Summary
Proteins with bacterial immunoglobulin-like (Big) domains, such as the Yersinia pseudotuberculosis invasin and Escherichia coli intimin, are surface-expressed proteins that mediate host mammalian cell invasion or attachment. Here, we report the identification and characterization of a new family of Big domain proteins, referred to as Lig (leptospiral Ig-like) proteins, in pathogenic Leptospira. Screening of L. interrogans and L. kirschneri expression libraries with sera from leptospirosis patients identified 13 lambda phage clones that encode tandem repeats of the 90 amino acid Big domain. Two lig genes, designated ligA and ligB, and one pseudo-gene, ligC, were identified. The ligA and ligB genes encode amino-terminal lipoprotein signal peptides followed by 10 or 11 Big domain repeats and, in the case of ligB, a unique carboxy-terminal non-repeat domain. The organization of ligC is similar to that of ligB but contains mutations that disrupt the reading frame. The lig sequences are present in pathogenic but not saprophytic Leptospira species. LigA and LigB are expressed by a variety of virulent leptospiral strains. Loss of Lig protein and RNA transcript expression is correlated with the observed loss of virulence during culture attenuation of pathogenic strains. High-pressure freeze substitution followed by immunocytochemical electron microscopy confirmed that the Lig proteins were localized to the bacterial surface. Immunoblot studies with patient sera found that the Lig proteins are a major antigen recognized during the acute host infection. These observations demonstrate that the Lig proteins are a newly identified surface protein of pathogenic Leptospira, which by analogy to other bacterial immunoglobulin superfamily virulence factors, may play a role in host cell attachment and invasion during leptospiral pathogenesis.
Introduction
Leptospirosis is the most widespread zoonosis in the world and has emerged as an important public health problem in large urban centres of developing countries (Ko et al., 1999; Levett, 2001). Its severe disease form, known as Weil’s syndrome, is an acute febrile illness associated with multiorgan system complications including jaundice, renal failure, meningitis and pulmonary haemorrhage, with a mortality rate that may exceed 15% (Faine et al., 1999; Marotto et al., 1999). Leptospirosis is caused by spirochetes belonging to the genus Leptospira, which includes pathogenic and saprophytic species. Pathogenic Leptospira are highly motile and invasive organisms (Merien et al., 1997; Barocchi et al., 2002) which rapidly disseminate to target organs after entering the host, usually through abrasions in the skin or mucous membranes (Arean, 1962; Faine et al., 1999). These organisms may be cleared by the humoral immune response, but in carriers they have the ability to colonize and persist in the kidney tubules. The rat (Rattus norvegicus) is the principal reservoir associated with urban epidemics of leptospirosis (Ko et al., 1999). Urine excreted by infected animals contains viable Leptospira, which can survive for days to weeks in soil or water (Chang et al., 1948; Hellstrom and Marshall, 1978). Flooding following heavy rainfall is frequently associated with large outbreaks of leptospirosis (Trevejo et al., 1998; Ko et al., 1999).
To date, few leptospiral factors that contribute to the pathogenesis of disease have been identified. The periplasmic endoflagella involved in propeller movement of the bacterium is an important factor in tissue penetration and motility through highly viscous fluids (Greenberg and Canale-Parola, 1977; Trueba et al., 1992). Moreover, chemotaxis for haemoglobin has been demonstrated, which indicates that Leptospira may be attracted to abraded skin surfaces (Yuri et al., 1993). Many pathogenic Leptospira secrete sphingomyelinase C (SphA) and pore-forming haemolysins (SphH), possibly associated with the haemolytic anaemia observed in leptospirosis patients (Trowbridge et al., 1981; Segers et al., 1992; Lee et al., 2002). Leptospiral lipopolysaccharide (LPS) is a major outer membrane component recognized by Toll-like receptor 2 (TLR2) on macrophages (Werts et al., 2001). The acquired immune response to the carbohydrate component of leptospiral LPS confers serovar-specific immunity and underlies the serological classification of the genus Leptospira into >200 serovars (Jost et al., 1989; de la Peña-Moctezuma et al., 2001).
The ability of pathogenic leptospires to penetrate, disseminate and persist in mammalian host tissues appears to rely on the ability of these organisms to attach to eukaryotic cells and extracellular matrix proteins (Tsuchimoto et al., 1984; Vinh et al., 1984; Ballard et al., 1986; Thomas and Higbie, 1990). The variety of host cells recognized by Leptospira suggests the presence of several adhesins. However, the only putative leptospiral adhesin identified to date is a virulence-associated leptospiral surface protein that binds purified fibronectin (Merien et al., 2000). In a recent report, pathogenic leptospires were distinguished from saprophytic organisms by their ability to rapidly translocate through a polarized MDCK monolayer without disrupting tight junctions (Barocchi et al., 2002). Rapid translocation across tissue barriers may be a mechanism used by this pathogen to invade host organs.
Surface-exposed moieties or structures are presumably the factors that mediate adherence of the leptospiral pathogen to mammalian host cells. Like other spirochetes, leptospires have a large repertoire of lipoproteins, a subgroup of which may be involved in mediating host cell interactions, as in the case of Borrelia lipoproteins (Haake, 2000). A leptospiral integral membrane protein, OmpL1, lipoproteins, LipL41 and LipL32, and a peripheral membrane protein P31LipL45 have been identified, some of which have been shown to be surface-exposed and expressed during host infection (Haake et al., 1993; 2000; Shang et al., 1996; Barnett et al., 1999; Matsunaga et al., 2002). More recently, a L. interrogans protein, LigA, was identified which has tandem repeats of the bacterial immunoglobulin-like (Big) domain (Palaniappan et al., 2002). This repeat motif is found in bacterial virulence factors that mediate mammalian host cell adherence and invasion, such as Escherichia coli intimin (Luo et al., 2000) and Yersinia pseudotuberculosis invasin (Hamburger et al., 1999). However, the sequence of LigA consists entirely of tandem Big repeats and does not have a C-terminal non-repeat domain that, by analogy to the structure of intimin and invasin, would serve as the binding domain to host cell-associated receptors. In addition, the surface expression of LigA and its association with virulence were not characterized (Palaniappan et al., 2002).
Analysis of leptospiral antigens targeted by the humoral immune response to naturally acquired infection is an effective approach to identify proteins expressed during infection. Antibodies to the infection-associated proteins OmpL1, LipL41, LipL32, P31LipL45 (Qlp42), and LigA are generated during human or animal infection (Flannery et al., 2001; Guerreiro et al., 2001; Nally et al., 2001; Palaniappan et al., 2002). In this report, we used sera from leptospirosis patients as a tool to identify DNA sequences from Leptospira genomic libraries which encode for proteins expressed during host infection. This approach allowed us to determine that there are in fact two intact leptospiral genes, ligA and ligB, and one disrupted gene, ligC, that encode domains belonging to the bacterial Ig-like superfamily. The ligB and ligC genes encode a C-terminal non-repeat domain following the tandem Big domain repeats, an organization which is similar to that of intimin and invasin. Furthermore, we found that LigA and LigB were surface-exposed lipoproteins whose expression correlated with the virulence of Leptospira strains. Together, these findings indicate that these newly identified surface lipoproteins may play an analogous role to intimin and invasin in mediating host cell interactions during leptospiral pathogenesis.
Results
Identification of leptospiral ligA, ligB and ligC genes
Pooled sera from leptospirosis patients were used to identify DNA sequences that encode host-expressed leptospiral proteins from λ genomic expression libraries of two virulent strains, L. kirschneri serovar grippotyphosa, strain RM52 and L. interrogans serovar copenhageni, strain Fiocruz L1-130. Antibody screening yielded 98 and 27 reactive clones from L. kirschneri and L. interrogans libraries, respectively. Triage of λ clones with antisera against characterized leptospiral proteins and sequencing of DNA inserts identified 70 clones with inserts encoding heat shock proteins GroEL and DnaK (Ballard et al., 1993), three clones containing the gene encoding the lipoprotein LipL41 (Shang et al., 1996), one clone with an insert encoding the putative inner membrane lipoprotein LipL31 (Haake and Matsunaga, 2002), and one encoding a putative 28 kDa lipoprotein. Thirteen clones had inserts that encoded imperfect tandem repeat sequences. A query of the repeat sequences against the Pfam 7.7b database (Bateman et al., 2002) identified each of these 90 residue repeats as Big2 (PF02368), a family of bacterial immunoglobulin-like (Big) domains. In addition, the cloned insert sequences encoded polypeptides that were homologous to the recently described LigA protein from L. interrogans serovar pomona type kennewicki (Palaniappan et al., 2002).
Assembly of overlapping cloned sequences identified two novel leptospiral big genes, designated ligB and ligC, distinct from ligA. Of the recombinant clones, two L. kirschneri and three L. interrogans clones encoded LigA, six L. kirschneri clones encoded LigC, whereas one L. interrogans clone encoded LigB. Because none of the cloned sequences contained a complete open reading frame, inverse PCR of flanking DNA was performed to obtain the full-length nucleotide sequence. Primers derived from the L. interrogans ligB and L. kirschneri ligC sequences allowed for PCR amplification of the corresponding genes in L. kirschneri and L. interrogans respectively.
Sequence features of the ligA, ligB, and ligC genes
From the assembled sequences, ligA (3675 bp in both L. interrogans and L. kirschneri) and ligB (5673 bp in L. interrogans and 5661 bp in L. kirschneri) have open reading frames predicted to encode polypeptides of 128 and 212 kDa respectively. The ligC gene (5871 bp in L. interrogans and 5865 bp in L. kirschneri) appears to be a pseudogene in both L. interrogans and L. kirschneri. In L. interrogans, the ligC open reading frame is interrupted at codon 499 by a TAA stop codon. Alignment of the L. interrogans and L. kirschneri ligC nucleotide sequences reveals an extra thymine at nucleotide 1008 (codon 336) in the L. kirschneri gene. This frameshift mutation results in a TAG stop codon downstream at codon 347. The predicted size of LigC, correcting for the stop and frame-shift mutations, is 210 kDa, similar to that predicted for LigB. LigA, LigB and LigC orthologues from L. kirschneri and L. interrogans had 90.7–94.9% amino acid sequence identity whereas the L. kirschneri and L. interrogans LigA sequences were 80.5% and 84.5% identical to that of L. interrogans type kennewicki LigA respectively (Palaniappan et al., 2002). Horizontal transfer of DNA or convergent evolution may account for the closer relatedness of the L. interrogans strain Fiocruz L1-130 ligA gene to the L. kirschneri strain RM52 ligA gene than to that of L. interrogans type kennewicki.
The ligA and ligB genes are predicted to encode lipoproteins based on the identification of a 17 amino acid N-terminal signal peptide and lipoprotein signal peptidase cleavage site that conforms to the spirochetal lipobox (Haake, 2000). Following the signal peptide cleavage sites, the Lig proteins contain 10–11 repeats (Fig. 1D). Analysis of the primary amino acid sequences with the motif discovery tool MEME v3.0 (Bailey and Elkan, 1994) revealed two types of motifs within the 90 residue Big2 repeat sequences. Motif 1 is comprised of 52 residues, whereas Motif 2 is 24 residues in length (Fig. 1A and B). The repetitive nature of these motifs, along with their symmetrical placement within the Big domain, correlates with the predicted three-dimensional β-immunoglobulin sandwich structure of the Lig proteins (Fig. 1C).
Fig. 1.
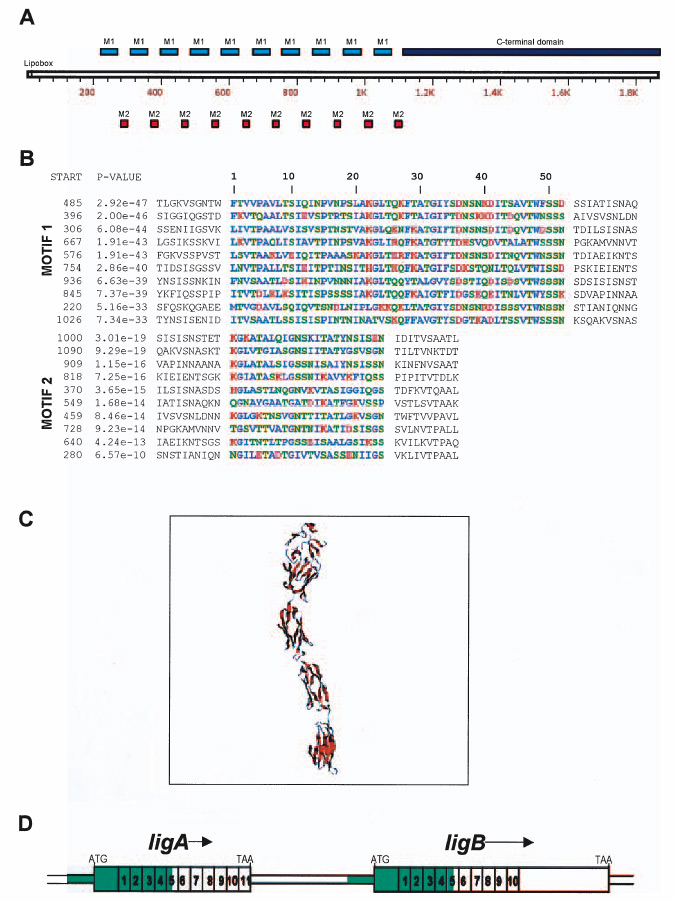
A. Schematic representation of LigB. Motif 1 (blue box) and Motif 2 (red box) comprise the 10 Big2 repetitive domains from residue 220 through 1109, whereas the C-terminus (dark blue box) is located between 1111 and 1890.
B. MEME Motif Discovery analysis of the Big2 repetitive domains of LigB. L. interrogans Lig proteins contain ~90mer repeats composed of two motifs of length 52 residues (M1) and 24 residues (M2), respectively. Residues are coloured based on their chemical properties: hydrophobic, basic, acidic and polar uncharged. The sites are listed in order of decreasing statistical significance (P-value). The P-values and start sites of the motifs are indicated. The P-value of a site is computed from the match score of the site with the position specific scoring matrix for the motif and represents the probability of a random string (generated from the background letter frequencies) having the same match score or higher.
C. Comparative modelling of L. interrogans LigB repetitive domains (residues 493–917).
D. Genetic structure of the ligAB locus. The 11 and 10 Big2 domains encoded by ligA and ligB, respectively, are numbered. Two additional N-terminal Big2 domains with poor E-values (>10−10) are not shown. The >2 kb segment of DNA sequence identity between the 5′ portions of ligA and ligB is indicated in green.
Like the Big superfamily proteins intimin and invasin, ligB and ligC encode a C-terminal, non-repeat domain (771 and 789 residues, respectively) following the Big tandem repeat sequences. In contrast, LigA is a smaller truncated form consisting entirely of Big repeat domains. Whereas the amino acid sequence identity between C-terminal domains encoded by ligB and ligC is 51%, these domains do not have significant homology with proteins deposited in the NCBI database.
Inverse PCR with primers annealing to the 3′ end of the ligA gene (Table 2) demonstrated that the ligB gene is located <1.5 kb downstream of the truncated ligA gene. Both genes are transcribed in the same orientation (Fig. 1D). Open reading frames larger than 100 nucleotides are absent between ligA and ligB. Sequencing and Southern blot analyses indicate that ligC is not linked to the ligA-ligB locus (data not shown). Although L. kirschneri and L. interrogans sequences of ligA and ligB are homologous with >91% DNA sequence identity, there is considerable divergence between the L. kirschneri and L. interrogans ligA-ligB intergenic regions (943 bp and 1347 bp in length, respectively).
Table 2.
Nucleotide sequence of inverse PCR primers.
| Gene | Primer sequence 5′ to 3′ | Co-ordinatesa | Restriction enzymeb |
|---|---|---|---|
| ligA 5′ end | CAGATATTCTTACCGTTTCCAATACA | 1883–1908 | NsiI |
| ATCCGATCAGATTTTTGCATCAAGTC | −209–(−234) | ||
| ligA 3′ end | CCGCTTTCTGTAGGTTCCTCTAAAAT | 3562–3587 | HincII |
| CGGTAGCGGTCAGTTGTAGTGTAAGA | 3175–3150 | ||
| ligB 3′ end | ATATCCGGAATGAATTTTGGTGTAAA | 5569–5594 | EcoRI |
| ATTTTCAAGATTTGTTCTCCAGATTT | 5487–5462 | ||
| LigC 5′ end | TCCTGCAAATCCAAGCGTAGCCAATG | 1054–1079 | XhoI |
| GTAAGAACTGCGGAAGTGACGTTTACTA | 1038–1011 | ||
| LigC 3′ end | ACCGGACTCTTGTTCTAACTGGGTGG | 5209–5234 | BglII |
| ATCCGATTCCGTCGTTGCTTGACTTT | 4868–4843 |
Relative to A in ATG start codon.
Leptospiral DNA digested with restriction enzyme before treatment with T4 ligase and PCR.
Interestingly, the 5′ portion of the ligA sequence is identical to the 5′ portion of the ligB sequence (Fig. 1D). This region of sequence identity begins 266 bp (L. interrogans) or 538 bp (L. kirschneri) upstream from the start codon of ligA and ligB and extends 1890 bp into the open reading frame sequence of both genes. Consequently, the first 630 amino acid residues of LigA and LigB, which includes the first four and part of the fifth Big2 repeat domain, are identical. In addition, there are four tandem imperfect 38-nucleotide repeats within the duplicated region upstream of the L. kirschneri ligA and ligB genes. The second and third repeats are identical. The first and fourth repeats differ from the other two repeats by two and six nucleotides respectively. The corresponding regions of the ligA and ligB genes in L. interrogans contain a single copy of a similar 39-nucleotide sequence that differs from the fourth repeat in L. kirschneri by two nucleotides. These small repeat sequences were not found in the 3′ flanking regions of either ligA or ligB. Several homopolymeric tracts were found in the lig genes. In L. kirschneri, ligA has a run of 8As, ligB has a run of 8As and a run of 9As, whereas ligC has a run of 8Ts. In L. interrogans, ligB has a run of 9As, whereas ligC has two runs of 8Ts and a run of 6 Gs. However, changes in the length of homopolymeric tracts would not correct the frameshift and stop codon mutations of the ligC genes of L. kirschneri and L. interrogans respectively.
Distribution of lig genes in the genus Leptospira
Southern blot studies with ligA, ligB, and ligC-specific probes confirmed that L. interrogans Fiocruz L1-130 has a single copy of each lig gene (Fig. 2A, middle lanes). Further analysis found that specific lig sequences were not detected in the saprophytic, non-pathogenic L. biflexa Patoc 1 (Fig. 2A, right lanes) whereas the L. kirschneri (data not shown) and L. interrogans genomes contained the three genes encoding the Lig repetitive domain.
Fig. 2.
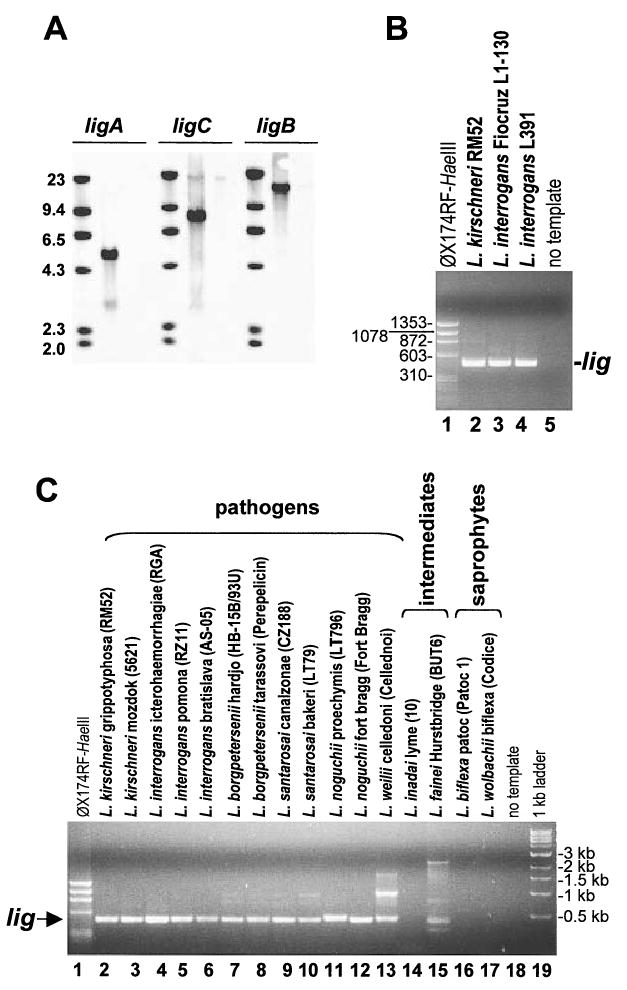
Detection of lig sequences in Leptospira species.
A. Southern blot analysis. Genomic DNA from L. interrogans Fiocruz L1-130 (middle lane) and L. biflexa Patoc 1 (right lane) were digested with NsiI and PacI and probed in Southern blots with ligA, ligB and ligC specific probes.
B and C. PCR analysis. Leptospiral DNA was subjected to PCR analysis by degenerate primers designed to anneal to all lig genes in L. kirschneri RM52 and L. interrogans Fiocruz L1-130. The expected size of the PCR product is −0.5 kb.
To examine the distribution of lig genes in a larger collection of pathogenic and saprophytic strains of Leptospira spp., a pair of degenerate PCR primers was designed that annealed to all six lig genes in L. interrogans Fiocruz L1-130 and L. kirschneri RM52. These primers amplified lig sequences in L. kirschneri RM52 and L. interrogans Fiocruz L1-130 (Fig. 2B, lanes 2 and 3, respectively) and in 12 strains belonging to six pathogenic Leptospira species (Fig. 2B, lane 4; Fig. 2C, lanes 2–13). Leptospira fainei and L. inadai are considered ‘intermediate’ pathogens and are phylogenetically distinct from typical pathogenic species (Perolat et al., 1998). Polymerase chain reaction analysis yielded a product of the expected 500 bp size for a L. fainei strain, but not for a L. inadai strain (Fig. 2C, lanes 14 and 15). Polymerase chain reaction amplification of the L. fainei and L. weilii DNA yielded higher molecular weight products that may represent primers annealing to related sequences within the lig genes. Furthermore, lig sequences appear to be restricted to pathogenic strains since they were not detected in the two saprophytic strains that were evaluated (Fig. 2C, lanes 16–17).
Expression of Lig proteins in virulent and culture-attenuated Leptospira strains
Leptospires lose their virulence phenotype with sequential in vitro culture passages (Haake et al., 1991). Therefore, expression of virulence factors may be downregulated during culture attenuation. In an earlier report, antisera to LigA of L. interrogans type kennewicki failed to react with antigens obtained from in vitro cultured leptospires (Palaniappan et al., 2002). However, it was not determined whether the lack of antibody reactivity was due to the absence of detectable LigA expression in virulent strains or due to prolonged culture and virulence attenuation in the study strains.
To examine this issue, we performed immunoblot analysis with polyclonal antibodies raised against unique portions of recombinant LigA, LigB and LigC fragments and extracts from virulent, low-passage and culture-attenuated, high-passage (>200 passages) isolates of L. kirschneri strain RM52. Loss of virulence in high-passage isolates was documented by the failure of large (≥107) inoculum doses to produce a lethal infection in the hamster model (Haake et al., 1991). In experiments with low-passage isolates, antibody against the last three Big2 repeats of LigA (amino acid residues 936–1224) reacted with a protein band with similar Mr (~130 kDa) to the predicted molecular weight for LigA (128 kDa) (Fig. 3A, lane1). Likewise, antibody against the C-terminal segment of LigB (residues 1113–1886) reacted with a >200 kDa protein band, which is close to the predicted molecular weights for LigB (212 kDa) and LigC, corrected for mutations (210 kDa) (Fig. 3B, lane 1). Anti-LigC antibodies (Table 1) did not react with a protein band >50 kDa in low-passage isolate extracts (data not shown), indicating that a high molecular weight ligC product is not expressed and that LigB is the >200 kDa protein detected in immunoblots with anti-LigB antibody. To confirm the expression of LigA and LigB, we raised antisera against residues 342 through 1224 of LigA, which includes a portion (residues 342–630) of the region of sequence identity with LigB. This antiserum reacted with ~130 and >200 kDa protein bands as expected (Fig. 3C, lane 1).
Fig. 3.
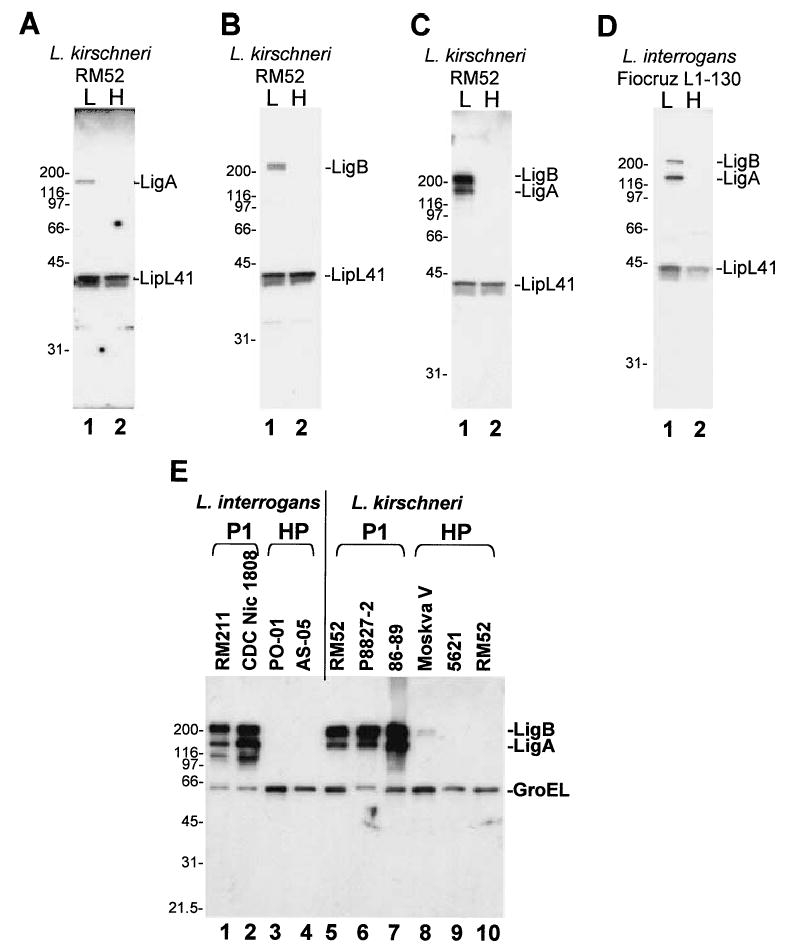
Western blot analysis of LigA and LigB expression in low- and high-passage Leptospira.
A and B. Immunoblots of low (9 passages) and high-passage (>200 passages) isolates of L. kirschneri RM52 were probed with LipL41 antiserum (1:10 000), and either LigA antiserum (A, 1:2000) or LigB antiserum (B, 1:2000). L, low passage; H, high passage.
C and D. Immunoblots of L. kirschneri RM52 or L. interrogans Fiocruz L1-130 (passage 25 and passage 69) were probed with LipL41 (1:10 000) and LigA/B repeat antisera (1:5000).
E. An immunoblot of L. interrogans and L. kirschneri isolates was probed with LigA/B repeat antisera (1:5000) and GroEL antisera (1:30 000). Strain names are shown above each lane. P1, passage 1; HP, high passage. The relative mobility (kDa) of molecular mass standards (Bio-Rad) is shown on the left of each panel.
Table 1.
Plasmids used to express recombinant Lig proteins in E. coli.
| Protein | Source | Tag | Amino acid co-ordinates | Plasmid name |
|---|---|---|---|---|
| LigA | RM52 | His6 | 936–1224 | p204orf1B |
| LigA | RM52 | His6 | 342–1224 | p204orf1 |
| LigB | RM52 | MBP | 20–581 | pROY1 |
| LigB | RM52 | MBP | 579–1126 | pROY2 |
| LigB | RM52 | MBP | 296–847 | pROY3 |
| LigB | RM52 | MBP | 1113–1886 | pJIM520 |
| LigB | RM52 | His6 | 1113–1886 | pJIM518 |
| LigC | RM52 | His6 | 621–1060 | p249orf1 |
| LigB | L1-130 | His6 | 131–649 | pAE-ligBrep |
Loss of LigA and LigB expression correlates with loss of virulence during culture attenuation since antibodies to Lig proteins did not react with a protein band in extracts of high-passage L. kirschneri (Fig. 3A–C, lanes 2). This phenomenon appears to be specific for LigA and LigB, as expression of most leptospiral proteins, including LipL41 (Fig. 3A–C), is conserved in both low and high-passage isolates (Haake et al., 1991). DNA sequencing of the promoter regions upstream of ligA and ligB revealed no differences between virulent and attenuated forms of L. kirschneri strain RM52, ruling out changes in these regions as an explanation for the observed loss of LigA and LigB expression.
Immunoblot analysis was performed with strain Fiocruz L1-130 to determine whether these observations could be extended to L. interrogans. Figure 3D shows that a virulent, low-passage but not attenuated, high-passage (45 passages) isolates of this strain expressed LigA and LigB. The attenuated virulence of the high-passage Fiocruz L1-130 isolate was demonstrated when a large inoculum of 108 leptospires failed to produce a lethal infection. Additional low- and high-passage L. kirschneri and L. interrogans isolates were examined in immunoblots shown in Fig. 3E. GroEL was detected by anti-GroEL antibodies in low and high-passage strains whereas significant levels of LigA and LigB were found only in low-passage isolates. Although a small amount of LigB was detected in high passage L. kirschneri strain Moskva V, the quantity detected was markedly less than that in the low-passage isolate. The additional ~110 kDa band observed in the L. interrogans strains appears to be a proteolytic breakdown product produced when the sample is boiled repeatedly rather than expression of an additional Lig protein. Neither LigA nor LigB was detected by immunoblot analysis in the saprophytic strain, L. biflexa Patoc 1 (data not shown).
Reverse transcriptase-polymerase chain reaction analyses indicate that variation in lig transcript levels in low-and high-passage strains may account for the loss of LigA and LigB expression during culture attenuation (Fig. 4). For assays with RNA extracts of L. kirschneri strain RM52, PCR primers were used to amplify sequences corresponding to a ~0.5 kb region near the 3′ ends of the transcripts. Amplified products were not observed in control reactions lacking reverse transcriptase, indicating that extracts were not contaminated with DNA. The ligA and ligB transcripts were detected in low-passage strains while these transcripts were absent or detected at reduced levels in high-passage strains (Fig. 4). The lipl45 gene expresses the P31LipL45 protein, whose levels are unaffected by in vitro passaging and culture attenuation (Matsunaga et al., 2002). Differences in ligA and ligB transcript levels do not appear to be due to differential extraction of leptospiral RNA since equivalent levels of lipl45 transcripts were detected in high and low-passage strains (Fig. 4, lanes 14 and 16). The ligC transcript was not observed in either low- or high-passage L. kirschneri, supporting the conclusion that ligC is not expressed as a gene product.
Fig. 4.
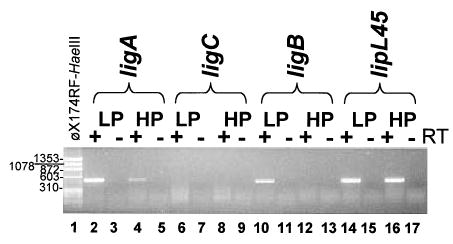
RT-PCR analysis of lig transcripts in low- and high-passage L. kirschneri RM52. Total RNA from low- (passage 4) and high-passage L. kirschneri RM52 was analysed by RT-PCR with primers specific for each lig gene. +, reverse transcriptase present in the reaction; −, reverse transcriptase omitted.
Early recognition of the Lig repeats by the immune system
At least one Lig protein is expressed during human infection since patient sera were used to identify lig clones from the λ genomic expression library. This finding was confirmed in immunoblot studies demonstrating that pooled patient sera specifically reacted with purified recombinant proteins corresponding to repeats one through five of LigB (Fig. 5, lane A3), repeats two through seven of LigB (data not shown), and repeats five through 10 of LigB (data not shown). In contrast, serum from healthy human volunteers did not react with any recombinant LigB proteins (Fig. 5, lane B3, and data not shown). These data suggest that LigB is expressed during infection; however, we can not exclude the possibility that the positive reaction in these blots is due to cross-reactivity to an antigenically related protein, such as LigA. Anti-recombinant LigB IgM antibody was detected in more than 95% of individual leptospirosis patients during their acute illness, suggesting that Lig proteins are expressed early in the course of infection (data not shown). In addition to human leptospirosis, Lig proteins are expressed during natural infection of animal reservoirs: anti-LigB antibodies were found in sera from captured domestic rat (Rattus norvegicus) reservoirs (Fig. 5, lane C3), but not in sera from captured culture-negative rats (lane D3). In contrast to the findings observed with natural infection, multiple immunizations of laboratory rats with extracts of in vitro cultured low-passage strain Fiocruz L1-130 did not produce detectable serum antibodies against recombinant LigB (Fig. 5, lane E3). This immunization protocol produced strong antibody responses to more than 20 other leptospiral proteins found in whole extracts (Fig. 5, lane E1), as well as to recombinant LipL32 (Fig. 5, lane E2), a protein previously found to be expressed in vitro and during host infection (Haake et al., 2000).
Fig. 5.
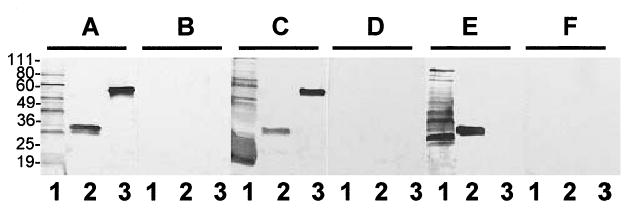
Reactivity of sera from leptospirosis patients and Rattus norvegicus reservoirs with recombinant LigB. Membranes were prepared from SDS-PAGE of whole L. interrogans extract (108 organisms per lane, lanes 1); recombinant L. interrogans lipoprotein, LipL32 (1 μg per lane, lanes 2) and recombinant protein containing repeat domains 1–5 of L. interrogans LigB (1 μg per lane, lanes 3). Membranes were probed with sera from leptospirosis patients (A); healthy individuals (B); captured R. norvegicus from which L. interrogans was isolated (C); captured culture-negative R. norvegicus (D); laboratory rats immunized with whole-cell preparations of in vitro cultured low-passage L. interrogans (E); and laboratory rats obtained prior to immunization (F). The relative mobility (kDa) of molecular mass standards (Invitrogen) is shown on the left.
Surface localization of Lig proteins in immunocytochemical electron microscopy and cell fractionation
Immunoelectron microcopy (IEM) studies demonstrated that the C-terminal non-repeat (Fig. 6E and F) and Big repeat (data not shown) domains of Lig proteins are present on the surface of virulent, low passage leptospires. In whole-cell IEM assays (Fig. 6A–C), live L. kirschneri RM52 were settled onto EM grids, incubated with antibody, and labelled with gold-conjugated anti-rabbit secondary antibody. Gold particles were present on leptospires incubated with antibodies against the LigB C-terminal domain (32 ± 9 particles per bacterium, mean ± standard deviation) (Fig. 6E). Staining was similar to that observed when leptospires were incubated with antibodies against the leptospiral surface moiety, LPS (64 ± 6 particles per bacterium) (Fig. 6A). Assay conditions appeared to preserve the integrity of the leptospiral membranes as bacteria incubated with antibodies against the cytoplasmic heat shock protein, GroEL, showed low levels of background staining (5 ± 2 particles per bacterium) (Fig. 6C). Furthermore, IEM of thin-sectioned L. kirschneri strain RM52 demonstrated that antibodies against the LigB C-terminal non-repeat domain (Fig. 6F) and LPS (Fig. 6B) labelled the external surface of the outer membrane. Labelling with anti-GroEL antibodies was restricted to the cytoplasm of leptospires. Similar results were obtained for whole-cell and thin-section IEMs with L. interrogans Fiocruz L1-130 (data not shown). Indirect immunofluorescence assays were performed with live, intact leptospires as a third method to confirm the surface localization of Lig proteins. Polyclonal antibodies raised against the first to fifth Big repeat domains of LigA and LigB bound to the surface of the virulent, low passage L. interrogans Fiocruz L1-130 as did antibodies to the outer membrane lipoprotein LipL41 (data not shown). Surface labelling was not detected in control assays with antibodies against the inner membrane lipoprotein LipL31 (Haake and Matsunaga, 2002), and GroEL. Anti-LigA/B antibodies did not label the surface of culture attenuated, high passage L. interrogans Fiocruz L1-130. The loss of LigA/B surface expression during culture attenuation of virulence appeared to be specific as anti-LipL41 antibodies strongly labelled these leptospires.
Fig. 6.
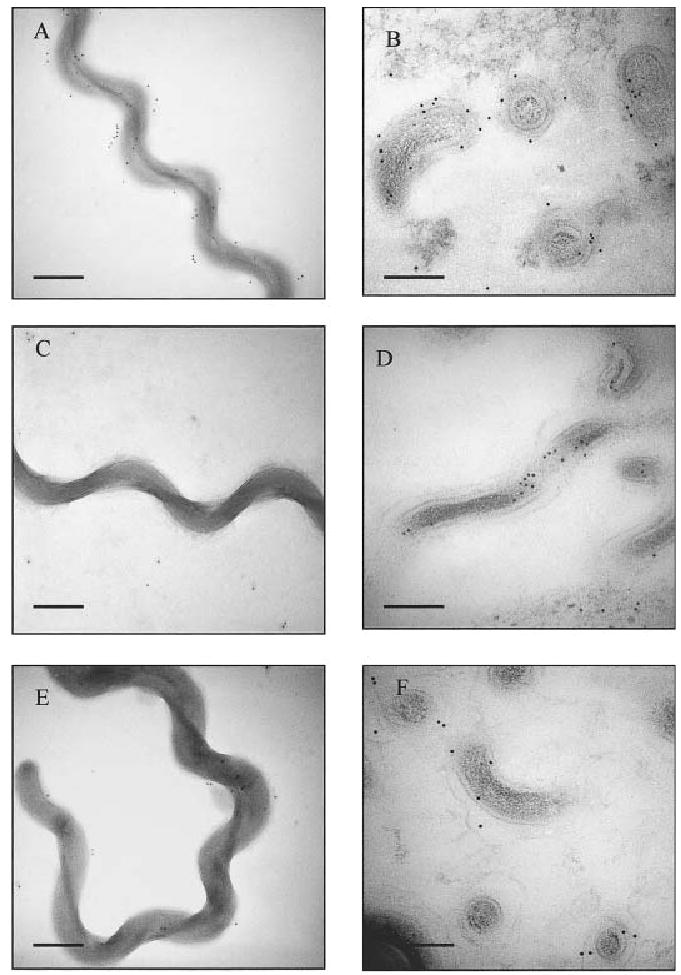
Immunoelectron microscopy of whole-cell (A, C and E) and thin-section (B, D, F) preparations of L. kirschneri strain RM52. Preparations were incubated with anti-LPS (A, B), anti-GroEL (C, D), and anti-LigB antibody (E, F), followed by anti-rabbit secondary antibody conjugated to 10 nm gold particles. Bars represent 100 nm.
The unique cellular architecture of spirochetes, in which the peptidoglycan is associated with the inner instead of the outer membrane, permits solubilization of the outer membrane with the non-ionic detergents Triton X-114 and Triton X-100 (Haake et al., 1991; 1998; Zuerner et al., 1991). The periplasmic cylinder (PC) and the Triton X-100-soluble fractions of virulent L. kirschneri were analysed by immunoblotting (Fig. 7). The fractions were probed with antibody to the inner membrane protein LipL31, which was found in the PC fraction, as expected (Haake and Matsunaga, 2002). Both LigA and LigB were partially solubilized by Triton X-100, consistent with the surface exposure demonstrated by immunogold electron microscopy. Incomplete solubilization of LigA and LigB suggests that the Lig proteins may be distributed between the inner and outer membranes.
Fig. 7.
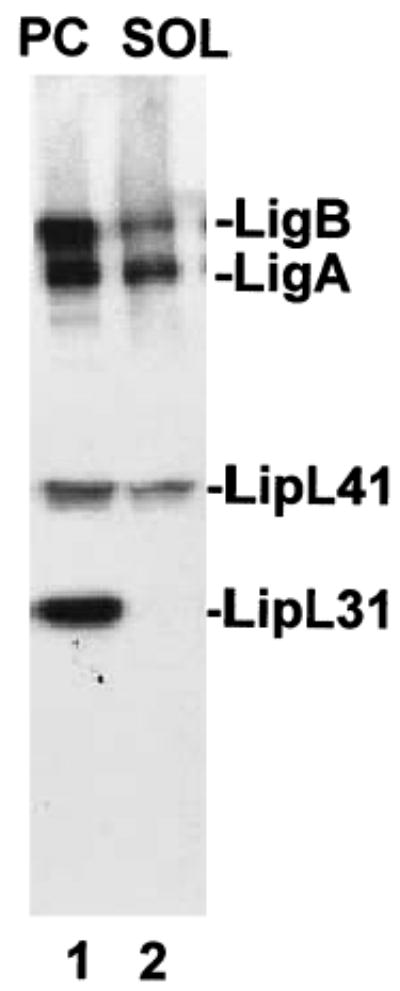
Triton X-100 extraction of Lig proteins from L. kirschneri RM52. Leptospires were incubated for 2 h in MEM (ATCC) at 37°C. Bacteria were then washed once in 1× PBS with 5 mM MgCl2 and extracted with 1% Triton X-100. The insoluble protoplasmic cylinder (PC) was pelleted by centrifugation. The protoplasmic cylinder and the Triton-soluble (SOL) fractions were probed in a Western blot using LigA/B repeat (1:5000), LipL41 (1:10 000), and LipL31 (1:8000) antisera. LipL31 is a marker for the leptospiral inner membrane (Haake and Matsunaga, 2002).
Discussion
In this study, we characterized a new family of leptospiral surface-exposed proteins we refer to as Lig (leptospiral Ig-like), which may play a role in the pathogenesis of leptospirosis. Our results are consistent with LigA and LigB being expressed in the infected host (Fig. 5) and demonstrate that the lig genes are found only in pathogenic Leptospira species (Fig. 2). More importantly, LigA and LigB expression is correlated with virulence. The loss of the ability to produce lethal infection in hamsters by high-passage L. interrogans strain Fiocruz L1-130 (this study) and L. kirschneri strain RM52 (Haake et al., 1991) is associated with loss of LigA and LigB expression (Fig. 3). Whereas the vast majority of leptospiral proteins expressed in virulent L. kirschneri RM52 is also expressed in the culture-attenuated form of the same strain (Haake et al., 1991), LigA and LigB expression is specifically lost during culture attenuation, suggesting that the latter proteins are closely tied to the pathogenesis of leptospirosis. The molecular basis for the loss of Lig protein expression with culture passage is unknown. Changes in the number of the 38-nucleotide repeat in the promoter region may affect transcription. However, sequence analysis of the promoter regions, including the four 38-nucleotide tandem repeats, and the first 380 nucleotides of ligA and ligB of both virulent and culture-attenuated L. kirschneri revealed no sequence differences. Alternatively, several sites consisting of runs of at least six identical nucleotides are present within the lig genes, raising the possibility of the loss of lig expression by slipped-strand mispairing.
Our studies suggest that Lig expression is upregulated during infection of the mammalian host. Rats immunized with killed virulent L. interrogans Fiocruz L1-130, which expresses LigA and LigB in vitro (Fig. 3D), failed to produce detectable antibody against recombinant Lig protein (Fig. 5E). In contrast, anti-Lig antibody was detected in rats infected with live L. interrogans (Fig. 5C). Similarly, lig sequences were not found among 104 clones obtained from a leptospiral expression library probed against serum from rabbits immunized with killed virulent L. kirschneri RM52 (Matsunaga et al., 2002), whereas in this study, 10% (13 of 125) of the clones isolated from expression libraries which reacted with leptospirosis patient sera had lig gene sequences. Upregulation of Lig expression during infection could also explain the inability of Palaniappan et al. (2002) to detect LigA in cultivated L. interrogans subtype kennewicki by immunoblot analysis while detecting expression of this moiety in leptospires from infected hamster kidneys by immunohistochemical methods.
The Lig proteins are new members of the bacterial immunoglobulin-like (Big) superfamily, some of which serve as adhesins and invasion-mediating determinants in other bacterial pathogens. Invasin of Yersinia pseudotuberculosis and intimin of enteropathogenic E. coli are two examples of virulence factors in this family. The structural organization and localization of Lig proteins share common themes to those observed for intimin and invasin. Lig proteins are presumed to be anchored to the outer membrane with the same N-terminal orientation as observed for intimin and invasin. However, LigA and LigB are predicted lipoproteins, which are anchored to the outer membrane via fatty acids attached to the amino-terminal cysteine residue rather than a transmembrane domain. The sequence of the amino termini of LigA and LigB largely conforms to the spirochetal lipobox, as previously defined (Haake, 2000). As with intimin and invasin, Lig proteins have tandem Big repeats that follow the membrane anchor. Computer modelling of Big2 domains from Lig proteins found significant sequence identity with Big domains from intimin and invasin, indicating that Lig Big2 domains adopt a folding structure resembling those observed for Big1 and 2 domains in these proteins (Fig. 1C).
The role of Big repeat domains in Lig proteins is unclear but they may serve a similar function as observed in intimin and invasion. Big domain repeats in these virulence factors are believed to act as linkers that project the receptor binding C-terminal domain from away from the bacterial surface therefore providing higher accessibility for interaction (Hamburger et al., 1999; Luo et al., 2000). In addition, one of the four Big domains of Y. pseudotuberculosis invasin mediates efficient invasion by promoting homotypic interaction of invasin subunits (Dersch and Isberg, 2000). Finally, LigB has a large C-terminal domains (771 residues) following the Big domain repeats that, by analogy with invasin and intimin (106 and 99 residue C-terminal domains, respectively), may have a host cell binding function. LigA lacks the C-terminal domain found in LigB. Interestingly, ligA is located <2 kb upstream of ligB, and the finding that ligA and ligB have identical sequences in the 5′ regions raises the question whether these genes arose from a duplication event. Together, these findings suggest that LigB may be a more plausible candidate among the Lig family to serve as an adhesin. As a caveat, the possibility cannot be excluded that Big2 domains in LigA play a role in host cell receptor binding.
The sequences of the L. kirschneri and L. interrogans ligC genes have frameshift and stop codons, respectively, which interrupt the open reading frame. Introduction of early stop codons in bacterial mRNA often destabilize the entire message (Nilsson et al., 1987; Grunberg-Manago, 1999). Reverse transcriptase PCR and immunoblot studies could not detect expression of ligC at the RNA and protein level, providing additional evidence that ligC is a pseudogene. Nevertheless, the possibility that a truncated ~50 kDa or ~35 kDa protein is expressed from the open reading frames at the 5′ end of the L. kirschneri or L. interrogans ligC transcript, respectively, cannot be excluded.
Adherence of leptospires to host mammalian cells plays an essential role in the pathogenesis of leptospirosis. Host cell entry may play a role in the rapid dissemination of leptospires as L. interrogans has been shown to rapidly translocate across polarized cell monolayers (Thomas and Higbie, 1990; Barocchi et al., 2002). Electron microscopy studies found that leptospires tightly adhere to host cell plasma membrane during the translocation process, suggesting that this type of association may be a required early step for translocation (Barocchi et al., 2002). In addition, leptospires attach tightly to the luminal surface of the renal epithelium in order to establish persistent carriage in animal reservoirs (Marshall, 1976).
Proteins that mediate these host cell interactions are expected to be exposed on the leptospiral cell surface. In this study, imunoelectron microscopy and immunofluorescence microscopy of L. kirschenri and L. interrogans demonstrated that the Lig proteins are surface-exposed (Fig. 6, data not shown). Furthermore, surface expression of LigB proteins, as detected by immunofluorescent microscopy, disappeared as virulent strains were culture attenuated (data not shown). Previous studies have shown that virulent leptospires, in contrast to culture-attenuated forms of the same strains, attach more readily to tissue culture cells and extracellular matrix proteins (Ballard et al., 1986; Ito and Yanagawa, 1987). Merien et al. (1997) observed the same phenomenon with respect to the ability of virulent and culture attenuated leptospires to invade Vero cells in vitro. A 36 kDa surface-exposed protein potentially involved in fibronectin binding has been identified in virulent L. interrogans but is absent in the culture-attenuated form of that strain (Merien et al., 2000). The correlation of LigA and LigB expression and leptospiral adhesion with virulence and their structural similarity with invasin and intimin suggests that LigA and/or LigB may be another leptospiral adhesin.
In conclusion, we have identified a new family of Big proteins in Leptospira that may play a role in pathogenesis. The evidence that LigA and B proteins are virulence factors is that (i) they are found only in pathogenic Leptospira species; (ii) they are detected in virulent but not in culture-attenuated strains of L. kirschneri and L. interrogans; and (iii) like the Big proteins intimin and invasin of known enteric bacterial pathogens, LigB, and perhaps LigA, is surface exposed. Further studies are needed to determine the role of LigB and the other members of the Lig family in the attachment and invasion of Leptospira spp. to host tissues.
Experimental procedures
Bacterial strains and media
All leptospiral strains were cultivated at 30°C in EMJH medium supplemented with 1% rabbit serum (Johnson and Harris, 1967) (Faine, 1982). Leptospira interrogans serovar copenhageni, strain Fiocruz L1-130 was isolated from a patient during an outbreak of leptospirosis in Salvador, Brazil (Ko et al., 1999; Barocchi et al., 2001). The low passage clinical isolate Fiocruz L1-130 had an LD50 of 104 in the hamster model of virulence (Barocchi et al., 2002). Forty-five in vitro passages were performed to obtain a culture-attenuated strain for which an inoculum of 108 organisms of this high passage strain did not induce death or clinical disease in hamsters. All other leptospiral strains, including virulent and culture-attenuated L. kirschneri serovar grippotyphosa, strain RM52, were obtained from the National Leptospirosis Reference Center (National Animal Disease Center, Agricultural Research Service, U.S. Department of Agriculture, Ames, Iowa) and are described in a recent study (Matsunaga et al., 2002). Experimental protocols involving hamsters were approved by the VA Greater Los Angeles Healthcare System Animal Research Committee.
Escherichia coli strains were grown in LB supplemented with 100 μg ml−1 ampicillin (Sigma) or 100 μg ml−1 carbenicillin (ICN) where appropriate. λ bacteriophage expression libraries were screened in Escherichia coli XL1-Blue MRF’ (Stratagene) and E. coli XL1-Blue (Clontech). Escherichia coli SOLR and E. coli BM25.8 (Stratagene) allowed for in vivo excision of plasmid sequences from the λ expression vectors. Expression of His6-tagged and MBP-tagged recombinant proteins were done in E. coli BLR(DE3)/pLysS (Novagen) and E. coli TB1 (New England Biolabs) respectively.
Isolation of genes encoding leptospiral protein antigens
Genomic DNA prepared from low-passage L. interrogans serovar copenhageni, strain Fiocruz L1-130 and L. kirschneri serovar grippotyphosa, strain RM52 were partially digested with Tsp509I (New England Biolabs) and ligated to λZap II (Stratagene) and _TriplEx arms (Clontech) respectively. Recombinant phage DNA was packaged into λ heads following the instructions provided in the Gigapack III Gold Packaging Extract (Stratagene).
Sera from 10 to 20 leptospirosis patients identified during an urban outbreak in Brazil were pooled and adsorbed three times with lysates of E. coli XL1-Blue to remove antibodies to E. coli antigens (Gruber and Zingales, 1995). A titre of 1:100 to 1:500 of the adsorbed sera was used to probe the genomic expression libraries as described (Matsunaga et al., 2002). Reactive phage clones were purified by a round of plaque purification. The plasmid DNA was excised from the λ bacteriophage sequence by in vivo recombination in E. coli SOLR (Stratagene) or E. coli BM25.8 (Clontech) as instructed.
Gel electrophoresis and immunoblots
Electrophoresis and Western blots were performed as described (Guerreiro et al., 2001). Serum titres are indicated in the figure legends.
Oligodeoxynucleotides and DNA sequencing
The sequence of the first and last 500–700 nucleotides of each insert was obtained with vector-specific primers that anneal adjacent to the insert. The sequence of the upstream primer is 5′-TCCGAGATCTGGACGAGC-3′ (Clontech), and the sequence of the downstream primer is 5′-TAATACGACT CACTATAGGG-3′ (T7 promoter). The sequence of the remainder of each insert was obtained from plasmid clones deleted of restriction fragments extending from inside the insert into the multicloning sites or from intact plasmid clones with custom-designed gene-specific primers. Oligodeoxynucleotides were synthesized by Invitrogen. The sequencing reactions were performed by the Yale/Keck Core DNA Sequencing Facility, the UC Berkeley Core DNA Sequencing Facility, and Instituto Butantan. The lig sequences were assembled with Sequencer 4.0.5 for Windows (Genecodes).
Polymerase chain reaction
The termini of the lig genes and their flanking regions were amplified by inverse PCR prior to sequencing. The 5′ and 3′ends of each lig sequence were subjected to separate inverse PCR reactions (Table 2). Leptospiral DNA was digested with an appropriate restriction enzyme (Table 2), treated with T4 DNA ligase, and amplified with Taq DNA polymerase (Qiagen). The primers listed in Table 2 (except for the ligA upstream primer) were designed to anneal near the ends of the lig sequences assembled from the clones obtained from the L. kirschneri expression library. After the duplicated segment was revealed by comparing the sequence of the 5′-most ligA clone with the inverse PCR product generated by the ligA 3′ end primers, the 5′ end of ligA and its promoter region were amplified with the sense primer 5′-ATTGTAATTTTTCTGATGGTCGTCAAAC-3′, which based on the sequence upstream of ligB was predicted to hybridize 265–238 nucleotides upstream of ligA, and the anti-sense primer 5′-CGGTAGCGGTCAGTTGTAGTGTAAGA-3′(positions 3175–3150 within ligA). Sequence upstream of the duplicated region of ligA was obtained by inverse PCR with the primers shown in Table 2 (ligA 5′ end).
To design PCR primers for the detection of lig sequences (Fig. 2B and C), the nucleotide sequences of all six lig genes in L. interrogans and L. kirschneri were aligned with MegAlign (DNAStar). Segments of near sequence identity were used to design the degenerate primers 5′-(G/C)AAAGTTG(T/C) (A/G)(T/C)G(T/G)CTTGGCC-3′ and 5′-(G/C)(A/T)ACC (A/G)TC(C/T)GAAAA(A/G)AT(A/T)CC-3′. About fifty nano-grams of leptospiral genomic DNA was amplified with Taq DNA polymerase (Qiagen) with an initial denaturation at 94°C for 1 min, then 35 cycles of 94°C for 30 s, 49°C for 30 s, and 72°C for 30 s, and a final 1 min extension at 72°C. The reactions were separated in a 1.4% agarose gel in TAE buffer (Fisher Biotech).
Plasmid constructions
Standard recombinant DNA techniques were used to construct all expression plasmids. The His6 expression vectors pRSETA and pRSETC were purchased from Invitrogen, and pMAL-c2x was purchased from New England Biolabs. pAE expression vector, a hybrid of pET and pRSET expression vectors, was kindly provided as a gift from Paulo Lee Ho (Instituto Butantan). The 2.8 kb KpnI-HindIII fragment of plasmid clone 204 harbouring codons 342 through 1224 of L. kirschneri ligA was ligated to pRSETC to create p204orf1. All other expression plasmids were constructed by PCR with lig-specific primers with restriction sites introduced at their 5′ends (Table 1). Polymerase chain reactions were performed with Pfu Turbo DNA polymerase (Stratagene) during 30 cycles of amplification with L. kirschneri genomic DNA or 15 cycles of amplification with plasmid DNA. Polymerase chain reaction products were purified with the QIAquick PCR Purification Kit (Qiagen), subsequently gel-purified with the Zymoclean Gel DNA Recovery Kit (Zymo Research), digested with the appropriate restriction enzymes, and ligated to a His6 expression vector with T4 DNA ligase (New England Biolabs).
Expression and purification of recombinant Lig
Expression plasmids were transformed into E. coli BLR(DE3)/pLysS. Transformed bacteria were grown at 37°C in 500 ml LB containing 100 μg ml−1 of carbenicillin to an OD600 of 0.4. IPTG was then added to a final concentration of 0.5 mM, and induction was continued at 37°C for 2.5–3 h. Except for p204orf1B (Table 1), all plasmids expressed insoluble His6-tagged recombinant protein. Ni2+-nitrilotriacetic acid (NTA) affinity columns were used to purify His6-tagged recombinant proteins according to the instructions of the manufacturer (Qiagen).
The C-terminal domain of LigB fused to the maltose binding protein was expressed with pJIM520 (Table 1) transformed into E. coli TB1. Transformed bacteria were grown at 30°C in 500 ml containing 2 g litre−1 glucose and 100 μg ml−1 ampicillin to an OD600 of 0.5. Expression of the recombinant protein was then induced for 2.5 h with 0.5 mM IPTG. Bacteria were harvested, and the recombinant protein was purified by amylose resin as described in the instruction manual (New England Biolabs).
Animal sera
Purified recombinant His6-tagged Lig proteins were introduced into preparative 10% or 12% SDS-PAGE. The full-length recombinant protein was excised from the gel and used to immunize New Zealand White rabbits (Harlan Sprague Dawley) as previously described (Matsunaga et al., 2002). The MBP-LigB fusion protein was dialysed against PBS with a Float-A-Lyzer (Spectra/Por) and concentrated with the Centricon YM-10 spin column with a molecular weight cut-off of 10 000. Immunization protocols were approved by the VA Greater Los Angeles Healthcare System Animal Research Committee. Rabbit antibody to LipL41 and GroEL were prepared as previously described (Matsunaga et al., 2002; Shang et al., 1996). Domestic rats (Rattus norvegicus) were captured from urban communities from outbreaks in Brazil (Barocchi et al., 2001). Western blot analysis for Fig. 5C was done with sera from culture-positive rats identified as reservoirs for L. interrogans.
Reverse transcriptase PCR
Leptospira kirschneri strain RM52 was grown to late exponential phase. Total RNA was extracted from 1 × 1010 leptospiral cells by the hot-phenol method and resuspended in water following ethanol precipitation (Case et al., 1990). DNA was digested with the DNA-free kit from Ambion. RT-PCR was performed with ~100 ng leptospiral RNA and Omniscript RT as described (Qiagen). Transcripts were hybridized with the following primers:
ligA, 5′-CGCAGAAATTTTAGAGGAACCTACAG-3′;
ligC, 5′-TTTGACTCCAAGACGCAGAGGATGAT-3′;
ligB, 5′-ATTTTCAAGATTTGTTCTCCAGATTT-3′;
lipl45, 5′-ATTACTTCTTGAACATCTGCTTGAT-3′.
Following reverse transcription, the following primers were added to the reactions for DNA PCR:
ligA, 5′-CTGCTACGCTTGTTGACATAGAAGTA-3′;
ligC, 5′-TAGAACCAACACGAAATGGCACAACA-3′;
ligB, 5′-ATCCGAAGTGGCATAACTCTCCTCAT-3′;
lipl45, 5′-TGAAAAGAACATTACCAGCGTTGTA-3′.
Assembly of the reaction mixtures was completed in 10× PCR Buffer, dNTPs, and Taq DNA polymerase, as instructed by the manufacturer (Qiagen). Polymerase chain reaction was performed in a Techne Progene thermocycler. An initial denaturation step of 95°C for 1 min was followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 53°C for 30 s, and extension at 72°C for 30 s. A final 72°C incubation for 30 s was then performed. Polymerase chain reaction products of 500 bp for ligA, 479 bp for ligC, 440 bp for ligB, and 438 bp for lipl45 were expected.
Southern blot analysis
Genomic DNA was extracted from leptospiral strains with the Blood and Cell Culture kit (Qiagen) from 500 ml of 7-day cultures. Approximately 3 μg of DNA was digested with 5–20 units of NsiI (NEB) overnight in a final volume of 50 μl. DNA was then purified with phenol:chloroform: isoamyl and precipitated with 100% cold ethanol and 3 M sodium acetate pH 5.2 and washed with 70% ethanol. Purified DNA was then re-digested with 5–20 units of PacI overnight in a final volume of 25 μl. The double digested DNA was separated in a 0.8% agarose gel at 20 V overnight. The gel was then incubated twice for 30 min in denaturing buffer (1.5 M NaCl, 0.5 N NaOH), and twice for 30 min in neutralization buffer (1 M Tris 1.5 M NaCl, pH 7.4). Genomic DNA was transferred onto a positively charged nylon membrane (Roche Molecular Biochemicals) according to the method described by Southern (Southern, 1975).
Probes were synthesized with the PCR Dig-labelling kit (Roche). Reactions were assembled according to the manufacturer in a final volume of 50 μl. Temperature cycles for the amplification were 94°C for 5 min, 94°C for 30 s, 57°C for 30 s, and 72°C for 1 min, with a final extension time of 7 min after 35 cycles of amplification. Primers were designed against the L. interrogans Fiocruz L1-130 lig gene sequences and are as follows: for the LigB repetitive domain 4–6 LigB_395 GATTTTAAAGTTACACAAGC, LigB_573 AAACCG GACTACTTACCTTTCC, for the C terminal specific probes: LigA.2078p TTACGGCTACAGGTATTTTTACG, LigA.2691p ATTGGAAGATTTCCAAGTAACC, LigB.5071p CATAACTC TCCTCATAACA, LigB.5548p TATGTAGAGATAAGATCC, LigC.5121p TATCTACGCTGCAAATGG, LigC.5865p TTGTT GGCGATACGTCCG.
The UV cross-linked membrane was subjected to prehybridization at 42°C for 1 h in Dig Easy Hybridization solution (Roche). Before hybridization, the Dig labelled probes were boiled for 10 min and rapidly transferred to ice for 5 min. The denatured probes were mixed with hybridization solution and incubated overnight with the membrane at 42°C. Following hybridization, the membranes were washed twice for 5 min at room temperature with 2× SSC (0.3 M NaCl, 30 mM sodium citrate), 0.1% SDS. The membranes were then washed twice for 30 min at 42°C with 0.1× SSC, 0.1% SDS. Membranes were exposed for 1–3 min to Biomax ML film (Eastman Kodak) for the detection of chemiluminescent products.
Computational analysis of bacterial-Ig like repeats and structural modelling
Lig sequences from both L. interrogans and L. kirschneri were used to search the nr NCBI database (http://www.ncbi.nlm.nih.gov), and the Pfam 7.7b database (http://pfam.wustl.edu). Schematic representation of the LigB protein was drawn with DSGene (Accelrys). Detection of motifs was based on the MEME algorithm (http://meme.sdsc.edu/meme/website/intro.html), whereas structural modelling was performed by a web based iterative search method 3-D PSSM (http://www.sbg.bio.ic.ac.uk/~3dpssm). Briefly, PSI-BLAST generated a weighted position specific scoring matrix (PSSM), which then searched a structural database for remote homologues. This method combined secondary structure and solvation potentials to recognize structural relationships.
Detergent fractionation
Leptospira kirschneri strain RM52 was fractionated by solubilization with Triton X-100. 0.5–1 × 109 leptospiral cells were washed once in 0.5 ml 1× PBS-5 mM MgCl2 and extracted in 1% Triton X-100 (Calbiochem), 20 mM Tris-HCl (pH 8), 150 mM NaCl, 2 mM EDTA, and 0.5% protease inhibitor cocktail (cat. # P8849 Sigma) for 30 min at 4°C. The insoluble protoplasmic cylinder was removed by centrifugation at 10 000 g for 10 min The soluble phase was precipitated with acetone before electrophoresis.
Immunoelectron microscopy
Leptospira interrogans (low-passage 4) and L. kirschneri (low-passage 6) were grown to log phase and centrifuged at 3000 r.p.m. for 5 min. Bacteria were resuspended in 0.1% gluteraldehyde in 1 × PBS and processed by High Pressure Freezing (Bal-Tec HPM010) followed by Freeze substitution (Leica EMAFS Automatic Freeze Substitution System). Samples were embedded in LRWhite and thin sections were placed on carbon coated nickel grids. Thin sections were incubated with 1:50 or 1:100 primary antibody in blocking buffer (1× PBS 0.1% Fish gelatin, and Tween 20), and washed with 1× PBS-Tween 20 rinse. Secondary gold conjugated goat anti-rabbit (IgG H + l) or goat anti-mouse (IgG H + l) were used at a dilution of 1:25. Grids were rinsed in 1× PBS, fixed with 0.5% gluteraldehyde, rinsed five times with distilled water, and stained in 2% Methanolic Uranyl Acetate and Lead Citrate. Sections were examined with the FEI Tec-nai 12 Electron microscope.
Immunofluorescence-labelling assays
To detect surface labelling of live, intact leptospires, cultures were centrifuged at 5000 r.p.m. for 5 min and resuspended in EMJH media. After a second washing step, 20 μl of a suspension of 2 × 109 organisms per ml were placed on poly l-lysine (Sigma) coated slides and incubated with antisera (1:50 to 1:100 dilution) to leptospiral recombinant proteins for 60 min in a humidified chamber. The slides were gently washed with EMJH media, air-dried, fixed with methanol for 10 min and then washed with 1× PBS with 10% goat serum. Antibody-labelled leptospires were incubated for 30 min with goat anti-rat Ig-FITC conjugated antibodies (Sigma), diluted 1:200. Air-dried slides were washed and mounted with a 1× PBS-glycerol solution with p-phenylene-diamine (Sigma). Labelled organisms were visualized by fluorescence microscopy.
Accession numbers
The nucleotide sequence of L. kirschneri RM52 ligA and ligB was deposited in GenBank under the accession number AY190126. The ligC sequence was deposited under accession number AY190127. The nucleotide sequence of L. interrogans Fiocruz L1-130 ligA was deposited under accession number AY221109.
Acknowledgments
We thank Betty Liu, Erin Hurley, Roya Saisan and Paul Cullen for providing technical assistance. We would also like to thank Rudy Hartskeerl for the use of the anti-LPS monoclonal antibody (F71C2), Ben Adler for the leptospiral His6-GroEL-expressing plasmid and Paulo Lee Ho for providing the pAE expression plasmid. In addition, we thank Kent McDonald, Reena Zalpuri and Gordon Vrdoljak at the UC Berkeley Electron Microscope lab for their help in preparing the EM samples for immunocytochemistry and Elizabeth Martins and Márcia Gamberini at Instituto Butantan for their help in sequencing cloned L. interrogans sequences.
This work was supported by Public Health Service grants AI-34431 (to D.A.H.) and AI-01605 (to A.I.K.) from the National Institute of Allergy and Infectious Diseases; Public Health Service grants TW-00905 and TW-00919 from the Fogarty Program in International Research and Training in Emerging Infectious Diseases; VA Medical Research Funds (to J.M. and D.A.H.); and grant 09224–7 from Biomanguinhos, the Oswaldo Cruz Foundation, Brazilian Ministry of Health (to A.I.K.).
References
- Arean VM. The pathologic anatomy and pathogenesis of fatal human leptospirosis (Weil’s disease) Am J Pathol. 1962;40:393–423. [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Ballard SA, Williamson M, Adler B, Vinh T, Faine S. Interactions of virulent and avirulent leptospires with primary cultures of renal epithelial cells. J Med Microbiol. 1986;21:59–67. doi: 10.1099/00222615-21-1-59. [DOI] [PubMed] [Google Scholar]
- Ballard SA, Segers RPAM, Bleumink-Pluym N, Fyfe J, Faine S, Adler B. Molecular analysis of the hsp (groE) operon of Leptospira interrogans serovar copenhageni. Mol Microbiol. 1993;8:739–751. doi: 10.1111/j.1365-2958.1993.tb01617.x. [DOI] [PubMed] [Google Scholar]
- Ballard SA, Go M, Segers RP, Adler B. Molecular analysis of the dnaK locus of Leptospira interrogans serovar copenhageni. Gene. 1998;216:21–29. doi: 10.1016/s0378-1119(98)00329-1. [DOI] [PubMed] [Google Scholar]
- Barnett JK, Barnett D, Bolin CA, Summers TA, Wagar EA, Cheville NF, et al. Expression and distribution of leptospiral outer membrane components during renal infection of hamsters. Infect Immun. 1999;67:853–861. doi: 10.1128/iai.67.2.853-861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barocchi MA, Ko AI, Ferrer SR, Faria MT, Reis MG, Riley LW. Identification of new repetitive element in Leptospira interrogans serovar copenhageni and its application to PCR-based differentiation of Leptospira serogroups. J Clin Microbiol. 2001;39:191–195. doi: 10.1128/JCM.39.1.191-195.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barocchi MA, Ko AI, Reis MG, McDonald KL, Riley LW. Rapid translocation of polarized MDCK cell monolayers by Leptospira interrogans, an invasive but nonintracellular pathogen. Infect Immun. 2002;70:6926–6932. doi: 10.1128/IAI.70.12.6926-6932.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, et al. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case CC, Simons EL, Simons RW. The IS10 transposase mRNA is destabilized during antisense RNA control. EMBO J. 1990;9:1259–1266. doi: 10.1002/j.1460-2075.1990.tb08234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SL, Buckingham M, Taylor MP. Studies on Leptospira icterohaemorrhagiae. IV. Survival in water and sewage: destruction in water by halogen compounds, synthetic detergents, and heat. J Infect Dis. 1948;82:256–266. doi: 10.1093/infdis/82.3.256. [DOI] [PubMed] [Google Scholar]
- Dersch P, Isberg RR. An immunoglobulin superfamily-like domain unique to the Yersinia pseudotuberculosis invasin protein is required for stimulation of bacterial uptake via integrin receptors. Infect Immun. 2000;68:2930–2938. doi: 10.1128/iai.68.5.2930-2938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faine, S. (1982) Guidelines for the Control of Leptospirosis. Geneva: World Health Organization.
- Faine, S., Adler, B., Bolin, C., and Perolat, P. (1999) Leptospira and Leptospirosis. Melbourne, Australia: MediSci.
- Flannery B, Costa D, Carvalho FP, Guerreiro H, Matsunaga J, Da Silva ED, et al. Evaluation of recombinant leptospira antigen-based enzyme-linked immunosorbent assays for the serodiagnosis of leptospirosis. J Clin Microbiol. 2001;39:3303–3310. doi: 10.1128/JCM.39.9.3303-3310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg EP, Canale-Parola E. Motility of flagellated bacteria in viscous environments. J Bacteriol. 1977;132:356–358. doi: 10.1128/jb.132.1.356-358.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A, Zingales B. Alternative method to remove antibacterial antibodies from antisera used for screening of expression libraries. Biotechniques. 1995;19 (28):30. [PubMed] [Google Scholar]
- Grunberg-Manago M. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu Rev Genet. 1999;33:193–227. doi: 10.1146/annurev.genet.33.1.193. [DOI] [PubMed] [Google Scholar]
- Guerreiro H, Croda J, Flannery B, Mazel M, Matsunaga J, Galvao Reis M, et al. Leptospiral proteins recognized during the humoral immune response to leptospirosis in humans. Infect Immun. 2001;69:4958–4968. doi: 10.1128/IAI.69.8.4958-4968.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA. Spirochetal lipoproteins and pathogenesis. Microbiology. 2000;146:1491–1504. doi: 10.1099/00221287-146-7-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Matsunaga J. Characterization of the leptospiral outer membrane and description of three novel leptospiral membrane proteins. Infect Immun. 2002;70:4936–4945. doi: 10.1128/IAI.70.9.4936-4945.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Walker EM, Blanco DR, Bolin CA, Miller MN, Lovett MA. Changes in the surface of Leptospira interrogans serovar grippotyphosa during in vitro cultivation. Infect Immun. 1991;59:1131–1140. doi: 10.1128/iai.59.3.1131-1140.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Champion CI, Martinich C, Shang ES, Blanco DR, Miller JN, Lovett MA. Molecular cloning and sequence analysis of the gene encoding OmpL1, a transmembrane outer membrane protein of pathogenic Leptospira spp. J Bacteriol. 1993;175:4225–4234. doi: 10.1128/jb.175.13.4225-4234.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Martinich C, Summers TA, Shang ES, Pruetz JD, McCoy AM, et al. Characterization of leptospiral outer membrane lipoprotein LipL36: Down-regulation associated with late log-phase growth and mammalian infection. Infect Immun. 1998;66:1579–1587. doi: 10.1128/iai.66.4.1579-1587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Chao G, Zuerner RL, Barnett JK, Barnett D, Mazel M, et al. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect Immun. 2000;68:2276–2285. doi: 10.1128/iai.68.4.2276-2285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger ZA, Brown MS, sberg RR, Bjorkman PJ. Crystal structure of invasin: a bacterial integrin-binding protein. Science. 1999;286:291–295. doi: 10.1126/science.286.5438.291. [DOI] [PubMed] [Google Scholar]
- Hellstrom JS, Marshall RB. Survival of Leptospira interrogans serovar pomona in an acidic soil under simulated New Zealand field conditions. Res Vet Sci. 1978;25:29–33. [PubMed] [Google Scholar]
- Ito T, Yanagawa R. Leptospiral attachment to extracellular matrix of mouse fibroblast (L929) cells. Vet Microbiol. 1987;15:89–96. doi: 10.1016/0378-1135(87)90133-7. [DOI] [PubMed] [Google Scholar]
- Johnson RC, Harris VG. Differentiation of pathogenic and saprophytic letospires. I. Growth at low temperatures. J Bacteriol. 1967;94:27–31. doi: 10.1128/jb.94.1.27-31.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost BH, Adler B, Faine S. Experimental immunisation of hamsters with lipopolysaccharide antigens of Leptospira interrogans. J Med Microbiol. 1989;29:115–120. doi: 10.1099/00222615-29-2-115. [DOI] [PubMed] [Google Scholar]
- Ko AI, Galvao Reis M, Ribeiro Dourado CM, Johnson WD, Jr, Riley LW, Group TSLS. Urban epidemic of severe leptospirosis in Brazil. Lancet. 1999;354:820–825. doi: 10.1016/s0140-6736(99)80012-9. [DOI] [PubMed] [Google Scholar]
- Lee SH, Kim S, Park SC, Kim MJ. Cytotoxic activities of Leptospira interrogans hemolysin SphH as a pore-forming protein on mammalian cells. Infect Immun. 2002;70:315–322. doi: 10.1128/IAI.70.1.315-322.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Frey EA, Pfuetzner RA, Creagh AL, Knoechel DG, Haynes CA, et al. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature. 2000;405:1073–1077. doi: 10.1038/35016618. [DOI] [PubMed] [Google Scholar]
- Marotto PC, Nascimento CM, Eluf-Neto J, Marotto MS, Andrade L, Sztajnbok J, Seguro AC. Acute lung injury in leptospirosis: clinical and laboratory features, outcome, and factors associated with mortality. Clin Infect Dis. 1999;29:1561–1563. doi: 10.1086/313501. [DOI] [PubMed] [Google Scholar]
- Marshall RB. The route of entry of leptospires into the kidney tubule. J Med Microbiol. 1976;9:149–152. doi: 10.1099/00222615-9-2-149. [DOI] [PubMed] [Google Scholar]
- Matsunaga J, Young TA, Barnett JK, Barnett D, Bolin CA, Haake DA. Novel 45-kilodalton leptospiral protein that is processed to a 31- kilodalton growth-phase-regulated peripheral membrane protein. Infect Immun. 2002;70:323–334. doi: 10.1128/IAI.70.1.323-334.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merien F, Baranton G, Perolat P. Invasion of Vero cells and induction of apoptosis in macrophages by pathogenic Leptospira interrogans are correlated with virulence. Infect Immun. 1997;65:729–738. doi: 10.1128/iai.65.2.729-738.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merien F, Truccolo J, Baranton G, Perolat P. Identification of a 36-kDa fibronectin-binding protein expressed by a virulent variant of Leptospira interrogans serovar icterohaemorrhagiae. FEMS Microbiol Lett. 2000;185:17–22. doi: 10.1111/j.1574-6968.2000.tb09034.x. [DOI] [PubMed] [Google Scholar]
- Nally JE, Timoney JF, Stevenson B. Temperature-regulated protein synthesis by Leptospira interrogans. Infect Immun. 2001;69:400–404. doi: 10.1128/IAI.69.1.400-404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson G, Belasco JG, Cohen SN, von Gabain A. Effect of premature termination of translation on mRNA stability depends on the site of ribosome release. Proc Natl Acad Sci USA. 1987;84:4890–4894. doi: 10.1073/pnas.84.14.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniappan RU, Chang YF, Jusuf SS, Artiushin S, Timoney JF, McDonough SP, et al. Cloning and molecular characterization of an immunogenic LigA protein of Leptospira interrogans. Infect Immun. 2002;70:5924–5930. doi: 10.1128/IAI.70.11.5924-5930.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Peña-Moctezuma A, Bulach DM, Adler B. Genetic differences among the LPS biosynthetic loci of serovars of Leptospira interrogans and Leptospira borgpetersenii. FEMS Immunol Medical Microbiol. 2001;31:73–81. doi: 10.1111/j.1574-695X.2001.tb01589.x. [DOI] [PubMed] [Google Scholar]
- Perolat P, Chappel RJ, Adler B, Baranton G, Bulach DM, Billinghurst ML, et al. Leptospira fainei sp. nov. , isolated from pigs in Australia. Int J Syst Bacteriol. 1998;48(Part 3):851–858. doi: 10.1099/00207713-48-3-851. [DOI] [PubMed] [Google Scholar]
- Segers RP, van Gestel JA, van Eys GJ, van der Zeijst BA, Gaastra W. Presence of putative sphin-gomyelinase genes among members of the family Leptospiraceae. Infect Immun. 1992;60:1707–1710. doi: 10.1128/iai.60.4.1707-1710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang ES, Summers TA, Haake DA. Molecular cloning and sequence analysis of the gene encoding LipL41, a surface-exposed lipoprotein of pathogenic Leptospira species. Infect Immun. 1996;64:2322–2330. doi: 10.1128/iai.64.6.2322-2330.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thomas DD, Higbie LM. In vitro association of leptospires with host cells. Infect Immun. 1990;58:581–585. doi: 10.1128/iai.58.3.581-585.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevejo RT, Rigau-Perez JG, Ashford DA, McClure EM, Jarquin-Gonzalez C, Amador JJ, et al. Epidemic leptospirosis associated with pulmonary hemorrhage-Nicaragua, 1995. J Infect Dis. 1998;178:1457–1463. doi: 10.1086/314424. [DOI] [PubMed] [Google Scholar]
- Trowbridge AA, Green JB, Bonnett JD, Shohet SB, Ponnappa BD, McCombs WB. Hemolytic anemia associated with leptospirosis. Morphologic and lipid studies. Am J Clin Pathol. 1981;76:493–498. doi: 10.1093/ajcp/76.4.493. [DOI] [PubMed] [Google Scholar]
- Trueba GA, Bolin CA, Zuerner RL. Characterization of the periplasmic flagellum proteins of Leptospira interrogans. J Bacteriol. 1992;174:4761–4768. doi: 10.1128/jb.174.14.4761-4768.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto M, Niikura M, Ono E, Kida H, Yanagawa R. Leptospiral attachment to cultured cells. Zentralbl Bakteriol Mikrobiol Hyg [a] 1984;258:268–274. doi: 10.1016/s0176-6724(84)80044-9. [DOI] [PubMed] [Google Scholar]
- Vinh T, Faine S, Adler B. Adhesion of leptospires to mouse fibroblasts (L929) and its enhancement by specific antibody. J Med Microbiol. 1984;18:73–85. doi: 10.1099/00222615-18-1-73. [DOI] [PubMed] [Google Scholar]
- Werts C, Tapping RI, Mathison JC, Chuang TH, Kravchenko V, Saint Girons I, et al. Leptospiral lipopolysaccharide activates cells through a TLR2-dependent mechanism. Nat Immunol. 2001;2:346–352. doi: 10.1038/86354. [DOI] [PubMed] [Google Scholar]
- Yuri K, Takamoto Y, Okada M, Hiramune T, Kikuchi N, Yanagawa R. Chemotaxis of leptospires to hemoglobin in relation to virulence. Infect Immun. 1993;61:2270–2272. doi: 10.1128/iai.61.5.2270-2272.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuerner RL, Knudtson W, Bolin CA, Trueba G. Characterization of outer membrane and secreted proteins of Leptospira interrogans serovar pomona. Microb Pathog. 1991;10:311–322. doi: 10.1016/0882-4010(91)90014-2. [DOI] [PubMed] [Google Scholar]


