Abstract
Import of proteins with a PTS1 (peroxisomal targeting signal 1) into the Leishmania glycosomal organelle involves docking of a PTS1-laden LdPEX5 [Leishmania donovani PEX5 (peroxin 5)] receptor to LdPEX14 on the surface of the glycosomal membrane. In higher eukaryotes, the PEX5–PEX14 interaction is mediated by a conserved diaromatic WXXXY/F motif. Site-directed and deletion mutageneses of the three WXXXY/F repeats in LdPEX5 did not abolish the LdPEX5–LdPEX14 association. Analysis of the equilibrium dissociation constant (Kd) revealed that ldpex5-W53A (Trp53→Ala), ldpex5-W293A, ldpex5-W176,293A and ldpex5-W53,176,293A mutant receptors were capable of binding LdPEX14 with affinities comparable with wild-type LdPEX5. That the diaromatic motifs were not required for the LdPEX5–LdPEX14 interaction was further verified by deletion analysis that showed that ldpex5 deletion mutants or ldpex5 fragments lacking the WXXXY/F motifs retained LdPEX14 binding activity. Mapping studies of LdPEX5 indicated that the necessary elements required for LdPEX14 association were localized to a region between residues 290 and 323. Finally, mutational analysis of LdPEX14 confirmed that residues 23–63, which encompass the conserved signature sequence AX2FLX7SPX6FLKGKGL/V present in all PEX14 proteins, are essential for LdPEX5 binding.
Keywords: glycosome, Leishmania, mutagenesis, peroxin (PEX), peroxisomal targeting signal 1 (PTS1), protein–protein interaction
Abbreviations: ABS, adult bovine serum; ABTS, 2,2′-azinobis-(3-ethylbenzothiazoline-6-sulphonic acid); DTT, dithiothreitol; ITC, isothermal titration calorimetry; PEX, peroxin; LdPEX5, Leishmania donovani PEX5; PTS, peroxisomal targeting signal; SH3 domain, Src homology 3 domain; TPR, tetratricopeptide repeat; XPRT, xanthine phosphoribosyltransferase
INTRODUCTION
The human protozoan pathogens Leishmania, Trypanosoma brucei and T. cruzi are nucleated cells that diverged early from the main eukaryotic cell lineage [1]. These organisms have a number of unique biochemical and structural features that include trans-splicing [2], polycistronic mRNAs [3], RNA editing [4], kinetoplastid DNA [5] and the presence of a subcellular organelle called a glycosome [6], which is evolutionarily related to peroxisomes of higher eukaryotic cells. These organelles share a number of architectural attributes that include a single phospholipid bilayer surrounding the microbody and an electron-dense protein matrix, and both are devoid of nucleic acids and protein translational machinery [6]. Glycosomes from the kinetoplastids Leishmania and trypanosomes are distinguished by the presence of a multiplicity of vital metabolic and biosynthetic pathways that include glycolysis, purine salvage and pyrimidine biosynthesis [7,8]; however they lack the classical peroxisomal marker enzyme, catalase [8].
The targeting of matrix proteins to the glycosome, as in the peroxisome, is dependent on two major types of topogenic signals designated peroxisomal targeting signal 1 and 2 (PTS1 and PTS2) [9,10]. PTS1, which is found on a preponderance of matrix proteins, consists of a C-terminal tripeptide with the sequence Ser-Lys-Leu or Ala-Lys-Leu or a conserved variant of these sequences [10]. PTS2 proteins are less abundant and generally contain the consensus motif R/K-L/I/V/-X5-H/Q-A/L located proximal to the N-terminus [10]. Biogenesis of the peroxisome, glycosome and glyoxysome is dependent on a family of soluble and membrane-associated proteins designated PEXs (peroxins) that are involved in sorting, targeting and translocation of polypeptides into these microbodies. Nascent PTS1 and PTS2 polypeptides synthesized on cytosolic ribosomes are selectively bound by the mobile cytosolic receptors PEX5 or PEX7 respectively [9,10]. These PEX5–PTS1 and PEX7–PTS2 complexes converge at the peroxisome-like microbody membrane where they dock to a receptor containing two core components, PEX13 and PEX14 [11–13]. A number of models for the import of PTS1 proteins into these organelles propose the recycling of PEX5 between the cytosolic and peroxisomal matrix by differentially binding to PEX13 or PEX14. These models postulate that cargo-laden PEX5 receptors preferentially bind PEX14; after translocations and unloading of the cargo proteins into the lumen of the peroxisome, these receptors preferentially associate with the membrane protein PEX13 that shuttles the PEX5 back into the cytosolic compartment [12,14,15]. In the case of mammalian and yeast PEX5, interactions with PEX13 and PEX14 have been shown to be mediated by a WXXXY/F pentapeptide, a motif that is conserved among all PEX5 receptors [16,17]. Mutations that alter either of the aromatic residues in this motif dramatically compromise the PEX5–PEX13 or PEX5–PEX14 interaction [14,16,18]. Mutational analysis of PEX14 has also shown that the association with the WXXXY/F pentapeptide repeat on PEX5 is mediated by an N-terminal region that contains a conserved signature motif that is a characteristic feature of all PEX14 proteins [19,21]. However, the exact nature of this protein–protein interaction is unclear. Although the WXXXY/F motif is known to be important for PEX5 docking to PEX13, the molecular mechanisms accounting for this interaction are not well defined. Three-dimensional structures of the yeast PEX5–PEX13 complex have suggested that this binary complex is stabilized by the C-terminal SH3 domain (Src homology 3 domain) of PEX13 binding to the WXXXY/F motif, which is a non-classical PXXP ligand for the SH3 domain [15,20]. In contrast, experiments with the mammalian system have shown that the PEX13 SH3 motif is not essential since the human PEX5–PEX13 interaction involves an N-terminal region of PEX13 [14].
The targeting and import of PTS1 proteins into the Leishmania glycosome is dependent on the two proteins LdPEX5 and LdPEX14 [17,21]. LdPEX5, like other PEX5 proteins, is a bidomain molecule consisting of a conserved C-terminal domain composed of seven TPRs (tetratricopeptide repeats) and a divergent N-terminal region, which aside from the three conserved WXXXY/F motifs, shows no significant sequence homology with other PEX5 proteins [17]. The N-terminal portion of LdPEX5 is also known to be important for LdPEX5 oligomerization [17,22] and for interaction with LdPEX14 [21]. Analysis of LdPEX14 has revealed that, with the exception of an N-terminal signature motif [21], this protein shares very limited sequence homology with other PEX14 proteins. Moreover, unlike other PEX14 homologues, LdPEX14 is a soluble peripheral membrane-associated protein that is anchored to the cytosolic face of the glycosomal membrane, an orientation that is consistent with the protein forming a docking complex that permits the association of PTS1-loaded LdPEX5 receptor. However, little is known about the molecular mechanisms involved in the LdPEX5–LdPEX14 interaction. In the present study, we show by site-directed mutagenesis and biophysical techniques that none of the conserved WXXXY/F motifs in LdPEX5 are essential for LdPEX14 binding. These studies also demonstrate that the N-terminal signature motif on LdPEX14 is critical for LdPEX5–LdPEX14 binding.
MATERIALS AND METHODS
Materials
All restriction endonucleases and DNA-modifying enzymes were procured from either Invitrogen Life Technologies (Grand Island, NY, U.S.A.) or Roche Molecular Biochemicals (Indianapolis, IN, U.S.A.). Secondary horseradish peroxidase-conjugated antibodies were obtained from Sigma–Aldrich (St. Louis, MO, U.S.A.). Chitin beads were purchased from New England Biolabs (Beverly, MA, U.S.A.) and S-protein beads were purchased from Novagen (Madison, WI, U.S.A.). All other reagents were of the highest quality commercially available.
LdPEX5 mutants
Site-directed mutants were created using the QuikChange PCR method (Stratagene, La Jolla, CA, U.S.A.) with the Pwo polymerase (Roche Molecular Biochemicals) and the pTYB12-LdPEX5 construct as a template. Primer pairs 5′-GCGGCTCAGGCAGCACAGAAT-3′ (SW53A) and 5′-ATTCTGTGCTGCCTGAGCCGC-3′ (AW53A), 5′-CAGCAACAAGCTAGCACCGACTAC-3′ (SW176A) and 5′-GTAGTCGGTGCTAGCTTGTTGCTG-3′ (AW176A) and 5′-GTCGAGGACGCAGCGCAGGAG-3′ (SW293A) and 5′-CTCCTGCGCTGCGTCCTCGAC-3′ (AW293A) were used to introduce the W53A (Trp53→Ala), W176A and W293A mutations respectively.
The triple-mutant ldpex5-W53,176,293A (lower-case letters are used to denote mutant proteins) was constructed by replacing the ClaI/SacI fragment in the pTYB12-ldpex5 W53A construct with the corresponding fragment from the pTYB12-ldpex5-W176,293A. All constructs were verified by automated DNA sequence analysis.
The NdeI/NotI fragment containing the LdPEX5 open reading frame was subcloned from the pBAce-LdPEX5 vector [17] into the corresponding sites of the pTYB12 vector (New England Biolabs) to generate a CBD (chitin-binding domain)–LdPEX5 fusion construct. The expression vector for ldpex5-(203–391) was created by subcloning the EcoRI/XhoI fragment from the pET30b(+)-NT-ldpex5-His6 vector [17] into the corresponding sites of the pTYB12 vector to create pTYB12-ldpex5-203–391. pTYB12-ldpex5-203–269, encoding ldpex5-(203–269), was generated by digesting pTYB12-ldpex5-203–391 with AatII and XhoI, then filling in the overhangs with T4 DNA polymerase and religating with T4 DNA ligase. The pTYB12-ldpex5-290–391 vector, encoding ldpex5-(290–391), was constructed by digesting pTYB12-ldpex5-203–391 with EcoRI/AatII, eliminating the 3′-overhangs with T4 DNA polymerase with a mixture of dATP, dTTP and dGTP and religating with T4 DNA ligase. The pTYB12-ldpex5-203–347 vector was generated from the pTYB12-ldpex5-203–391 construct using a site-directed mutagenesis approach to introduce a stop codon immediately downstream of the codon encoding Phe347. The pTYB12-ldpex5-Δ181–313 construct, encoding Δ181–313-ldpex5, a protein lacking residues 181–313, was generated from pTYB12-LdPEX5 by deletion mutagenesis using the QuikChange and Pwo polymerase method with primers SW176A and AW176A. The pTYB12-ldpex5-268–303 vector, encoding ldpex5-(268–303), was created by PCR amplifying the corresponding fragment using Pwo polymerase with the sense primer 5′-AGAATTCCATATGACGTCTCCGGAGAAC-3′ (M268s) and antisense primer 5′-GAATTCTTAGCGTTCCTGCATCTCCGC-3′ containing NdeI and EcoRI restriction sites (underlined) respectively, with 25 cycles of denaturation at 95 °C for 30 s, annealing at 54 °C for 45 s and extension at 68 °C for 1 min. The pTYB12-ldpex5-268–323 vector was generated by amplifying the coding region for residues 268–323 using the M268s primer and the antisense primer 5′-GAACATGTACTGGTTGTTAGG-3′ (this primer also contains a P320A point mutation) with Pwo polymerase and 25 cycles of 95 °C for 30 s, 54 °C for 45 s, and extension at 68 °C for 1 min. The PCR fragment was digested with NdeI and cloned into the pTYB12 vector that was prepared by digesting with XhoI, then treating with T4 DNA polymerase to fill in the ends, followed by NdeI digestion. The pTYB12-ldpex5-1–270 vector was generated by digesting pTYB12-LdPEX5 with AatII, to remove a 60 bp fragment, then re-ligating with T4 ligase. This resulted in a frameshift mutation that introduced a termination codon 48 bp downstream of the AatII site. The fragment encoding His6–ldpex5-(283–625) was produced by PCR amplification of a 1026 bp fragment using the sense primer 5′-AATGTACATATGGATATGGCCGCGAACGAC-3′ containing an NdeI restriction site (underlined) and the antisense primer 5′-CGCGGATCCTTAGACGTGGCCCTCAAGTCC-3′ containing a BamHI restriction site (underlined). PCRs were performed with Pwo polymerase using 20 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s and extension at 68 °C for 90 s using pTYB12-LdPEX5 as the template. The PCR fragment was cloned into NdeI/BamHI sites of the pET15b vector (Novagen). All constructs were verified by automated DNA sequence analysis.
Expression and purification of LdPEX5 proteins
Escherichia coli ER2566 cells (New England Biolabs) transformed with pTYB12-LdPEX5 were grown in Luria broth with 50 μg/ml ampicillin to an absorbance A600 of 1.0 and then induced with 0.5 mM isopropyl thiogalactoside for 5 h at 25 °C with vigorous shaking. Bacterial cultures (1 litre) were harvested and then the cell pellet was resuspended in 30 ml of 40 mM Tris/HCl (pH 8.0) containing an EDTA-free mini-tab protease inhibitor cocktail (Roche Molecular Biochemicals) and cells were lysed by two passes through a French press. Lysates were clarified by centrifugation and NaCl was added to the supernatant to a final concentration of 500 mM before loading on to a chitin column (1.5 cm×8 cm; New England Biolabs) equilibrated with 40 mM Tris/HCl (pH 8.0) and 500 mM NaCl (buffer I). The column was washed first with 30 column vol. of buffer I and then with 2 column vol. of buffer I containing 50 mM DTT (dithiothreitol). Intein cleavage of LdPEX5 protein was achieved by resuspending the chitin resin in 1.5 column vol. of buffer I containing 50 mM DTT and incubating at 4 °C for 40 h. Column eluate containing LdPEX5 was dialysed against 40 mM Tris/HCl (pH 8.0), 100 mM NaCl and 1 mM DTT, concentrated to 4–10 mg/ml in a Biomax 5K NMWL centrifugal filter (Millipore, Bedford, MA, U.S.A.), aliquots were made, and stored at −80 °C. For the ldpex5-(268–303) and ldpex5-(268–323) peptides, the fusion protein cleavage was performed in 40 mM Tris/HCl (pH 8.0) containing 50 mM DTT at 4 °C for 40 h. The chitin column eluate was loaded on to a Q-Sepharose column (1 cm×5 cm) equilibrated with 40 mM Tris (pH 8.0). The column was washed with 10 vol. of equilibration buffer (50 mM Tris, pH 8.0) to remove the DTT and the ldpex5-(268–303) peptide was eluted with 4 vol. of 0.1% trifluoroacetic acid, and concentrated by freeze-drying. The integrity of ldpex5-(268–303) was verified by reversed-phase HPLC and the mass was confirmed by surface-enhanced laser desorption time-of-flight MS. All ldpex5 site-directed mutant proteins and ldpex5 protein fragments expressed using the pTYB12 vector were prepared using the method that was employed for the wild-type LdPEX5.
NT-ldpex5–His6 and His6–CT-ldpex5 (where NT and CT stand for N- and C-terminal respectively) were overexpressed and purified as described in [17]. For His6–ldpex5-(283–625) protein, E. coli ER2566 cultures (1 litre) transformed with pET15b-His6-ldpex5-283–625 were grown to an A600∼0.7 in Luria broth supplemented with 50 μg/ml ampicillin, and protein expression was induced by adding 0.7 mM isopropyl thiogalactoside and incubating the cultures for 5 h at 25 °C. Bacterial cell pellets were resuspended in 30 ml of a buffer (40 mM Tris/HCl and 500 mM NaCl, pH 8.0) containing a protease inhibitor cocktail and lysed by two passes through a French press. His6–ldpex5-(283–625) was purified by affinity chromatography on an Ni2+-nitrilotriacetate matrix (Qiagen, Valencia, CA, U.S.A.) according to the manufacturer's instructions. All protein concentrations were measured at 280 nm by the method of Gill and Von Hippel [23].
Expression and purification of LdPEX14 proteins
His6/S–LdPEX14 and His6/S–ldpex14-(1–120) containing His6 and S-tags were expressed as described in [21]. LdPEX14 truncation mutants His6/S–ldpex14-(24–464), His6/S–ldpex14-(44–464) and His6/S–ldpex14-(64–464) were constructed by PCR amplification of the corresponding fragment using the sense primers containing an NcoI restriction site (underlined), 5′-CATGCCATGGCTTCGTCGGAACTGGACGCT-3′ (PEX14-23NT), 5′-CATGCCATGGCACGCGTGCGGCGCTCGCCG-3′ (PEX1443NT) and 5′-CATGCCATGGCAGATGAACAGATAAAGTAC3′ (PEX14-63NT), and the antisense primer containing a BamHI restriction site (underlined), 5′-CGGGATCCTTAGCCAATCGACATCGG-3′ (PEX14 stop), to obtain the corresponding open reading frames. The PCR fragments were cloned into the NcoI and BamHI sites of the pET30b(+) vector to generate recombinant LdPEX14 proteins that all contained an N-terminal His6 tag and an S-peptide tag. All constructs were verified by automated DNA sequence analysis.
LdPEX5–LdPEX14 interaction
For pull-down experiments, 10 μg of His6/S–LdPEX14 or His6/S–ldpex14-(1–120) was mixed with 8 μg of LdPEX5, ldpex5-(1–270), Δ181–313-ldpex5, Δ269–291-ldpex5, ldpex5-W53A, ldpex5-W293A, ldpex5-W176,293A, ldpex5-W53,176,293A, His6–CT-ldpex5 or His6–ldpex5-(283–625) in 30 μl of TS buffer (40 mM Tris and 500 mM NaCl, pH 8.0) and then incubated with 35 μl of packed S-protein agarose beads (Novagen) for 30 min at 25 °C with occasional mixing. Supernatants were removed and the beads were washed three times with 400 μl of TS containing 1% Triton X-100 and then washed three times with 500 μl of TS to remove unbound proteins. Proteins bound to the S-protein beads were analysed by Coomassie Blue-stained SDS/PAGE.
For competition pull-down assays, 10 μg of His6/S–ldpex14-(1–120) was loaded on to 35 μl of S-protein beads and mixed either with no competitor protein or with 100 μg of ldpex5-(268–303), 64 μg of ldpex5-(268–323), 40 μg of ldpex5-(290–391) or 40 μg of ldpex5-(203–347) preincubated for 25 min at 25 °C, followed by the addition of 4 μg of LdPEX5 to each test tube and incubating the mixture at 25 °C for another 25 min. Unbound proteins were removed by washing the S-protein beads with TS buffer as described above and bound proteins were analysed by SDS/PAGE.
For ELISA-based LdPEX5–LdPEX14 interaction assays, 96-well microtitre plates (Packard BioScience, Groningen, The Netherlands) were coated with 1 μg/well His6/S–LdPEX14, His6/S–ldpex14-(1–120), His6/S–ldpex14-(24–464), His6/S–ldpex14-(44–464), His6/S–ldpex14-(64–464), XPRT (xanthine phosphoribosyltransferase), xprtΔAKL, a mutant lacking the PTS1 signal [17], or BSA in 100 μl of PBS for 16 h at 4 °C. Unbound protein was removed and plates were blocked with 200 μl of 2% milk powder in PBS for 45 min at 25 °C. Microtitre plates were rinsed and incubated with increasing concentrations of LdPEX5 proteins (0.4–860 nM) diluted in 100 μl of 2% (v/v) ABS (adult bovine serum), 0.05% Tween 20 and PBS for 2 h at 25 °C. Unbound proteins were removed by washing the wells four times with 300 μl of PBS/0.05% Tween 20 and bound LdPEX5 was measured by indirect ELISA using anti-LdPEX5 rabbit antisera (1:5000) and goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (1:5000; Sigma–Aldrich) diluted in 2% ABS, 0.05% Tween 20 and PBS. ELISAs were developed using the chromogenic substrate ABTS [2,2′-azinobis-(3-ethylbenzothiazoline-6-sulphonic acid)] and, after a 15 min incubation at 25 °C, plates were read on a Benchmark microplate reader (Bio-Rad Laboratories) at 405 nm. All binding experiments were performed in triplicate and the data were analysed with the ORIGIN software package (Originlab Corporation, Northhampton, MA, U.S.A.).
LdPEX5–LdXPRT interaction
High-binding flat-bottom 96-well microtitre plates (Packard BioScience) were coated with 1 μg/well purified LdXPRT in 100 μl of PBS for 16 h at 4 °C. Unbound protein was removed by washing twice with PBS and plates were blocked with 200 μl of 2% milk powder in PBS for 45 min at 25 °C. Plates were washed with PBS/0.05% Tween 20 and incubated for 2 h at 25 °C with various concentrations of LdPEX5 or ldpex5 mutants (0.4–850 nM) diluted in 100 μl of PBS/0.05% Tween 20/2% ABS in the absence or presence of 850 nM His6/S–LdPEX14. Plates were washed four times with 200 μl of PBS/0.05% Tween 20 and bound LdPEX5 was quantified by indirect ELISA using LdPEX5-specific rabbit antisera (1:5000) and goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (1:5000; Sigma) diluted in PBS/0.05% Tween 20/2% ABS. ELISAs were developed using the chromogenic substrate ABTS. Colour development was measured on a Benchmark microplate reader (Bio-Rad Laboratories) at 405 nm and the data were analysed with ORIGIN software.
Chromatographic analysis of LdPEX5 proteins
Size-exclusion chromatography was performed on a Beckman Coulter 32 Karat HPLC system equipped with a Bio-Sil SEC 250 column (300 mm×7.8 mm or 600 mm×7.8 mm; Bio-Rad Laboratories). LdPEX5 protein (25–50 μg) was injected and the column was developed with 25 mM Tris and 120 mM NaCl (pH 7.5) at a flow rate of 0.25 ml/min. Protein elution was monitored at 280 nm. Columns were calibrated with a standard protein mixture containing thyroglobulin dimer (660 kDa), thyroglobulin monomer (330 kDa), bovine IgG (160 kDa), ovalbumin (45 kDa), equine myoglobin (17 kDa) and vitamin B12 (1.3 kDa; Bio-Rad Laboratories).
The 35 amino acid ldpex5-(268–303) fragment was measured by injecting samples on to an HP Lichrospher 100 RP8 column equilibrated with 0.1% trifluoroacetic acid, and the column was developed with a 0–50% (v/v) acetonitrile gradient for 30 min at a flow rate of 1.0 ml/min. Column effluent was monitored at 225 nm. Analysis was performed on a Beckman Coulter 32 Karat HPLC system.
ITC (isothermal titration calorimetry)
ITC experiments were performed at 30 °C on a MicroCal VP-ITC calorimeter (MicroCal, Northampton, MA, U.S.A.). Purified His6/S–ldpex14-(1–120) and ldpex5-(203–391) were dialysed exhaustively against 40 mM sodium phosphate (pH 7.5), 120 mM NaCl and 3 mM 2-mercaptoethanol (SSME buffer). The reference cell of the calorimeter was filled with degassed SSME buffer and the sample cell was filled with 1.43 ml of 30 μM ldpex5-(203–391) diluted in SSME buffer. Typically, 30–40 5 μl injections of a 1.0 mM His6/S–ldpex14-(1–120) solution were made at 6 min intervals. The heat evolved per injection was determined by integrating the area under the peak and plotted as a function of the mole ratio of His6/S–ldpex14-(1–120): ldpex5-(203–391), and best-fit curve analysis was performed using ORIGIN software to determine His6/S–ldpex14-(1–120): ldpex5-(203–391) binding constant (Kd).
RESULTS
Mutagenesis of the WXXXY/F motifs does not abolish the LdPEX5–LdPEX14 interaction
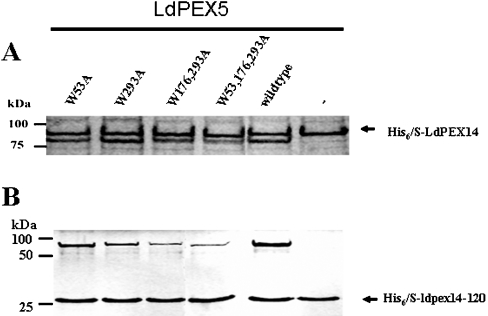
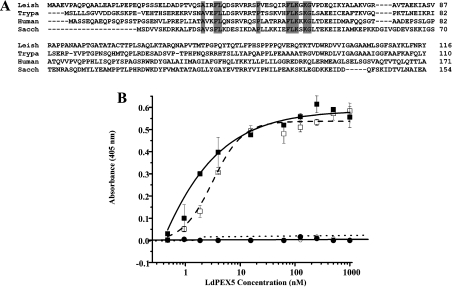
LdPEX5 contains three WXXXY/F pentapeptide repeats with the sequences W53AQNF57, W176STDY180 and W293AQEY297 (Figure 1, shaded regions). To ascertain the importance of these repeats for the LdPEX5–LdPEX14 interaction, a series of single, double and triple mutations consisting of W53A, W293A, W176,293A and W53,176,293A were introduced into the full-length LdPEX5 sequence and the effect of these mutations on His6/S–LdPEX14 or His6/S–ldpex14-(1–120) association was assessed by pull-down assays [21]. As shown in Figure 2, all ldpex5 mutants were capable of binding His6/S–LdPEX14 and His6/S–ldpex14-(1–120). However, the amount of ldpex5-W176,293A and ldpex5-W53,176,293A that bound to the S-protein beads was reduced in comparison with wild-type LdPEX5, suggesting that the double (W176,293A) and triple (W53,176,293A) mutations altered the ability of these mutants to bind His6/S–LdPEX14. In the absence of His6/S–LdPEX14 or His6/S–ldpex14-(1–120), none of the LdPEX5 proteins bound to the S-protein beads.
Figure 1. Multiple sequence alignment of PEX5 proteins.
A partial sequence alignment of the N-terminal region of the L. donovani (Leish), T. brucei (Trypa), human and S. cerevisiae (Sacch) PEX5 was performed using the CLUSTAL_X computer program [35]. The WXXXY/F motifs in the LdPEX5 are designated by the grey shaded boxes. The black bar delineates the first three TPRs that form part of the PTS1-binding pocket.
Figure 2. LdPEX14–LdPEX5 interaction.
The interaction of His6/S–LdPEX14 (A) or His6/S–ldpex14-(1–120) (His6/S–ldpex14-120) (B) with either the wild-type LdPEX5 or ldpex5 WXXXY/F site-directed mutants was assessed by pull-down assays using S-protein agarose beads. LdPEX14 proteins were mixed with 10 μg of purified, ldpex5-W53A, ldpex5-W293A, ldpex5-W176,293A, ldpex5-W53,176,293A or wild-type LdPEX5 or with no LdPEX5 and the mixture was added to S-protein agarose. Beads were stringently washed with 1% Triton X-100 and 500 mM NaCl in TBS (Tris-buffered saline; 50 mM Tris/HCl, pH 8.0 and 150 mM NaCl) and bound proteins were analysed by Coomassie Blue-stained SDS/PAGE.
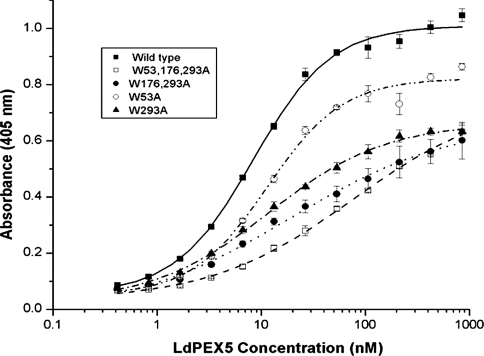
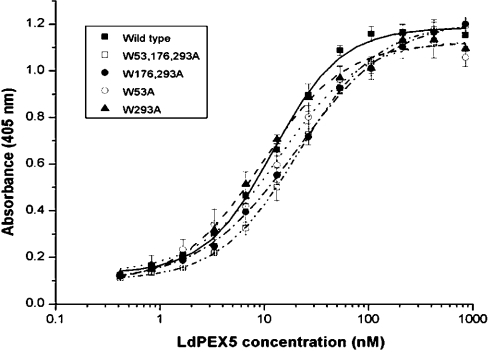
To characterize further the interaction of the site-directed ldpex5 mutant proteins with His6/S–LdPEX14 and His6/S–ldpex14-(1–120), we employed a modified ELISA using anti-LdPEX5 polyclonal antisera to measure the equilibrium dissociation constant Kd for the LdPEX5–LdPEX14 interaction [24]. Saturable kinetics with Kd values of approx. 9 and 16 nM were observed for the association of LdPEX5 with His6/S–LdPEX14 or His6/S–ldpex14-(1–120) non-covalently immobilized on the microtitre plates (Figure 3 and Table 1). Quantitative analysis of the interaction of His6/S–LdPEX14 with the mutant ldpex5 proteins revealed that these proteins also exhibited saturable binding kinetics (Figure 3); however, the amount of ldpex5 protein binding to His6/S–LdPEX14 at the saturation point (Bmax) was notably reduced by approx. 25% for ldpex5-W53A and 40% for ldpex5-W293A. However, with the concentrations of ldpex5 used in these experiments, saturable binding was not observed with the mutants ldpex5-W176,293A and ldpex5-W53,176,293A. Since polyclonal anti-LdPEX5 antisera were used in these experiments, it is likely that the diminished binding with the latter ldpex5 mutants was due to decreased binding to LdPEX14 as ELISAs performed with LdPEX5 and ldpex5 proteins directly immobilized on microtitre plates showed comparable immunoreactivities with the LdPEX5 antisera (results not shown). The Bmax values observed with the WXXXY/F mutants are consistent with the results obtained with the pull-down experiments (Figure 2). Interestingly, the Kd values measured for ldpex5-W53A, ldpex5-W293A and ldpex5-W176,293A were approx. 16 nM, a value comparable with that of LdPEX5 (Table 1). The fact that the Kd value for these proteins was not markedly altered suggests that the WXXXY/F pentapeptide motifs do not constitute a direct LdPEX14-binding domain. The most significant change in ldpex5 binding affinity for His6/S–LdPEX14 was with the triple-mutant ldpex5-W53,176,293A, which had a measured Kd value of approx. 108 nM.
Figure 3. Determination of the LdPEX14–LdPEX5 equilibrium dissociation constant.
Microtitre plates were coated with His6/S–LdPEX14 and then incubated with increasing amounts of LdPEX5 (■), ldpex5-W53A (○), ldpex5-W293A (▲), ldpex5-W176,293A (●), or ldpex5-W53,176,293A (□). Bound LdPEX5 or ldpex5 proteins were quantified by an indirect ELISA using anti-LdPEX5 antisera. Each assay was performed in triplicate and the average absorbance values were plotted as a function of the log of the LdPEX5 or ldpex5 concentration using the ORIGIN 7.0 software. Kd values were determined as the protein concentration that gave half the maximal LdPEX5 or ldpex5 binding.
Table 1. Equilibrium dissociation constants for the LdPEX5–LdPEX14 interaction.
Numbers in parentheses indicate the number of times the experiment was performed. ND, not determined.
| Kd (nM) | ||
|---|---|---|
| LdPEX5 | LdPEX14 | LdPEX14-(1–120) |
| Wild type | 16±8 (5) | 9.0±0.6 (2) |
| W53A | 16±5 (4) | 18±12 (2) |
| W293A | 16±6 (3) | 15±11 (2) |
| W176,293A | 25±12 (4) | 10±2 (2) |
| W53,176,293A | 108±49 (3) | ND |
Mapping the LdPEX5–LdPEX14 interaction domain
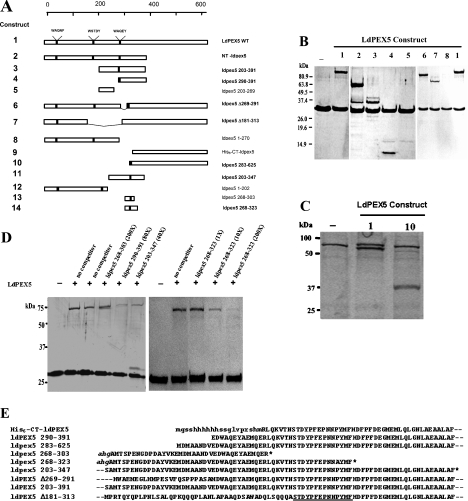
Previous studies with NT-ldpex5–His6, a fragment encompassing residues 1–391, showed that this region formed a tight interaction with His6/S–LdPEX14 [17]. Studies with ldpex5-(1–202) and ldpex5-(1–270) showed that, in pull-down assays and ELISAs, neither of these fragments was capable of binding His6/S–LdPEX14 (Figure 4 and Table 2). These observations are consistent with site-directed mutagenesis studies showing that replacement of the tryptophan residue in the W53AQNF57 and W176STDY180 repeats did not affect the LdPEX14 interaction affinity. These results suggest that the LdPEX14 recognition domain is localized to a region between residues 270 and 391 on LdPEX5. Similar experiments with smaller ldpex5 fragments encompassing residues 203–269, 203–347, 203–391 and 290–391 showed that only ldpex5-(203–347), ldpex5-(203–391) and ldpex5-(290–391) associated with His6/S–ldpex14-(1–120) in a pull-down assay (Figure 4). Analysis of these interactions by ELISA revealed that ldpex5-(203–391) and ldpex5-(290–391) bound His6/S–LdPEX14 with an affinity of 192±31 and 116±34 nM respectively. These Kd values were further validated by isothermal titration microcalorimetry [25]. Using this thermodynamic approach, a Kd of 204±22 nM was measured for the association of ldpex5-(203–391) with His6/S–ldpex14-(1–120), which is in good agreement with the ELISA data.
Figure 4. Mapping of the LdPEX14-binding domain.
(A) A schematic representation of the ldpex5 constructs that were overexpressed, purified and tested in pull-down or ELISA-based assays in order to map the LdPEX14 interaction motif. Constructs designated in boldface were shown to have LdPEX14 binding activity. (B) Coomassie Blue-stained SDS/PAGE analysis of S-protein agarose pull-down assays performed with His6/S–ldpex14-(1–120). For these assays, 10 μg of His6/S–ldpex14-(1–120) was mixed with no LdPEX5 or 10 μg of LdPEX5, NT-ldpex5, ldpex5-(203–391), ldpex5-(290–391), ldpex5-(203–269), Δ269–291-ldpex5, Δ181–313-ldpex5 and ldpex5-(1–270). The LdPEX5 constructs indicated by the numbers above each gel lane correspond to the LdPEX5 structures shown in (A). (C) For these assays, 10 μg of His6/S–LdPEX14 was mixed with no LdPEX5, 10 μg of LdPEX5 or 10 μg of ldpex5-(283–625). Numbers above each lane correspond to the LdPEX5 constructs illustrated in (A). (D) LdPEX5 competition assays. S-protein agarose beads were incubated with 10 μg of His6/S–ldpex14-(1–120) alone or with 4 μg of LdPEX5 in the presence and absence of increasing concentrations of competitor peptides ldpex5-(268–303), ldpex5-(290–391), ldpex5-(203–347) and ldpex5-(268–323) at molar excess ranging from 1- to 200-fold. Proteins bound to the S-protein beads were analysed by SDS/PAGE. (E) Partial alignment of overlapping sequences of ldpex5 mutants and ldpex5 fragments used to map the LdPEX14-binding motif. The amino acid sequence retained in the ldpex5 constructs with LdPEX14 binding activity is designated by the underline. Sequences in lower-case letters are derived from the expression vector. Broken lines preceding and following the sequence indicate that this portion of the protein sequence extends beyond the residues shown. The asterisk indicates the C-terminus of the ldpex5 fragment.
Table 2. Equilibrium dissociation constants for the ldpex5–LdPEX14 interaction.
Constructs correspond to the schematic diagram in Figure 4(A). Numbers in parentheses indicate the number of times the experiment was performed.
| LdPEX5 | LdPEX5 construct | Kd (nM) |
|---|---|---|
| ldpex5-(203–391) | 3 | 192±31 (2) |
| ldpex5-(290–391) | 4 | 116±34 (1) |
| ldpex5-(203–269) | 5 | No binding (2) |
| ldpex5-Δ181–313 | 7 | 69±16 (4) |
| ldpex5-(1–270) | 8 | No binding (3) |
| ldpex5-(283–625) | 10 | 111±57 (1) |
| ldpex5-(1–202) | 12 | No binding (4) |
| ldpex5-(268–303) | 13 | No binding (2) |
Alignment of ldpex5-(203–391), ldpex5-(203–347) and ldpex5-(290–391) (Figure 4) showed that these fragments retained the pentapeptide repeat W293AQEY297. Although mutagenesis studies indicated that this motif was not important for LdPEX14 binding, this was further confirmed using pull-down and competition assays with the fragments ldpex5-(268–303) and ldpex5-(268-323). No interaction between ldpex5-(268–303) and His6/S–ldpex14-(1–120) was detected as a 200-fold molar excess of ldpex5-(268–303) did not out-compete LdPEX5 binding to His6/S–ldpex14-(1–120) (Figure 4D). In contrast, competition assays using either a 10- or 200-fold molar excess of ldpex5-(268–323) resulted in a dramatic decrease in LdPEX5 bound to His6/S–ldpex14-(1–120), confirming that this peptide contained elements capable of out-competing LdPEX5 binding. Direct analysis of pull-down assays with ldpex5-(268–323) showed no detectable peptide bound to His6/S–ldpex14-(1–120), suggesting that this protein–protein interaction may be relatively weak and dissociates from His6/S–ldpex14-(1–120) during the stringent wash step. Competition experiments using a 40- or 80-fold molar excess of ldpex5-(203–347) and ldpex5-(290–391) respectively showed that these fragments reduced LdPEX5 binding by approx. 5–10-fold (Figure 4D). Moreover, both LdPEX5 and ldpex5-(203–347) were observed in pull-down assays with His6/S–ldpex14-(1–120) (Figure 4D). Experiments with ldpex5-Δ269–291, a mutant lacking the 30 amino acids immediately upstream of the third WXXXY/F repeat, did not alter the LdPEX14 binding activity (Figure 4B). Pull-down experiments with ldpex5-Δ181–313, a protein lacking repeats W176STDY180 and W293AQEY297, showed that this internal deletion mutant still bound His6/S–LdPEX14 with a Kd value of approx. 69 nM (Table 2 and Figure 4B).
Previous studies with His6–CT-ldpex5 established that this portion of LdPEX5 was not sufficient for interaction with His6/S–LdPEX14 [21]. In contrast, the mutant ldpex5-(283–625) generated in the present study revealed that extending the N-terminal region by 20 amino acids restored the His6/S–LdPEX14 binding activity and the Kd value for this interaction was 111±57 nM.
In contrast with wild-type LdPEX5, the various ldpex5 fragments (Table 2) exhibited 4–8 times higher Kd values. This is not surprising since these fragments may not retain a conformation that is fully competent to favour optimal LdPEX14 association. Analysis in silico with several secondary-structure predictive algorithms (http://www.expasy.org) suggests that the LdPEX5 region spanning residues 290–323 has a high propensity for random coil structure. The differences in binding affinity may also be due to an avidity effect since LdPEX5 has been shown to exist either as a tetramer or dimer in the absence or presence of PTS-1 ligand [27].
Mutagenesis of WXXXY/F motifs does not alter LdPEX5 quaternary structure
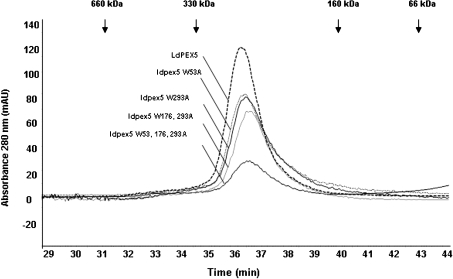
In the absence of a PTS1 ligand, LdPEX5 forms a homotetrameric structure [17]. Size-exclusion chromatography of ldpex5-W53A, ldpex5-W293A, ldpex5-W176,293A and ldpex5-W53,176,293A revealed that these mutant proteins co-eluted with the wild-type LdPEX5 as a single peak with an apparent molecular mass of approx. 270 kDa, a molecular mass consistent with the idea that these proteins form tetrameric structures (Figure 5).
Figure 5. Size-exclusion chromatography analysis of the ldpex5 mutants.
Wild-type LdPEX5 and site-directed mutant ldpex5-W53A, ldpex5-W293A, ldpex5-W176,293A, ldpex5-W53,176,293A proteins were purified to homogeneity as fusion proteins using the New England Biolabs IMPACT system. Intein fusion proteins were cleaved using DTT, dialysed and concentrated. The oligomeric state of these proteins was determined by loading 25–50 μg of purified protein on to a Bio-Sil 250 column (7.8 mm×600 mm) in 25 mM Tris/HCl (pH 7.5) and 120 mM NaCl at a flow rate of 0.25 ml/min. Column eluate was monitored spectrophotometrically at 280 nm. Arrows indicate the elution positions of the standard proteins.
Mutagenesis of the LdPEX5 WXXXY/F motifs does not alter PTS1 binding
To investigate if the mutagenesis of the WXXXY/F repeats affected the capacity of ldpex5-W53A, ldpex5-W293A, ldpex5-W176,293A and ldpex5-W53,176,293A to bind the PTS1 topogenic signal, we analysed the affinity of interaction between these receptor proteins and the model PTS1 protein XPRT from Leishmania donovani [26]. As shown in Figure 6, all LdPEX5/ldpex5 proteins exhibited comparable saturation kinetics with Kd values in the range 10–19 nM for the LdPEX5/ldpex5–XPRT interaction (Table 3). The binding of LdPEX5 in these modified ELISAs has been previously demonstrated to be dependent on the AKL tripeptide [17]. In control experiments performed with ldxprt ΔAKL, a variant of LdXPRT that lacks the AKL PTS1 signal sequence [17], no significant binding of LdPEX5 or ldpex5 mutant receptors was observed. Similarly, no appreciable binding of LdPEX5 was detected using BSA as a control protein.
Figure 6. LdPEX5–XPRT interaction.
Microtitre plates were coated with recombinant L. donovani XPRT and then incubated with increasing concentrations of one among wild-type LdPEX5 (■), ldpex5-W53A (○), ldpex5-W293A (▲), ldpex5-W176,293A (●) and ldpex5-W53,176,293A (□). The amount of LdPEX5 bound to the XPRT was determined by indirect ELISA using anti-LdPEX5 antisera (see the Materials and methods section). Binding experiments were performed in triplicate and absorbance values were averaged and plotted as a function of the log of the LdPEX5 or ldpex5 concentration. Data were fitted to a sigmoidal curve using the ORIGIN 7.0 computer program and Kd values are the LdPEX5 concentrations that gave half the maximal LdPEX5 or ldpex5 binding.
Table 3. Equilibrium dissociation constants for LdPEX5–LdXPRT interaction.
Numbers in parentheses indicate the number of times the experiment was performed.
| Kd (nM) | ||
|---|---|---|
| LdPEX5 | LdXPRT | LdXPRT in the presence of LdPEX14 |
| Wild type | 10±2 (6) | 92±31 (4) |
| W53A | 11±2 (2) | 57±9 (1) |
| W293A | 19±5 (3) | 90±17 (3) |
| W176,293A | 14±1 (4) | 65±12 (3) |
| W53,176,293A | 12±7 (4) | 56±16 (3) |
Recently, the binding of His6/S–LdPEX14 to the LdPEX5–LdXPRT complex was demonstrated to reduce dramatically the LdPEX5–XPRT interaction affinity [27]. As shown in Table 3, the ldpex5 Trp→Ala mutants, like wild-type LdPEX5, exhibited approx. 5–9-fold decrease in affinity for the LdPEX5–LdXPRT association, suggesting that these mutations did not affect the capacity of His6/S–LdPEX14 to alter the ldpex5 affinity for the PTS1 signal.
The conserved LdPEX14 signature sequence is critical for LdPEX5 interaction
Multiple sequence analysis of PEX14 revealed a conserved signature sequence AX2FLX7SPX6FLKGKGL/V (Figure 7). Deletion mapping of the LdPEX14 N-terminal region revealed that mutant proteins lacking the first 23 amino acids [His6/S–ldpex14-(24–464)] were capable of binding LdPEX5 with affinities comparable with wild-type LdPEX14 (Figure 7B), although the binding kinetics for His6/S–ldpex14-(24–464) appears to have shifted from a sigmoidal to a hyperbolic function. This observation suggests that the extended N-terminal sequence found on LdPEX14 adopts a configuration that may influence the association with LdPEX5. However, removal of the first 43 amino acids, which deleted a putative helix containing one of the conserved Phe-Leu dyads, or removal of the first 63 residues, which eliminated the entire signature sequence, resulted in a complete loss of LdPEX5 binding activity.
Figure 7. Mapping the LdPEX5 binding domain on LdPEX14.
(A) The multiple sequence alignment of the N-terminal region of the L. donovani (Leish), T. brucei (Trypa), human and S. cerevisiae (Sacch) PEX14 protein illustrates the conserved signature sequence (boldface). (B) Binding assay for the interaction of LdPEX5 with LdPEX14 N-terminal truncation mutants. Microtitre plates were coated with 1 μg/well wild-type LdPEX14 (□), ldpex14-(24–464) (■), ldpex14-(44–464) (○) or ldpex14-(64–464) (●), blocked with 2% milk powder in PBS 0.05% Tween 20 and incubated with various concentrations of LdPEX5 (0.4–860 nM). Amount of bound LdPEX5 was determined by indirect ELISA using anti-LdPEX5 rabbit antisera and goat anti-rabbit horseradish peroxidase-conjugated secondary antibody.
DISCUSSION
A critical step in the import of PTS1 proteins into peroxisome-like microbodies involves the interaction of the PTS1-laden PEX5 receptor with the microbody membrane-associated docking complex containing the PEX14 [9,10,28]. The interaction of PEX5 with PEX14 and PEX13 in higher eukaryotes has been previously shown to be dependent on WXXXY/F sequence motifs, which are conserved among all PEX5 proteins [9]. Although the Leishmania PEX5 contains three WXXXY/F pentapeptide repeats, here we show that these repeats are not required for the LdPEX5–LdPEX14 association. This argument is supported by several lines of evidence; first, sequential ablation of all three WXXXY/F repeats by site-directed mutagenesis failed to abolish completely the LdPEX5–LdPEX14 interaction. This contrasts with previous reports for the human and Saccharomyces cerevisiae PEX5 showing that replacement of the tryptophan or tyrosine residue of the WXXXY/F motif with an alanine was sufficient to cause a complete loss of binding to PEX14 or PEX13 [10,18,19]. Secondly, in vitro binding studies with ldpex5 truncation mutants encompassing residues 1–202 and 1–270, regions containing the repeats W53AQNF57 and W176STDY180, showed that these fragments were not capable of binding His6/S–LdPEX14 or His6/S–ldpex14-(1–120). Finally, experiments with ldpex5-(268–303), a 35 amino acid peptide encompassing the W293AQEY297 motif, showed no detectable binding to His6/S–LdPEX14. Moreover, competition assays showed that a large excess of this peptide was not sufficient to disrupt LdPEX5 binding to His6/S–ldpex14-(1–120).
In the present study, we show that, for LdPEX5, the motif involved in the interaction with LdPEX14 contrasts markedly with recent reports on the human and T. brucei PEX5, which demonstrated that short synthetic peptides spanning the WXXXY/F repeats were capable of binding PEX14. In the human system, peptides corresponding to all seven pentapeptide repeats exhibited PEX14 binding activity, whereas only two of the three WXXXY/F peptides derived from TbPEX5 bound TbPEX14 [16,18]. The Kd values measured with the TbPEX5 peptides were approx. 150–180 nM [18], which are considerably higher than the Kd values obtained in similar studies with human WXXXY/F peptides that had binding constants of approx. 20 nM. These differences may not be surprising since the flanking sequences, in addition to the sequence of the pentapeptide motif itself, have been proposed to be critical elements that affect the binding affinities and dictate whether a WXXXY/F motif interacts specifically with PEX13 or PEX14 [11–16,20]. Recent studies with an N-terminal fragment of T. brucei tbpex14 (residues 1–166) have shown that this fragment was capable of binding LdPEX5 (A. Jardim and K. P. Madrid, unpublished work). The tbpex14 fragment was also assayed for its ability to bind the mutants ldpex5-W53A, ldpex5-W293A, ldpex5-W176,293A and ldpex5-W53,176,293A in pull-down assays and a similar binding pattern comparable with that of Figure 2(B) was observed, suggesting that TbPEX14 may be capable of binding other motifs in addition to the WXXXY/F (results not shown).
Multiple sequence alignments of PEX5 proteins, together with partial three-dimensional structures of the human and T. brucei PEX5 TPR domains [29,30], indicate that the first TPR motif in LdPEX5 is predicted to start at residue 328 (Figure 1); it is therefore tempting to propose that the LdPEX14-binding site may be situated between residues 310 and 327, and indeed ldpex5-(268–323), a fragment spanning this region, was capable of competing with LdPEX5 for His6/S–ldpex14-(1–120) binding, whereas the peptide ldpex5-(268–303) failed to displace LdPEX5 binding even in the presence of a large excess of this peptide. Of note, however, was the finding that His6–CT-ldpex5, a fragment spanning residues 303–625, showed no LdPEX14 binding activity. This finding was not unanticipated since the crystal structure of the T. brucei PEX5 [30] predicts that this region, which exhibits a high degree of sequence identity with residues 309–322 of LdPEX5, forms a random coil structure that wraps around the TPR helix bundle, resulting in a compact arrangement that is stabilized by an extensive network of hydrogen bonds [30]. A similar compact packing architecture may be inferred for His6–CT-ldpex5, since partial proteolysis experiments have shown that this C-terminal portion of LdPEX5 is more resistant to proteolytic degradation [17]. This tight packing organization most probably shrouds the LdPEX14 interaction domain, which explains the lack of binding with His6–CT-ldpex5. Interestingly, extension of His6–CT-ldpex5 by 20 residues to create the ldpex 283–625 mutant protein led to restoration of LdPEX14 binding activity. These additional amino acids make up part of a coiled-coil motif encompassing residues 277–310, a region which is important for LdPEX5 oligomerization [27], a structure not found in the T. brucei, human or yeast PEX5. It is feasible that a protein–protein interaction between ldpex5-(283–625) subunits mediated by this coiled-coil motif may induce a conformational change that exposes the putative LdPEX14-binding motif S310TDYPFEPNNPYMFHDFP327. Another possibility for the absence of LdPEX14 binding activity observed with His6–CT-ldpex5 is that a truncation close to the LdPEX14-binding motif may lead to a disordered structure near the N-terminus, resulting in a weak binding affinity.
The PEX14 protein family is poorly conserved across phylogeny sharing only approx. 10% sequence identity. The only significant feature retained among the PEX14 proteins is the signature sequence AX2FLX7SPX6FLKGKGL/V. Mutagenesis studies in the Arabidopsis PEX14 [19] and T. brucei PEX14 [18] have demonstrated that this sequence is critical for binding the WXXXY/F pentapeptide and mediating the PEX5–PEX14 interaction. Since LdPEX5 is postulated to have a novel LdPEX14-binding motif, it was essential to verify that the LdPEX5–LdPEX14 interaction was dependent on the conserved PEX14 signature sequence. Progressive N-terminal deletions showed that removal of the first 23 amino acids had no effect on LdPEX5–LdPEX14 interaction. However, removal of the first 43 or 63 residue mutations that eliminated part or the entire PEX14 signature motif resulted in complete disruption of the LdPEX5–LdPEX14 association. The PEX14 signature sequence in the Arabidopsis PEX14 has been suggested to form a groove that binds the amphipathic α-helical structure formed by the WXXXY/F motifs [19]. It is not known whether the novel LdPEX5 motif, STDYPFEPNNPYMFHDFP, which has a PXXP pattern reminiscent of a polyproline type II helix, interacts with the signature motif in LdPEX14 in an analogous fashion [31–33]. Three-dimensional structures of the yeast PEX13 complexed with PEX5 have confirmed that the non-classical WXXXY/F motif can be bound by the SH3 domain [15,20]. Crystal structures of the Fyn tyrosine kinase SH3 [33] or Csk-SH3 [31] domains complexed with the PXXP peptide, of the HIV-1 Nef or the proline-enriched tyrosine phosphatase respectively, reveal that these high-affinity interactions are stabilized by hydrophobic and electrostatic interactions formed when the polyproline helices insert into a hydrophobic pocket in the SH3 domain.
The measured Kd for the LdPEX5–LdPEX14 interaction in the absence of a PTS1 ligand was approx. 16 nM, which is comparable with the 1.0–3.5 nM determined for the human PEX5–PEX14 interaction [22]. Using SDS/PAGE analysis, it was previously suggested that the LdPEX5–LdPEX14 interaction affinity was in the μM range. However, recent isothermal titration microcalorimetry experiments suggest that LdPEX5–LdPEX14 interaction appears to be a more complex phenomenon since two Kd values in the nM and μM range have been measured for this protein–protein interaction (K. P. Madrid, S. Wang and A. Jardim, unpublished work).
Mutagenesis of the WXXXY/F motifs in LdPEX5 had no significant effect on the LdPEX5–LdPEX14 protein–protein interaction or on its ability to bind PTS1 ligands. Since the diaromatic motifs are not required for mediating the docking of a PTS1-laden LdPEX5 receptor with LdPEX14 on the glycosomal membrane, it is possible that these pentapeptide repeats may be important for association with a putative kinetoplastid homologue or other components of the glycosome biogenesis machinery [14,34].
Acknowledgments
We acknowledge the excellent technical assistance of Ms S. Boyd in the creation and purification of the ldpex5 mutant proteins. A.J. was supported by a grant from the Natural Sciences and Engineering Research Council of Canada and this grant was used to support K.P.M. and Ms S. Boyd.
References
- 1.Dacks J. B., Doolittle W. F. Reconstructing/deconstructing the earliest eukaryotes: how comparative genomics can help. Cell (Cambridge, Mass.) 2001;107:419–425. doi: 10.1016/s0092-8674(01)00584-0. [DOI] [PubMed] [Google Scholar]
- 2.Zeiner G. M., Sturm N. R., Campbell D. A. The Leishmania tarentolae spliced leader contains determinants for association with polysomes. J. Biol. Chem. 2003;278:38269–38275. doi: 10.1074/jbc.M304295200. [DOI] [PubMed] [Google Scholar]
- 3.Muhich M. L., Boothroyd J. C. Polycistronic transcripts in trypanosomes and their accumulation during heat shock: evidence for a precursor role in mRNA synthesis. Mol. Cell. Biol. 1988;8:3837–3846. doi: 10.1128/mcb.8.9.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kable M. L., Heidmann S., Stuart K. D. RNA editing: getting U into RNA. Trends Biochem. Sci. 1997;22:162–168. doi: 10.1016/s0968-0004(97)01041-4. [DOI] [PubMed] [Google Scholar]
- 5.Englund P. T., Hajduk S. L., Marini J. C. The molecular biology of trypanosomes. Annu. Rev. Biochem. 1982;51:695–726. doi: 10.1146/annurev.bi.51.070182.003403. [DOI] [PubMed] [Google Scholar]
- 6.Opperdoes F. R. The glycosome of trypanosomes and Leishmania. Biochem. Soc. Trans. 1990;18:729–731. doi: 10.1042/bst0180729. [DOI] [PubMed] [Google Scholar]
- 7.Hannaert V., Bringaud F., Opperdoes F. R., Michels P. A. M. Evolution of energy metabolism and its compartmentation in Kinetoplastida. Kinetoplastid Biol. Dis. 2003;2:11–41. doi: 10.1186/1475-9292-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Opperdoes F. R., Baudhuin P., Coppens I., De Roe C., Edwards S. W., Weijers P. J., Misset O. Purification, morphometric analysis, and characterization of the glycosomes (microbodies) of the protozoan hemoflagellate Trypanosoma brucei. J. Cell Biol. 1984;98:1178–1184. doi: 10.1083/jcb.98.4.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purdue P. E., Lazarow P. B. Peroxisome biogenesis. Annu. Rev. Cell Dev. Biol. 2001;17:701–752. doi: 10.1146/annurev.cellbio.17.1.701. [DOI] [PubMed] [Google Scholar]
- 10.Subramani S. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol. Rev. 1998;78:171–188. doi: 10.1152/physrev.1998.78.1.171. [DOI] [PubMed] [Google Scholar]
- 11.Otera H., Harano T., Honsho M., Ghaedi K., Mukai S., Tanaka A., Kawai A., Shimizu N., Fujiki Y. The mammalian peroxin Pex5pL, the longer isoform of the mobile peroxisome targeting signal (PTS) type 1 transporter, translocates the Pex7p·PTS2 protein complex into peroxisomes via its initial docking site, Pex14p. J. Biol. Chem. 2000;275:21703–21714. doi: 10.1074/jbc.M000720200. [DOI] [PubMed] [Google Scholar]
- 12.Urquhart A. J., Kennedy D., Gould S. J., Crane D. I. Interaction of Pex5p, the type 1 peroxisome targeting signal receptor, with the peroxisomal membrane proteins Pex14p and Pex13p. J. Biol. Chem. 2000;275:4127–4136. doi: 10.1074/jbc.275.6.4127. [DOI] [PubMed] [Google Scholar]
- 13.Albertini M., Rehling P., Erdmann R., Girzalsky W., Kiel J. A., Veenhuis M., Kunau W. H. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell (Cambridge, Mass.) 1997;89:83–92. doi: 10.1016/s0092-8674(00)80185-3. [DOI] [PubMed] [Google Scholar]
- 14.Otera H., Setoguchi K., Hamasaki M., Kumashiro T., Shimizu N., Fujiki Y. Peroxisomal targeting signal receptor Pex5p interacts with cargoes and import machinery components in a spatiotemporally differentiated manner: conserved Pex5p WXXXF/Y motifs are critical for matrix protein import. Mol. Cell. Biol. 2002;22:1639–1655. doi: 10.1128/MCB.22.6.1639-1655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pires J. R., Hong X., Brockmann C., Volkmer-Engert R., Schneider-Mergener J., Oschkinat H., Erdmann R. The ScPex13p SH3 domain exposes two distinct binding sites for Pex5p and Pex14p. J. Mol. Biol. 2003;326:1427–1435. doi: 10.1016/s0022-2836(03)00039-1. [DOI] [PubMed] [Google Scholar]
- 16.Saidowsky J., Dodt G., Kirchberg K., Wegner A., Nastainczyk W., Kunau W. H., Schliebs W. The di-aromatic pentapeptide repeats of the human peroxisome import receptor PEX5 are separate high affinity binding sites for the peroxisomal membrane protein PEX14. J. Biol. Chem. 2001;276:34524–34529. doi: 10.1074/jbc.M104647200. [DOI] [PubMed] [Google Scholar]
- 17.Jardim A., Liu W., Zheleznova E., Ullman B. Peroxisomal targeting signal-1 receptor protein PEX5 from Leishmania donovani. Molecular, biochemical, and immunocytochemical characterization. J. Biol. Chem. 2000;275:13637–13644. doi: 10.1074/jbc.275.18.13637. [DOI] [PubMed] [Google Scholar]
- 18.Choe J., Moyersoen J., Roach C., Carter T. L., Fan E., Michels P. A., Hol W. G. Analysis of the sequence motifs responsible for the interactions of peroxins 14 and 5, which are involved in glycosome biogenesis in Trypanosoma brucei. Biochemistry. 2003;42:10915–10922. doi: 10.1021/bi034248n. [DOI] [PubMed] [Google Scholar]
- 19.Nito K., Hayashi M., Nishimura M. Direct interaction and determination of binding domains among peroxisomal import factors in Arabidopsis thaliana. Plant Cell Physiol. 2002;43:355–366. doi: 10.1093/pcp/pcf057. [DOI] [PubMed] [Google Scholar]
- 20.Douangamath A., Filipp F. V., Klein A. T., Barnett P., Zou P., Voorn-Brouwer T., Vega M. C., Mayans O. M., Sattler M., Distel B., et al. Topography for independent binding of alpha-helical and PPII-helical ligands to a peroxisomal SH3 domain. Mol. Cell. 2002;10:1007–1017. doi: 10.1016/s1097-2765(02)00749-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jardim A., Rager N., Liu W., Ullman B. Peroxisomal targeting protein 14 (PEX14) from Leishmania donovani. Molecular, biochemical, and immunocytochemical characterization. Mol. Biochem. Parasitol. 2002;124:51–62. doi: 10.1016/s0166-6851(02)00164-0. [DOI] [PubMed] [Google Scholar]
- 22.Schliebs W., Saidowsky J., Agianian B., Dodt G., Herberg F. W., Kunau W. H. Recombinant human peroxisomal targeting signal receptor PEX5. Structural basis for interaction of PEX5 with PEX14. J. Biol. Chem. 1999;274:5666–5673. doi: 10.1074/jbc.274.9.5666. [DOI] [PubMed] [Google Scholar]
- 23.Gill S. C., von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 24.Molloy D. P., Barral P. M., Bremner K. H., Gallimore P. H., Grand R. J. Structural determinants in adenovirus 12 E1A involved in the interaction with C-terminal binding protein 1. Virology. 2000;277:156–166. doi: 10.1006/viro.2000.0580. [DOI] [PubMed] [Google Scholar]
- 25.Cliff M. J., Ladbury J. E. A survey of the year 2002 literature on applications of isothermal titration calorimetry. J. Mol. Recognit. 2003;16:383–391. doi: 10.1002/jmr.648. [DOI] [PubMed] [Google Scholar]
- 26.Jardim A., Bergeson S. E., Shih S., Carter N., Lucas R. W., Merlin G., Myler P. J., Stuart K., Ullman B. Xanthine phosphoribosyltransferase from Leishmania donovani. Molecular cloning, biochemical characterization, and genetic analysis. J. Biol. Chem. 1999;274:34403–34410. doi: 10.1074/jbc.274.48.34403. [DOI] [PubMed] [Google Scholar]
- 27.Madrid K. P., De Crescenzo G., Wang S., Jardim A. Modulation of the Leishmania donovani peroxin 5 quaternary structure by peroxisomal targeting signal 1 ligands. Mol. Cell. Biol. 2004;24:731–744. doi: 10.1128/MCB.24.17.7331-7344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Walque S., Kiel J. A., Veenhuis M., Opperdoes F. R., Michels P. A. Cloning and analysis of the PTS-1 receptor in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;104:106–119. doi: 10.1016/s0166-6851(99)00144-9. [DOI] [PubMed] [Google Scholar]
- 29.Gatto G. J., Jr, Geisbrecht B. V., Gould S. J., Berg J. M. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 2002;7:1091–1095. doi: 10.1038/81930. [DOI] [PubMed] [Google Scholar]
- 30.Kumar A., Roach C., Hirsh I. S., Turley S., deWalque S., Michels P. A., Hol W. G. An unexpected extended conformation for the third TPR motif of the peroxin PEX5 from Trypanosoma brucei. J. Mol. Biol. 2001;307:271–282. doi: 10.1006/jmbi.2000.4465. [DOI] [PubMed] [Google Scholar]
- 31.Ghose R., Shekhtman A., Goger M. J., Ji H., Cowburn D. A novel, specific interaction involving the Csk SH3 domain and its natural ligand. Nat. Struct. Biol. 2001;8:998–1004. doi: 10.1038/nsb1101-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kay B. K., Williamson M. P., Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 33.Lee C.-H., Saksela K., Mirza U. A., Chait B. T., Kuriyan J. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell (Cambridge, Mass.) 1996;85:931–942. doi: 10.1016/s0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- 34.Bottger G., Barnett P., Klein A. T., Kragt A., Tabak H. F., Distel B. Saccharomyces cerevisiae PTS1 receptor Pex5p interacts with the SH3 domain of the peroxisomal membrane protein Pex13p in an unconventional, non-PXXP-related manner. Mol. Biol. Cell. 2000;11:3963–3976. doi: 10.1091/mbc.11.11.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]