Abstract
Mutations in the human genes encoding WNK1 [with no K (lysine) protein kinase-1] and the related protein kinase WNK4 are the cause of Gordon's hypertension syndrome. Little is known about the molecular mechanism by which WNK isoforms regulate cellular processes. We immunoprecipitated WNK1 from extracts of rat testis and found that it was specifically associated with a protein kinase of the STE20 family termed ‘STE20/SPS1-related proline/alanine-rich kinase’ (SPAK). We demonstrated that WNK1 and WNK4 both interacted with SPAK as well as a closely related kinase, termed ‘oxidative stress response kinase-1’ (OSR1). Wildtype (wt) but not catalytically inactive WNK1 and WNK4 phosphorylated SPAK and OSR1 to a much greater extent than with other substrates utilized previously, such as myelin basic protein and claudin-4. Phosphorylation by WNK1 or WNK4 markedly increased SPAK and OSR1 activity. Phosphopeptide mapping studies demonstrated that WNK1 phosphorylated kinase-inactive SPAK and OSR1 at an equivalent residue located within the T-loop of the catalytic domain (Thr233 in SPAK, Thr185 in OSR1) and a serine residue located within a C-terminal non-catalytic region (Ser373 in SPAK, Ser325 in OSR1). Mutation of Thr185 to alanine prevented the activation of OSR1 by WNK1, whereas mutation of Thr185 to glutamic acid (to mimic phosphorylation) increased the basal activity of OSR1 over 20-fold and prevented further activation by WNK1. Mutation of Ser325 in OSR1 to alanine or glutamic acid did not affect the basal activity of OSR1 or its ability to be activated by WNK1. These findings suggest that WNK isoforms operate as protein kinases that activate SPAK and OSR1 by phosphorylating the T-loops of these enzymes, resulting in their activation. Our analysis also describes the first facile assay that can be employed to quantitatively assess WNK1 and WNK4 activity.
Keywords: Gordon's syndrome, hypertension, mass spectrometry (MS), Na+–K+–2Cl− co-transporter-1 (NKCC1), protein kinase, pseudohypoaldosteronism type II (PHAII)
Abbreviations: DTT, dithiothreitol; ERK, extracellular-signal-regulated kinase; GST, glutathione S-transferase; H2A, histone 2A; ki, kinase-inactive; MALDI-TOF, matrix-assisted laser-desorption ionization–time-of-flight; MBP, myelin basic protein; NKCC1, Na+–K+–2Cl− co-transporter-1; OSR1, oxidative stress response kinase-1; PHAII, pseudohypoaldosteronism type II; SPAK, STE20/SPS1-related proline/alanine-rich kinase; WNK, with no K (lysine) protein kinase; WNK1-(CT), WNK1 C-terminus; wt, wild-type
INTRODUCTION
The WNK [with no K (lysine) protein kinase] family of protein kinases comprise four members (WNK1, WNK2, WNK3 and WNK4) and were originally identified as distinctive serine/threonine protein kinases that lack a conserved lysine residue normally found in subdomain II of the catalytic domain [1,2]. Subsequent studies identified mutations in the genes encoding WNK1 and WNK4 in families with an inherited hypertension and hyperkalaemia (elevated plasma K+) disorder called pseudohypoaldosteronism type II (PHAII, also known as Gordon's syndrome) [3]. WNK isoforms are large protein kinases (WNK1, 2382 residues; WNK4, 1243 residues) in which the catalytic domain is located at the N-terminus (residues 221–479 for WNK1 and 174–432 for WNK4). Apart from two putative coiled-coil domains, the remainder of the WNK polypeptides possess no obvious structural features. Mutations in the WNK1 gene found in PHAII subjects are deletions in intron-1, which reportedly elevate the expression of the WNK1 protein, indicating that hypertension could result from increased expression of WNK1 [3]. Consistent with this notion, mice lacking one allele of WNK1, which were presumed to possess decreased levels of WNK1 protein, had lower blood pressure [4]. The WNK1 knockout is an embryonic lethal, indicating that WNK1 is also required for normal development [4]. Thus far the mutations in the WNK4 gene found in PHAII subjects lie distal to both of the putative coiled-coil domains [3,5].
Most functional studies on WNK1 and WNK4 have focused on the overexpression of these enzymes and monitoring of the effects that this has on co-expressed ion channels or co-transporters in Xenopus oocytes or paracellular ion flux through tight junctions in epithelial cells [6–8]. For example, overexpression of WNK4 markedly inhibited Na+ flux mediated by the Na+–Cl− co-transporter, by reducing its level at the plasma membrane [9,10]. Overexpression of WNK4 in MDCK (Madin–Darby canine kidney) epithelial cells increased paracellular ion flux through tight junctions, an effect that was postulated to result from phosphorylation of claudins, a family of transmembrane tight-junction proteins [11]. It was recently reported that WNK1 interacted with, and phosphorylated, synaptotagmin-2, a Ca2+ sensor, which regulates endocytosis and exocytosis and may affect trafficking of ion channels to the plasma membrane [12]. In the present study we have analysed the proteins that interact with WNK1, and this has led to the identification of two related protein kinases of the STE20 family that are phosphorylated and activated by WNK isoforms.
MATERIALS AND METHODS
Immunoprecipitation of endogenous WNK1
The WNK1-(CT) (WNK1 C-terminus) and pre-immune IgG antibodies were covalently coupled to Protein G–Sepharose in a ratio of 1 mg of antibody to 1 ml of resin using a dimethyl pimelimidate cross-linking procedure [13]. As a pre-clearing step, 50 mg of rat testis lysate was incubated at 4 °C for 20 min on a rolling shaker with 0.5 ml of Protein G–Sepharose. The supernatant was then incubated at 4 °C for 1 h on a rolling shaker with 50 μl of WNK1-(CT) or IgG–Protein G–Sepharose-conjugated antibodies. The immunoprecipitates were washed four times with 10 ml of Lysis Buffer containing 0.15 M NaCl and lacking DTT (dithiothreitol) and twice with 10 ml of 10 mM Tris/HCl (pH 8)/0.1 mM EGTA. The resin was resuspended in a total volume of 0.1 ml and 30 μl of NuPAGE® LDS Sample Preparation Buffer [purchased from Invitrogen and containing 0.14 M Tris, 2% (w/v) LDS (lithium dodecyl sulphate) and 10% (v/v) glycerol, final pH 8.5] in the absence of DTT. The samples were filtered through a 0.44-μm-pore-size Spin-X filter tube (Corning), DTT was added to a final concentration of 10 mM, then samples were heated for 5 min at 70 °C and concentrated by speed-vacuum centrifugation to 30 μl. Samples were alkylated for 30 min at room temperature using 50 mM 4-vinylpyridine in 10 mM NH4HCO3 and then subjected to electrophoresis on a 4–12% (w/v) polyacrylamide gel using Mops as a running buffer. The gel was stained with colloidal Coomassie Blue (Invitrogen). The bands were excised, washed and digested with trypsin as described previously [14]. Peptides were analysed by combined MALDI-TOF (matrix-assisted laser-desorption ionization–time-of-flight) and MALDI-TOF/TOF MS analysis on an Applied Biosystems 4700 ToF/ToF Proteomics Analyser using 5 mg/ml α-cyanocinnamic acid in 10 mM ammonium phosphate as the matrix or by liquid chromatography–MS on an Applied Biosystems 4000 Q-TRAP instrument. The Celera Discovery System (Applied Biosystems) Rodent database was searched using the Mascot search algorithm (http://www.matrixscience.com [15]).
Expression and purification of GST (glutathione S-transferase) fusion proteins in HEK-293 cells
Dishes (10 cm diameter) of HEK-293 cells were transfected with 5–10 μg of the pEBG-6P or pEBG-2T constructs alone or together with the indicated pCMV5 constructs using the polyethylenimine method [16]. At 36 h post-transfection the cells were lysed in 0.5 ml of ice-cold Lysis Buffer and the clarified lysates were incubated for 1 h on a rotating platform with glutathione–Sepharose 4B (10 μl/10-cm-diameter dish of lysate) previously equilibrated in Lysis Buffer. The beads were washed four times with Lysis Buffer containing 0.15 M NaCl and twice with Buffer A [50 mM Tris/HCl (pH 7.5)/0.1 EGTA/1 mM DTT] containing 0.27 M sucrose. The resin was incubated in a 1:1 slurry of Buffer A containing 0.27 M sucrose and 20 mM glutathione to elute the GST-fusion proteins. The beads were then removed by filtration through a 0.44-μm-pore-size Spin-X filter tube (Corning) and the eluate divided into aliquots, snap-frozen in liquid N2 and stored at −80 °C.
Expression of WNK1-(1–661), SPAK (oxidative stress response kinase 1), OSR1 (STE/SPS1-related proline/alanine-rich kinase), NKCC1-(1–260) [Na+–K+-2Cl− co-transporter 1-(1–260)] and claudin-4 in Escherichia coli
All pGEX-6P-1 constructs were transformed into BL21 E. coli cells and a 0.5-litre culture was grown at 37 °C in Luria Broth containing 100 μg/ml ampicillin and 30 μg/ml chloramphenicol until the D600 (attenuance at 600 nm) was 0.8. Isopropyl β-D-thiogalactopyranoside (‘IPTG’; 30 μM) was then added. The cells were cultured for a further 8–16 h at 26 °C, resuspended in 12.5 ml of ice-cold Lysis Buffer and frozen in liquid N2. After thawing, 12.5 ml of Lysis Buffer containing 1 mg/ml chicken egg lysozyme and 10 units/ml of DNase1 were added. Samples were gently agitated at 4 °C for 20 min and then sonicated briefly. Lysates were centrifuged at 4 °C for 15 min at 26 000 g and incubated with 0.5 ml of glutathione–Sepharose for 1 h. The resin was washed in Lysis Buffer containing 0.5 M NaCl, followed by Buffer A containing 0.27 M sucrose. Proteins were either eluted by the addition of 20 mM glutathione or by incubating the resin overnight with GST-PreScission™ Protease (30 μg/ml of slurry; for source, go to http://www.BiochemJ.org/bj/391/bj3910017add.htm) Eluted protein was stored in aliquots in −80 °C.
In vitro WNK1 and WNK4 phosphorylation reactions
Assays were set up in a total volume of 25 μl of Buffer A containing 0.1 μM kinase {GST–WNK1-(1–661), GST–[D368A]-WNK1-(1–661), GST–WNK4-(1–593) or GST–[K186A/D321A]WNK4-(1–593)}, 5 μM of substrate {[D212A]SPAK, [D164A]OSR1, MBP (myelin basic protein), claudin-4 or H2A (histone 2A)}, 10 mM MgCl2 and 0.1 mM [γ-32P]ATP (≈300 c.p.m./pmol). After incubation for 30 min at 30 °C, incorporation of phosphate was determined after electrophoresis of samples on a NuPAGE® Bis-Tris/10%-polyacrylamide gels and autoradiography of the dried Coomassie Blue-stained gels. The proteins bands corresponding to SPAK, OSR1, MBP, claudin-4 and H2A were excised, and phosphate incorporation was quantified on a Wallac liquid-scintillation counter.
Activation of SPAK/OSR1 by WNK1/WNK4
The activation assays mixtures were set up in a total volume of 25 μl of Buffer A containing 0.25 μM kinase {GST–WNK1-(1–661), GST–[D368A]WNK1-(1–661), GST–WNK4-(1–593) or GST–[K186A/D321A]WNK4-(1–593)}, 5 μM substrate (GST–SPAK, [D212A]-GST–SPAK, GST–OSR1 or [D164A]-GST–OSR1), 10 mM MgCl2 and 0.1 μM non-radioactive ATP. After incubation for 40 min at 30 °C, 5 μl of the activation assay mix was transferred to 20 μl of a solution containing 6.25 μM NKCC1-(1–260), 10 mM MgCl2 and 0.1 μM [γ-32P]ATP (≈300 c.p.m./pmol). After incubation for 20 min at 30 °C, incorporation of phosphate was determined after electrophoresis of samples on a NuPAGE® Bis-Tris/10%-polyacrylamide gel and autoradiography of the dried Coomassie Blue-stained gels. The protein bands corresponding to NKCC1 were excised and phosphate incorporation was quantified on a Wallac liquid-scintillation counter. It should be noted that NKCC1-(1–260) (in which the GST tag has been removed) migrates with an apparent molecular mass of 40 kDa on our gel system. Electrospray MS confirmed that the molecular mass of the NKCC1-(1–260) fragment corresponded to the theoretical mass (results not shown).
Mapping phosphorylation sites on SPAK and OSR1
The phosphorylation assays were set up in a 25 μl volume of Buffer A containing 2 μM GST–WNK1-(1–661), 10 μM [D212A]-SPAK or [D164A]OSR1, 10 mM MgCl2, 0.1 μM [γ-32P]ATP (≈2000 c.p.m./pmol). After incubation for 40 min at 30 °C, incorporation of phosphate was determined after electrophoresis of samples on a NuPAGE® Bis-Tris 10%-polyacrylamide gel and autoradiography of Coomassie Blue-stained gels. The protein bands corresponding to SPAK and OSR1 were excised, and phosphate incorporation was quantified on a Wallac liquid-scintillation counter. After tryptic digestion, more than 86% of the 32P radioactivity incorporated into the gel band was recovered and the samples were chromatographed on a reverse-phase Vydac 218TP5215 C18 HPLC column as described in the legend of Figure 5 (below). Peptide analysis of the fractions corresponding to the major 32P-containing peaks was performed using an Applied Biosystems 4700 Proteomics Analyser (MALDI-TOF/TOF) and solid-phase Edman degradation on an Applied Biosystems 494C sequenator of the peptide coupled to Sequelon-AA membrane (Milligen Corp., Bedford, MA, U.S.A.) as described previously [17].
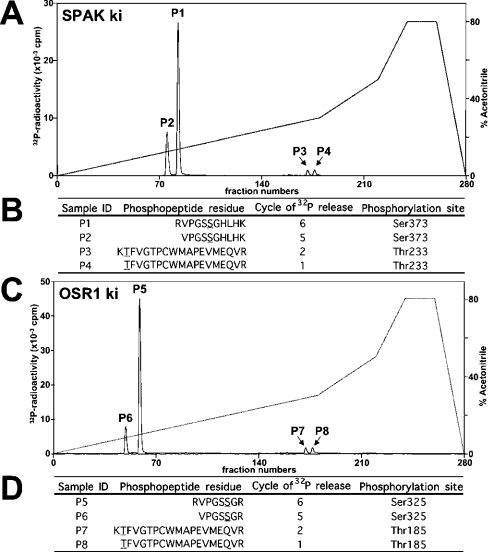
Figure 5. Analysis of phosphorylation of SPAK and OSR1.
(A) ki [D212A]SPAK was phosphorylated by wt WNK1-(1–661) for 40 min under conditions in which phosphorylation was maximal (results not shown). The 32P-labelled SPAK was isolated by PAGE, digested with trypsin and the resulting peptides were chromatographed on a C18 column. Fractions containing the major 32P-labelled peptides are marked. (B) The indicated peptides were analysed by MALDI-TOF/TOF-MS as described in the Materials and methods section. The site of phosphorylation within each peptide was determined by solid-phase Edman sequencing in which 32P radioactivity was measured after each cycle of Edman degradation. The cycle number in which 32P radioactivity was released is indicated. The deduced amino acid sequences of peptides P1, P2, P3, and P4 are indicated; the phosphorylated residues are underlined. (C and D) As above, except that ki [D164A]OSR1 was employed.
Supplementary materials and methods
Supplementary materials and methods, including the topics Materials, Antibodies, General methods and buffers, DNA constructs, Immunoblotting, Multiple sequence alignment and supplementary references can be accessed by going to http://www.BiochemJ.org/bj/391/bj3910017add.htm.
RESULTS
Analysis of WNK1-interacting proteins
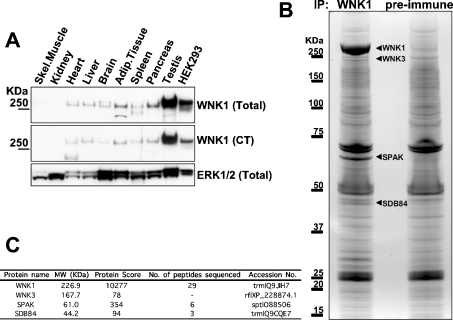
To identify a tissue in which WNK1 was highly expressed, we immunoblotted adult rat tissue extracts with two anti-WNK1 antibodies (Figure 1A). We found that WNK1 was expressed in several tissues, but was most abundant in testis. We were unable to detect significant expression of WNK1 in skeletal-muscle or kidney extracts (Figure 1A). WNK1 was also expressed at a high level in HEK-293 epithelial cells, albeit at a lower level than in testis. In order to identify WNK1-binding proteins, we immunoprecipitated endogenous WNK1 from 50 mg of testis extract and, in parallel experiments, performed a control immunoprecipitation using a pre-immune antibody. The purified preparations were subjected to electrophoresis on a polyacrylamide gel, which was then stained with colloidal Coomassie Blue (Figure 1B). The identities of the major bands in the WNK1 and the control immunoprecipitation were established by tryptic-peptide MS fingerprinting procedures (Figure 1C; results not shown). In the WNK1 immunoprecipitation, the major 250 kDa band that was not present in the control immunoprecipitates was identified as WNK1. In addition to WNK1, one major colloidal Coomassie Blue-stained band at ≈65 kDa was also only observed in the WNK1, and not in the control immunoprecipitation, and this was identified as SPAK, a STE20-like kinase [18]. Interestingly, Gagnon and co-workers have also recently reported that WNK4 can interact with SPAK in a yeast two-hybrid system [19]. Two other more minor bands that appeared specifically associated with WNK1 immunoprecipitate were identified as WNK3 (≈220 kDa) [2] and SDB84 antigen (≈45 kDa) [20]. WNK3 is unlikely to be directly immunoprecipitated with the WNK1 antibody raised against the C-terminal peptide of WNK1, as the sequence to which the antibody was raised is not conserved in WNK3. However, as WNK1 and WNK4 have been reported to oligomerize [21], WNK3 might also bind WNK1. We cloned SDB84, but were unable to demonstrate that it interacted with WNK1 when overexpressed in HEK-293 cells, suggesting that it may not interact with WNK1 directly. We were also unable to demonstrate that WNK1 phosphorylated recombinant SDB84 in a standard in vitro kinase assay (results not shown).
Figure 1. Analysis of WNK1-binding proteins.
(A) Rat tissue extracts (40 μg of protein) were immunoblotted with the indicated antibodies. Similar findings were obtained in at least two separate experiments. (B) Rat testis extracts were subjected to immunoprecipitation with WNK1 or pre-immune antibody. The immunoprecipitates were subjected to PAGE and the protein bands visualized following colloidal Coomassie Blue staining. The major bands observed in the WNK1, but not the pre-immune, purification are indicated. (C) These bands, as well as the corresponding region in the preimmune sample, were excised from the gel, digested in-gel with trypsin, and the identities of the proteins were determined by tryptic-peptide mass-spectral fingerprinting as described in the Materials and methods section. Mascot protein score where a value of >63 is considered significant (P<0.05), number (No.) of peptides sequenced by MALDI-TOF/TOF-MS/MS (SPAK and SDB84) and liquid chromatography–tandem MS (WNK1) and the accession numbers for each protein identified are indicated. Tryptic peptides derived from WNK1, WNK3, SPAK and SDB84 were present in the WNK1, but not in the preimmune, immunoprecipitate.
Specific interaction of WNK1 and WNK4 with SPAK and OSR1
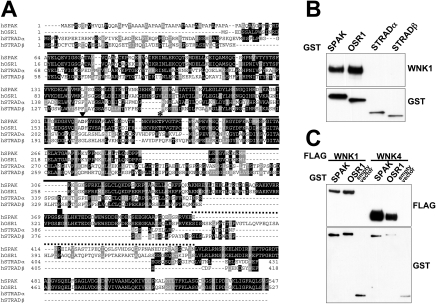
SPAK belongs to a group of four highly related STE20 family kinases, the other members being OSR1 [22], STRADα and STRADβ [23,24] (Figure 2A). SPAK and OSR1 are reported to be active kinases and are 68% identical in sequence with one another (89% in the catalytic domain). STRADα and STRADβ are catalytically inactive pseudokinases that interact with, and activate, the LKB1 tumour suppressor protein kinase [23,24] and are ≈30% identical in their pseudokinase domains with the catalytic domain of SPAK and OSR1. In order to analyse the binding of WNK1 to these STE20 family kinases, we expressed SPAK, OSR1, STRADα and STRADβ as GST fusion proteins in HEK-293 cells and, following affinity purification, tested whether they were associated with endogenously expressed WNK1. Both SPAK and OSR1, but neither isoform of STRAD, interacted with WNK1 (Figure 2B). As described in the Introduction, mutations in WNK4, like those found in WNK1, lead to Gordon's syndrome, suggesting that WNK1 and WNK4 may possess similar physiological functions and/or interacting partners. We therefore sought to determine whether WNK4 could also interact with SPAK and OSR1. We overexpressed WNK4 with SPAK and OSR1 in HEK-293 cells, as endogenous WNK4 is not detectable in these cells, and found that WNK4 interacted with SPAK and OSR1 (Figure 2C).
Figure 2. Interaction of WNK1/WNK4 with SPAK and OSR1.
(A) Multiple sequence alignment of the indicated STE20 human kinases. Identical residues are highlighted in black and similar residues are highlighted in grey. The kinase domain is marked with a continuous line, regions of non-similarity between SPAK and OSR1 are marked with a dotted line, the Mg2+-binding aspartic acid residue mutated to inactivate SPAK and OSR1 is marked with a triangle, the T-loop threonine residue phosphorylated by WNK1/WNK4 is marked with an asterisk, and the non-catalytic serine residue phosphorylated by WNK1/WNK4 with a square. (B) HEK-293 cells were transfected with constructs encoding the indicated GST-fusion proteins. At 36 h post-transfection, the GST-fusion proteins were affinity-purified and immunoblotted with GST antibodies or WNK1 antibody to detect endogenously associated WNK1. (C) HEK-293 cells were co-transfected with the indicated combinations of FLAG-tagged WNK1/WNK4, GST–SPAK, GST–OSR1 or empty pEBG2T vector. GST-fusion proteins were affinity-purified and immunoblotted with FLAG antibody to detect WNK1 or WNK4 expression or GST antibody to detect SPAK, OSR1 or GST expression.
Phosphorylation of SPAK/OSR1 by WNK1/WNK4
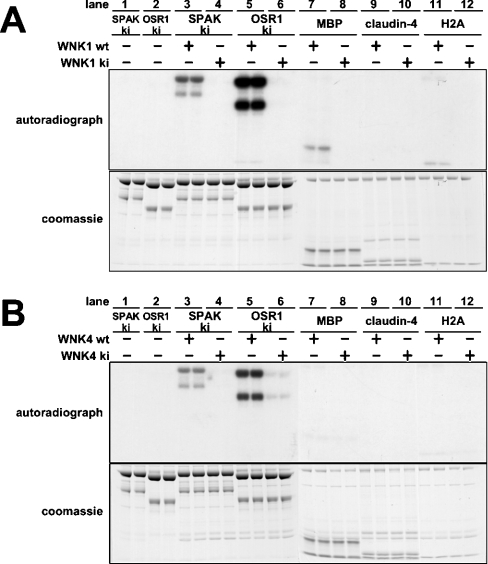
We next tested whether an N-terminal fragment of WNK1, encompassing residues 1–661, the largest catalytically active fragment of WNK1 that we have thus far been able to express in a soluble form [25], could phosphorylate SPAK and OSR1. We found that WNK1-(1–661), expressed in E. coli (Figure 3A) or HEK-293 cells (results not shown), was capable of phosphorylating catalytically inactive mutants of [D212A]SPAK (Figure 3A, lane 3) or [D164A]OSR1 (Figure 3A, lane 5). A catalytically inactive mutant of WNK1, namely [D368A]WNK1-(1–661), did not phosphorylate SPAK or OSR1, suggesting that wt (wild-type) WNK1 directly phosphorylates SPAK (Figure 3A; cf. lanes 3 and 4) and OSR1 (Figure 3A; cf. lanes 5 and 6). Under the conditions employed, in which all substrates were assayed at a concentration of 5 μM, SPAK and OSR1 were phosphorylated to a significantly greater extent than MBP, a previously utilized as an in vitro substrate [1], claudin-4, a previously reported WNK4 substrate [11], or H2A.
Figure 3. Phosphorylation of SPAK/OSR1 by WNK1 and WNK4.
(A) Kinase-active wt WNK1-(1–661) or kinase-inactive (ki) [D368A]WNK1-(1–661) was incubated with the indicated proteins (ki-[D212A]SPAK, ki [D164A]OSR1, MBP, claudin-4 and H2A) in the presence of Mg2+ and [γ-32P]ATP. Phosphorylation of protein substrates was determined after PAGE and autoradiography (upper panel) of the Coomassie Blue-stained bands (lower panel) corresponding to each substrate. It should be noted that recombinant SPAK and OSR1 migrate as a doublet band with the upper band migrating with the expected molecular mass for the full-length protein. We presume that the lower band is a proteolytic fragment lacking the C-terminal region, as these enzymes were expressed as N-terminal GST-fusion proteins. (B) As above, expect that kinase-active wt WNK4-(1–593) or ki [K186A/D321A]WNK4-(1–593) were employed. A double mutant of WNK4 was generated in order to ensure complete catalytic inactivation of the kinase. For both (A) and (B) each experimental condition was assayed in duplicate and similar results obtained in at least two different experiments.
To evaluate whether WNK4 could phosphorylate SPAK and OSR1, we expressed a fragment of WNK4-(1–593) in HEK-293 cells, as we could not express WNK4 in E. coli, and found that, like WNK1, it could phosphorylate SPAK and OSR1 (Figure 3B). A catalytically inactive mutant of WNK4, namely [K186A/D321A]-WNK4-(1–593), did not phosphorylate SPAK or OSR1. WNK4-(1–593) did not significantly phosphorylate MBP, Claudin-4 or H2A (Figure 3B).
Activation of SPAK/OSR1 by WNK1/WNK4
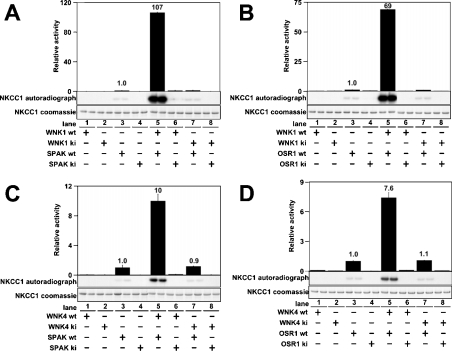
We next tested whether WNK isoforms could activate SPAK or OSR1. In order to assess the activity of SPAK and OSR1, we employed a fragment of NKCC1 encompassing the N-terminal intracellular domain (residues 1–260) that SPAK has been reported to phosphorylate [26]. Active WNK1, wt full-length SPAK or OSR1 and NKCC1-(1–260) employed in these studies were expressed in E. coli to ensure that they were not contaminated with any other mammalian kinase. WNK1, SPAK or OSR1 alone did not phosphorylate NKCC1-(1–260) significantly (Figures 4A and 4B, lanes 1 and 3). However, if SPAK or OSR1 (Figures 4A and 4B, lane 5) were incubated with active WNK1-(1–661), in the presence of Mg-ATP, NKCC1-(1–260) was markedly phosphorylated. The extent of phosphorylation was over 100-fold higher for SPAK and over 60-fold higher for OSR1 than background levels observed with SPAK or OSR1 alone. In contrast, a catalytically inactive mutant of WNK1 failed to activate SPAK and OSR1 (Figures 4A and 4B, lane 7). Moreover, catalytically inactive [D212A]SPAK or [D164A]OSR1 were not activated by WNK1-(1–661) (Figures 4A and 4B, lane 6). wt WNK4-(1–593), but not catalytically inactive WNK4-(1–593), was also capable of activating SPAK (Figure 4C) and OSR1 (Figure 4D).
Figure 4. Activation of SPAK/OSR1 by WNK1 and WNK4.
(A) The indicated combinations of wt or ki WNK1-(1–661) were tested for their ability to activate wt SPAK or ki [D212A]SPAK. Activity of SPAK was assayed employing [1-260]NKCC1 as a substrate. Phosphorylation of NKCC1 was quantified after PAGE and autoradiography (middle panel) of the Coomassie Blue-stained band of NKCC1-(1–260) (lower panel). The phosphorylation of NKCC1 was also measured as 32P radioactivity by Cerenkov counting (upper panel). The results are plotted as means±S.D. for a duplicate experiment relative to the phosphorylation obtained with wt SPAK alone. (B) As above, except that wt OSR1 or ki [D164A]OSR1 were employed. (C and D) As above, except that wt WNK4-(1–593) or ki [K186A/D321A]WNK4-(1–593) were employed.
Residues in SPAK and OSR1 that are phosphorylated by WNK1
To map the WNK1 phosphorylation sites in SPAK and OSR1, the catalytically inactive mutants [D212A]SPAK and [D164A]OSR1 were phosphorylated by WNK1-(1–661). Under these conditions, SPAK and OSR1 were phosphorylated to 0.20 and 0.36 mol of phosphate/mol respectively. We were unable to phosphorylate the SPAK and OSR1 to a higher stoichiometry, indicating that a significant proportion of the recombinant enzymes may be in a conformation that is not phosphorylated. 32P-labelled SPAK (Figure 5A) and OSR1 (Figure 5C) were digested with trypsin and chromatographed on a C18 column to isolate 32P-labelled phosphopeptides. The analysis of SPAK (Figure 5A) revealed one major phosphopeptide (P1) and three minor phosphopeptides (P2, P3 and P4). A similar profile of one major (P5) and three minor (P6, P7 and P8) phosphopeptides was obtained for OSR1 (Figure 5C). MS and solid-phase Edman sequencing of the SPAK phosphopeptides established the identity of P1 and P2 as peptides phosphorylated at Ser373 and P3 and P4 as peptides phosphorylated at Thr233 (Figure 5A, lower panel). The OSR1 P5 and P6 peptides were phosphorylated at Ser325 (the residue equivalent to Ser373 of SPAK) and P7 and P8 peptides were phosphorylated at Thr185 (the residue equivalent to Thr233 of SPAK) (Figure 5D). Thr233/Thr185 are located within the kinase domain T-loop, whereas Ser373/Ser325 lie in a conserved C-terminal non-catalytic region of SPAK and OSR1 (Figure 2A).
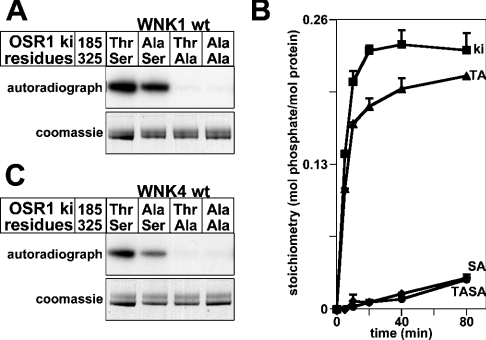
We next assessed how mutation of Thr185 and Ser325 in OSR1 affected phosphorylation by WNK1. Mutation of Thr185 moderately decreased phosphorylation of OSR1 by WNK1, whereas mutation of Ser325 virtually abolished phosphorylation of OSR1 by WNK1 (Figures 6A and 6B). Similar results were obtained employing WNK4 (Figure 6C), indicating that WNK4 phosphorylates the same residues on OSR1 that are phosphorylated by WNK1.
Figure 6. Analysis of phosphorylation of OSR1 by WNK1 and WNK4.
(A) The indicated mutants of the ki [D164A]OSR1 were phosphorylated by wt WNK1-(1–661) in the presence of Mg2+ and [γ-32P]ATP. Phosphorylation of OSR1 was determined after PAGE and autoradiography (upper panel) of the Coomassie Blue-stained band (lower panel) corresponding to OSR1. (B) As above, except that phosphorylation reactions were terminated at the indicated time points and the stoichiometry of 32P phosphorylation of the indicated mutants of ki OSR1 was determined by Cerenkov counting. The results are plotted as means±S.D. for an experiment performed in duplicate. Abbreviations: ki, kinase-inactive [D164A]OSR1; TA, [D164A/T185A]OSR1; SA, [D164A/S325A]OSR1; TASA, [D164A/T185A/S325A]OSR1. (C) As in (A), except that WNK4-(1–593) was employed.
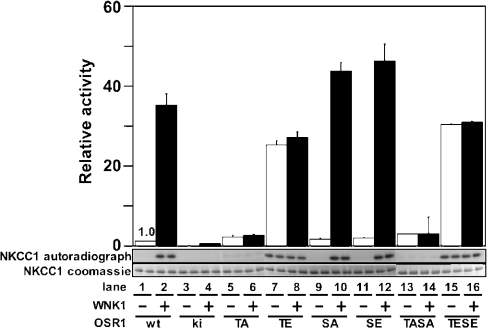
Activation of OSR1 requires Thr185 but not Ser325 phosphorylation
In order to determine the importance of the phosphorylation of Thr185 and Ser325 in regulating the activation of OSR1 by WNK1, we mutated these residues to either alanine (to prevent phosphorylation) or glutamic acid (to mimic phosphorylation) and determined how this affected the activation of OSR1 by WNK1. Mutation of Thr185 to alanine prevented WNK1 from activating OSR1 (Figure 7, lane 6). Consistent with Thr185 phosphorylation mediating activation of OSR1, mutation of Thr185 to glutamic acid markedly enhanced OSR1 activity (Figure 7, lane 7) and this mutant could not be activated further by phosphorylation with WNK1 (Figure 7, lane 8). Mutation of Ser325 to alanine or glutamic acid had no effect on the basal activity of OSR1 or on the ability of WNK1 to activate OSR1 (Figure 7, lanes 9–12). Double OSR1 mutants in which both Thr185 and Ser325 were altered to either alanine (Figure 7, lanes 13 and 14) or glutamic acid (Figure 7, lanes 15 and 16) had similar properties to the single Thr185 mutants.
Figure 7. Analysis of activation of OSR1 by WNK1.
The indicated mutants of OSR1 were incubated in the absence (−) or presence (+) of WNK1-(1–661) in the presence of Mg2+ and [γ-32P]ATP. After this incubation, the activity of OSR1 was assayed employing NKCC1-(1–260) as a substrate. Phosphorylation of NKCC1 was quantified after PAGE and autoradiography (middle panel) of the Coomassie Blue-stained band of NKCC1-(1–260) (lower panel). The phosphorylation of NKCC1 was also measured as 32P radioactivity by Cerenkov counting (upper panel). The results are plotted as means±S.D. for a duplicate experiment relative to the phosphorylation obtained with wt OSR1 alone. Abbreviations: ki, kinase-inactive [D164A]OSR1; TA, [T185A]OSR1; TE, [T185E]OSR1; SA, [S325A]OSR1; SE, [S325E]OSR1; TASA, [T185A/S325A]OSR1; TESE, [T185E/S325E]OSR1.
DISCUSSION
We report here that endogenously expressed WNK1 in testis is associated with SPAK and that WNK1 and WNK4 can phosphorylate and activate SPAK and OSR1 in vitro. WNK isoforms are likely to have a very restricted substrate specificity, as they failed to phosphorylate significantly any of ≈50 proteins or ≈500 synthetic peptides that we had previously tested (results not shown). It is a common feature of upstream kinases, such as Raf, MEK [MAPK (mitogen-activated protein kinase)/ERK (extracellular-signal-regulated kinase) kinase], PDK1 (3-phosphoinositide-dependent kinase 1) and LKB1, that they tend to possess highly restricted substrate specificities. Our results also indicate that activation of OSR1 by WNK1 is mediated by the phosphorylation of Thr185 within its T-loop. This is based on the finding that OSR1 is phosphorylated at Thr185 by WNK1, mutation of Thr185 to alanine abolishes activation of OSR1 by WNK1 and that mutation of Thr185 to glutamic acid is sufficient to activate OSR1 in the absence of WNK1. Thus the mechanism of activation of OSR1 is similar to that of many other kinases and is mediated by phosphorylation of the T-loop residue [27]. It is likely that SPAK will be activated by WNK1 in a similar manner to OSR1.
Interestingly, the major site that WNK1 phosphorylates in SPAK and OSR1 is not the T-loop Thr, but a Ser residue located in a C-terminal non-catalytic region of the enzyme. Both phosphorylation sites on WNK1 and WNK4, as well as the residues surrounding them, are conserved in mammalian, Xenopus (frog), Drosophila (fruitfly) and Caenorhabditis elegans (nematode worm) homologues of SPAK/OSR1. The role that phosphorylation of the serine residue plays is unknown, as its mutation does not affect the basal activity of OSR1, nor does it affect its ability to be activated by WNK1. Further analysis is therefore required to define the role that the serine phosphorylation site plays in regulating SPAK and OSR1 function.
We have also observed consistently that OSR1 is phosphorylated more efficiently by WNK1 and WNK4 than SPAK (Figure 3), despite the residues surrounding the sites of phosphorylation being almost identical in SPAK and OSR1 (Figure 2A). Major differences between SPAK and OSR1 are the presence of a highly unusual proline-and-alanine-rich motif within the first 50 amino acid residues of SPAK and a region spanning ≈40 amino acids towards the C-terminal region (Figure 2A) that might affect its folding when expressed in E. coli and/or its ability to interact with WNK isoforms. SPAK has also been reported to interact with various cytoskeletal components [28] that could potentially play a scaffolding role to facilitate the phosphorylation of SPAK by WNK isoforms. We have been unable to express recombinant forms of full-length non-degraded WNK1 and WNK4, owing to the large size of these enzymes, and therefore it is possible that the full-length kinases would phosphorylate SPAK and OSR1 more efficiently than the catalytic fragments of WNK1 and WNK4 employed in the present study.
The analysis of WNK1 and WNK4 activity has been hampered by the lack of a sensitive and quantitative in vitro assay. Previous studies relied upon measurement of the autophosphorylation activity of WNK1/WNK4 and its ability to poorly phosphorylate MBP and histone. The finding that WNK isoforms phosphorylate and activate OSR1 and SPAK can readily be adapted to provide a sensitive and accurate method for assessing WNK isoform activity in vitro. Moreover, as WNK1+/− mice possess lower blood pressure [4], and mutations in the WNK1 gene that increase its expression lead to Gordon's hypertension syndrome in humans [3], drugs which inhibit WNK1 activity might be useful in the treatment of this disorder and of hypertension in general. The assays that we have developed in the present study could be deployed in a screen to identify small-molecule inhibitors of WNK1.
SPAK was previously demonstrated to phosphorylate and activate the NKCC1 co-transporter [26]. NKKC1 was also shown to interact with SPAK and OSR1 and to localize SPAK to the apical membrane of choroid-plexus epithelial cells [29]. In addition to directly activating NKCC1 by phosphorylation, SPAK was suggested to play a scaffolding role in regulating NKCC1 function, perhaps by controlling its phosphorylation by other kinases [28]. Recently, WNK4 was reported to interact with SPAK in a yeast two-hybrid screen, and evidence was presented that co-expression of WNK4 and SPAK in Xenopus oocytes increased potassium uptake through the NKCC1 transporter [19]. That study did not address the question as to whether WNK4 could phosphorylate and activate SPAK, but the finding that stimulation of NKCC1 activity was only observed following overexpression of wt WNK4 and SPAK, but not by overexpression of inactive WNK4 or SPAK [19], is consistent with the notion that WNK4 could activate NKCC1 through its ability to phosphorylate and activate SPAK. Our findings also indicate that, in addition to WNK4 activating SPAK, WNK1 can also mediate the activation of SPAK, as well as OSR1, and could therefore play a similar role in regulating NKCC1 activity.
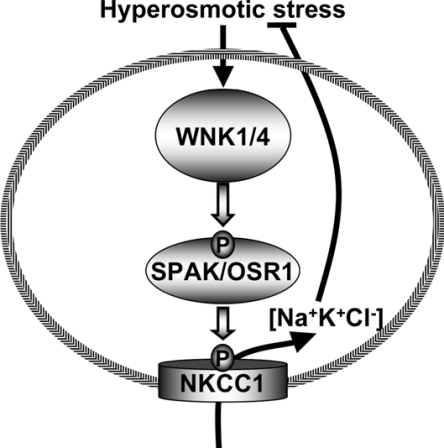
The rate of autophosphorylation of endogenously expressed WNK1 is increased ≈5-fold by hypertonic stress in kidney epithelial cells and in breast- and colon-cancer cell lines [30]. These conditions are well known to stimulate NKCC1 activity by inducing its phosphorylation, leading to increased uptake of Na+ and K+ ions to maintain cell volume [31,32]. As summarized in Figure 8, we suggest that hyperosmotic stress induces the activation of WNK isoforms through an uncharacterized mechanism, leading to the phosphorylation and activation of SPAK and OSR1, which then phosphorylate and stimulate the activity of the NKCC1 co-transporter. Significant further studies are required to validate this model. In particular, it will be important to establish that phosphorylation and activation of SPAK and/or OSR1, as well as NKCC1, is dependent on WNK activity in vivo. This may not be a trivial task, owing to the presence of four distinct isoforms of WNK in mammalian cells. It will also be important to investigate the effect of mutating the sites on SPAK/OSR1 phosphorylated by WNK1, of the ability of WNK isoforms to stimulate NKCC1 activity, and also to determine the role phosphorylation of the serine residue phosphorylated by WNKs plays in controlling SPAK/OSR1 function. It would also be interesting to investigate whether any mutations in the SPAK and OSR1 protein kinases are found in humans with familial hypertension syndrome and whether SPAK and OSR1 kinases are hyperactive in subjects with Gordon's syndrome.
Figure 8. Summary of the mechanism by which hyperosmotic stresses may stimulate NKCC1 co-transporter activity.
Online data
Acknowledgments
We thank Steven L. Roberds (Pfizer, St Louis, MO, U.S.A.) for support and discussion, David Campbell for MS analysis, Agnieszka Kieloch and Gail Fraser for technical assistance, Kei Sakamoto for providing rat tissue extracts, the Sequencing Service (School of Life Sciences, University of Dundee, Dundee, Scotland, U.K.) for DNA sequencing, the Post Genomics and Molecular Interactions Centre for Mass Spectrometry facilities (School of Life Sciences, University of Dundee) and the protein production and antibody purification teams [Division of Signal Transduction Therapy (DSTT), University of Dundee] co-ordinated by Hilary McLauchlan and James Hastie for the expression and purification of antibodies. A. C. V. is the recipient of a Pfizer-sponsored studentship. D. R. A. is supported by the Association for International Cancer Research, Diabetes UK, the Medical Research Council and the Moffat Charitable Trust.
References
- 1.Xu B., English J. M., Wilsbacher J. L., Stippec S., Goldsmith E. J., Cobb M. H. WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J. Biol. Chem. 2000;275:16795–16801. doi: 10.1074/jbc.275.22.16795. [DOI] [PubMed] [Google Scholar]
- 2.Verissimo F., Jordan P. WNK kinases, a novel protein kinase subfamily in multi-cellular organisms. Oncogene. 2001;20:5562–5569. doi: 10.1038/sj.onc.1204726. [DOI] [PubMed] [Google Scholar]
- 3.Wilson F. H., Disse-Nicodeme S., Choate K. A., Ishikawa K., Nelson-Williams C., Desitter I., Gunel M., Milford D. V., Lipkin G. W., Achard J. M., et al. Human hypertension caused by mutations in WNK kinases. Science. 2001;293:1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- 4.Zambrowicz B. P., Abuin A., Ramirez-Solis R., Richter L. J., Piggott J., BeltrandelRio H., Buxton E. C., Edwards J., Finch R. A., Friddle C. J., et al. Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14109–14114. doi: 10.1073/pnas.2336103100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamide K., Takiuchi S., Tanaka C., Miwa Y., Yoshii M., Horio T., Mannami T., Kokubo Y., Tomoike H., Kawano Y., Miyata T. Three novel missense mutations of WNK4, a kinase mutated in inherited hypertension, in Japanese hypertensives: implication of clinical phenotypes. Am. J. Hypertens. 2004;17:446–449. doi: 10.1016/j.amjhyper.2003.12.020. [DOI] [PubMed] [Google Scholar]
- 6.Kahle K. T., Wilson F. H., Lifton R. P. Regulation of diverse ion transport pathways by WNK4 kinase: a novel molecular switch. Trends Endocrinol. Metab. 2005;16:98–103. doi: 10.1016/j.tem.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 7.Cope G., Golbang A., O'Shaughnessy K. M. WNK kinases and the control of blood pressure. Pharmacol. Ther. 2005;106:221–231. doi: 10.1016/j.pharmthera.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Gamba G. Role of WNK kinases in regulating tubular salt and potassium transport and in the development of hypertension. Am. J. Physiol. Renal Physiol. 2005;288:F245–F252. doi: 10.1152/ajprenal.00311.2004. [DOI] [PubMed] [Google Scholar]
- 9.Kahle K. T., Wilson F. H., Leng Q., Lalioti M. D., O'Connell A. D., Dong K., Rapson A. K., MacGregor G. G., Giebisch G., Hebert S. C., Lifton R. P. WNK4 regulates the balance between renal NaCl reabsorption and K+ secretion. Nat. Genet. 2003;35:372–376. doi: 10.1038/ng1271. [DOI] [PubMed] [Google Scholar]
- 10.Yang C. L., Angell J., Mitchell R., Ellison D. H. WNK kinases regulate thiazide-sensitive Na–Cl cotransport. J. Clin. Invest. 2003;111:1039–1045. doi: 10.1172/JCI17443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamauchi K., Rai T., Kobayashi K., Sohara E., Suzuki T., Itoh T., Suda S., Hayama A., Sasaki S., Uchida S. Disease-causing mutant WNK4 increases paracellular chloride permeability and phosphorylates claudins. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4690–4694. doi: 10.1073/pnas.0306924101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee B. H., Min X., Heise C. J., Xu B. E., Chen S., Shu H., Luby-Phelps K., Goldsmith E. J., Cobb M. H. WNK1 phosphorylates synaptotagmin 2 and modulates its membrane binding. Mol. Cell. 2004;15:741–751. doi: 10.1016/j.molcel.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 13.Harlow E., Lane D. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. Antibodies: a Laboratory Manual. [Google Scholar]
- 14.Woods Y. L., Rena G., Morrice N., Barthel A., Becker W., Guo S., Unterman T. G., Cohen P. The kinase DYRK1A phosphorylates the transcription factor FKHR at Ser329 in vitro, a novel in vivo phosphorylation site. Biochem. J. 2001;355:597–607. doi: 10.1042/bj3550597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 16.Durocher Y., Perret S., Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell D. G., Morrice N. A. Identification of protein phosphorylation sites by a combination of mass spectrometry and solid phase Edman sequencing. J. Biomol. Techn. 2002;13:121–132. [PMC free article] [PubMed] [Google Scholar]
- 18.Ushiro H., Tsutsumi T., Suzuki K., Kayahara T., Nakano K. Molecular cloning and characterization of a novel Ste20-related protein kinase enriched in neurons and transporting epithelia. Arch. Biochem. Biophys. 1998;355:233–240. doi: 10.1006/abbi.1998.0736. [DOI] [PubMed] [Google Scholar]
- 19.Gagnon K. B., England R., Delpire E. Volume sensitivity of cation–chloride cotransporters is modulated by the interaction of two kinases: SPAK and WNK4. Am. J. Physiol. Cell Physiol. 2005 doi: 10.1152/ajpcell.00037.2005. 10.1152/ajpcell.0037.2005. [DOI] [PubMed]
- 20.Scanlan M. J., Gout I., Gordon C. M., Williamson B., Stockert E., Gure A. O., Jager D., Chen Y. T., Mackay A., O'Hare M. J., Old L. J. Humoral immunity to human breast cancer: antigen definition and quantitative analysis of mRNA expression. Cancer Immun. 2001;1:4. [PubMed] [Google Scholar]
- 21.Yang C. L., Zhu X., Wang Z., Subramanya A. R., Ellison D. H. Mechanisms of WNK1 and WNK4 interaction in the regulation of thiazide-sensitive NaCl cotransport. J. Clin. Invest. 2005;115:1379–1387. doi: 10.1172/JCI22452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamari M., Daigo Y., Nakamura Y. Isolation and characterization of a novel serine threonine kinase gene on chromosome 3p22–21.3. J. Hum. Genet. 1999;44:116–120. doi: 10.1007/s100380050121. [DOI] [PubMed] [Google Scholar]
- 23.Baas A. F., Boudeau J., Sapkota G. P., Smit L., Medema R., Morrice N. A., Alessi D. R., Clevers H. C. Activation of the tumour suppressor kinase LKB1 by the STE20-like pseudokinase STRAD. EMBO J. 2003;22:3062–3072. doi: 10.1093/emboj/cdg292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boudeau J., Baas A. F., Deak M., Morrice N. A., Kieloch A., Schutkowski M., Prescott A. R., Clevers H. C., Alessi D. R. MO25α/β interact with STRAD α/β enhancing their ability to bind, activate and localize LKB1 in the cytoplasm. EMBO J. 2003;22:5102–5114. doi: 10.1093/emboj/cdg490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vitari A. C., Deak M., Collins B. J., Morrice N., Prescott A. R., Phelan A., Humphreys S., Alessi D. R. WNK1, the kinase mutated in an inherited high-blood-pressure syndrome, is a novel PKB (protein kinase B)/Akt substrate. Biochem. J. 2004;378:257–268. doi: 10.1042/BJ20031692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dowd B. F., Forbush B. PASK (proline-alanine-rich STE20-related kinase), a regulatory kinase of the Na–K–Cl cotransporter (NKCC1) J. Biol. Chem. 2003;278:27347–27353. doi: 10.1074/jbc.M301899200. [DOI] [PubMed] [Google Scholar]
- 27.Nolen B., Taylor S., Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 28.Piechotta K., Garbarini N., England R., Delpire E. Characterization of the interaction of the stress kinase SPAK with the Na+-K+-2Cl− cotransporter in the nervous system: evidence for a scaffolding role of the kinase. J. Biol. Chem. 2003;278:52848–52856. doi: 10.1074/jbc.M309436200. [DOI] [PubMed] [Google Scholar]
- 29.Piechotta K., Lu J., Delpire E. Cation chloride cotransporters interact with the stress-related kinases Ste20-related proline-alanine-rich kinase (SPAK) and oxidative stress response 1 (OSR1) J. Biol. Chem. 2002;277:50812–50819. doi: 10.1074/jbc.M208108200. [DOI] [PubMed] [Google Scholar]
- 30.Lenertz L. Y., Lee B. H., Min X., Xu B. E., Wedin K., Earnest S., Goldsmith E. J., Cobb M. H. Properties of WNK1 and implications for other family members. J. Biol. Chem. 2005;280:26653–26658. doi: 10.1074/jbc.M502598200. [DOI] [PubMed] [Google Scholar]
- 31.Kurihara K., Moore-Hoon M. L., Saitoh M., Turner R. J. Characterization of a phosphorylation event resulting in upregulation of the salivary Na+-K+-2Cl− cotransporter. Am. J. Physiol. 1999;277:C1184–C1193. doi: 10.1152/ajpcell.1999.277.6.C1184. [DOI] [PubMed] [Google Scholar]
- 32.Lytle C., Forbush B., 3rd The Na–K–Cl cotransport protein of shark rectal gland. II. Regulation by direct phosphorylation. J. Biol. Chem. 1992;267:25438–25443. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.