Abstract
Recently, we described a 160 kDa protein (designated AS160, for Akt substrate of 160 kDa) with a predicted Rab GAP (GTPase-activating protein) domain that is phosphorylated on multiple sites by the protein kinase Akt. Phosphorylation of AS160 in adipocytes is required for insulin-stimulated translocation of the glucose transporter GLUT4 to the plasma membrane. The aim of the present study was to determine whether AS160 is in fact a GAP for Rabs, and, if so, what its specificity is. We first identified a group of 16 Rabs in a preparation of intracellular vesicles containing GLUT4 by MS. We then prepared the recombinant GAP domain of AS160 and examined its activity against many of these Rabs, as well as several others. The GAP domain was active against Rabs 2A, 8A, 10 and 14. There was no significant activity against 14 other Rabs. GAP activity was further validated by the finding that the recombinant GAP domain with the predicted catalytic arginine residue replaced by lysine was inactive. Finally, it was found by immunoblotting that Rabs 2A, 8A and 14 are present in GLUT4 vesicles. These results indicate that AS160 is a Rab GAP, and suggest novel Rabs that may participate in GLUT4 translocation.
Keywords: Akt, Akt substrate of 160 kDa (AS160), GTPase-activating protein (GAP), GLUT4, Rab
Abbreviations: AS160, Akt substrate of 160 kDa; GAP, GTPase-activating protein; GAP R/K, GAP domain with a R973K replacement; GST, glutathione S-transferase; IRAP, insulin-regulated aminopeptidase
INTRODUCTION
Insulin treatment of fat and muscle cells causes a rapid increase in glucose transport. This increase is due to a rise in glucose transporters of the GLUT4 type at the cell surface. This elevation occurs as the result of insulin-stimulated movement of intracellular vesicles containing GLUT4 to the plasma membrane and fusion therewith, a process known as GLUT4 translocation (reviewed in [1]). There is considerable evidence that a signalling pathway necessary for GLUT4 translocation is the one that proceeds from the insulin receptor to the activation of the protein kinase Akt [1–4]. However, there is less information about the connection between Akt activation and GLUT4 translocation. Recently, we described a 160 kDa Akt substrate protein that has the properties expected to mediate this connection [5,6]. This protein, which has been designated AS160 (Akt substrate of 160 kDa), has a predicted GAP (GTPase-activating protein) domain towards members of the Rab protein family.
Rabs are small G-proteins that in their GTP-bound form participate in vesicle movement and fusion (reviewed in [7]). The GAP for a Rab stimulates the typically low intrinsic GTPase activity of the Rab to generate the inactive GDP-bound form of the Rab. Our finding that AS160 is phosphorylated by insulin-activated Akt has led to the following hypothesis: phosphorylation of AS160 inhibits its GAP activity; consequently, the GTP form of a Rab(s) required for GLUT4 translocation is elevated, and thus translocation is triggered [6]. In agreement with this proposal, insulin-stimulated GLUT4 translocation in adipocytes was blocked by expression of a mutant of AS160 lacking Akt phosphorylation sites [6,8]. Presumably, this non-phosphorylatable mutant of AS160 continued to function as a GAP in the presence of insulin. This blockage required a functional GAP domain. The non-phosphorylatable mutant was not effective when the catalytic arginine residue in the GAP domain was mutated to lysine.
Although the studies summarized above are consistent with AS160 being a Rab GAP, to date there has been no demonstration that AS160 has this activity. In the present study we show that the recombinant GAP domain of AS160 is functional towards a small number of Rabs, and that these Rabs are present in vesicles containing GLUT4.
EXPERIMENTAL
Plasmids
The following plasmids were generously provided by the following individuals: pGEX-Rab1A (human), William Balch (Department of Molecular and Cell Biology, Scripps Research Institute, La Jolla, CA, U.S.A.); pGEX-Rab4A, -5A and -6 (all human), Guangpu Li (Department of Biochemistry and Molecular Biology, University of Oklahoma Health Sciences Center, Oklahoma City, OK, U.S.A.); pGEX-Rab7 (dog), Angela Wandinger-Ness (Department of Pathology, University of New Mexico Health Sciences Center, Albuquerque, NM, U.S.A.); pGEX-Rab11A (dog), Marino Zerial (Max-Planck Institute for Molecular Cell Biology and Genetics, Dresden, Germany); pQE-Rab11B (mouse), James Goldenring (Department of Surgery, Vanderbilt University School of Medicine, Nashville, TN, U.S.A.); pGEX-Rab21 (human), Jack Fransen (Department of Cell Biology, University of Nijmegen, Nijmegen, Netherlands). The pGEX plasmids for Rabs 3A (mouse), 4B (human), 8A (dog), 10 (human), 14 (mouse), 18 (human), 27A (mouse), 27B (mouse), and 35 (human) were prepared by PCR amplification of the Rab coding sequences with templates from several sources, followed by ligation into the pGEX vector. The plasmids encoding N-terminal FLAG-tagged AS160 and the AS160 R973K mutant are as described previously [6]. The pGEX plasmids for the GAP domain of AS160 and the R973K mutant GAP were generated by PCR amplification of the DNA encoding GAP domain and ligation into the vector. These comprised amino acids 865–1299 of human AS160 (GI code 7662198).
Antibodies
Anti-Rab2 antibody was purchased from Santa Cruz Biotechnology (catalogue number sc307). Anti-Rab8 antibody was obtained from BD Transduction Laboratories (catalogue number 610844); this antibody reacts with both Rabs 8A and 8B (J. Peränen, unpublished work). The antibodies against the C-terminus of GLUT4, the C-terminus of AS160 and the cytoplasmic domain of IRAP (insulin-regulated aminopeptidase) are as described previously [6,9,10]. An antibody against a peptide corresponding to amino acids 167–183 of mouse Rab14 was generated by immunization of rabbits with the conjugated peptide (Biosource International), followed by affinity purification on the immobilized peptide.
GLUT4 vesicles
The method for isolation of GLUT4 vesicles was as described in [11], with slight modifications. 3T3-L1 adipocytes in serum-free medium were treated with 160 nM insulin or not for 30 min. The cells, on a 10-cm diameter plate, were washed with PBS and then with buffer A (1 mM EDTA, 225 mM sucrose and 20 mM Hepes, pH 7.4). The cells were then scraped into 1 ml of buffer A with protease inhibitors (10 μM each of leupeptin, pepstatin and EP475) and homogenized at 20 °C. Subsequent steps were carried out at 4 °C. The homogenate was centrifuged at 16000 g for 15 min, and the supernatant centrifuged again at 48000 g for 15 min. The supernatant from the second centrifugation, which is a low-density microsome/cytosol fraction, was made up to a concentration of 100 mM in NaCl and then incubated for 2 h with anti-GLUT4 antibody or control rabbit immunoglobulin bound to Pansorbin (Calbiochem) (10 μg of antibody for 4 μl of Pansorbin per ml). The adsorbent was then washed several times with buffer A/100 mM NaCl and the bound vesicles were solubilized with 0.5% nona(ethylene glycol) dodecyl ether in buffer A/100 mM NaCl. For identification of the Rabs by MS, SDS samples of the solubilized GLUT4 and control vesicles, each from three 10-cm diameter plates of unstimulated adipocytes, were separated on a short gradient gel, and the gel region containing the Rabs (21–38 kDa section) was excised. Gel slices were subjected to in-gel tryptic digestion, and the tryptic peptides were sequenced by microcapillary liquid-chromatography tandem MS, as described in [6]. For immunoblotting, samples of the vesicles were separated by SDS/PAGE, and immunoblotted for specific proteins as described in [5].
Recombinant proteins
The pGEX-Rab plasmids were introduced into Escherichia coli BL21 cells or, in the case of GST–Rab14, into the E. coli Rosetta BL21 strain (Novagen). Bacterial cultures (200 ml) were grown to a D600 of approx. 0.5, and then induced with 0.1 mM isopropyl β-D-thiogalactoside for 6 h at 25 °C or overnight at 15 °C. The bacteria were pelleted, resuspended in 10 ml of 2.5 mM MgCl2/PBS with protease inhibitors (Roche, catalogue number 11836170), and lysed in a French press. The lysate was centrifuged at 23000 g for 45 min, and the supernatant was mixed with 300 μl of glutathione beads (Pierce) for 1 h at 4 °C. The beads were washed with 2.5 mM MgCl2/PBS, and the GST (glutathione S-transferase)–Rab fusion protein was eluted three times with 300 μl of 10 mM glutathione, 1 mM dithiothreitol, 2.5 mM MgCl2 and 20 mM Tris/HCl, pH 8.0. The procedure for expression of the GST–GAP domain was similar, except that the bacteria were lysed in PBS, 5 mM mercaptoethanol and 10% glycerol with protease inhibitors, and the GST-fusion protein was eluted with 20 mM glutathione, 5 mM mercaptoethanol, 150 mM NaCl, 10% glycerol and 100 mM Tris/HCl, pH 8.5. Rab11B, which was His-tagged, was expressed in the E. coli XL-1 Blue strain and purified on an immobilized nickel column (Qiagen), according to the manufacturer's instructions. Purified recombinant human untagged Rab2A was a gift from Ellen Tisdale (Department of Pharmacology, Wayne State University School of Medicine, Detroit, MI, U.S.A.).
Recombinant FLAG-tagged AS160 and the R973K mutant were generated by transfecting HEK-293F cells with the AS160 plasmids described above. Each 10-cm diameter plate was transfected with 10 μg of plasmid using Lipofectamine 2000 (Invitrogen). After 24 h, the cells were serum starved for 2 h, and treated with 75 μM LY294002 (phosphoinositide 3-kinase inhibitor) for the final 45 min before lysis in order to deactivate any activated Akt [12]. The cells were then lysed in 2 ml of 1.5% nona(ethylene glycol) dodecyl ether, 150 mM NaCl and 40 mM Hepes, pH 7.4, with protease inhibitors (10 μM each of pepstatin, leupeptin, aprotinin and EP475), the lysate was cleared by centrifugation at 12000 g for 15 min, and the AS160 was adsorbed on to 10 μl of anti-FLAG beads (Sigma), which were then washed thoroughly with 150 mM NaCl/40 mM Hepes, pH 7.5. This procedure yielded approx. 15 μg of AS160 per 10-cm diameter plate.
In order to estimate the purity and concentration of the recombinant proteins, samples were run on SDS/PAGE along with known amounts of standard proteins and stained with Coomassie Blue. The desired protein was the predominant component in the preparation of each recombinant protein.
Assays for Rab GTP loading and GAP activity
The loading procedure involved the exchange of added [α-32P]-GTP with the bound unlabelled GTP and GDP (mainly GDP) on the recombinant GST–Rab, facilitated by complexing Mg2+ ion with EDTA. Then the bound [α-32P]GTP is fixed on the Rab protein by the addition of Mg2+ ion, and binding is measured by separation of the [α-32P]GTP complex on a nitrocellulose filter [13,14]. The GST–Rab fusion protein (1 μM) was incubated with [α-32P]GTP (0.1 μM) in 2 mM EDTA, 10 mM glutathione, 1 mM dithiothreitol, 200 μg/ml BSA and 20 mM Tris/HCl, pH 8.0, in a volume of 43.5 μl at 30 °C for 30 min. Then 2.5 μl of 200 mM MgCl2 was added, and this addition was followed by 4 μl of the buffer for the recombinant GST–GAP domain, so that the conditions for the loading assay were similar to those for the GAP assay (see below). Aliquots (10 μl) of the mixture were filtered through 24 mm nitrocellulose filters (Millipore, HAWP type), and the filters washed with ice-cold 10 mM MgCl2, 2 mM EDTA, 50 mM NaCl and 20 mM Tris/HCl, pH 8.0. The percentage of bound GTP was calculated from radioactivity measured on the filter and the input radioactivity. In the case of Rab10, the percentage of bound GTP was relatively low (approx. 15%). Consequently, for Rab10, after loading the mixture was passed through a small gel-filtration spin column (Pierce, catalogue number 89849) to remove some of the unbound GTP, and then filtered through the nitrocellulose filter. With this modification, 40% of the [α-32P]GTP from the spin column was bound to the Rab10.
The GAP assay was carried out as descibed in [13]. The GST–Rab protein was loaded with [α-32P]GTP, as described above. Immediately after the addition of MgCl2, a 10 μl aliquot was removed as the zero time point. Then the GST–GAP domain was added as a 4 μl aliquot, so that the final concentration was 0.4 μM unless stated otherwise, and incubation was continued at 30 °C, with 10 μl aliquots removed at various times. The samples were immediately placed in 20 μl of 0.2% SDS, 5 mM EDTA, 5 mM GDP and 5 mM GTP at 65 °C, and these were held at 65 °C for 2 min. Samples (5 μl) were spotted on to a polyethyleneimine cellulose plate (Selecto Scientific, catalogue number 10078), and TLC was carried out in 0.75 M potassium phosphate, pH 3.5. The radioactivity in the GTP and GDP spots was measured by phosphor imaging.
RESULTS
Rabs in GLUT4 vesicles by MS
The mammalian family of Rabs has 60 different members [7,15]. Each of these is believed to participate in a specific trafficking step, for example, from the recycling endosomes to the plasma membrane. However, the site of action has only been clearly established for a small fraction of the 60 Rabs. Since it seems likely that the Rab(s) involved in GLUT4 translocation is located on GLUT4 vesicles, we initially narrowed the number of Rabs to examine by determining the Rab content of GLUT4 vesicles by MS.
GLUT4 vesicles were isolated from the low-density microsome fraction of 3T3-L1 adipocytes by immunoadsorption with an antibody against GLUT4, and negative control immunoadsorption was also carried out with non-specific rabbit immunoglobulin. The vesicle proteins were separated by SDS/PAGE on a short gel, and the proteins in the 21–38 kDa region were identified by microcapillary liquid-chromatography tandem MS. This region encompasses the sizes of almost all the 60 Rabs, which typically have a mass of approx. 25 kDa [15].
Using this approach, tryptic peptides from the following Rabs were found in GLUT4 vesicles: 1A, 1B, 2A, 3A or 3D, 4B, 5A, 5B, 5C, 6A or 6B, 7, 8A or 8B, 10, 11B, 14, 18 and 35 (results not shown). Some of the Rabs are listed as two possibilities, such as Rabs 3A or 3B, because the peptides found were ones common to both members of the Rab subfamily. In addition, it should be noted that in some cases both peptides common to a Rab subfamily (for example, Rabs 2A and 2B) and ones specific to a member of the subfamily (for example, Rab2A) were found. We have listed only the specific member, although it is possible that some common peptides come from another member of the subfamily. In previous studies GLUT4 vesicles have been found to contain Rab4 and Rab11 by immunoblotting [16,17].
Surprisingly, the negative control for the GLUT4 vesicle preparation also yielded peptides from all the Rabs present in the GLUT4 vesicles, with the exception of Rabs 1A, 1B, 5A and 35. There are probably two reasons for large number of Rabs common to both the GLUT4 vesicles and the negative control. Firstly, a small amount of GLUT4 vesicles may have bound to the non-specific adsorbent. Second, some non-GLUT4 vesicles may have bound to the anti-GLUT4 adsorbent, as well as to the non-specific adsorbent. The enormous sensitivity of microcapillary liquid-chromatography tandem MS then resulted in detection of these impurities. Unfortunately, MS as used here does not give a measure of amounts, only of identity, and so no reliable comparison can be made between the amounts of a Rab in the GLUT4 vesicle preparation and the negative control. Thus it is likely that this group of Rabs found in the GLUT4 vesicle preparation includes most, if not all, the Rabs in vesicles with GLUT4, as well as some Rabs that are not in such vesicles. Despite this ambiguity, these results served the intended purpose in that they provided the basis for the selection of Rabs to test as substrates for the AS160 GAP domain.
GAP activity of the AS160 GAP domain
We prepared a GST-fusion protein of the AS160 GAP domain and GST-fusion proteins of one or more members of each subfamily of Rab found in the GLUT4 vesicle preparation, as well as of several other Rabs implicated in exocytosis in other systems. The activity of the GAP domain was assayed by measuring its effect on the rate of conversion of the GTP form of each GST–Rab into the GDP form. This assay required that the recombinant GST–Rab be functional in binding GTP. Table 1 lists the Rabs used in this study and summarizes the results from the assay for [α-32P]GTP loading of each of the Rabs. Each Rab specifically bound GTP, and thus was at least partially active in GTP binding. The percentage of the [α-32P]GTP bound ranged from a high value of 70% to a low one of 25%.
Table 1. GTP loading of Rabs.
The percentage of total [α-32P]GTP that was bound to the Rab protein was measured as described in the Experimental section. In the case of Rab5A the GTP binding was done for 1 min, rather than the usual 30 min, because at longer times considerable hydrolysis of the GTP to GDP occurred. In the case of Rab10, the value is that after removal of a portion of the unbound GTP by gel filtration (see the Experimental section).
| Rab | GTP bound (%) |
|---|---|
| 1A | 43 |
| 2A | 50 |
| 3A | 43 |
| 4A | 58 |
| 4B | 57 |
| 5A | 45 |
| 6 | 29 |
| 7 | 48 |
| 8A | 50 |
| 10 | 40 |
| 11A | 48 |
| 11B | 33 |
| 14 | 42 |
| 18 | 25 |
| 21 | 43 |
| 27A | 27 |
| 27B | 70 |
| 35 | 33 |
The rate of conversion of the [α-32P]GTP form of the Rab into the [α-32P]GDP form was measured in the presence and absence of the GAP domain through separation of the GTP from GDP by TLC. Figure 1 shows the method for Rab14, one of the Rabs against which the AS160 GAP domain was active. In order to be certain that the GTPase activity was due to the GAP domain, we generated a point mutant of the GST–GAP domain in which Arg973, which is predicted to be required for activity [6], was mutated to lysine. This mutation is the same one that affects AS160 function in vivo (see the Introduction section). As expected, this mutated GAP domain (designated GAP R/K) showed no activity against Rab14 (Figure 1).
Figure 1. Rab GAP assay with AS160 GST–GAP domain and GST–Rab14.
(A) TLC separation of α32P GDP and GTP in assays with Rab14 alone, Rab14 plus the GAP domain, and Rab14 plus the R/K mutant of the GAP domain. (B) The data from (A) are presented as the percentage of the total radioactivity in GDP versus time for Rab14 (●), Rab14 plus the GAP domain (■), and Rab14 plus the R/K mutant GAP domain (▲). For further details, see the Experimental section.
In another type of control we found that neither the GST–GAP domain nor GST–GAP R/K exhibited significant hydrolytic activity against [α-32P]GTP alone (that is, not bound to a Rab) (results not shown). Since GST-fusion proteins are known to exist as dimers, we also considered whether the activity of the GAP domain against Rab14 could be dependent upon formation of a heterodimer comprising one subunit of GST–GAP and one subunit of GST–Rab14. In order to test this possibility, we carried out the assay in the presence of GST, rather than BSA, as the carrier protein, at a concentration such that the GST was in 10-fold molar excess over the combined GST–GAP and GST–Rab14. If heterodimerization were required for GAP activity, the GST would be expected to inhibit the activity markedly, since it should be preferred as the partner in the heterodimers due to its higher concentration. However, the presence of excess GST did not inhibit the GAP activity (results not shown). In addition, we directly compared the activity of the recombinant GST–GAP domain with that of recombinant GAP domain without GST against Rab14, and found no difference (results not shown). The recombinant GAP domain without GST was prepared from recombinant GST–GAP domain bound to immobilized glutathione by thrombin cleavage at the thrombin site in the linker between the GST and GAP domain.
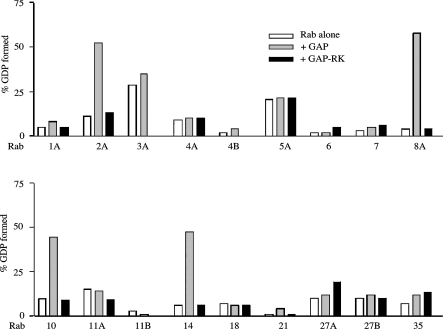
Figure 2 summarizes the results from assays of the GAP domain against the Rabs in Table 1. The GAP domain markedly stimulated the hydrolysis of [α-32P]GTP bound to Rabs 2A, 8A, 10 and 14. In each of these cases the GAP R/K construct exhibited no significant activity. It is worth noting that the intrinsic GTPase activity of the Rabs varied considerably. For example, after correction for the fraction of GTP bound to the Rab (Table 1), Rab7 showed only 4% of hydrolysis of its bound GTP in 15 min at 30 °C, whereas Rab3A showed 60% hydrolysis of its bound GTP in 15 min.
Figure 2. Activity of AS160 GAP domain against the 18 Rab proteins.
The GAP assay was carried out as described in the Experimental section and shown in Figure 1. The data show the increase in the percentage of the total radioactivity in GDP between 0 and 15 min for the Rab alone and the Rab plus the AS160 GAP domain. In addition, for any Rab that appeared to be a GAP substrate, as well as most of the others, the assay was also performed with the GAP R/K mutant. In the case of Rab5A, the data are for 3 min, rather than 15 min, due to the high intrinsic GTPase activity of this Rab.
At the concentration of the GST–GAP domain used in Figure 2, the activities toward Rabs 2A, 8A, 10 and 14 were such that the percentages of the [α-32P]GTP hydrolysed in the initial 15 min period of the assay were approximately the same as the percentages that were bound to the Rab (Table 1). Thus, since most of the Rab-bound GTP was hydrolysed, the assay under these conditions did not yield an accurate measure of the GAP activity toward each Rab. In order to measure the activity more accurately, we performed the assay with lower concentrations of the GST–GAP domain with Rabs 2A, 8A, 10, and 14 (Figure 3). At the lower concentrations of the GST–GAP domain, the rate of GTP hydrolysis for each Rab became more linear with time and approximately proportional to the concentration of the GAP domain.
Figure 3. Dependence of the GAP activity upon the concentration of the AS160 GST–GAP domain.
The GAP assay was carried out with Rabs 2A, 8A, 10 and 14, as described in the Experimental section and shown in Figure 1. The concentration of the GST–GAP domain in the assay was: none (■), 0.04 μM (◆), 0.13 μM (●) and 0.4 μM (▲).
In an effort to assess the GAP activity of full-length AS160, we generated recombinant full-length FLAG-tagged AS160 and the corresponding inactive R973K mutant in HEK-293F cells, and isolated these proteins from non-ionic detergent lysates on anti-FLAG beads. These proteins were then used in the GAP assay at the same molar concentration at which the GST–GAP domain had been used, with Rabs 2A, 8A, 10 and 14 as substrates. Unfortunately, full-length AS160 exhibited no significant activity above that of the R973K mutant (results not shown). As it was not possible to release the FLAG-tagged AS160 from the anti-FLAG beads in native form, the GAP assay was performed with the full-length proteins attached to the beads with frequent mixing. This difference from the normal assay did not account for the absence of GAP activity, since in a control assay the same molar amount of the GST–GAP domain attached to glutathione beads showed the expected activity against Rab14. The reason for the lack of activity of the recombinant full-length AS160 remains unknown; possibly this large protein (1299 amino acids) denatured during isolation procedures.
Rabs 2, 8 and 14 in GLUT4 vesicles
The finding of activity against Rabs 2A, 8A, 10 and 14 made it of interest to determine whether these Rabs were actually present in vesicles containing GLUT4. GLUT4 vesicles and control immunoadsorbates were prepared from the low-density microsomal fraction of unstimulated and insulin-stimulated 3T3-L1 adipocytes. These were immunoblotted with antibodies against Rabs 2, 8 and 14, as well as with an antibody against IRAP. IRAP is a membrane aminopeptidase that co-localizes with GLUT4 and translocates to the plasma membrane in response to insulin in the same way as GLUT4 [18]. It is more convenient for immunoblotting in an experiment of this type than is GLUT4 itself, which is of the same size as the antibody heavy chain. Unfortunately, no antibody against Rab10 was available.
Figure 4 shows the results of this experiment. The IRAP blot shows that GLUT4 vesicles were adsorbed by the anti-GLUT4 adsorbent, and at the load used here not detectably by the control adsorbent (but see below). In addition, as has been shown previously [18], the amount of IRAP in the vesicles isolated from insulin-treated cells was approx. 50% of that in the vesicles from unstimulated cells. This decrease is due to the translocation of IRAP to the plasma membrane. Rab2 was present in the GLUT4 vesicles, but not the control. Rab8 was enriched in the GLUT4 vesicles relative to the control, which also had Rab8 present. Rab14 was present in the GLUT4 vesicles, and the control also showed a faint Rab14 signal. Unlike IRAP, the amounts of Rabs 2 and 8 in the GLUT4 vesicles did not decrease in response to insulin. This finding suggests that Rabs 2 and 8 occur in subfractions of GLUT4 vesicles that are not mobilized by insulin. In the case of Rab2 these vesicles may be ones containing GLUT4 in the biosynthetic pathway, since Rab2 is known to participate in trafficking between the endoplasmic reticulum and the Golgi [19]. The effect of insulin on the relative amount of Rab14 in the GLUT4 vesicles was variable. In the experiment shown in Figure 4, the amount of Rab14 in the GLUT4 vesicles from insulin-treated cells was 50% of that in the vesicles from untreated cells, but in two other replicate experiments the amounts of Rab14 in the GLUT4 vesicles from insulin-treated cells were 75 and 100% of that of the Rab14 in the vesicles from untreated cells (results not shown). The reason for this variability is not known, and no conclusion can be drawn concerning whether Rab14 is in GLUT4 vesicles mobilized by insulin.
Figure 4. Rabs in GLUT4 vesicles.
GLUT4 vesicles (G4V) and control adsorbates (CV) were prepared from unstimulated (B) and insulin-treated (I) 3T3-L1 adipocytes, as described in the Experimental section. SDS samples of the vesicles were immunoblotted for IRAP, Rabs 2, 8 and 14, and AS160. The 1× loads were derived from 0.15% of a 10-cm diameter plate for IRAP, 2% of a plate for Rabs 2, 8 and 14, and 4% of a plate for AS160. Two repetitions of this experiment gave similar results, except for variable relative amounts of Rab14 in the insulin-treated GLUT4 vesicles (see the text).
The presence of smaller amounts of Rabs 8 and 14 in the control immunoadsorbates of vesicles is probably the result of non-specific binding of GLUT4 and possibly other vesicles. When larger amounts of the control vesicles than those used in Figure 4 were immunoblotted for IRAP, a weak IRAP signal was detected (results not shown). This result indicates that small amounts of the GLUT4 vesicles bound to the adsorbent with control antibody. These findings are consistent with the detection of many Rabs in the control vesicle immunoadsorbate by MS, as described above.
AS160 was also found in the GLUT4 vesicles by immunoblotting (Figure 4). We previously reported that AS160 was not detectable in the vesicles by this method [6], but recently with a more sensitive blotting procedure we have detected it. Although there is some AS160 in the GLUT4 vesicles, we have found by immunofluorescence that most of the AS160 in 3T3-L1 adipocytes is not colocalized with GLUT4 (S. Kane and G. E. Lienhard, unpublished work).
DISCUSSION
The results of the present study show that the GAP domain of AS160 is functional as a Rab GAP. The GAP domain of AS160 was assayed against a group of 18 Rabs. To our knowledge, this survey is the largest one with any Rab GAP that has been reported. The GAP domain showed activity against Rabs 2A, 8A, 10 and 14. It is notable that in a neighbour-joining tree, defining the relationships of the 60 mammalian Rabs, Rabs 2A and 14 are closely related, as are Rabs 8A and 10 [15]. On the other hand, these two pairs are not closely related. Moreover, Rabs 4A and 4B, which are not substrates, are also closely related to Rabs 2A and 14. Activity of the AS160 GAP domain with diverse Rabs is not unprecedented. Several yeast Rab GAPs each show activity against several Rabs in vitro [20]. One such yeast Rab GAP, Gyp1p, has been demonstrated to have greater specificity in vivo due to its localization in the vicinity of its substrate, Ypt1p [20]. The same situation is likely to be the case for AS160.
The magnitude of the activity of the AS160 GAP domain is slightly lower than that reported for other Rab GAPs, but is in the same range. For example, with GST–Rab14 as the substrate, the rate of hydrolysis of the bound [α-32P]GTP in the assay was approx. 3% per min with 130 nM GST–GAP domain (Figure 3). On the assumption that the low concentration of the GTP complex of GST–Rab14 (approx. 50 nM) was not saturating, this value corresponds to a kcat/Km value of 0.2 min−1·μM−1. Only three other mammalian Rab GAPs of the catalytic-arginine-type have been characterized. These are RNTre [21] and PR17 [22], two Rab5 GAPs, and GAPCenA [23], a Rab6 GAP. From the data in the literature with recombinant forms of these GAPs and their Rab substrates, the kcat/Km values are approx. 2.5, 1.2 and 0.4 min−1·μM−1 for RNTre, PR17 and GAPCenA respectively. A group of yeast Rab GAPs have been characterized [24]. The kcat/Km values for five of these range from 1.1 to 42 min−1·μM−1. Thus the activity of the AS160 GAP domain is between 0.5 and 50% of the values for these other Rab GAPs. Its slightly low activity may be due to a failure of a portion of the GST–GAP domain to fold correctly or to the use of GST forms of the Rabs in the assay.
The results from immunoblotting show that the GLUT4 vesicles contain Rabs 2A, 8A or 8B, and 14. In addition, the vesicles may contain Rab10. Since these Rabs are substrates for the AS160 GAP domain, one or more may be the Rab(s) on which AS160 acts in vivo to control GLUT4 translocation. On the basis of information in the literature, the more likely candidates are Rabs 8, 10 and 14. Rab2 is a less likely candidate, since there is strong evidence that it functions in trafficking between the endoplasmic reticulum and the Golgi [19]. Rab8 has been implicated in trafficking from the trans-Golgi network and recycling endosomes to the plasma membrane [25]. Moreover, Rab8 regulates actin organization [26] and also the movement of at least one type of vesicle (melanosomes) on actin filaments [27]. These properties of Rab8 are consistent with a role in GLUT4 translocation, since the GLUT4 vesicles move from the trans-Golgi network/recycling endosome region to the plasma membrane [1,28], and GLUT4 translocation requires actin remodelling [1]. In addition, it is worth noting that Rab8 is the mammalian Rab that is most similar to Sec4p, the yeast Rab that interacts with the yeast exocyst. The exocyst is a plasma membrane complex to which vesicles dock prior to fusion (reviewed in [29]). The mammalian Rab that interacts with the mammalian exocyst is not known, but the sequence similarity between Rab8 and Sec4p suggests that it could be Rab8. Two recent studies present evidence that the mammalian exocyst plays a role in the docking of GLUT4 vesicles to the plasma membrane [30,31].
Much less is known about the functions of Rab10 and Rab14 than Rab8. Rab10 has been localized to the perinuclear region of cells, a region where GLUT4 vesicles also reside [1,32]. Rab14 has been localized to both the Golgi and the endosomes [33], and has been postulated to be involved in trafficking between these two organelles. In this regard GLUT4 has been found to cycle through a subdomain of the trans-Golgi network, and this trafficking pathway has been hypothesized to participate in insulin-regulated GLUT4 translocation [28].
In conclusion, the present study supports the hypothesis that AS160 is a functional Rab GAP, and has yielded several Rabs that are candidates to be the Rab(s) that regulates GLUT4 translocation. In the future it will be important to determine which, if any, of these in vitro Rab substrates is a substrate for AS160 in vivo, and then to define the role in GLUT4 translocation of the Rab(s) that is the in vivo substrate.
Acknowledgments
We thank those scientists noted in the Experimental section for their gifts of plasmids and proteins. This work was support by grant DK025336 from the National Institutes of Health to G. E. L.
References
- 1.Watson R. T., Kanzaki M., Pessin J. E. Regulated membrane trafficking of the insulin-responsive glucose transporter 4 in adipocytes. Endocrine Rev. 2004;25:177–204. doi: 10.1210/er.2003-0011. [DOI] [PubMed] [Google Scholar]
- 2.Bae S. S., Cho H., Mu J., Birnbaum M. J. Isoform-specific regulation of insulin-dependent glucose uptake by Akt/protein B kinase. J. Biol. Chem. 2003;278:49530–49536. doi: 10.1074/jbc.M306782200. [DOI] [PubMed] [Google Scholar]
- 3.Jiang Z. Y., Zhou Q. L., Coleman K. A., Chouinard M., Boese Q., Czech M. P. Insulin signaling through Akt/protein kinase B analyzed by small interfering RNA-mediated gene silencing. Proc. Natl. Acad. U.S.A. 2003;100:7569–7574. doi: 10.1073/pnas.1332633100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katome T., Obata T., Matsushima R., Masuyama N., Cantley L. C., Gotoh Y., Kishi K., Shiota H., Ebina Y. Use of RNA interference-mediated gene silencing and adenoviral overexpression to elucidate the roles of Akt/protein kinase B isoforms in insulin actions. J. Biol. Chem. 2003;278:28312–28323. doi: 10.1074/jbc.M302094200. [DOI] [PubMed] [Google Scholar]
- 5.Kane S., Sano H., Liu S. C. H., Asara J. M., Lane W. S., Garner C. C., Lienhard G. E. A method to identify serine kinase substrates. J. Biol. Chem. 2002;277:22115–22118. doi: 10.1074/jbc.C200198200. [DOI] [PubMed] [Google Scholar]
- 6.Sano H., Kane S., Sano E., Mîinea C. P., Asara J. M., Lane W. S., Garner C. W., Lienhard G. E. Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J. Biol. Chem. 2003;278:14599–14602. doi: 10.1074/jbc.C300063200. [DOI] [PubMed] [Google Scholar]
- 7.Zerial M., McBride H. Rabs as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001;2:107–119. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 8.Zeigerer A., McBrayer M. K., McGraw T. E. Insulin stimulation of GLUT4 exocytosis, but not its inhibition of endocytosis, is dependent on Rab GAP AS160. Mol. Biol. Cell. 2004;15:4406–4415. doi: 10.1091/mbc.E04-04-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calderhead D. M., Kitagawa K., Tanner L. I., Holman G. D., Lienhard G. E. Insulin regulation of the two glucose transporters in 3T3-L1 adipocytes. J. Biol. Chem. 1990;265:13800–13808. [PubMed] [Google Scholar]
- 10.Keller S. R., Scott H. M., Mastick C. C., Aebersold R., Lienhard G. E. Cloning and characterization of a novel insulin-regulated membrane aminopeptidase from Glut4 vesicles. J. Biol. Chem. 1995;270:23612–23618. doi: 10.1074/jbc.270.40.23612. [DOI] [PubMed] [Google Scholar]
- 11.Cain C. C., Trimble W. S., Lienhard G. E. Members of the VAMP family of synaptic vesicle proteins are components of glucose transporter-containing vesicles from rat adipocytes. J. Biol. Chem. 1992;267:11681–11684. [PubMed] [Google Scholar]
- 12.Basu S., Totty N. F., Irwin M. S., Sudol M., Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol. Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 13.Liu K., Li G. Catalytic domain of the p120 Ras GAP binds to Rab5 and stimulates its GTPase activity. J. Biol. Chem. 1998;273:10087–10090. doi: 10.1074/jbc.273.17.10087. [DOI] [PubMed] [Google Scholar]
- 14.Ridley A. J., Self A. J., Kasmi F., Patterson H. F., Hall A., Marshall C. J., Ellis C. Rho family GTPase activating proteins p190, bcr, and rhoGAP show distinct specificities. EMBO J. 1993;12:5151–5160. doi: 10.1002/j.1460-2075.1993.tb06210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pereira-Leal J. B., Seabra M. C. Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- 16.Cormont M., Tanti J.-F., Zahraoui A., van Obberhegen E., Tavitian A., Le Marchand-Brustel Y. Insulin and okadaic acid induce Rab4 redistribution in adipocytes. J. Biol. Chem. 1993;268:19491–19497. [PubMed] [Google Scholar]
- 17.Kessler A., Tomas E., Immler D., Meyer H. E., Zorzano A., Eckel J. Rab 11 is associated with GLUT4-containing vesicles and redistributes in response to insulin. Diabetologia. 2000;43:1518–1527. doi: 10.1007/s001250051563. [DOI] [PubMed] [Google Scholar]
- 18.Ross S. A., Scott H. M., Morris N. J., Leung W.-Y., Mao F., Lienhard G. E., Keller S. R. Characterization of the insulin-regulated membrane aminopeptidase in 3T3-L1 adipocytes. J. Biol. Chem. 1996;271:3328–3332. doi: 10.1074/jbc.271.6.3328. [DOI] [PubMed] [Google Scholar]
- 19.Tisdale E. J., Balch W. E. Rab2 is essential for maturation of pre-Golgi intermediates. J. Biol. Chem. 1996;271:29372–29379. doi: 10.1074/jbc.271.46.29372. [DOI] [PubMed] [Google Scholar]
- 20.Du L.-L., Novick P. Yeast rab GTPase-activating protein Gyp1p localizes to the Golgi apparatus and is a negative regulator of Ypt1p. Mol. Biol. Cell. 2001;12:1215–1226. doi: 10.1091/mbc.12.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanzetti L., Rybin V., Malabarba M. G., Christoforidis S., Scita G., Zerial M., Di Fiore P. P. The Eps8 protein coordinates EFG receptor signaling through Rac and trafficking through Rab 5. Nature (London) 2000;408:374–377. doi: 10.1038/35042605. [DOI] [PubMed] [Google Scholar]
- 22.Pei L., Peng Y., Yang Y., Ling X. B., van Eyndhoven W. G., Nguyen K. C. Q., Rubin M., Hoey T., Powers S., Li J. PRC17, a novel oncogene encoding a Rab GTPase-activating protein, is amplified in prostate cancer. Cancer Res. 2002;62:5420–5424. [PubMed] [Google Scholar]
- 23.Cuif M.-H., Possmayer F., Zander H., Bordes N., Jollivet F., Coudel-Courteille A., Janoueix-Lerosey I., Langsley G., Bornens M., Gould B. Characterization of GAPCenA, a GTPase activating protein for Rab6, part of which associates with the centrosome. EMBO J. 1999;18:1772–1782. doi: 10.1093/emboj/18.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Will E., Gallwitz D. Biochemical characterization of Gyp6p, a Ypt/Rab-specific GTPase-activating protein from yeast. J. Biol. Chem. 2001;276:12135–12139. doi: 10.1074/jbc.M011451200. [DOI] [PubMed] [Google Scholar]
- 25.Ang A. L., Fölsch H., Koivisto U.-M., Pypaert M., Mellman I. The Rab8 GTPase selectively regulates AP-1B-dependent basolateral transport in polarized Madin–Darby canine kidney cells. J. Cell Biol. 2003;163:339–350. doi: 10.1083/jcb.200307046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peränen J., Auvinen P., Virta H., Wepf R., Simons K. Rab8 promotes polarized membrane transport through reorganization of actin and microtubules in fibroblasts. J. Cell Biol. 1996;135:153–167. doi: 10.1083/jcb.135.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chabrillat M. L., Wilhelm C., Wasmeier C., Sviderskaya E. V., Louvard D., Coudrier E. Rab8 regulates the actin-based movement of melanosomes. Mol. Biol. Cell. 2005;16:1640–1650. doi: 10.1091/mbc.E04-09-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shewan A. M., van Dam E. M., Martin S., Luen T. B., Hong W., Bryant N. J., James D. E. GLUT4 recycles via a trans-Golgi network (TGN) subdomain enriched in syntaxins 6 and 16 but not TGN38: involvement of an acidic targeting motif. Mol. Biol. Cell. 2003;14:973–986. doi: 10.1091/mbc.E02-06-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu S.-C., TerBush D., Abraham M., Guo W. The exocyst complex in polarized exocytosis. Int. Rev. Cytol. 2004;233:243–265. doi: 10.1016/S0074-7696(04)33006-8. [DOI] [PubMed] [Google Scholar]
- 30.Inoue M., Chang L., Hwang J., Chiang S.-H., Saltiel A. R. The exocyst complex is required for targeting of Glut4 to the plasma membrane by insulin. Nature (London) 2003;422:629–633. doi: 10.1038/nature01533. [DOI] [PubMed] [Google Scholar]
- 31.Ewart M.-A., Clarke M., Kane S., Chamberlain L. H., Gould G. W. Evidence for a role of the exocyst in insulin-stimulated Glut4 trafficking in 3T3-L1 adipocytes. J. Biol. Chem. 2005;280:3812–3816. doi: 10.1074/jbc.M409928200. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y. T., Holcomb C., Moore H. P. Expression and localization of two low molecular weight GTP-binding proteins, Rab8 and Rab10, by epitope tag. Proc. Natl. Acad. Sci. U.S.A. 1993;90:6508–6512. doi: 10.1073/pnas.90.14.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jununtula J. R., De Maziére A. M., Peden A. A., Ervin K. E., Advani R. J., van Dijk S. M., Klumperman J., Scheller R. H. Rab14 is involved in membrane trafficking between the Golgi complex and endosomes. Mol. Biol. Cell. 2004;15:2218–2229. doi: 10.1091/mbc.E03-10-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]