
The poly(A)-limiting element (PLE) is a conserved sequence that restricts the length of the poly(A) tail to <20 nt. This study compared the translation of PLE-containing short poly(A) mRNA expressed in cells with translation in vitro of mRNAs with varying length poly(A) tails. In transfected cells, PLE-containing mRNA had a <20-nt poly(A) and accumulated to a level 20% higher than a matching control without a PLE. It was translated as well as the matching control mRNA with long poly(A) and showed equivalent binding to polysomes. Translation in a HeLa cell cytoplasmic extract was used to examine the impact of the PLE in the context of varying length poly(A) tails. Here the overall translation of +PLE mRNA was less than control mRNA with the same length poly(A), and the PLE did not overcome the effect of a short poly(A) tail. Because poly(A)-binding protein (PABP) is a dominant effector of poly(A)-dependent translation we reasoned excess PABP in our extract might overwhelm PLE regulation of translation. This was confirmed by experiments where PABP was inactivated with poly(rA) or Paip2, and the effect of both treatments was reversed by addition of recombinant PABP. These data indicate that the PLE functionally substitutes for bound PABP to stimulate translation of short poly(A) mRNA.
Keywords: poly(A)-limiting element, poly(A), translation, translational control, poly(A)-binding protein
INTRODUCTION
The translation of most mRNAs is regulated at initiation (Arava et al. 2003), which in turn depends on two ubiquitous features: the 5′ cap and the 3′ poly(A) tail (Raught et al. 2000; Mangus et al. 2003). The current view of translation initiation is based on the circular polysome model (Sachs 2000), in which eIF4G brings together the 5′ cap bound by eIF4E and the 3′ poly(A) tail bound by poly(A)-binding protein (PABP). Initiation commences with the binding of eIF3 and the met-tRNA-bound 43S ribosome subunit to this complex. Most vertebrate mRNAs exit the nucleus with a poly(A) tail of ~200 residues (Wahle and Keller 1996), and this undergoes progressive shortening in the cytoplasm until it reaches a limit of 10–15 residues (Tharun and Parker 1997), following which it is no longer associated with eIF4E or PABP (Tharun and Parker 2001) and instead becomes associated with Dcp1p, Dcp2p, and the cytoplasmic Lsm1p-7p proteins in discrete sites of mRNA degradation termed processing bodies, or P-bodies (Tharun and Parker 2001; Sheth and Parker 2003).
The minimum length poly(A) bound by PABP is 12 residues, and on longer tracts PABP binds approximately every 25 residues (Sachs 1987). The notion that poly(A) is a length-dependent enhancer of translation is supported by experiments performed in vitro using cell extracts (Preiss et al. 1998; Bergamini et al. 2000; Raught et al. 2000) and in electroporated cells (Gallie 1991), where, compared to A0, A50 increased luciferase translation 11-fold in tobacco protoplasts and 156-fold in CHO cells. A similar stimulation of cap-dependent translation was observed for PABP–MS2 fusion proteins binding to reporter mRNAs in which poly(A) was replaced with multiple MS2 binding sites (Gray et al. 2000). The ability of tethered domains of PABP to stimulate translation initiation suggests that poly(A) length-dependent enhancement of translation results from the increased availability of PABP bound to the mRNA 3′ end. More recent work with PABP-depleted extracts proved that PABP is an initiation factor that works by enhancing the formation of both the 48S initiation complex and 60S subunit joining (Kahvejian et al. 2005).
While the results described above appear to be generally applicable to most polyadenylated mRNAs, our laboratory identified an exception to this. Serum albumin is the most abundant protein produced by Xenopus liver, and cytoplasmic albumin mRNA sediments almost exclusively with heavy polysome fractions (Cunningham et al. 2001; Yang and Schoenberg 2004), a characteristic of efficiently translated mRNAs. Nevertheless, this mRNA has a remarkably short, discrete 17-nt poly(A) tail (Schoenberg et al. 1989), which is present both on the mature mRNA and nuclear pre-mRNA (Rao et al. 1996). Further, our laboratory (Gu et al. 1999) and others (Choi and Hagedorn 2003) found that this short poly(A) phenotype is shared by a number of mRNAs. Albumin pre-mRNA contains two functionally redundant poly(A)-limiting elements (PLEs) in the last exon that restrict the length of the poly(A) tail to <20 nt even in the context of a strong synthetic polyadenylation element polyadenylation element (SPA) (Das Gupta et al. 1998).
Given the link between poly(A) tail length, translation, and translocation of deadenylated mRNAs to P-bodies for decay, one might anticipate that PLE-containing mRNAs would be unstable. However, we recently showed that β-globin mRNA with a PLE is as stable as control mRNA with a long poly(A) tail (Peng et al. 2005). The PLE also increases the efficiency of pre-mRNA 3′ processing, resulting in greater accumulation of this mRNA with a <20-nt poly(A) tail. The stability of PLE-containing mRNA and the association of albumin mRNA with polysomes suggested that these mRNAs are efficiently translated in spite of their short poly(A) tails. Since this seemed to contradict the generally accepted view of poly(A) and translation we looked directly at the impact of the PLE on translation. In transfected cells, ~20% more luciferase mRNA and enzyme activity was observed for +PLE mRNA compared to a control mRNA that lacked the element. The majority of both control and +PLE mRNA were bound by two or more ribosomes; however, more of the control mRNA with long poly(A) sedimented with heavy polysomes. Enhanced translation of PLE-containing mRNA with short poly(A) mRNA was not seen in vitro until extracts were depleted of PABP by addition of excess poly(rA) or Paip2. This effect was overcome by addition of recombinant PABP, indicating that excess PABP in the extracts overwhelmed the action of the PLE as a translational enhancer. We discuss this phenomenon in light of new concepts for regulating translation.
RESULTS
PLE-containing mRNA is efficiently translated in vivo
Previous work characterizing the function of the PLE (Das Gupta et al. 1998, 2001; Gu et al. 1999; Gu and Schoenberg 2003) used a human β-globin reporter gene driven by the CMV promoter in which the 3′ UTR was replaced with a multiple cloning site for the PLE and a strong synthetic polyadenylation element (SPA) (Levitt et al. 1989). For the current study the intron-containing β-globin gene in those constructs was replaced with an intronless firefly (Photinus pyralis) luciferase gene. We previously showed that PLE regulation of poly(A) tail length was unaffected by the absence of either upstream intron in the β-globin reporter genes (Das Gupta et al. 2001). The results in Figure 1 show that the PLE also restricted the length of the poly(A) tail to <20 nt on intronless luciferase mRNA. In this experiment cells were transfected with plasmids that lacked (control) or contained a PLE (+PLE) upstream of the polyadenylation site, and the length of the poly(A) tail on luciferase mRNA was determined using the RT-PCR-based LM-PAT assay developed by Salles et al. (1999). As seen previously (Das Gupta et al. 1998, 2001), the <20-nt poly(A) tail on PLE-containing mRNA appeared as a discrete group of bands. Interestingly, the long poly(A) tail on control mRNA showed a periodicity of sizes corresponding to the 20–25-nt packing density of poly(A)-binding protein on poly(A) (Sachs 1987).
FIGURE 1.
The PLE imparts a short poly(A) tail on luciferase mRNA. The sequence corresponding to PLE B (shown above the figure) was inserted downstream of the firefly luciferase gene, 12 bp upstream of a minimal synthetic polyadenylation element (SPA; Das Gupta et al. 1998) in a plasmid with transcription controlled by the CMV promoter. Poly(A) tail length of mRNA expressed in transiently transfected cells was determined by PAT assay (Salles et al. 1999) using an oligo(dT) primer-adapter to prime reverse transcription and a 5′ [32P]-labeled upstream primer for luciferase. The amplified products were separated on a denaturing 6% polyacrylamide/urea gel and visualized by PhosphorImager analysis. The lane marked M contains size markers of [32P]-labeled HinfI digested φX174 DNA.
Recent work from our laboratory found that the PLE also enhances the rate of pre-mRNA 3′ processing, resulting in a 50% increase in the accumulation of a β-globin reporter mRNA (Peng et al. 2005). That report also shows that +PLE mRNA with a <20-nt poly(A) tail decays at the same rate as control mRNA with long poly(A). Although we did not examine the 3′ processing efficiency and mRNA decay in this study, adding a PLE increased the accumulation of intronless luciferase mRNA by 20% (Fig. 2A) and this was matched by a similar increase in luciferase expression (Fig. 2B). We conclude that +PLE luciferase mRNA with a <20-nt poly(A) is at least as stable and well translated as control mRNA with a long poly(A) tail.
FIGURE 2.
Impact of a PLE on mRNA and protein expression in vivo. Cells were transfected with firefly luciferase (Fluc) constructs without (control) and with (+PLE) a PLE together with a plasmid expressing Renilla luciferase (Rluc). (A) RNA extracted 24 h after transfection was analyzed by Northern blot using [32P]-labeled probes to each mRNA. Radioactivity was visualized by PhosphorImager analysis. (B) Luciferase activity was measured in cytoplasmic extracts 24 h after transfection. The data are presented as the ratio of firefly (Fluc) to Renilla (Rluc) activity and show the mean ± standard deviation for triplicate determinations. Shown are typical results from three independent experiments.
PLE-containing mRNA is efficiently recruited to polyribosomes
Translation is primarily regulated at initiation (Arava et al. 2003), and the efficiency of this process is generally determined by polysome profile analysis to assess the degree of ribosome loading onto mRNA. In the experiment in Figure 3, cytoplasmic extracts from cells transfected with the luciferase expression plasmids used in Figure 2 were separated on a 10%–45% linear sucrose density gradient, and RNA extracted from even-numbered fractions was analyzed by RNase protection assay. The overall pattern showed that 19% of the control mRNA with long poly(A) was on the heaviest polysomes compared to 10% of +PLE mRNA. This was mirrored by a greater amount of +PLE mRNA (14%) associated with 80S monosomes compared to 10% of control mRNA. Overall, 53% of +PLE mRNA sedimented with two or more bound ribosomes compared to 57% of control mRNA. The similarity between these data and those in Figure 2 indicate that the PLE must functionally substitute for long poly(A) in stimulating translation in vivo.
FIGURE 3.
Polysome profile analysis of PLE-containing mRNA. Cos-1 cells were transfected with firefly luciferase expression plasmids without (control) or with a PLE (+PLE). Post-nuclear extracts were applied to a linear 10%–45% sucrose gradient and centrifuged at 225,000g for 2 h at 4°C in the direction indicated by the arrow. (A) Fractions of 0.25 mL were collected from the bottom and the absorbance profile at 254 nm is shown. (B) The sedimentation of control and PLE-containing (+PLE) luciferase mRNA was determined by RNase protection assay of RNA recovered from even-numbered fractions, and radioactivity was determined by PhosphorImager analysis. (C) The radioactivity in each RPA fraction in B was normalized to the total counts to yield a percent distribution of luciferase mRNA across the gradient.
In vitro translation of PLE-containing mRNAs
To evaluate the relationship between the presence of a PLE and the length of the poly(A) tail on translation efficiency we prepared a series of plasmid vectors that transcribe luciferase ±PLE mRNA with varying length poly(A) tails (Table 1). These mRNAs either had a 5′ 7mGpppG cap, an inactive 5′ ApppG or no cap, no 3′ poly(A) or 3′ poly(A) up to 98 residues. The spatial relationship of the PLE with respect to the luciferase coding sequence and the poly(A) tail was the same as that for mRNAs expressed in vivo in Figures 1–3. Translation of these mRNAs was evaluated using a cap- and poly(A)-dependent in vitro translation system prepared from HeLa cytoplasmic extracts prepared and optimized as described by Thoma et al. (2004b). In each sample the amount of firefly luciferase produced was normalized to a capped Renilla luciferase mRNA control.
TABLE 1.
Capped and polyadenylated mRNAs
| Poly(A) length | ||||||
| luc mRNA | 5′ end | A0 | A14 | A20 | A54 | A98 |
| Control | ApppG | X | X | |||
| 7mGpppG | X | X | X | X | X | |
| +PLE | ApppG | X | X | |||
| 7mGpppG | X | X | X | X | X |
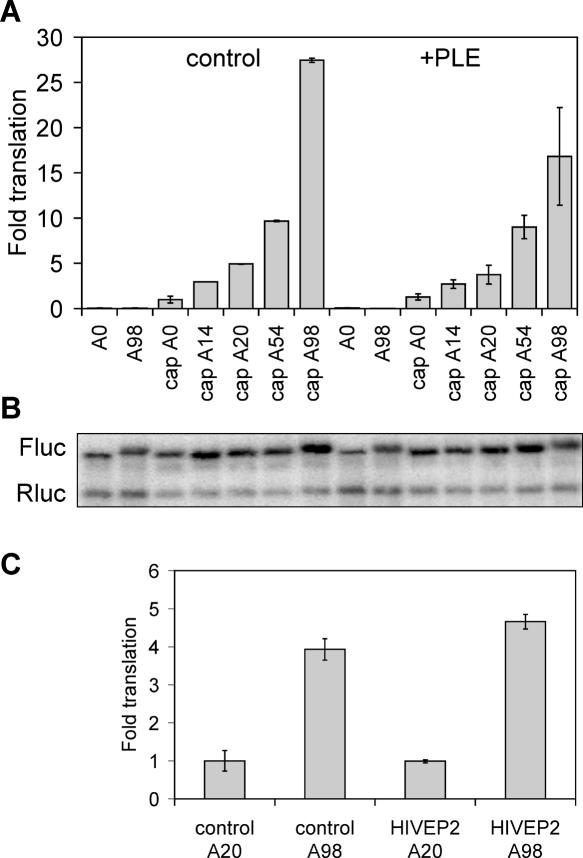
Luciferase activity was not observed for mRNA with 5′ ApppG regardless of the presence of a poly(A) tail (Fig. 4A, lanes 1 [A0] and 2 [A98]). In contrast, a small amount of translation was observed for A0 mRNA bearing a 5′ 7mGpppG cap (Fig. 4A, lane 3). In this and subsequent experiments, the normalized value for capped A0 mRNA was arbitrarily set to 1. Translation of capped A14 and A20 mRNAs were 2.5- and fivefold greater, respectively, than that of capped A0 mRNA, even though the length of each of these poly(A) tails is sufficient to bind only one molecule of PABP. Luciferase expression increased with increasing length poly(A), with an overall fivefold increase between A20 and A98, and a 25-fold increase comparing capped A0 to capped A98. These results are in good agreement with results obtained by others using a similar translation system (Bergamini et al. 2000), and they confirm the applicability of this system to study the impact of the PLE on translation.
FIGURE 4.
Impact of the PLE on translation in vitro. (A) The mRNAs shown in Table 1 were translated in a HeLa cytoplasmic extract for 1 h together with a control Renilla luciferase mRNA. A0 and A98 mRNA have 5′ ApppG, and mRNAs labeled “cap” have 5′ 7mGpppG. The fold translation shown in the upper panel represents firefly luciferase activity normalized to Renilla luciferase activity, and each bar consists of the mean ± SD for triplicate determinations. To facilitate comparison between the individual mRNAs, the value for capped A0 mRNA was arbitrarily set to 1. (B) RNA extracted from each pooled set of reactions was analyzed by Northern blot as in Figure 1 using a mixed probe for firefly (Fluc) and Renilla (Rluc) luciferase. (C) The PLE identified in HIV-EP2 mRNA (HIVEP2) was inserted into A20 or A98 firefly luciferase mRNA in place of PLE B from albumin mRNA. These capped mRNAs were translated in triplicate and compared to translation of control A20 and A98 mRNA together with Renilla luciferase control. In this experiment the translation of capped A20 mRNA normalized to translation of Renilla luciferase mRNA was arbitrarily set to 1 and results with the other mRNAs were normalized to this. The bars represent the mean ± SD for triplicate determinations.
Based on in vivo results in Figures 1–3 we anticipated that capped +PLE mRNA with A14 or A20 would be translated as efficiently in vitro as control mRNA with A98. However, the translation of PLE-containing mRNAs with A0, A14, A20, and A54 was essentially the same as that of the matching controls. RNA recovered from each translation reaction was analyzed by Northern blot to determine whether the lower than expected translation seen in Figure 4A was due to deadenylation or differential degradation of mRNAs during the incubation period (Fig. 4B). Significantly less firefly luciferase mRNA was only seen with A0 mRNA with 5′ ApppG, a result that is consistent with previous observations on mRNA stability in cytoplasmic extracts (Bergman et al. 2004). A comparison of the electrophoretic mobility of each of the polyadenylated mRNAs with that of A0 mRNA showed no evidence for significant deadenylation during the reaction. Thus, the inability of the PLE to enhance the in vitro translation of short poly(A) mRNA was not due to enhanced degradation or deadenylation.
The PLE used here came from Xenopus albumin mRNA. To rule out the possibility that the lack of PLE stimulation in vitro was due to a species difference in the source of the element, we examined translation of luciferase bearing the PLE from human HIV-EP2 mRNA (Gu et al. 1999). The choice of PLE made no difference in translation efficiency (Fig. 4C); the same degree of translation was seen in A20 mRNA regardless of the presence of the element, and lengthening poly(A) to A98 had a similar effect on the translation for each mRNA.
Translational activity of PLE-containing mRNA transfected directly into cells
In the experiment in Figure 5 the mRNAs listed in Table 1 were transfected directly into cells together with a capped, polyadenylated Renilla luciferase control mRNA. Much like the results observed in vitro this yielded little translation from uncapped A0 or A98 mRNAs (Fig. 5A). Compared to capped A0 mRNA, A14 and A20 stimulated the translation of capped control mRNA sixfold, translation increased another twofold when poly(A) was increased to A54, and there was an overall 17-fold increase in luciferase expression between control A0 and A98. Although the fold increase in translation observed here was somewhat lower, these data are in good agreement with those seen in Figure 4A and confirm previous work showing poly(A) stimulates translation of capped mRNAs in transfected cells (Gallie 1991).
FIGURE 5.
Impact of the PLE on translation of mRNA transfected into cultured cells. (A) mRNAs shown in Table 1 were transfected together with Renilla luciferase mRNA into LM(tk–) cells. Extracts prepared 1 h after transfection were analyzed for luciferase activity as in Figure 4, with the results for capped A0 control mRNA arbitrarily set to 1. The bars represent the mean ± SD for triplicate determinations. (B) RNA extracted from RNA-transfected cells was analyzed as in Figure 1 by Northern blot using a mixed probe for firefly (Fluc) and Renilla (Rluc) luciferase mRNA.
As observed in vitro, adding a PLE to A14 or A20 mRNA did not increase luciferase expression beyond that seen for the matching controls, and a similar increase in translation was seen with increasing poly(A) tail length to A54 and A98. With the exception of uncapped +PLE A98 mRNA equivalent amounts of each mRNA were recovered from transfected cells, indicating that the observed differences in luciferase expression did not result from differences in mRNA stability. Together with results obtained in vitro, these data indicate that with respect to the efficiency of initiating translation there is something fundamentally different between +PLE mRNA that has been transcribed and processed within a cell versus mRNA that has been transcribed in vitro.
Impact of inhibiting PABP on the translation of PLE-containing mRNA
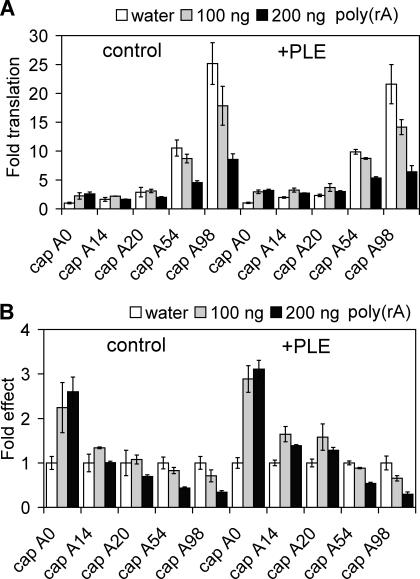
Cells contain an excess of PABP over poly(A) (Gorlach et al. 1994), and one possible explanation for our results was that this might overwhelm the function of some other PLE-binding factor in stimulating translation of short poly(A) mRNA. While it was not possible to address this in vivo, one can manipulate PABP function in vitro. This was first accomplished by adding excess poly(A) to the translation mixture to titrate poly(A)-binding proteins (Fig. 6A). In this experiment, capped ±PLE mRNAs with different length poly(A) tails were translated in HeLa cytoplasmic extract that was supplemented with 0, 100, or 200 ng of poly(rA). Results with control mRNA in the left panel of Figure 6A show that, as expected, the addition of poly(rA) inhibited the translation of polyadenylated mRNA in a concentration-dependent manner. Poly(rA) addition also inhibited the translation of polyadenylated +PLE mRNA (Fig. 6A, right panel); however, a different picture emerged when the relative amount of translation for each mRNA was normalized to translation in the absence of added poly(rA) (Fig. 6B). Here, both 100 and 200 ng of poly(rA) stimulated the translation of capped A0 control and PLE-containing luciferase mRNA two- to threefold. This trans stimulation of A0 mRNA translation by poly(rA) was observed previously in ribosome-depleted rabbit reticulocyte lysate (Borman et al. 2002), and it disappeared at poly(A) lengths ≥54 nt for both control and PLE-containing mRNA. More interesting was the effect of poly(rA) on translation of A14 and A20 mRNAs, where in three independent experiments, PLE-containing mRNA was translated 30%–50% more efficiently than control mRNA.
FIGURE 6.
Impact of poly(rA) competition for poly(A)-binding proteins on the translation of PLE-containing mRNAs. In vitro translation was performed as in Figure 4 without (white bars) or with addition of 100 ng (gray bars) or 200 ng (black bars) of poly(rA) to each reaction. (A) The activity from capped control A0 luciferase mRNA with no added poly(rA) was arbitrarily set to 1, and the values shown are the fold difference in translation for each of the indicated mRNAs in the absence or presence of poly(rA). The bars represent the mean ± SD for triplicate determinations. (B) To determine the impact of poly(rA) on the translation of each individual mRNA, the values obtained in A with added poly(rA) were normalized to the value obtained for each buffer control. For comparison the buffer control for each mRNA was set to 1. The difference between control A20 versus +PLE A20 mRNA was statistically significant, with p < 0.01 by Student’s T-test.
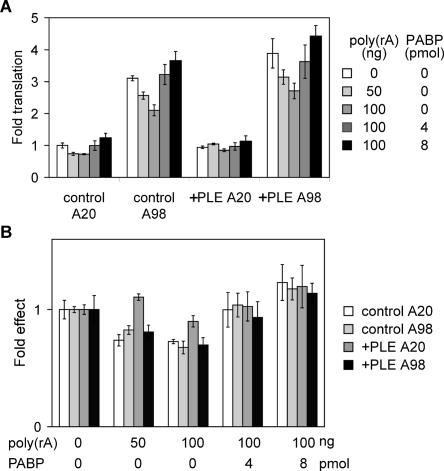
Next, recombinant PABP was added back to these reactions to confirm that poly(rA) acted by titrating PABP (Fig. 7A). In this experiment, capped A20 or A98 luciferase mRNAs without (control) or with a PLE (+PLE) were translated in reactions containing 0, 50, or 100 ng of poly(rA), plus 0, 4, or 8 pmol of bacterially expressed, His-tagged hPABP. Recombinant hPABP effectively overcame the inhibitory effect of poly(rA) on the translation of both control and +PLE A98 mRNA, thus confirming that poly(rA) was acting by titrating PABP. However, enhanced translation of +PLE A20 mRNA was evident when the results of each reaction with added poly(rA) ± hPABP were normalized to that of buffer alone (Fig. 7B). These data indicate that +PLE mRNA with a short poly(A) tail is translated more efficiently under conditions of limiting PABP, and this effect is reversed by adding recombinant hPABP to the mixture.
FIGURE 7.
Impact of PABP on poly(rA) inhibition of poly(A)-dependent translation. (A) Capped control and +PLE mRNAs with 20- (A20) and 98- (A98) nt poly(A) were translated as in Figure 6 in reactions containing 0, 50, or 100 ng of poly(rA). The reactions containing 100 ng of poly(rA) were also supplemented with 4 or 8 pmol of recombinant hPABP (dark gray and black bars, respectively). Each bar represents the mean ± SD for triplicate determinations, and similar results were obtained for three independent repeats of this experiment. (B) The fold effect of added PABP was determined as in Figure 6B by normalizing results for each mRNA to its corresponding buffer control. In each case the buffer control was arbitrarily set to 1. The difference between control A20 versus +PLE A20 mRNA was statistically significant, with p < 0.01 by Student’s T-test.
PABP is an essential translation initiation factor (Kahvejian et al. 2005), and the long poly(A) tail stimulates translation initiation by recruiting PABP. Paip2 is a selective repressor of PABP-dependent translation (Khaleghpour et al. 2001), and immobilized Paip2 can be used to deplete PABP from cytoplasmic extracts (Svitkin and Sonenberg 2004) without affecting the levels of eIF4E, eIF4A, and eIF4G (Svitkin and Sonenberg 2004). To explore further the relationship between PABP and the translation of +PLE mRNA we examined the impact of Paip2 on the translation of A20 and A98 mRNAs with or without a PLE (Fig. 8A). As observed for poly(rA) in Figure 7, Paip2 inhibited translation in a concentration-dependent manner, with the greatest effects observed for both control and +PLE A98 mRNAs. The addition of recombinant hPABP reversed the inhibitory effect of Paip2, returning the translation of each mRNA to that of the buffer control when both proteins were present in equal amounts. Preferential translation of +PLE A20 mRNA was again apparent when the results obtained for each mRNA in the presence of Paip2 ± hPABP were normalized to the respective buffer controls (Fig. 8B). Together the results in Figures 7 and 8 confirm that PABP in the translation extract overwhelmed the action of any trans-acting factor involved in PLE stimulation of translation initiation, and the latter became apparent only when PABP was inactivated. Ideally we would like to reduce PABP to a level that is sufficient to act synergistically with the PLE to stimulate translation of A20 mRNA as seen in vivo, but this has proven difficult to achieve.
FIGURE 8.
Impact of Paip2 inhibition of PABP on the translation of PLE-containing mRNAs. Capped control and +PLE mRNAs with 20-(A20) and 98- (A98) nt poly(A) were incubated on ice for 30 min with buffer (white bars) or 4 pmol or 8 pmol (light gray bars) of Gst-Paip2. Prior to the start of the reaction, 4 or 8 pmol of recombinant hPABP (dark gray and black bars, respectively) were added to mixtures containing 8 pmol of Gst-Paip2. Each bar represents the mean ± SD for triplicate determinations, and similar results were obtained for three independent repeats of this experiment. (B) The impact of Paip2 and added PABP on translation of each individual mRNA was determined as in Figure 7B by normalizing each data set to the values obtained for the buffer control, which for each mRNA was set to 1. The difference between control A20 versus +PLE A20 mRNA both with added Gst-Paip2 and Gst-Paip2+PABP was statistically significant, with p < 0.01 by Student’s T-test.
DISCUSSION
The long poly(A) tail on most vertebrate mRNAs acts as an enhancer of translation initiation by recruiting multiple copies of PABP to the 3′ end of the mRNA (Gray et al. 2000; Raught et al. 2000; Sachs 2000; Kahvejian et al. 2005). However, Xenopus serum albumin mRNA has a <20-nt poly(A) tail (Schoenberg et al. 1989; Rao et al. 1996) and is one of the most abundantly expressed liver proteins. We previously identified and characterized a poly(A)-limiting element (PLE) (Das Gupta et al. 1998) whose presence on this mRNA was responsible for imparting a short poly(A) tail. Database searches performed before completion of any metazoan genomes identified several PLE-containing mRNAs (Gu et al. 1999), and we have identified several more candidates. Another laboratory identified a number of mRNAs with short poly(A) when they compared mRNA recovered by binding to eIF4E versus selection on oligo(dT) (Choi and Hagedorn 2003); however, these do not have sequence elements related to the PLE, suggesting that the short poly(A) mRNA phenotype might be more widespread than previously thought. Note that we previously showed that the PLE regulates poly(A) tail length on nuclear pre-mRNA (Rao et al. 1996). A similar analysis was not performed for the mRNAs identified by Choi and Hagedorn (2003), so it is unclear whether the short poly(A) phenotype they observed was due to regulation of poly(A) addition or deadenylation.
The purpose of the current study was to determine whether the PLE in the context of a short poly(A) tail functionally substitutes for a long poly(A) tail in stimulating translation. To test this in vivo a PLE was inserted upstream of AAUAAA in a plasmid vector expressing firefly luciferase. Results in Figures 1 and 2 demonstrate that PLE-containing luciferase mRNA has a <20-nt poly(A) tail, that this mRNA was expressed at slightly greater levels than control, and that it was translated as efficiently as control mRNA with long poly(A). The polysome profile for PLE-containing luciferase mRNA was similar to that of control mRNA with a long poly(A) tail (Fig. 3), although the latter showed a higher proportion of mRNA in the heaviest polysome fractions. There is ample evidence that poly(A) acts as an enhancer of translation initiation by bringing PABP into the initiation complex (Munroe and Jacobson 1990; Tarun et al. 1997; Fraser et al. 1999; Gray et al. 2000; Mangus et al. 2003; Thoma et al. 2004a; Kahvejian et al. 2005). Therefore, the most reasonable conclusion for the similar translation of +PLE mRNA mRNA with a <20-nt poly(A) tail and control mRNA with a long poly(A) tail is that a cellular PLE-binding factor functionally substitutes for PABP.
This notion is supported by results obtained in vitro under conditions where PABP was inactivated by adding excess poly(rA) (Figs. 6, 7) or Paip2 (Fig. 8) to an in vitro translation system from exponentially growing HeLa cells that accurately recapitulates cap- and poly(A) length-dependent translation (Thoma et al. 2004b). In this system +PLE mRNA with ≤20-nt poly(A) showed the same translation as control mRNA with the same length poly(A) tails, and never approximated the degree of translation seen with a 98-nt poly(A) tail (A98). However, +PLE mRNA with 20-nt poly(A) showed better translation than A98 mRNA under conditions in which PABP was functionally inactivated by addition of excess poly(rA) or Gst-Paip2. Based on this we conclude that in intact cells a PLE-binding factor functionally substitutes for bound poly(A) in stimulating translation, and this is only poorly replicated in vitro or with mRNAs directly transfected into cells, most likely because of competition with PABP.
A combination of both conventional and affinity chromatography identified U2AF (Gu and Schoenberg 2003), La autoantigen, hnRNP A1 and SF3b (J. Peng, unpubl.) as proteins capable of binding the PLE. Although U2AF modulates PLE-directed control of poly(A) length (Gu and Schoenberg 2003), neither it nor any of the other proteins noted above has any demonstrable effect on the in vitro translation of A20 +PLE mRNA (data not shown). This leaves open the question of the identity of the trans-acting factor(s) responsible for PLE control of both poly(A) length and PLE stimulation of translation. One possibility is that the PLE is a target for regulation by one or more micro RNAs. A recent report (Lewis et al. 2005) identified thousands of mRNAs as potential miRNA targets, and a search of the miRNA registry identified substantial base-pairing for PLE A of albumin mRNA with human and mouse miR-338 and miR-325, albumin mRNA PLE B with human and mouse miR-17-3p, and the PLE from HIV-EP2 mRNA with human and mouse miR-136. While these pairings do not match the “seed” for nucleotides 2–7, they are nevertheless quite extensive, covering 10–14 nt. Involvement of one or more miRNAs would explain a number of previous observations. For example, the two PLEs in albumin mRNA are 21 and 23 nt, respectively, yet they share only limited sequence identity and act independently to limit poly(A) length to <20 nt (Das Gupta et al. 1998). It was difficult to see how this fit with binding by one or more proteins, and experiments are in progress to test whether the identified miRNAs are involved in PLE regulation of poly(A) tail length. Although miRNAs were first identified as translational repressors (Bartel 2004), to the best of our knowledge there are no data to indicate they cannot function as translational enhancers. The requirement for miRNAs to assemble onto mRNA in the context of an miRNP might also explain why PLE regulation of poly(A) tail length and translation are poorly replicated in vitro and in RNA-transfected cells, and why increased translation in vitro of +PLE mRNA with a 20-nt poly(A) tail is only observed under conditions where a competing translation factor (PABP) has been removed.
Finally, a recurring question is why cells would possess a mechanism like the PLE to restrict the length of the poly(A) tail during nuclear processing. Different cells possess different miRNAs, and the quantity of each can change. Perhaps the PLE (or some other PLE-like element; Choi and Hagedorn 2003) acts in concert with miRNAs to regulate translation by mimicking the effect of PABP (Kahvejian et al. 2005), enhancing translation in the context of a matching miRNA, and repressing translation in its absence.
MATERIALS AND METHODS
Plasmids constructs
The construction of the plasmids CMV-glo-SPA and CMV-glo-PLE B-SPA were described previously (Das Gupta et al. 1998, 2001). To prepare CMV-luc-SPA, CMV-glo-SPA was digested with Asp718 and end-filled with Klenow fragment of DNA polymerase followed by digestion with NcoI to remove the globin cassette, leaving intact the CMV promoter and SPA 3′ processing element. The luciferase gene was obtained by digesting pGtetOβAc.luc3 (generously provided by Jose Garcia-Sanz) with XbaI followed by end-filling using Klenow fragment of DNA polymerase and digestion with NcoI. The recovered luc gene was cloned into the CMV promoter-containing constructs prepared above. PLE B was inserted into CMV-luc-SPA through the XbaI site to generate CMV-luc-PLEB-SPA. Plasmid pcDNARluc was the ligation product of pcDNA3 and Renilla luciferase fragment from pRLTK (Promega) through HindIII and XbaI sites.
The plasmids used for generating luciferase transcripts with different poly(A) tail lengths in the in vitro translation experiments were constructed as following: LM(tk–) cells were transfected with CMV-glo-SPA, and the fragments containing part of β-globin exon3 with different poly(A) tail lengths were amplified by RT-PCR with primers XG1 (5′-GGCAACGTGCTGGTCTGTGT) and XG4 (5′-GGGGATCCGCGGT15), then cloned into the BamH1 site of pGEM3Z. DNA sequence analysis was used to identify clones with 14- and 54-residue poly(A) tails. Next, the entire β-globin exon 3 was amplified by PCR from plasmid CMV-glo-SPA with Joy18 (5′-CCTGGGCAACGTGCTGGTCT) and Joy26 (5′-CG GAAGCTTCTAATGAAAATAAAGATCT), then inserted into the A14 and A54 constructs made above through XbaI and HincII sites. Full-length β-globin cDNA was generated by RT-PCR with primers SP6gloC (5′-GAATACAAGCTAGCTTGCTT) and HG35 (5′-TC TTTGCCAAAGTGATGGGC) from RNA of LM(tk–) cells that were transfected with CMV-glo-SPA. This was inserted into the plasmids with A14 and A54 digested with PstI and BstXI that were treated with T4 DNA polymerase to remove the 3′ overhang of the PstI site. To create plasmids pGLuc-A14, A54 and pGLuc-PLEB-A14, A54, β-globin cDNA was replaced with an NcoI and BglII digest CMV-luc-SPA and CMV-luc-PLEB-SPA containing the luciferase gene. To construct pGluc-PLEB-A98, T3LucA98 (kindly provided by Nahum Sonenberg, McGill University) was digested with EcoRI, end-filled with Klenow fragment of DNA polymerase, then digested with BamHI. This was used to generate an A98 fragment that was used to replace A14 from pGLuc-PLEB-A14 that was removed by digestion with BglII followed by end-filling with Klenow fragment of DNA polymerase and digestion with BamHI. To generate pGLucA98 the PLE was removed by digestion with XbaI, followed by religation of the plasmid. To construct pGluc-A20 and pGluc-PLEB-A20, primers JP23 (5′-GCGGT20AATAAAGATCTTT TAT) and JP25 (5′-CT TTATTA20CCGCGGATC) were annealed and end-filled with Klenow fragment of DNA polymerase, the A20 fragment was then used to replace A14 from pGLuc-A14 and pGLuc-PLEB-A14 through BglII and SstII sites. Plasmid pGlucA78 was used to clone pGRLucA78 in which firefly luciferase was replaced with an NheI to Xba1 fragment containing Renilla luciferase.
Cell culture and transfection
Cos-1, Cos-7, and LM(tk–) cells were maintained in Dulbecco’s Minimal Essential Medium (DMEM) with 10% fetal bovine serum (FBS) and 2 mM L-glutamine. Hela S3 cells were grown in DMEM with 10% FBS and 4 mM L-glutamine. To grow Hela S3 cells in suspension, cells grown attached were trypsinized and seeded to a250-mL spinner flask at 5–8 × 104 cells/mL, which was used to inoculate a 1-L flask. For experiments using linear sucrose density gradients to evaluate polysome loading, 3.5 × 106 Cos-1 cells were seeded in 150-mm dishes the day before transfection. Cells were transfected with 10 μg plasmid DNA and 30 μL Fugene-6 (Roche) following the manufacturer’s instructions. In RNA transfection experiments, 1.6 × 104 LM(tk–) cells were seeded in 96-well plates the day before transfection; 160 ng (240 pmol) of each firefly luciferase transcript and 40 ng (120 pmol) GRLA78 transcript were used for transfection with 0.4 μL lipofectamine 2000 reagent (Invitrogen) following the manufacturer’s instructions. Cells were harvested 1 h after transfection, washed twice with 100 μL phosphate-buffered saline (PBS), and lysed in 100 μL 1× passive lysis buffer (Promega). Luciferase activities were measured by Promega Dual Luciferase Assay kit with 10 μL cell lysate. To extract RNA, 8 × 104 cells were seeded and transfected with 800 ng of each firefly luciferase transcript, 200 ng GRLA78 transcript, and 2 μL lipofectamine 2000 reagent. Total cell RNA was extracted with 500 μL Trizol reagent 1 h after transfection. One-tenth of the extracted RNA was used for Northern blot analysis.
Northern blot, RNase protection, and poly(A) length assay
RNA was separated on 1% agarose gel and transferred to nylon membrane. The probes were synthesized with an Invitrogen Random Primer kit. The template for firefly luciferase was a 491-bp PpuMI and XbaI fragment of CMV-luc-SPA and the template for Renilla luciferase was a 947-bp NheI and XbaI fragment of pRLTK. The probe used for firefly luciferase was a transcript corresponding to the first 156 nt of the coding region. For unknown reasons this probe yields a doublet RNase protection product. The experiments were performed using the Ambion RPAIII kit following the standard protocol from the manufacturer. Poly(A) tail length was assayed as described previously (Das Gupta et al. 1998) using the upstream primer BVLuc1 (5′-TCGACGCAAGAAAAATCAGAGAGAT).
Polysome profile analysis
Polysome profile analysis of transfected Cos-1 cells used the approach described by Davydova et al. (1997). Briefly, 3.5 × 106 cells on a 150-mm plate were washed two times with ice-cold PBS and scraped off in 1 mL PBS. The cell pellet was collected by centrifugation at 200g for 2 min and resuspended in 400 μL of lysis buffer that contained 20 mM HEPES/KOH (pH 7.5), 100 mM KCl, 10 mM MgCl2, 1 mM DTT, 0.25% (v/v) NP-40, 1 mM PMSF, 100 μg/mL cycloheximide, 100 units/mL RNaseOut (Invitrogen), and 25 μL/mL protease inhibitor cocktail (Sigma). The cells were lysed with five strokes of a syringe with a 25-gauge needle. Cells debris and nuclei were removed by centrifugation at 15,000g for 5 min at 4°C in a refrigerated microcentrifuge. This was applied to a linear 10–45% sucrose gradient and centrifuged at 225,000g for 2 h at 4°C in a Sorvall TH641 rotor. The linear gradients consisted of 12 mL 10–45% sucrose prepared in lysis buffer. Fractions of 0.25 mL were collected from the bottom of the gradient and RNA was extracted from each even-numbered fractions with 1 mL TRIzol reagent (Invitrogen).
Preparation of HeLa cytoplasmic extract
HeLa S3 cells were grown in 1-L spinner flasks until cell density reached 4–6 × 105 cells/mL, at which time they were harvested by centrifugation at 200g for 10 min at 4°C in a Sorvall HS-4 rotor. The cell pellet was washed three times with ice-cold PBS at 10× volume of the pellet and resuspended in a volume of hypotonic MC buffer equal to the volume of the pellet. MC buffer contained 10 mM HEPES-KOH (pH 7.4), 10 mM potassium acetate, 0.5 mM magnesium acetate, 5 mM DTT, 1 mM PMSF, and 25 μL/mL protease inhibitor cocktail (Sigma). Cells were swollen on ice for 5–10 min, then lysed using 25–30 strokes of a Dounce homogenizer (B pestle). The cell debris and nuclei were removed by centrifugation at 15,000g for 20 min at 4°C in an refrigerated microcentrifuge, and the supernatant HeLa cell cytoplasmic extract (referred to as S15) was frozen in aliquots at −80°C.
Preparation of transcripts for in vitro translation
7mGpppG capped luciferase transcripts with A14, A20, or A54 were prepared by linearizing plasmid DNA with SstII followed by in vitro transcription using the Ambion mMessage mMachine SP6 kit following the manufacturer’s instructions. Luciferase expression plasmids with A98 were linearized by digestion with BamHI followed by digestion with mung bean nuclease treated before transcription. A0 transcripts were prepared from A14 plasmids that were digested with BglII, which cleaves between the 3′ end and the poly(A) tail. To prepare RNA with 5′ ApppG instead of a 7mGpppG cap, transcription was performed with the Ambion SP6 MegaScript kit with 4 μL 20 mM ApppG (Ambion) and 0.4 μL of 75 mM GTP added to each reaction. For all RNAs transcription was performed overnight at 37°C, followed by treatment with DNase I to remove plasmid DNA and precipitation with LiCl.
In vitro translation
A typical 10-μL reaction contained 4 μl Hela S15 cytoplasmic extract, 2 μL of RNA (15 fmol/μL), 2 μL water, and 2 μL 5× translation buffer (30 mM HEPES-KOH at pH 7.4, 330 mM KCl, 5 mM MgCl2, 5 mM ATP, 0.25 mM GTP, 125 μM bovine liver tRNA, 50 μM amino acid mix [Promega], 21 mM β-mercaptoethanol, 50 mM creatine phosphate, 125 ng/μL creatine phosphokinase, 1.2 mM spermidine) and 4 units of RNaseOut (Invitrogen). Each translation reaction was preformed in triplicate. The reaction mixture was incubated at 37°C for 1 h, and 2 μL were then used to assay luciferase activity. The remaining reaction mixtures for each transcript were combined and 20 μL were used to extract RNA using 200 μL Trizol reagent. One-tenth of the recovered RNA was used for the Northern blot. In experiments using recombinant PABP and Paip2, the purified protein was dialyzed against MC buffer, and the amount indicated in each of the figures was added in a 2-μL volume to the reaction in place of water. Recombinant GST-Paip2 (kindly provided by Nahum Sonenberg) was expressed in Escherichia coli BL21(DE3)pLysS strain and purified using Amersham’s GSTrap FF column according to the manufacturer’s instruction. Recombinant human PABP (hPABP-(His)6), also obtained from Nahum Sonenberg, was expressed in the same E. coli strain, and purified on a HiTrap chelating column charged with NiCl2. Protein was eluted with a gradient of 50–500 mM imadazole with individual fractions analyzed by SDS-PAGE for hPABP recovery. In the poly(A) competition experiment, 2 μL poly(A) potassium salt (Sigma) with the amount indicated in each figure was added to the reaction to replace water.
Acknowledgments
We thank Yuri Svitkin and Nahum Sonenberg for plasmids expressing Gst-Paip2, recombinant hPABP, and luciferase mRNA with 98-residue poly(A) tail. We also thank Nahum Sonenberg and members of the Schoenberg laboratory for their helpful comments. This work was supported by PHS grants R01 GM55407 and R01 GM38277 to D.R.S., and support for core facilities was provided by PHS grant P30 CA16058 to the Ohio State University Comprehensive Cancer Center.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2470905.
REFERENCES
- Arava, Y., Wang, Y., Storey, J.D., Liu, C.L., Brown, P.O., and Herschlag, D. 2003. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 100: 3889–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bergamini, G., Preiss, T., and Hentze, M.W. 2000. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 6: 1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman, N., Milone, J., Bates, E.J., Opyrchal, M., Bellofatto, V., and Wilusz, J. 2004. Analysis of RNA exonucleolytic activities in cellular extracts. In mRNA processing and metabolism; Methods and protocols (ed. D.R. Schoenberg), pp. 193–211. Humana Press, Totowa, NJ.
- Borman, A.M., Michel, Y.M., Malnou, C.E., and Kean, K.M. 2002. Free poly(A) stimulates capped mRNA translation in vitro through the eIF4G-poly(A)-binding protein interaction. J. Biol. Chem. 277: 36818–36824. [DOI] [PubMed] [Google Scholar]
- Choi, Y.H. and Hagedorn, C.H. 2003. Purifying mRNAs with a high-affinity eIF4E mutant identifies the short 3′ poly(A) end phenotype. Proc. Natl. Acad. Sci. 100: 7033–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, K.S., Hanson, M.N., and Schoenberg, D.R. 2001. Polysomal ribonuclease 1 exists in a latent form on polysomes prior to estrogen activation of mRNA decay. Nucleic Acids Res. 29: 1156–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Gupta, J., Gu, H., Chernokalskaya, E., Gao, X., and Schoenberg, D.R. 1998. Identification of two cis-acting elements that independently regulate the length of poly(A) on Xenopus albumin pre-mRNA. RNA 4: 766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Gupta, J., Gu, H., and Schoenberg, D.R. 2001. Position and sequence requirements for poly(A) length regulation by the poly(A) limiting element. RNA 7: 1034–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydova, E.K., Evdokimova, V.M., Ovchinnikov, L.P., and Hershey, J.W. 1997. Overexpression in COS cells of p50, the major core protein associated with mRNA, results in translation inhibition. Nucleic Acids Res. 25: 2911–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, C.S., Pain, V.M., and Morley, S.J. 1999. The association of initiation factor 4F with poly(A)-binding protein is enhanced in serum-stimulated Xenopus kidney cells. J. Biol. Chem. 274: 196–204. [DOI] [PubMed] [Google Scholar]
- Gallie, D.R. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes & Dev. 5: 2108–2116. [DOI] [PubMed] [Google Scholar]
- Gorlach, M., Burd, C.G., and Dreyfuss, G. 1994. The mRNA poly(A)-binding protein—Localization, abundance, and RNA-binding specificity. Exp. Cell Res. 211: 400–407. [DOI] [PubMed] [Google Scholar]
- Gray, N.K., Coller, J.M., Dickson, K.S., and Wickens, M. 2000. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 19: 4723–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, H. and Schoenberg, D.R. 2003. U2AF modulates poly(A) length control by the poly(A)-limiting element. Nucleic Acids Res. 31: 6264–6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, H., Das Gupta, J., and Schoenberg, D.R. 1999. The poly(A)-limiting element is a conserved cis-acting sequence that regulates the length of poly(A) on nuclear pre-mRNAs. Proc. Natl. Acad. Sci. 96: 8943–8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian, A., Svitkin, Y.V., Sukarieh, R., M’Boutchou, M.N., and Sonenberg, N. 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes & Dev. 19: 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour, K., Svitkin, Y.V., Craig, A.W., Demaria, C.T., Deo, R.C., Burley, S.K., and Sonenberg, N. 2001. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell 7: 205–216. [DOI] [PubMed] [Google Scholar]
- Levitt, N., Briggs, D., Gil, A., and Proudfoot, N.J. 1989. Definition of an efficient synthetic poly(A) site. Genes & Dev. 3: 1019–1025. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Burge, C.B., and Bartel, D.P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Mangus, D.A., Evans, M.C., and Jacobson, A. 2003. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4: 223.1–223.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe, D. and Jacobson, A. 1990. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol. Cell. Biol. 10: 3441–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., Murray, E.L., and Schoenberg, D.R. 2005. The poly(A)-limiting element enhances mRNA accumulation by increasing the efficiency of pre-mRNA 3′ processing. RNA 11: 958–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss, T., Muckenthaler, M., and Hentze, M.W. 1998. Poly(A)-tail-promoted translation in yeast: Implications for translational control. RNA 4: 1321–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, M.N., Chernokalskaya, E., and Schoenberg, D.R. 1996. Regulated nuclear polyadenylation of Xenopus albumin pre-mRNA. Nucleic Acids Res. 24: 4078–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught, B., Gingras, A.C., and Sonenberg, N. 2000. Regulation of ribosomal recruitment in eukaryotes. In Translational control of gene expression (eds. N. Sonenberg et al.), pp. 245–293. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sachs, A.B. 1987. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 7: 3268–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. Physical and functional interactions between the mRNA cap structure and the poly(A) tail. In Translational control of gene expression (eds. N. Sonenberg et al.), pp. 447–465. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Salles, F.J., Richards, W.G., and Strickland, S. 1999. Assaying the polyadenylation state of mRNAs. Methods 17: 38–45. [DOI] [PubMed] [Google Scholar]
- Schoenberg, D.R., Moskaitis, J.E., Smith Jr., J.H., and Pastori, R.L. 1989. Extranuclear estrogen-regulated destabilization of Xenopus laevis serum albumin mRNA. Mol. Endocrinol. 3: 805–814. [DOI] [PubMed] [Google Scholar]
- Sheth, U. and Parker, R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin, Y. and Sonenberg, N. 2004. An efficient system for cap- and poly(A)-dependent translation in vitro. In mRNA processing and metabolism; Methods and protocols (ed. D.R. Schoenberg), pp. 155–170. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Tarun Jr., S.Z., Wells, S.E., Deardorff, J.A., and Sachs, A.B. 1997. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl. Acad. Sci. 94: 9046–9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S. and Parker, R. 1997. Mechanisms of mRNA turnover in eukaryotic cells. In mRNA metabolism and post-transcriptional gene regulation (eds. J. Harford and D.R. Morris), pp. 181–200. Wiley, New York.
- ———. 2001. Targeting an mRNA for decapping: Displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell 8: 1075–1083. [DOI] [PubMed] [Google Scholar]
- Thoma, C., Bergamini, G., Galy, B., Hundsdoerfer, P., and Hentze, M.W. 2004a. Enhancement of IRES-mediated translation of the c-myc and BiP mRNAs by the poly(A) tail is independent of intact eIF4G and PABP. Mol. Cell 15: 925–935. [DOI] [PubMed] [Google Scholar]
- Thoma, C., Ostareck-Lederer, A., and Hentze, M.W. 2004b. A poly(A) tail-responsive in vitro system for cap- or IRES-driven translation from HeLa cells. In mRNA processing and metabolism; Methods and protocols (ed. D.R. Schoenberg), pp. 171–180. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Wahle, E. and Keller, W. 1996. The biochemistry of polyadenylation. Trends Biochem. Sci. 21: 247–250. [PubMed] [Google Scholar]
- Yang, F. and Schoenberg, D.R. 2004. Endonuclease-mediated mRNA decay involves the selective targeting of PMR1 to polyribosome-bound substrate mRNA. Mol. Cell 14: 435–445. [DOI] [PubMed] [Google Scholar]