Abstract
The LytC lysozyme of Streptococcus pneumoniae forms part of the autolytic system of this important pathogen. This enzyme is composed of a C-terminal CM (catalytic module), belonging to the GH25 family of glycosyl hydrolases, and an N-terminal CBM (choline-binding module), made of eleven homologous repeats, that specifically recognizes the choline residues that are present in pneumococcal teichoic and lipoteichoic acids. This arrangement inverts the general assembly pattern of the major pneumococcal autolysin, LytA, and the lytic enzymes encoded by pneumococcal bacteriophages that place the CBM (made of six repeats) at the C-terminus. In the present paper, a three-dimensional model of LytC built by homology modelling of each module and consistent with spectroscopic and hydrodynamic studies is shown. In addition, the putative catalytic-pair residues are identified. Despite the inversion in the modular arrangement, LytC and the bacteriophage-encoded Cpl-1 lysozyme most probably adopt a similar global fold. However, the distinct choline-binding ability and their substrate-binding surfaces may reflect a divergent evolution directed by the different roles played by them in the host (LytC) or in the bacteriophage (Cpl-1). The tight binding of LytC to the pneumococcal envelope, mediated by the acquisition of additional choline-binding repeats, could facilitate the regulation of the potentially suicidal activity of this autolysin. In contrast, a looser attachment of Cpl-1 to the cell wall and the establishment of more favourable interactions between its highly negatively charged catalytic surface and the positively charged chains of pneumococcal murein could enhance the lytic activity of the parasite-encoded enzyme and therefore liberation of the phage progeny.
Keywords: choline-binding protein, hydrodynamic particle, modular organization, pneumococcal lysin, secondary structure, structural modelling
Abbreviations: 3D, three-dimensional; CBM, choline-binding module; C-Cpl-1, choline-binding domain of Cpl-1; CM, catalytic module; GH, glycosyl hydrolase(s); TIM, triose phosphate isomerase
INTRODUCTION
GHs (glycosyl hydrolases) are a widespread group of enzymes that break down glycosidic bonds between carbohydrates. They are classified into over 100 sequence-unrelated families [1], whose members share a common general fold, active-site topology and stereochemical course [2]. The lysozymes encoded by Streptococcus pneumoniae [3] and its bacteriophages [4] cleave the β1-4-glycosydic bond between the N-acetylmuramoyl-N-glucosaminyl residues that form the polysaccharide chain of the bacterial cell wall. They are modular enzymes that harbour a CM (catalytic module) and a CBM (choline-binding module), which generally binds to the choline residues of the pneumococcal teichoic and lipoteichoic acid chains [5]. The CBM is made up of homologous repeats of approx. 20 amino acids (Pfam accession code PF01473). This modular organization is shared between all the pneumococcal murein hydrolases and enhances the enzymatic activity by several orders of magnitude [6]. The catalytic module of the pneumococcal cell wall lysozymes belongs to the GH25 family of GHs (chalaropsis-type lysozymes) [3] whose sequences and structures are unrelated to enzymes from the other three lysozyme families (GH22, GH23 and GH24) [7]. Only two members of the GH25 group have known 3D (three-dimensional) structure, i.e. Cpl-1 from pneumococcal bacteriophage Cp-1 [8] and cellosyl from Streptomyces coelicolor [9]. The catalytic residues that are involved in the cleavage of the scissile bond have been identified in the Cpl-1 lysozyme (Asp10 and Glu94) [8,10] and appear to be conserved within the GH25 family [8,9]. Their position is compatible with an inverting mechanism in which regeneration of the protonation states of the acid donor (Glu94) and the general base (Asp10) would be facilitated by low-energy hydrogen bonding to residues Asp92 and Asp182 respectively.
On the other hand, the crystal structures of the Cpl-1 lysozyme and C-LytA, the isolated CBM of the major pneumococcal LytA autolysin, showed that each sequence repeat forms a β-hairpin, followed by a loop and a coiled region [8,11]. The repeats tend to adopt a left-handed β-solenoid, although they can also arrange in an extended β-sheet [8,11]. Choline residues bind at the interface of two consecutive repeats of the β-solenoid in a cavity lined, primarily, by aromatic side chains [8,11].
Cpl-1 and LytC are representative examples of module shuffling. As in most pneumococcal murein hydrolases [5], the CM of Cpl-1 is located at the N-terminus, while, in LytC, it comprises the C-terminal region. Furthermore, the CBM of the former is made up of six homologous repeats (p1–p6) plus a C-terminal tail of 16 residues, while, in LytC, it comprises 11 repeating units (p1–p11), with a lower conservation of the consensus sequence [3]. The sequence differences between LytC and Cpl-1, together with their inverted modular arrangement, yield two lysozymes with very different catalytic efficiencies and temperature-activity profiles [3]. In the present paper, as a first approach towards the elucidation of the LytC structure, we propose a 3D model for the pneumococcal-encoded lysozyme based on homology-modelling strategies. This approach can give very reliable results, even when the sequence identity is lower than 40%, if the target and the template proteins belong to the same structural family and the homologous proteins of known structures show a high conservation of functional and structural features [12,13]. The proposed model is compatible with the hydrodynamic behaviour and the secondary structure of LytC, which have been characterized, respectively, by analytical ultracentrifugation and by CD and IR spectroscopy. The soundness of the validation tests was checked on the Cpl-1 lysozyme. Despite having an inverted modular arrangement, LytC and Cpl-1 can adopt similar overall structures whose specific features are discussed in relation to their functional roles.
EXPERIMENTAL
Protein purification and chemicals
LytC (55210 Da) and Cpl-1 (39249 Da) lysozymes were purified from Escherichia coli BL21(DE3)[pLCC14] and DH5α[pCIP100] cells respectively by affinity chromatography on DEAE-cellulose [14], followed by size-exclusion chromatography on a dextran–agarose column (Superdex 200; Amersham Biosciences) in the case of LytC. The purity of samples was analysed routinely by SDS/PAGE. Before use, proteins were dialysed extensively at 4 °C against the appropriate buffer and centrifuged for 5 min at 11600 g. Protein concentration was determined spectrophotometrically using molar absorption coefficients of 186107 M−1·cm−1 (LytC) and 97115 M−1·cm−1 (Cpl-1) at 280 nm. Choline concentration was measured by differential refractometry [15]. All reagents were from Sigma Chemical Co. and were of analytical grade.
Sedimentation velocity
Sedimentation velocity experiments were performed by centrifugation of 400 μl samples of LytC (2.4 μM) and Cpl-1 (5.2 μM) at 45000 rev./min and 4 °C in an Optima-XLA analytical ultracentrifuge (Beckman Instruments) using an An50Ti rotor. Radial concentration profiles were taken every 8 min at 280 nm. Sedimentation coefficients (s) were calculated from the ratio of the velocity of the solute boundary (Svedberg program) [16] and corrected to the standard values in water at 20 °C, s20,w [17]. The translational frictional coefficient (f) was determined from the protein molecular mass and the s20,w value. The frictional coefficient of the equivalent hydrated sphere (f0) was calculated assuming hydration coefficients (∂w) of 0.41 gwater/gprotein (LytC) and 0.39 gwater/gprotein (Cpl-1) estimated from the amino acid sequences [18]. A gross estimation of the hydrodynamic shape of the protein was obtained by generating a family of revolution ellipsoids compatible with the hydrodynamic properties from the f/f0 ratio [19].
CD
Far-UV CD spectra (average of four scans) were recorded at 4 °C (LytC) or 20 °C (Cpl-1) in a JASCO-810 spectropolarimeter (Jasco), equipped with a Peltier system for temperature control, using 0.02 cm path-length quartz cells and a scan rate of 20 nm·min−1. Addition of choline impaired data acquisition below 195 nm. The buffer spectrum was subtracted from the experimental data, and the corrected ellipticities were converted into mean residue ellipticities using average molecular masses per residue of 118.0 (LytC) and 115.8 (Cpl-1). Secondary-structural content was estimated by deconvolution of the experimental curves using the CONTIN (16 protein spectra data set) and CDNN (33 protein spectra data set) programs [20,21] that evaluate four conformations (α-helix, β-sheet, β-turn and remainder), although CDNN discriminates between parallel and antiparallel β-sheet. Structure assignment in CONTIN was based on the Kabsch and Sander method [22].
Fourier-transform IR spectroscopy
Infrared spectra were recorded at 20 °C in a Bruker IFS66 spectrophotometer equipped with a DTGS detector at 4 cm−1 resolution using the OPUS program. Protein samples were prepared as follows: 600 μg of protein equilibrated in 10 mM phosphate buffer, pH 8.0, in the absence or in the presence of 20 mM choline, were freeze-dried under a nitrogen stream, then were rehydrated by adding 30 μl of 2H2O, and maintained at 20 °C under vacuum for 2 h to ensure isotopic substitution of protein-exchangeable NH groups. Samples were assembled in a thermostatically controlled cell between two calcium fluoride windows with 50 μm spacers. For baselines, the process was repeated using the dialysis buffer. Data analysis (average of four spectra) was carried out following the previously described procedure [23]. Percentages of secondary structure were determined from the relative areas of the amide I band components, assuming that all they have equivalent molar absorbances [23].
Template searching and secondary-structure predictions
The identification of homologues for each module of LytC (Swiss-Prot code Q9Z4J8) in the protein databases was carried out using the Psi-BLAST server [24]. The secondary-structure prediction was performed at the PHD [25], PSIPRED [26] and Jpred [27] servers. Only those structural elements predicted with a reliability above 85.7% (score ≥6; range 0–9) by at least two of them were considered for the sequence-to-structure alignment.
Sequence-to-structure alignment and homology modelling of LytC
Sequence alignment based on the correlation between the secondary structure of the template and the predicted secondary structure of the target was performed with ClustalW [28], followed by a manual adjustment with SeaView [29] guided by the secondary-structure prediction. The alignment was formatted with JOY [30]. In the case of the CBM we used the LALIGN program [31] to compare the CBM of LytC and the template in order to find the best local alignments. Two overlapping fragments of the CBM of Cpl-1, which showed a significant similarity with the CBM of LytC, were thus identified.
3D models of each module containing all non-hydrogen atoms were obtained automatically from the best alignment using the method implemented in MODELLER [32]. The program deduces spatial restraints from the input template(s), subsequently applied to the model of the query protein. 3D models are obtained by optimization of the molecular pdf (probability density function), with simultaneous minimization of input-restraint violations. Initially, 15 models were generated for each input, but only that with the fewest restraint violations and the lowest energy was evaluated. Several cycles of realignment, modelling and validation were repeated until the model showed no further improvement.
The SVRs (structurally variable regions) were built with the standard procedure in MODELLER and they were, afterwards, refined. For short loops, MODELLER procedures yielded good results using the loop parent structures. Long loops were remodelled using the Search-Loop option in the SYBYL program software suite coupled database (Tripos, St. Louis, MO, U.S.A.). This database contains a large set of high-resolution protein structural fragments. A total of 25 loops clustered in families were identified, and the fragment that showed a lower number of van der Waals contacts and a higher sequence identity (≥25%) with each loop was selected for remodelling. The final model was subjected to an additional minimization procedure using the Tripos forcefield in order to avoid the unfavourable possible contacts. All models were validated using PROCHECK [34] and VERIFY 3D [35]. JOY and COMPARER [36] were used to generate structure-based alignments between the target and the template in every cycle of the modelling process.
RESULTS AND DISCUSSION
Homology modelling of LytC 3D structure
Template search and multiple sequence alignment
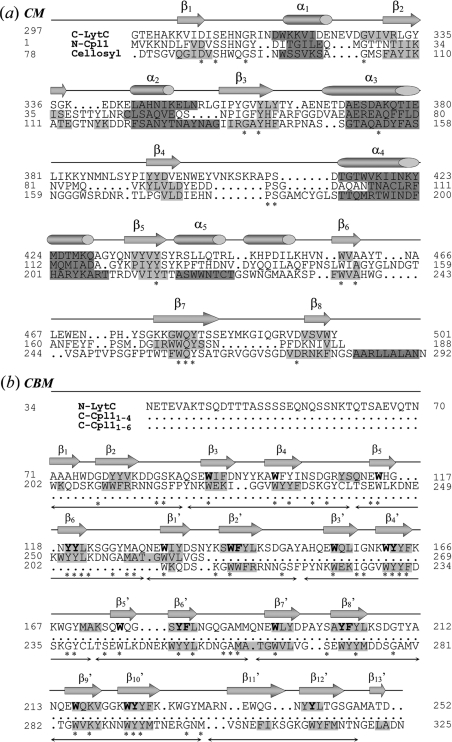
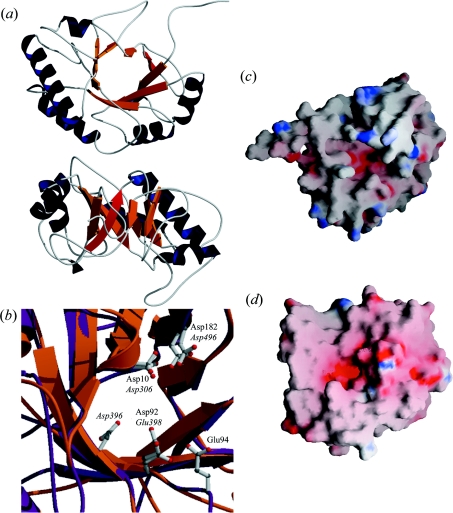
There are only two homologous 3D structures of the CM of LytC available in the PDB (Protein Data Bank), i.e. the catalytic module of Cpl-1 (Swiss-Prot accession number P15057; PDB code 1H09) [8] and cellosyl (Swiss-Prot accession number P25310; PDB code 1JFX) [9], that showed a sequence identity of 21.5% and 20% respectively. Figure 1(a) shows the sequential alignment of the target (CM of LytC) with the templates. The irregular (α/β)5β3 fold characteristic of cellosyl and the CM of Cpl-1, and the positioning of the potential catalytic residues (Asp306 and Glu398) at the end of β1 and in the loop connecting β4 to α4, seem to be conserved in LytC. The reliability of the predicted target structure is supported by the good correlation found between the structural elements present in the 3D structure of Cpl-1 and those predicted by the theoretical methods employed throughout the present work (see Figure 1a).
Figure 1. Sequence alignments of LytC modules with their structural templates.
(a) Sequence alignment of the CMs of LytC (C-LytC), Cpl-1 (N-Cpl1) and the cellosyl muramidase. Numbers at both sides correspond to residue numbers including the signal peptide. Strictly conserved residues are marked with an asterisk (*). Predicted elements of secondary structure in Cpl-1 and LytC are presented as dark (α-helix) and light (β-strands) grey-shaded regions. The same colour code is used for the secondary-structure elements present in the cellosyl 3D structure. Structural elements in the Cpl-1 crystal structure are depicted as cylinders (α-helix) and arrows (β-strands) over the sequence. (b) Alignment of the first nine repeats (p1–p9) of the CBM of LytC (N-LytC) with two fragments of the Cpl-1 CBM (C-Cpl11–4 and C-Cpl11–6). Symbols, colour coding and numbers are as in (a). Horizontal arrows under the sequence alignment indicate the regions that comprise each repeating unit. Conserved residues potentially involved in choline binding to LytC are in bold letters. β-Strands in the 3D structure of the Cpl-1 CBM are named βi (fragment p1–p4) and βi' (fragment p1–p6).
The search of close homologues for the whole CBM of LytC in several databases was unsuccessful, and only the structures of the CBM of Cpl-1 (C-Cpl-1) and C-LytA (PDB code 1HCX) [11] were considered to be significant hits. The identity was approx. 27% with both homologues, but the former was used as template, since its 3D structure contains a higher number of repeats. Using the LALIGN program, we identified two fragments of the CBM of Cpl-1, comprising repeats p1–p4 and p1–p6, which showed the best alignment with the N- and the C-terminal regions of the target (Figure 1b). Thus both fragments were used as templates for modelling the CBM of LytC. We found a good correlation between the predicted and the experimental distribution of secondary-structure elements in the CBM of Cpl-1 (Figure 1b). The aromatic residues that are characteristic of choline-binding repeats are conserved in the CBM of LytC, and the predicted secondary structure suggests a conservative folding of the module. Nevertheless, the higher variability in sequence and size of the LytC repeats yield a more irregular distribution of the structural elements. The conservation of the glycine residue in the segment connecting the two β-strands of each repeat will aid them to adopt the antiparallel disposal into the β-hairpin observed in the crystallographic structures [8,11].
3D modelling of LytC catalytic module
Three 3D models of the CM of LytC were built using cellosyl, C-Cpl-1 and both structures as templates. We selected, through the Ramachandran structural validation in the PROCHECK program and VERIFY 3D, the model using the structure of cellosyl as a template for the best model in terms of average stereochemical properties and energy profile. The 3D model showed that 97.9% of the residues were in allowed regions and only 2.1% were in the disallowed region. Moreover, the analysis of the template and the model with VERIFY 3D showed that their Z-scores were comparable (0.68 and 0.77 for the model and for cellosyl respectively). Remodelling of the loops, focused on the regions structurally different to the template [loops 1 (α1–β2), 3 (α3–β4) and 5 (α5–β6) or potentially involved in functionality loop 4 (β4–α5); see below], was performed using the Loop-Search option of SYBYL. No severely disallowed atomic contacts were detected upon re-evaluation of the final model. These results suggest that the proposed model is reasonable and represents an acceptable approximation of the real structure of the CM of LytC. The 3D superimposition of the model and template structures made by COMPARER and formatted by JOY is shown in Supplementary Figure 1(a) (available at http://www.BiochemJ.org/bj/391/bj3910041add.htm). The structural superimposition is made by aligning the pairs of residues selected automatically from the highest scoring local sequence alignment made by COMPARER, which identifies, as equivalent positions in the 3D structures of the model and the template, pairs of residues whose Cα atoms are located less than 3.0 Å (1 Å=0.1 nm) apart. The structurally corrected alignment generated in this way and the sequence-based one can show some differences in highly variable regions, as actually occurs in the loops of the CM connecting β4 to α4 and β6 to β7 (see Figure 1a, and Supplementary Figure 1a at http://www.BiochemJ.org/bj/391/bj3910041add.htm).
As shown in Figure 2(a), the model of the CM of LytC exhibits a TIM (triose phosphate isomerase) barrel-like (α/β)5β3 fold with the antiparallel orientation of the last β-strand being characteristic of the GH25 family [8,9]. The active site is at the C-terminal side of the β-barrel in a highly negatively charged cavity (Figure 2c). In agreement with sequence alignment (Figure 1a), the pair Asp306 and Asp496 superimpose with the Cpl-1 pair formed by Asp10 and Asp182 (Figure 2b), supporting the assignment of Asp306 as the general base that activates the nucleophilic water molecule. Although Glu398 (the putative donor) is slightly shifted in relation to the position of Glu94 in Cpl-1 (Figure 2b), they are both located (like Glu177 in cellosyl) at the end of the β4 strand and can be considered to be structurally conserved. Moreover, the distance of 9 Å between Glu398 and Asp306 is compatible with an inverting mechanism. The observation that the proton donor in all GHs with a TIM barrel structure characterized so far is a glutamic acid, usually located at the end of the β4 strand [37,38], gives additional support to the assignment of Glu398 as the proton donor. In addition, the position of Asp396 is consistent with its potential role as regulator of Glu398 protonation state. The shift of the Asp396 and Glu398 catalytic pair might result from a misalignment of sequences. However, the model derived from the alignment in which Asp396 and Glu398, and Asp92 and Glu94 were superimposed was significantly worse than the proposed model in relation to stereochemistry (7.3% residues in the disallowed region) and energetic profile.
Figure 2. Structural model of the catalytic module of LytC.
(a) Views from the N-terminal side of the barrel (top) and the region lacking α6–α8 helices (bottom). Helices and strands are coloured dark blue and orange respectively; antiparallel β8 is highlighted in red. (b) Superposition of the catalytic cavities of LytC (orange) and Cpl-1 (purple). Catalytic residues of Cpl-1 and putative catalytic residues in LytC are in ball-and-stick representation (C atoms, grey; N atoms, blue; O atoms, red). Labels indicate their position in the sequence of Cpl-1 (roman) and LytC (italic). (c and d) Surface electrostatic potentials of LytC and Cpl-1 respectively. Negatively charged regions are coloured red, and positive regions are blue.
As observed in cellosyl and Cpl-1, the catalytic cavity is layered by the side chains of conserved aromatic residues that can contribute, as in other carbohydrate-binding proteins, to the substrate specificity [39,40], and the shape of the active site pocket suggests that they could act as exoglycosidases.
3D modelling of the CBM
The 3D modelling of the CBM of LytC (p1–p9 repeats) was based on the alignment shown in Figure 1(b). Repeats p10 and p11 were not modelled because of their poor sequence conservation and the failure of protein-fold recognition methods to find a template for this region. The fact that prediction methods indicate low conservation of secondary structure in both repeats (results not shown) and the high content of polar residues, characteristic of linker regions [41], suggests that these repeats probably act as a linker holding the CM and the CBM of LytC in place.
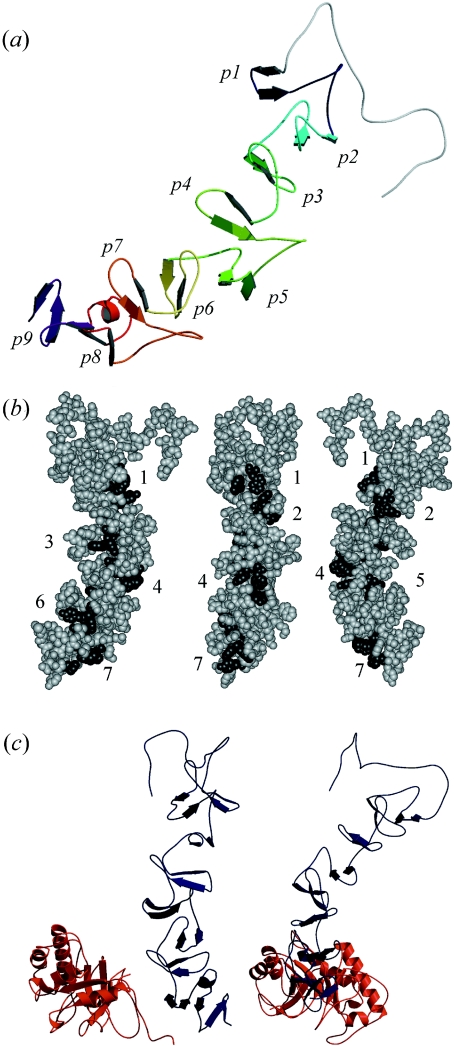
Figure 3 and Supplementary Figure 1(b) (available at http://www.BiochemJ.org/bj/391/bj3910041add.htm) show respectively the 3D model of the CBM of LytC and the JOY output with the final structural alignment of the model and the templates. The PROCHECK summary of the model showed that 97.4% residues were in allowed regions and only 1.6% were in the disallowed region. The average score of 0.67 obtained with VERIFY 3D is comparable with the value of 0.72 of the CBM of Cpl-1. According to the model, the CBM of LytC folds into a β-solenoid fold (Figure 3a), similar to those found in Cpl-1 and C-LytA structures [8,11]. Each repeat comprises a β-hairpin followed by a loop and a coiled region. Up to p8, the hairpin βn is oriented parallel to β(n+3) and is turned approx. 120° around the central axis with respect to the previous hairpin. Beyond p6, the CBM of LytC shows a small curvature, enhanced by a short helix at the end of the hairpin in p8, that modifies the orientation of p9. The choline-binding sites would be located at the interface of two consecutive repeats in the cavity lined by a tryptophan residue from the first strand of the upper β-hairpin and two aromatic side chains provided respectively by the second strand of both repeats [8,11]. The aromatic residues forming the choline-binding sites are structurally conserved, and the hydrophobic pocket is properly formed in the sites from p2 to p9 (Figure 3b). The solvent accessibility of the aromatic residues involved in choline recognition in the model of LytC and in the Cpl-1 structure are comparable, except for Phe181 at the interface of p6 and p7 (Figure 1b). Owing to the slight curvature of the CBM of LytC, the β-hairpin of p8 partially occludes the site. Nevertheless, a reliable characterization of the choline-binding sites available in LytC will require direct stoichiometric measurements. The presence of shorter repeats (17 amino acids) in LytC affects the loops connecting the β-hairpins and reduces the angle of 120° formed between two consecutive β-hairpins in the more regular modules, such as those of Cpl-1 and LytA. Thus the alternation of shorter (p3, p6 and p9) and longer repeats leads to an uneven distribution of choline-binding sites along the module surface (Figure 3b).
Figure 3. Structural models of the CBM of LytC and the whole enzyme.
(a) The CBM folds in a β-solenoid structure. β-Hairpins of repeating units appear differently coloured, from dark blue (p1) to purple (p9). (b) Disposition of the potential choline-binding sites. Numbers indicate the order of the hydrophobic cavities (dark grey) from the N- to the C-terminal end starting at the p2 repeat. Sites 2 and 5 are located between a short and a large repeat. (c) Different views of the model generated for the whole structure of LytC. The region comprising strands β6–β8 of the CM (red) interacts with repeats p7–p8 of the CBM (dark blue). The C-terminal side of the barrel, where the catalytic cavity is located, points towards the bottom of the Figure.
Overall fold of LytC
Inspection of the LytC structure suggests that the linker connecting the CM and the CBM could have arisen from the degeneration of canonical choline-binding repeats and would be composed of residues from the poorly conserved repeats p10 and p11. A tentative model for the entire molecule of LytC was obtained by structural superimposition of the 3D models of the CM and the CBM on the basis of their spatial orientation in Cpl-1 lysozyme (Figure 3c). The interacting surfaces of catalytic barrel (β6–β8) and the CBM (p7–p8 repeats) have a high content of aromatic and aliphatic residues, also observed in the equivalent regions of Cpl-1, but absent in the unimodular cellosyl muramidase, that would contribute to the intermodular interactions stabilizing the overall structure of LytC. Thermal denaturation experiments indicate that LytC is composed of two strongly interacting modules (B. Monterroso, J. L. Saiz, P. García, J. L. García and M. Menéndez, unpublished work). In order to test the proposed 3D model, the secondary structure and the hydrodynamic behaviour of LytC were characterized by CD and IR spectroscopies and analytical centrifugation respectively. The reliability of the analysis was checked by characterizing in parallel the Cpl-1 lysozyme.
Spectroscopic determination of secondary structure
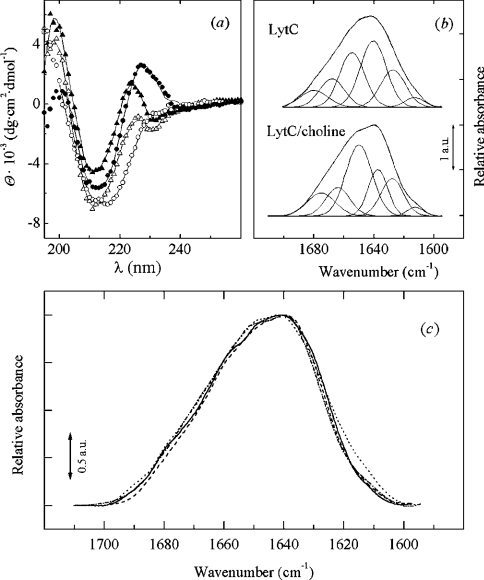
The far-UV region of the CD spectrum of pneumococcal murein hydrolases is characterized by the presence of a band in the region of 224–230 nm, which is sensitive to the interaction of choline with the tryptophan residues present at the binding site [15,42–44]. Interestingly, the presence of this band in the LytC spectrum is linked to choline binding (Figure 4a). This indicates that, in LytC, choline mediates a reorganization of the polypeptide chain that strongly affects the environment of the aromatic residues of the CBM. Table 1 summarizes the percentages of secondary structure estimated with CONTIN and CDNN, and solid lines in Figure 4(a) are the theoretical fits obtained by CONTIN. In general, there is a good correlation between the results obtained by both procedures. However, CONTIN slightly overestimates the β-turns in Cpl-1 considering the 3D structure [8], and the β-sheet percentage of LytC without choline is also higher than the CDNN estimate. Considering the aggregated state of LytC in the absence of choline (see below), this difference could reflect the specific capability of each method to overcome chiral effects unrelated to protein secondary structure [45]. The mean values from CDNN and CONTIN estimates are 10% α-helix, 46% β-strand and 18% β-turn for the monomer of LytC stabilized in the presence of choline, and 8% α-helix, 43% β-strand and 23% β-turn for Cpl-1 (average values of the free and choline-bound forms).
Figure 4. Spectroscopic data of LytC and Cpl-1.
(a) Far-UV CD spectra of LytC (13.4 μM; circles) and Cpl-1 (21.5 μM; triangles) lysozymes in the absence (open symbols) and in the presence (filled symbols) of 140 mM choline. Data were collected at 4 °C (LytC) and 20 °C (Cpl-1) in 20 mM phosphate buffer, pH 8.0. Solid lines are the theoretical curves calculated by CONTIN. (b) Decomposition of LytC amide I band in 2H2O. Parameters derived for the band components are shown in Table 2. (c) Amide I bands (2H2O) of LytC and Cpl-1 in arbitrary units.·····, LytC;·–·–·–, LytC–choline; ——, Cpl-1; – – –, Cpl-1–choline.
Table 1. Secondary structure estimated by analysis of far-UV CD spectra of LytC and Cpl-1.
Secondary-structure content was estimated using CDNN and CONTIN (values in parentheses) methods from spectra recorded in the absence (−) and presence (+) of choline. Errors in the estimated values are 1–6% for CDNN and 1–4% for CONTIN. Values are rounded off to the nearest integer.
| Cpl-1 (%) | LytC (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| Structural element | − | + | − | + | ||||
| α-Helix | 11 | (7) | 8 | (4) | 12 | (8) | 7 | (12) |
| β-Sheet | 40 | (42) | 48 | (42) | 37 | (52) | 45 | (47) |
| Antiparallel | 35 | 42 | 31 | 40 | ||||
| Parallel | 5 | 6 | 6 | 5 | ||||
| β-Turn | 17 | (30) | 15 | (31) | 18 | (17) | 17 | (19) |
| Random structure | 32 | (21) | 29 | (23) | 33 | (23) | 31 | (22) |
The unambiguous analysis of secondary-structure content may be hampered when the experimentally available CD spectra do not include data down to 185 nm. For this reason, the secondary structure of LytC and Cpl-1 was also characterized by IR spectroscopy. The amide I bands of LytC and Cpl-1 in 2H2O present two maxima at 1640 cm−1 and 1650 cm−1 (Figure 4c). In addition, Cpl-1 also presents a shoulder around 1660 cm−1. Choline binding induces a narrowing of the amide I band of LytC in the region around 1615 cm−1, and a slight decrease in the intensity of the spectra of both enzymes around 1650 cm−1. Figure 4(b) shows the resolved components derived from the analysis of the amide I band of LytC, and the corresponding parameters, i.e. band position and area percentage, for both proteins are shown in Table 2. The assignment of these bands has been carried out previously [23,46]. The bands around 1629 and 1637 cm−1 indicate the presence of extended structures [23,46], while those between 1660 cm−1 and 1680 cm−1 can be attributed primarily to β-turns [46], since the contribution of the high-frequency component of the β-sheet component (1672–1694 cm−1) would be at most 10% of the low-frequency component around 1630 cm−1 [23]. On the other hand, bands around 1642–1660 cm−1 are usually assigned to α-helix [23,46], although contributions from long loops [47] or structures with dihedral angles close to those found in the α-helix, such as the 310 helix, have been described around this frequency. Finally, the component centred around 1640 cm−1 is due to unordered segments and loops [23]. The presence of a component centred at 1659 cm−1 in the spectrum of Cpl-1 without choline could indicate some degree of overlap of the components from α-helix and long loops with those from the β-turns.
Table 2. Band frequencies, fractional areas and proposed structure assignments of the components derived from IR measurements of LytC and Cpl-1.
Spectra were recorded in 2H2O in the absence (−) or presence (+) of choline (10 mM phosphate, p2H 8.0). Values are rounded off to the nearest integer. Odd-numbered columns are values in cm−1, even-numbered columns are percentages. ↑υ and ↓υ indicate high- and low-frequency components respectively.
| LytC | Cpl-1 | |||||||
|---|---|---|---|---|---|---|---|---|
| − | + | − | + | Assignment | ||||
| 1680 | 9 | − | − | − | − | 1683 | 1 | β-Sheet (↑υ) and β-turns |
| − | − | 1675 | 13 | 1677 | 11 | 1673 | 12 | β-Sheet (↑υ) and β-turns |
| 1668 | 13 | 1664 | 13 | 1659* | 33 | 1660 | 18 | β-Turns |
| 1655 | 25 | 1650† | 37 | − | − | 1650† | 22 | α-Helix, 310 helix and long loops |
| 1640 | 32 | − | − | 1646 | 14 | − | − | Unordered structure |
| − | − | 1637 | 17 | 1637 | 24 | 1635 | 45 | β-Sheet (↓υ) |
| 1627 | 17 | 1628 | 17 | 1629 | 17 | − | − | β-Sheet (↓υ) |
| 1613‡ | 4 | 1612‡ | 3 | 1612‡ | 1 | 1614‡ | 2 | |
* Maximum position could indicate some overlap with the α-helix component.
† Maximum position could indicate some overlap with the component from unordered structures.
‡ Bands at around 1612 cm−1 in native proteins arise from amino acid side chains.
The simultaneous presence of bands at 1637 cm−1 and 1628 cm−1 in the spectra of LytC and Cpl-1 is consistent with the structure of Cpl-1 and the model of LytC. The contribution of the TIM-barrel β-strands would be centred around the values expected for the low-frequency component of large β-sheets (approx. 1629 cm−1), while the CBM double strands would account for the component at 1637 cm−1 [46]. Indeed, the appearance of the latter in the amide I band of the LytC–choline complex, with the simultaneous loss of the unordered segments signal at 1640 cm−1, suggests that formation of β-hairpins in the CBM of LytC could be to some degree mediated by the interaction with choline. The small differences found in the components of Cpl-1 with and without choline agree with the rather small variations found between the free and the choline-bound structures of Cpl-1 [8].
Taking together the results with and without choline, and assuming that the contribution of the high-frequency component of β-strands in the region of around 1672–1694 cm−1 can increase to 10% of the intensity of the low-frequency absorption, the secondary structure of Cpl-1 appears to be 15% α-helix (the difference between the intensity of the component around 1660 cm−1 in the Cpl-1–choline complex and the free enzyme), 47% β-strand and 27% β-turn. Analogously, IR data for the monomer of LytC stabilized by choline-binding are consistent with the following estimations for the secondary-structure composition: 25% α-helix (intensity of the component at 1655 cm−1 in the absence of choline), 37% β-strand and 23% β-turn.
It is worth mentioning the agreement between the IR and CD estimations of secondary structure for each protein, and the reasonable correlation found with the percentages derived from either the Cpl-1 3D structure (17% α-helix, 3% 310 helix, 12% parallel β-sheet, 23% antiparallel β-sheet and 22% β-turns) or the proposed 3D model of LytC (14% α-helix, 25% β-sheet and 25% β-turns), particularly if we consider that the length of strands and helices in the model could be underestimated.
Hydrodynamic properties of LytC and Cpl-1
The sedimentation velocity profiles of Cpl-1 showed an apparently single boundary both in the absence and in the presence of choline that can be described in terms of a single sedimenting species (results not shown) with s20,w of 3.28±0.01 S (free enzyme) and 4.46±0.01 S (Cpl-1–choline complex). These values are compatible with the molecular masses of the monomer and the dimer of Cpl-1 respectively. Thus choline regulates, as in the amidases with CBM composed of six repeats [42–44], the selfassociation of the phage-encoded lysozyme, inducing the dimerization of Cpl-1. In contrast, the complete sedimentation of LytC at very short times and sedimentation rates as low as 3000 rev./min revealed the formation of high-order molecular-mass aggregates that dissociate upon choline binding into a single species whose sedimentation coefficient (s20,w of 3.73±0.01 S) is compatible with the monomer molecular mass. Dissociation of LytC aggregates is parallel to the appearance of the band at 224 nm in the CD spectra. The model generated for LytC was tested further by calculating the expected s20,w value from the model co-ordinates using the bead modelling approach implemented in HYDROPRO [48] that yielded an excellent estimate of s20,w for Cpl-1 using the crystallographic co-ordinates (Table 3). The good agreement between the calculated (3.67 S) and the experimental (3.73 S) values of s20,w supports further the proposed model of LytC.
Table 3. Hydrodynamic parameters of LytC and Cpl-1.
Measurements were made in 20 mM phosphate buffer, pH 8.0, in the absence (Cpl-1 monomer) and in the presence of 60 mM (Cpl-1 dimer) and 140 mM choline (LytC monomer).
| LytC | Cpl-1 | ||
|---|---|---|---|
| Monomer | Monomer | Dimer | |
| s20,w (S) | 3.73±0.01 | 3.28±0.01 | 4.46±0.01 |
| sT20,w (S)* | 3.67±0.01 | 3.14±0.01 | − |
| f (g·s−1) | 6.83×10−8 | 5.47×10−8 | 7.82×10−8 |
| f/f0 | 1.41 | 1.27 | 1.44 |
| a/b (prolate)† | 4.6 | 2.9 | 5.0 |
| a/b (oblate)† | 5.0 | 3.1 | 5.5 |
* Calculated with HYDROPRO from the Cpl-1 structure and the 3D model of LytC.
† Axial ratio between length (a) and diameter (b) of the theoretical ellipsoids.
The translational frictional ratios (f/f0) derived from the sedimentation coefficients (Table 3) clearly deviate from the values expected for rigid spherical particles and are compatible with the prolate and oblate ellipsoids with the axial ratios reported in Table 3. Interestingly, the f/f0 ratio of the choline-bound forms of LytC (monomer) and Cpl-1 (dimer) are very close and suggest that both complexes can have similar hydrodynamic shapes. The hydrodynamic particle model that best agrees with the dimensions of Cpl-1 in the crystal structure [8] is an oblate ellipsoid of 25 Å×76 Å. Analogously, the dimensions (21 Å×88 Å) derived from the 3D model of LytC reasonably agree with the oblate ellipsoid model (20 Å×101 Å).
Structural comparison of Cpl-1 and LytC lysozymes
In spite of having an inverted disposition of their constituting modules, LytC and Cpl-1 seem to adopt a similar global fold. However, there are substantial differences between the 3D model generated for LytC and the crystal structure of Cpl-1, including the electrostatic potential of the catalytic surface (Figures 2c and 2d), the length of the loops at the C-terminal side of the β-strands (related in α/β barrels to substrate recognition [49]), the number of potential choline-binding sites (seven in LytC compared with four in the Cpl-1 dimer) and the shape of the active particle. Moreover, the structural changes promoted by choline in the host-encoded enzyme suggest that the conformation adopted by the CBM of LytC is strongly dependent on choline binding, as revealed CD and ultracentrifugation studies.
In contrast with LytC and cellosyl [9], Cpl-1 exhibits a broad negatively charged catalytic surface (Figures 3c and 3d) that would favour its interaction with a positively charged substrate (80% of glucosamine residues of pneumococcal peptidoglycan are deacetylated) [50]. Besides, its looser attachment to the bacterial cell wall (the choline IC50 is 15 times higher for LytC than for Cpl-1) will facilitate the diffusion of the protein through the bacterial envelope. These features might account for the higher specific activity of the parasite-encoded enzyme [3], especially if the active species is the Cpl-1 dimer bearing two catalytic centres.
Choline-induced self-association has been observed in all the pneumococcal lysins containing a CBM formed by six repeats located at the C-terminal extreme characterized to date [42–44]. Therefore the acquisition of additional choline-binding motifs by LytC may constitute an alternative adaptation mechanism leading to the enhancement of cell-wall-binding capability that reduces, at the same time, the curvature associated with the dimerization of CBM displayed by the crystal structure of C-LytA [11]. The lack of stabilizing contacts between sequence-distant repeats, a characteristic of the solenoid fold, would enhance the flexibility of the CBM of LytC and therefore its ability to selectively interact with those regions of the bacterial envelope where the complementarity of the target is maximal, providing a tight binding [51]. Indeed, shallow grooves in solenoids with small curvature are frequently found in structures that can bind extended and flexible structures (like those of teichoic and lipoteichoic acids) through multiple interactions [52]. High-affinity binding of peptidoglycan hydrolases has been proposed to be related to the bacterial regulation of the suicidal activity of the autolytic enzymes. Thus the acquisition of additional repeats by LytC and the uneven distribution of choline-binding sites in the CBM could reflect a divergent evolution toward an improved regulation of the autolytic activity of the pneumococcal lysozyme.
Concluding remarks
The family of pneumococcal choline-binding proteins, known as CBPs, represents one of the most paradigmatic examples that illustrate the theory of the modular evolution of proteins. Until very recently, it was assumed that the murein hydrolases encoded by Pneumococcus and its phages evolved following a general assembling pattern that placed the functional/catalytic domain at the N-terminus and the CBM at the C-terminus. However, the reversed order of these modules in the LytC lysozyme encoded by Pneumococcus confirmed further the versatility of this modular design and raised the question of why the bacteria had not taken advantage of this already existing gene to evolve its own autolytic enzyme. Lysozymes are extremely aggressive enzymes whose expression and activity can be tailored at the genetic level by natural selection to suit the prevailing physiological requirements. For the lytic bacteriophages, it is essential that a general and rapid destruction of the cell wall occur once the virions are formed, and Cpl-1 has probably evolved to fulfil this requirement for high efficiency, making it difficult for the host to adapt such an active enzyme. Thus Pneumococcus would have acquired a different, less active, version of this lytic gene to remodel the cell wall at its convenience and to carry out a slow autolysis at 30 °C. Our data provide evidence that, by increasing the number and variability of the repeats, LytC has acquired a more unstable, irregular and extended CBM whose conformation is extremely dependent on its binding to the choline-containing teichoic acids and provides tight binding to the cell wall. This fact may confine the activity of the host enzyme to specific areas of cell envelope, since complete dissociation from teichoic acids will be highly unfavourable. Furthermore, the catalytic activity can be also limited by steric boundaries, since the distinct binding patterns of LytC and Cpl-1 can result in a different orientation of the glycanic chain within the active site. Moreover, the dissimilarities found in the catalytic cavity can also contribute to reduce the activity of LytC relative to Cpl-1.
Online data
Acknowledgments
This work was supported by DGICYT (Dirección General de Investigación Científica y Técnica) Grants BIO2000-1307, BMC2000-1002, BIO2003-01952, BMC2003-00074 and by CAM Grant of the Program for Strategic Groups. We are grateful to Dr Germán Rivas for many helpful discussions, to Dr Concepción Domingo for assistance in IR experiments, to Dr Douglas Laurents for linguistic revision, and to Victoria López for technical support. B. M. was supported by a fellowship from the Spanish Ministerio de Ciencia y Tecnología.
References
- 1.Coutinho P. M., Henrissat B. Carbohydrate-active enzymes: an integrated database approach. In: Gilbert H. J., Davies G., Henrissat B., Svensson B., editors. Recent Advances in Carbohydrate Bioengineering. Cambridge: The Royal Society of Chemistry; 1999. pp. 3–12. [Google Scholar]
- 2.Gebler J., Gilkes N. R., Claeyssens M., Wilson D. B., Beguin P., Wakarchuk W. W., Kilburn D. G., Miller R. C., Warren R. A. J., Withers S. G. Stereoselective hydrolysis catalyzed by related β-1,4-Glucanases and β-1,4-xylanases. J. Biol. Chem. 1992;267:12559–12561. [PubMed] [Google Scholar]
- 3.García P., González M. P., García E., García J. L., López R. The molecular characterization of the first autolytic lysozyme of Streptococcus pneumoniae reveals evolutionary mobile domains. Mol. Microbiol. 1999;33:128–138. doi: 10.1046/j.1365-2958.1999.01455.x. [DOI] [PubMed] [Google Scholar]
- 4.García P., García J. L., García E., Sánchez-Puelles J. M., López R. Modular organization of the lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Gene. 1990;86:81–88. doi: 10.1016/0378-1119(90)90116-9. [DOI] [PubMed] [Google Scholar]
- 5.López R., García E., García P., García J. L. The pneumococcal cell wall degrading enzymes: a modular design to create new lysins? Microb. Drug Resist. 1997;3:199–211. doi: 10.1089/mdr.1997.3.199. [DOI] [PubMed] [Google Scholar]
- 6.Sanz J. M., Díaz E., García J. L. Studies on the structure and function of the N-terminal domain of the pneumococcal murein hydrolases. Mol. Microbiol. 1992;6:921–931. doi: 10.1111/j.1365-2958.1992.tb01542.x. [DOI] [PubMed] [Google Scholar]
- 7.Henrissat B., Bairoch A. New families in the classification of glycosylhydrolases based on amino acid sequence similarities. Biochem. J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hermoso J. A., Monterroso B., Albert A., Galán B., Ahrazem O., García P., Martínez-Ripoll M., García J. L., Menéndez M. Structural basis for selective recognition of pneumococcal cell wall by modular endolysin from phage Cp-1. Structure. 2003;11:1239–1249. doi: 10.1016/j.str.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Rau A., Hogg T., Marquardt R., Hilgenfeld R. A new lysozyme fold. J. Biol. Chem. 2001;276:31994–31999. doi: 10.1074/jbc.M102591200. [DOI] [PubMed] [Google Scholar]
- 10.Sanz J. M., García P., García J. L. Role of Asp-9 and Glu-37 in the active site of the pneumococcal Cpl-1 lysozyme: an evolutionary perspective of lysozyme mechanism. Biochemistry. 1992;31:8495–8499. doi: 10.1021/bi00151a016. [DOI] [PubMed] [Google Scholar]
- 11.Fernández-Tornero C., López R., García E., Giménez-Gallego G., Romero A. A novel solenoid fold in the cell wall anchoring domain of the pneumococcal virulence factor LytA. Nat. Struct. Biol. 2001;8:1020–1024. doi: 10.1038/nsb724. [DOI] [PubMed] [Google Scholar]
- 12.Tramontano A. Homology modelling with low sequence identity. Methods. 1998;14:293–300. doi: 10.1006/meth.1998.0585. [DOI] [PubMed] [Google Scholar]
- 13.Facchiano A. M., Struso P., Chiusano M. L., Caraglia M., Giuberti G., Marra M., Abbruzzese A., Colonna G. Homology modelling of the human eukaryotic initiation factor 5A (eIF-5A) Protein Eng. 2001;14:881–890. doi: 10.1093/protein/14.11.881. [DOI] [PubMed] [Google Scholar]
- 14.Sanz J. M., López R., García J. L. Structural requirements of choline derivatives for “conversion” of pneumococcal amidase. FEBS Lett. 1988;232:308–312. doi: 10.1016/0014-5793(88)80759-2. [DOI] [PubMed] [Google Scholar]
- 15.Medrano F. J., Gasset M., López-Zumel C., Usobiaga P., García J. L., Menéndez M. Structural characterization of the unligated and choline-bound forms of the major pneumococcal autolysin LytA amidase. J. Biol. Chem. 1996;271:29152–29161. doi: 10.1074/jbc.271.46.29152. [DOI] [PubMed] [Google Scholar]
- 16.Philo J. An improved function for fitting sedimentation velocity data for low-molecular weight solutes. Biophys. J. 1997;72:435–444. doi: 10.1016/S0006-3495(97)78684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Holde K. E. Physical Biochemistry. Englewood Cliffs: Prentice Hall; 1985. Sedimentation; pp. 110–136. [Google Scholar]
- 18.Kuntz I. D. Hydration of macromolecules IV. Polypeptide conformation in frozen solutions. J. Am. Chem. Soc. 1971;93:516–518. doi: 10.1021/ja00731a037. [DOI] [PubMed] [Google Scholar]
- 19.Waxman E., Laws W. R., Laue T. M., Nemerson Y., Ross J. B. A. Human factor VIIa and its complex with soluble tissue factor: evaluation of asymmetry and conformational dynamics by ultracentrifugation and fluorescence anisotropy decay methods. Biochemistry. 1993;32:3005–3012. doi: 10.1021/bi00063a011. [DOI] [PubMed] [Google Scholar]
- 20.Provencher S. W., Glöckner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 21.Böhm G., Muhr R., Jaenicke R. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 1992;5:191–195. doi: 10.1093/protein/5.3.191. [DOI] [PubMed] [Google Scholar]
- 22.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 23.Arrondo J. L. R., Muga A., Castresana J., Goñi F. M. Quantitative studies of the structure of proteins in solution by Fourier-transform infrared spectroscopy. Prog. Biophys. Mol. Biol. 1993;59:23–56. doi: 10.1016/0079-6107(93)90006-6. [DOI] [PubMed] [Google Scholar]
- 24.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and Psi-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rost B. PHD: predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 26.McGuffin L., Bryson K., Jones D. T. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 27.Cuff J. A., Barton C. J. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins Struct. Funct. Genet. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 28.Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galtier N., Gouy M., Gautier C. SeaView and Phylo_win, two graphic tools for sequence alignment and molecular phylogeny. CABIOS Comput. Appl. Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 30.Mizuguchi K., Deane C. M., Blundell T. M., Johnson M. S., Overington J. P. JOY: protein sequence structure representation and analysis. Bioinformatics. 1998;14:617–623. doi: 10.1093/bioinformatics/14.7.617. [DOI] [PubMed] [Google Scholar]
- 31.Pearson W. R., Wood T., Zhang Z., Miller W. Comparison of DNA sequences with protein sequences. Genomics. 1997;46:24–36. doi: 10.1006/geno.1997.4995. [DOI] [PubMed] [Google Scholar]
- 32.Sali A., Blundell T. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 33. Reference deleted.
- 34.Laskowski R. A., McArthur W., Moss D., Thornton J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 35.Luthy R., Bowie J. U., Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature (London) 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 36.Sali A., Blundell T. L. Definition of general topological equivalence in protein structures: a procedure involving comparison of properties and relationships thought simulated annealing and dynamic programming. J. Mol. Biol. 1990;212:403–428. doi: 10.1016/0022-2836(90)90134-8. [DOI] [PubMed] [Google Scholar]
- 37.van Roey P., Rao V., Plummer T. H., Jr, Tarentino A. L. Crystal structure of endo-β-N-acetylglucosaminidase F1, an α/β-Barrel enzyme adapted for a complex substrate. Biochemistry. 1994;33:13989–13996. doi: 10.1021/bi00251a005. [DOI] [PubMed] [Google Scholar]
- 38.Mark B. L., Vocadlo D. J., Knapp S., Triggs-Raine B. L., Withers S. G., James M. N. Crystallographic evidence for a substrate-assisted catalysis in a bacterial β-hexosaminidase. J. Biol. Chem. 2001;276:10330–10337. doi: 10.1074/jbc.M011067200. [DOI] [PubMed] [Google Scholar]
- 39.Simpson P. J., Bolam D. N., Cooper A., Ciruela A., Hazlewood G. P., Gilbert H. J., Williamson M. P. A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity. Structure. 1999;7:853–864. doi: 10.1016/s0969-2126(99)80108-7. [DOI] [PubMed] [Google Scholar]
- 40.Nagy T., Simpson P. J., Williamson M. P., Hazlewood G. P., Orosz L., Gilbert H. J. All three surface tryptophans in Type IIa cellulose binding domains play a pivotal role in binding both soluble and insoluble ligands. FEBS Lett. 1998;429:312–316. doi: 10.1016/s0014-5793(98)00625-5. [DOI] [PubMed] [Google Scholar]
- 41.Udwary D. W., Merski M., Townsend C. A. A method for prediction of the locations of linker regions within large multifunctional proteins, and application to a Type I polyketide synthase. J. Mol. Biol. 2002;323:585–598. doi: 10.1016/s0022-2836(02)00972-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sáiz J. L., López-Zumel C., Monterroso B., Varea J., Arrondo J. L. R., Iloro I., García J. L., Laynez J., Menéndez M. Characterization of Ejl, the cell-wall amidase coded by the pneumococcal bacteriophage Ej-1. Protein Sci. 2002;11:1788–1799. doi: 10.1110/ps.4680102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varea J., Sáiz J. L., López-Zumel C., Monterroso B., Medrano F. J., Arrondo J. L. R., Iloro I., Laynez J., García J. L., Menéndez M. Do sequence repeats play an equivalent role in the choline-binding module of pneumococcal LytA amidase? J. Biol. Chem. 2000;275:26842–26855. doi: 10.1074/jbc.M004379200. [DOI] [PubMed] [Google Scholar]
- 44.Varea J., Monterroso B., Sáiz J. L., López-Zumel C., García J. L., Laynez J., García P., Menéndez M. Structural and thermodynamic characterization of Pal, a phage natural chimeric lysin active against Pneumococci. J. Biol. Chem. 2004;279:43697–43707. doi: 10.1074/jbc.M407067200. [DOI] [PubMed] [Google Scholar]
- 45.Johnson W. C., Jr Protein secondary structure and circular dichroism: a practical guide. Proteins Struct. Funct. Genet. 1990;7:205–214. doi: 10.1002/prot.340070302. [DOI] [PubMed] [Google Scholar]
- 46.Barth A., Zscherp B. What vibrations tell us about proteins. Q. Rev. Biophys. 2002;35:369–430. doi: 10.1017/s0033583502003815. [DOI] [PubMed] [Google Scholar]
- 47.Wilder C. L., Friedrich A. D., Potts R. O., Daumy G. O., Francoeur M. L. Secondary structural analysis of two recombinant murine proteins, interleukins 1α and 1β: is infrared spectroscopy sufficient to assign structure? Biochemistry. 1992;31:27–31. doi: 10.1021/bi00116a006. [DOI] [PubMed] [Google Scholar]
- 48.García de la Torre J., Huertas M. L., Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Branden C., Tooze J. Introduction to Protein Structure. New York and London: Garland Publishing; 1991. Alpha/beta structures; pp. 43–56. [Google Scholar]
- 50.Vollmer W., Tomasz A. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J. Biol. Chem. 2000;275:20496–20501. doi: 10.1074/jbc.M910189199. [DOI] [PubMed] [Google Scholar]
- 51.Dyson H. J., Wright P. E. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 52.Kobe B., Kajava A. When protein folding is simplified to protein coiling: the continuum of solenoid protein structures. Trends Biochem. Sci. 2000;25:509–515. doi: 10.1016/s0968-0004(00)01667-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.