Abstract
The luminal domain of the type I transmembrane protein Ire1 senses endoplasmic reticulum stress by an undefined mechanism to up-regulate the signalling pathway for the unfolded protein response. Previously, we proposed that the luminal domain of yeast Ire1 is divided into five subregions, termed subregions I–V sequentially from the N-terminus. Ire1 lost activity when internal deletions of subregion II or IV were made. In the present paper, we show that partial proteolysis of a recombinant protein consisting of the Ire1 luminal domain suggests that subregions II–IV are tightly folded. We also show that a recombinant protein of subregions II–IV formed homodimers, and that this homodimer formation was impaired by an internal deletion of subregion IV. Furthermore, recombinant fragments of subregion IV exhibited a self-binding ability. Therefore, although its sequence is little conserved evolutionarily, subregion IV plays an essential role to promote Ire1 dimer formation.
Keywords: endoplasmic reticulum chaperone, immunoglobulin heavy-chain binding protein (BiP)/78 kDa glucose-regulated protein (GRP78), Ire1, partial proteolysis, stress sensor, unfolded protein response (UPR)
Abbreviations: aa, amino acid; BiP, immunoglobulin heavy-chain binding protein; CCD, charge-coupled device; DTT, dithiothreitol; ER, endoplasmic reticulum; GST, glutathione S-transferase; HA, haemagglutinin; mAb, monoclonal antibody; MBP, maltose-binding protein; rP, recombinant protein; SD, synthetic dextrose; UPR, unfolded protein response; UPRE, UPR element
INTRODUCTION
Accumulation of misfolded proteins in the ER (endoplasmic reticulum), so-called ER stress, activates a cytoprotective signalling cascade termed the UPR (unfolded protein response). Although it is a complicated branched signalling pathway in mammalian cells, UPR signalling is straightforward in the yeast Saccharomyces cerevisiae. ER stress promotes dimerization of the ER-resident type I transmembrane protein Ire1 [1–4], which is believed to lead to up-regulation of the kinase and RNase activities located in the cytosolic domain of this protein [3,5]. The target of this RNase activity is a precursor form of mRNA encoded by the HAC1 gene [6]. Upon ER stress, Ire1 promotes splicing of this RNA to produce the mature form that is translated into a functional transcription factor for induction of various genes, including those encoding ER-resident molecular chaperones and protein-folding catalysts [6–9]. In mammalian cells, there are two Ire1 paralogues, IRE1α and IRE1β [10–12], which appear to target RNAs and downstream signalling pathways differently [12–14].
Although the mechanism by which the luminal domain of Ire1 senses ER stress remains to be elucidated, several studies have partly uncovered functions of this domain. First, the luminal domain possesses dimer-forming ability [15,16], which is required for overall activation of the Ire1 protein [17]. Secondly, the ER-resident HSP70 (heat-shock protein 70) chaperone BiP (immunoglobulin heavy-chain binding protein) binds to Ire1 and represses its activity under non-stressed conditions [4,18,19]. Upon ER stress, BiP dissociates from Ire1 both in mammalian and yeast cells [4,19]. Furthermore, we have reported recently a comprehensive in vivo mutation study of the luminal domain of yeast Ire1 [20], which concluded by proposing the structure–function relationship schematically represented in Figure 1(A). In that report, the IRE1 gene was subjected to 10-aa (amino acid) deletion scanning, and phenotypes of the deletion mutant strains were analysed. This analysis predicted that the luminal domain is divided into five subregions, termed subregions I–V sequentially from the N-terminus. Ire1 lost UPR pathway-activating activity when internal 10-aa deletions of subregion II or IV were introduced. Although the BiP-binding site was assigned to a part of subregion V, deletions of subregion V preserved the ER-stress inducibility of Ire1. Based on these and other findings described in that report [20], we concluded that subregions II–IV constitute the core stress-sensing region, in which BiP is not involved. The predicted function of the core stress-sensing region is to promote dimerization of Ire1 and BiP release from subregion V.
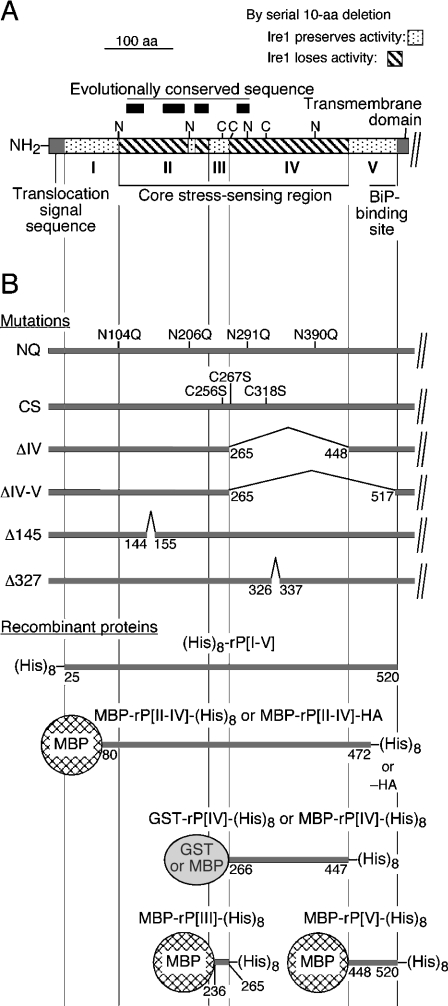
Figure 1. Predicted structure of the Ire1 luminal domain, and the mutations and recombinant proteins used in the present study.
(A) Structure of the proposed yeast Ire1 luminal domain [20]. Subregion I corresponds roughly to aa 25–104, subregion II to aa 105–235, subregion III to aa 236–265, subregion IV to aa 266–447, and subregion V to aa 448–520. The potential N-glycosylation sites and the cysteine residues are marked as N and C respectively. Positions of the conserved sequences indicated by an interspecies sequence alignment [17] are also shown. (B) Positions of the mutations, and structures of the recombinant proteins are presented.
The aim of the present study is to provide mainly in vitro experimental support for our prediction about the structure–function relationship of Ire1. For instance, we show the significance of subregion IV, which until now has been obscure. Liu et al. [16] reported that a chimaeric mutant of yeast Ire1, in which the full-length luminal domain was swapped for a region of human IRE1α corresponding only to subregion II, was functional. Moreover, the amino acid sequence of subregion IV is weakly conserved, whereas that of subregion II is highly conserved (see Figure 1A). However, the present in vitro and in vivo studies indicate that subregion IV possesses dimer-forming ability, which is required for dimer formation and activation of the overall Ire1 protein.
EXPERIMENTAL
Plasmids
Plasmid pRS315-IRE1-HA [20] is a yeast centromeric plasmid bearing the LEU2 selectable marker, and the yeast IRE1 gene fused with a C-terminal three-tandem copy of a HA (haemagglutinin)-tagging sequence which is expressed from its native promoter. To generate plasmid pRS315-Ire1(ΔC)-HA, a DNA fragment coding Met1–Lys585 [to number nucleotide and amino acid positions, we set the initiation methionine site of Ire1 (GenBank® accession number AAB68894; S. cerevisiae Ire1p) as position 1] of Ire1 was PCR-amplified from pRS315-IRE1-HA using the forward primer 5′-AGCACTGTCGACAATGCGTCTACTTCGAAGAAAC-3′ (the hybridizing coding sequence is underlined, and the attached SalI site is indicated in bold) and the reverse primer 5′-TTTAGCGCATGCGACTCAACTATGGGGATTTCCTTTTCAGGC-3′ (the hybridizing sequence is underlined, and the attached SphI site is indicated in bold), and the product was digested with SalI and SphI, and ligated into the same site of pRS315-IRE1-HA. PCR fragments carrying Δ145 and Δ327 mutations were generated by the overlap PCR technique [20], and ligated into pRS315-IRE1-HA in a similar way. pRS426-Ire1-FLAG is a yeast 2-μm plasmid bearing the URA3 selectable marker and the yeast IRE1 gene fused with a C-terminal three-tandem copy of FLAG-tagging sequence which is expressed from its native promoter [20]. The yeast URA3 2 μm plasmid pCZY1, carrying a fusion of the UPRE (UPR element)–CYC1 core promoter–lacZ, was a gift from Dr K. Mori (Department of Biophysics, Kyoto University, Kyoto, Japan). To generate plasmid pMBP-rP[II-IV]-(His)8, a DNA fragment coding Thr80–Ser472 of Ire1 was PCR-amplified from pRS315-IRE1-HA using the forward primer 5′-AACGGGGGATCCACTGCTGATAATCGACGTGCTAAC-3′ (the hybridizing sequence is underlined, and the attached BamHI site is indicated in bold) and the reverse primer 5′-CATCTCCTCGAGTCAgtgatggtgatggtgatggtgatgCGAATCAATCATCAGAGGTGGATA-3′ [the hybridizing sequence is underlined, and the attached sequence carried a His8-tag-coding sequence (indicated in lower-case) followed by the termination codon and the XhoI site (bold)], digested with BamHI and XhoI, and ligated with the BamHI/SalI-digested version of a bacterial expression vector for MBP (maltose-binding protein)-tagged proteins (pMAL-c2X; New England Biolabs). Plasmid pMBP-rP[II-IV]-HA was generated from pMBP-rP[II-IV]-(His)8 by replacement of the His8-tag coding sequence with that encoding three tandem copies of HA epitope. The Δ145 and Δ327 mutations were introduced into these plasmids using the overlap PCR technique as described previously [20]. Plasmid pYEX4T-1 (Clontech) is a yeast expression vector for GST (glutathione S-transferase)-tagged proteins. To generate plasmids pYEX-His and pYEX-MBP4, the GST-coding sequence on pYEX4T-1 was replaced with that encoding His8 residues and MBP respectively. DNA fragments corresponding to subregions III (Ile236–Glu265), IV (Cys266–Asp447) and V (His448–Leu520) were respectively PCR-amplified from pRS315-IRE1-HA (a His8-tag coding sequence was attached to the reverse primer), and cloned into either pYEX4T-1 or pYEX-MBP4. To generate plasmid pYEX-His-rP[I-V], a DNA fragment corresponding to the entire Ire1 luminal domain (Thr25–Leu520), was PCR-amplified from pRS315-IRE1-HA and cloned into pYEX-His. Construct details are available from K.K. upon request.
Yeast techniques
For the in vivo analysis shown in Figure 2, yeast cells were cultured at 30 °C in SD (synthetic dextrose) medium [21] supplemented with appropriate nutrients. To generate strains carrying the plasmid-borne mutant IRE1 genes (see Figure 1B for mutation positions), an in vivo gap repair technique was employed [22]. As described previously [20], DNA fragments corresponding to the Ire1 luminal domain were amplified using overlap PCR to introduce the desired mutations, mixed with linearized pRS315-IRE1-HA, and transformed into a Δire1 strain KMY1015 (MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 Δire1::TRP1) bearing pCZY1. Then transformants carrying the successfully gap-repaired plasmid were selected. Assays for cellular β-galactosidase activity were performed as described previously [18].
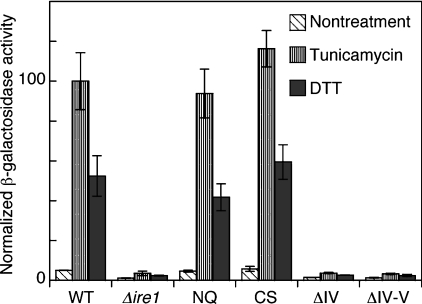
Figure 2. UPRE–lacZ reporter assay to monitor in vivo activity of the Ire1 mutants.
As described under the Experimental section, IRE1 mutations were generated on the IRE1 plasmid pRS315-IRE1-HA by in vivo gap repair in the yeast Δire1 strain KMY1015 carrying the UPRE–lacZ reporter plasmid pCZY1. For the wild-type Ire1 (WT) and empty vector (Δire1) controls, cells were respectively transformed with non-linearized pRS315-IRE1-HA and pPR315. Then cultures were treated at 30 °C with 2 μg/ml tunicamycin or 3 mM DTT for 4 h, and subjected to assays for cellular β-galactosidase activity. Each value is the average±S.D. for three independent transformants and was normalized to the tunicamycin-treated wild-type Ire1 control, which was set at 100.
Preparation of recombinant proteins of the yeast Ire1 luminal domain
An Escherichia coli strain named BL21 codon plus (DE3)-RIL (Stratagene) was used for bacterial expression of MBP–rP[II-IV]–His8 and MBP–rP[II-IV]–HA respectively from pMBP-rP[II-IV]-(His)8 and pMBP-rP[II-IV]-HA. After culturing at 37 °C in 2× YT (yeast extract/tryptone) medium [23] supplemented with 0.2% glucose, cells bearing the expression plasmids were transferred to 20 °C for 30 min and treated with 0.1 mM IPTG (isopropyl β-D-thiogalactoside) for 1 h. Then the cells were harvested and lysed by incubation in a buffer containing 50 mM Hepes, pH 7.0, 200 mM KCl, 5 mM MgCl2, protease inhibitors (2 mM PMSF, and 400 μg/ml each of aprotinin, pepstatin A and leupeptin), 1% (v/v) Triton X-100, 4 μg/ml of DNase I and 330 μg/ml of lysozyme on ice for 30 min, followed by sonication. After clarification of the lysates by ultracentrifugation at 30000 rev./min for 1 h in a Beckman SW40Ti rotor, the proteins were purified by affinity chromatography on amylose resin (New England Biolabs). The elution buffer contained 20 mM Hepes, pH 7.0, 100 mM KCl, 5 mM MgCl2, 20 mM maltose and 50% (v/v) glycerol. The same method was used for expression and purification of the Δ145 and Δ327 mutant versions of these proteins.
An S. cerevisiae strain named FY23 (MATa ura3-52 trp1-Δ63 leu2-Δ1) was used for cytosolic expression of GST–rP[IV]–His8, MBP–rP[III]–His8, MBP–rP[IV]–His8 and MBP–rP[V]–His8 from the above-mentioned derivatives of either pYEX4T-1 or pYEX-MBP4 under the control of the copper-inducible CUP1 promoter. For production of His8–rP[I-V], pYEX-His-rP[I-V] was introduced into FY23. Cells transformed with the expression plasmids were cultured at 30 °C in SD medium containing 0.5 mM CuSO4 for 2 h, harvested and lysed by glass-bead beating in a buffer containing 50 mM Hepes, pH 8.0, 300 mM KCl, 5 mM MgCl2, 10 mM imidazole, 1% (v/v) Triton X-100 and the above-mentioned protease inhibitors. After clarification of the lysates by ultracentrifugation at 100000 g for 1 h, the proteins were purified by nickel-chelate chromatography on Ni-NTA (Ni2+-nitrilotriacetate)–agarose (Qiagen). The elution buffer contained 50 mM Hepes, pH 8.0, 100 mM KCl, 5 mM MgCl2, 200 mM imidazole and 50% (v/v) glycerol.
Protein analyses
For partial proteolysis, 3 μg of the target protein (His8–rP[I-V]) dissolved in 15 μl of the elution buffer for the nickel-chelate chromatography was incubated for 40 min with 15 or 30 ng of trypsin (Roche) at 37 °C, 240 ng of endoproteinase Glu-C (Roche) at 25 °C, or 120 ng of endoproteinase Lys-C (Roche) at 37 °C. The resulting fragments were fractionated by standard reducing SDS/PAGE (10% polyacrylamide), and subjected to protein sequencing (ABI protein sequencer 476A, 492).
For native PAGE (5–20% polyacrylamide), 3 μg of proteins were diluted to 10 μl with Buffer A containing 20 mM Hepes, pH 7.0, 100 mM KCl, 5 mM MgCl2, mixed with 2 μl of commercially prepared loading buffer [0.125 M Tris/HCl, pH 6.8, 30% (v/v) glycerol, 0.01% (w/v) Bromophenol Blue (Daiichi Pure Chemicals, Tokyo, Japan)], and loaded on to a 4–20% polyacrylamide pre-cast gel (Daiichi Pure Chemicals). As the electrophoresis buffer, a commercially prepared [0.125 M Tris/0.192 M glycine, pH 8.4 (Daiichi Pure Chemicals)] Tris/glycine-based buffer was used.
The immunoprecipitation assays shown in Figure 4 were performed as described previously [20]. Western blot visualization of the protein bands was performed as described in [18], using mouse anti-HA mAb (monoclonal antibody) 12CA5 (Roche) or mouse anti-FLAG mAb M2 (Sigma–Aldrich) as primary antibody, and ECL® (enhanced chemiluminescence; Amersham Biosciences) signals were detected using a cooled CCD (charge-coupled device) camera system LAS-1000plus (Fuji Photo Film).
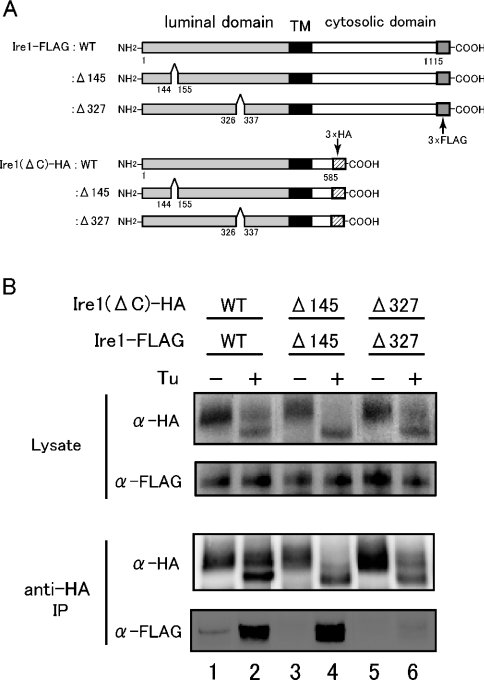
Figure 4. Immunoprecipitation assay to demonstrate the dimer-forming ability of core stress-sensing region in vivo.
(A) Diagram of constructs encoding full-length Ire1 or Ire1(ΔC), and its Δ145 and Δ327 mutants respectively. These proteins were C-terminally tagged with the 3× FLAG and 3× HA epitopes respectively. TM means transmembrane region (black). (B) A Δire1 strain KMY1015 was transformed with pRS315-Ire1(ΔC)-HA and pRS426-Ire1-FLAG, or their internal deletion mutants. The transformants carrying both plasmids were selected and cultured with (+) (lanes 2, 4 and 6) or without (−) (lanes 1, 3 and 5) 2 μg/ml tunicamycin (Tu) for 60 min, and subjected to anti-HA immunoprecipitation (IP) as described in the Experimental section. The cell lysates (equivalent to 3×106 cells) and immunoprecipitates (equivalent to 1×107 cells) were analysed by anti-HA (α-HA) and anti-FLAG (α-FLAG) Western blotting. Lanes 1 and 2, wild-type (WT) luminal region [Ire1(ΔC)–HA and Ire1–FLAG]; lanes 3 and 4, Δ145 mutants [Ire1(ΔC)–HA:Δ145 and Ire1–FLAG:Δ145]; lanes 5 and 6, Δ327 mutants [Ire1(ΔC)–HA:Δ327 and Ire1–FLAG:Δ327].
For pull-down assays shown in Figures 5 and 6, protein solutions were diluted to 200 μl with Buffer B [Buffer A supplemented with 1 mM DTT (dithiothreitol) and 0.1% (v/v) Triton X-100], and incubated with 10 μl of affinity beads [monoclonal anti-HA (Clone HA-7) agarose (Sigma–Aldrich) or glutathione–Sepharose 4B (Amersham Biosciences)] at 4 °C for 1 h. Then the beads were washed three times with Buffer B, and incubated with the differently tagged proteins in 200 μl of Buffer B for 1 h (at 30 °C for the experiment shown in Figure 5, and at 26.5 °C for that in Figure 6). After washing the beads three times with Buffer B, the bead-bound proteins were fractionated by standard reducing SDS/PAGE (10% polyacrylamide). Western blot visualization of the protein bands was performed as described in [18], using anti-His tag antibody (Amersham Biosciences) or anti-MBP antiserum (New England Biolabs) as primary antibody, and ECL® signals were detected using a cooled CCD camera system LAS-1000plus (Fuji Photo Film).
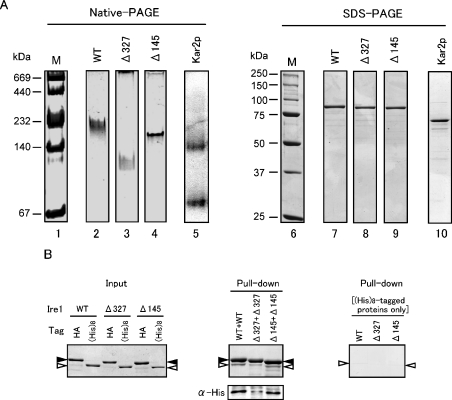
Figure 5. Dimer-forming ability of core-stress sensing region in vitro.
(A) Native PAGE analysis of the core-stress sensing region. Native PAGE (4–20% gradient gel, left-hand panel) or SDS/PAGE (10% gel, right-hand panel) of MBP–rP[II-IV]–His8 (marked WT) and its Δ327 and Δ145 mutant versions was performed as described in the Experimental section. The gel was stained with Coomassie Brilliant Blue except for lane 5 (immunoblotting with anti-Kar2 antibody) and photographed. Lanes 1 and 6, the High Molecular Weight Native Marker Kit (Amersham Biosciences) and Precision Protein standards (Bio-Rad) respectively, as molecular-mass markers (M) (sizes are indicated in kDa); lanes 2 and 7, MBP–rP[II-IV]–His8; lanes 3 and 8, Δ327 mutant; lanes 4 and 9, Δ145 mutant; lanes 5 and 10, yeast BiP protein. (B) In vitro pull-down assay to demonstrate the self-binding ability of the core stress-sensing region. Left-hand panel: 0.625 μg of the HA-tagged proteins {MBP–rP[II-IV]–HA (WT), and its Δ327 or Δ145 mutant} and 0.5 μg of the His8-tagged proteins {MBP–rP[II-IV]–His8 (WT), and its Δ327 or Δ145 mutant} were subjected to SDS/6% PAGE, and the gel was stained with Coomassie Blue. Positions of the HA-tagged and His8-tagged proteins were respectively marked by black and white arrowheads. Middle panel: 5 μg of the HA-tagged proteins and 4 μg of the His8-tagged proteins were subjected to the pull-down assay using anti-HA antibody–agarose beads as described in the Experimental section, and total (upper panel) or a 1/6 (lower panel) volume of the bead-bound proteins were fractionated by SDS/6% PAGE. Then the gels were stained with Coomassie Blue (upper panel) or subjected to anti-His tag Western blotting (lower panel). Positions of the HA-tagged and His8-tagged proteins were marked as in the left-hand panel. Right-hand panel: a similar experiment was performed as that shown in the middle panel, except for the absence of the HA-tagged proteins.
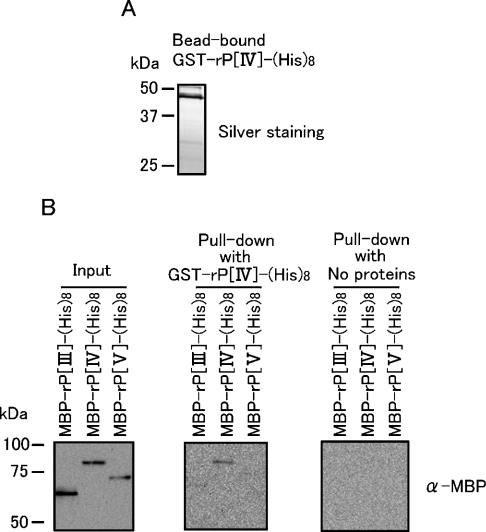
Figure 6. In vitro pull-down assay to demonstrate the self-binding ability of subregion IV.
(A) GST–rP[IV]–HA (500 ng) was incubated with 10 μl of glutathione–Sepharose beads, and the bead-bound protein was visualized by SDS/PAGE followed by silver staining. (B) Left-hand panel: 12.5 ng of the indicated proteins were visualized by anti-MBP Western blotting. Middle panel: the glutathione–Sepharose beads carrying GST–rP[IV]–HA and 400 ng of the MBP-tagged proteins were subjected to the in vitro pull-down assay as described in the Experimental section, and the pulled-down proteins were visualized by anti-MBP Western blotting. Right-hand panel: a similar experiment was performed as that shown in the middle panel, except for the absence of GST–rP[IV]–HA. Molecular-mass sizes are given in kDa.
RESULTS
The N-linked glycosyl chains and disulphide bonds are dispensable for regulation of yeast Ire1 in response to ER stress
Unlike the eukaryotic or prokaryotic cytosol, the ER possesses a specific ability to promote N-glycosylation and disulphide bond formation of proteins located on its luminal side. At the beginning of the present study, we experimentally confirmed that neither the N-glycosylation nor disulphide bond formation of the luminal domain of yeast Ire1 significantly influenced the activity of this protein. The aim of this experiment was to support our expectation that, in the later experiments, recombinant fragments of the Ire1 luminal domain expressed in the eukaryotic and prokaryotic cytosol would function as well as those that underwent native maturation in the ER.
As illustrated in Figure 1(A), there are four potential N-glycosylation sites (Asn-Xaa-Ser/Thr) and three cysteine residues in the luminal domain of yeast Ire1. The mutant CS, which is unable to undergo disulphide bond formation, carries replacements of all of the cysteine residues with serine. In the N-glycosylation minus mutant, NQ, N-glycosylation was blocked since asparagine-to-glutamine replacements were made at all of the potential N-glycosylation sites (Figure 1B). In Figure 2, the activity of Ire1 and its mutants in yeast cells was estimated by expression of a lacZ reporter that is controlled by UPRE, which is a promoter element positively regulated by the Ire1–Hac1 signalling pathway [24,25]. Genomic IRE1 gene knockout cells carrying the plasmid-borne wild-type or mutant IRE1 gene were cultured under non-stressed conditions or treated with the potent ER stressor, tunicamycin or DTT. Subsequently, these cells were subjected to β-galactosidase assays, which showed that both the NQ and CS mutant Ire1 proteins, as well as wild-type Ire1, functioned normally to up-regulate the UPR pathway in response to ER stress.
In vitro partial proteolysis shows tight folding of the core stress-sensing region
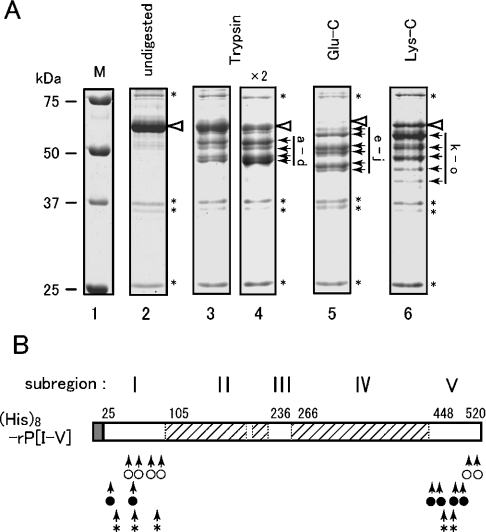
We then performed in vitro studies using recombinant proteins of the luminal domain of yeast Ire1. At first, a His8-tagged version of the entire luminal domain excluding the N-terminal translocation signal sequence {named His8–rP[I-V]; see Figure 1B} was expressed in yeast cytosol, and purified by nickel-chelate chromatography. In order to check the folding status, this protein was subjected to partial proteolysis analysis. As shown in Figure 3, His8–rP[I-V] was incubated with trypsin, or endoproteinase Glu-C or Lys-C, and then fractionated by SDS/PAGE. All of the resulting major fragments are marked by black arrows, and it should be noted that increasing the protease concentration did not yield new protein bands (Figure 3A, lanes 3 and 4, and results not shown). The molecular masses of these fragments were predicted from their mobility on SDS/PAGE, and a protein sequencer was used to determine their N-terminal cleavage sites (Table 1), which were all located in subregion I (aa 25–104). As shown in Table 1, the position of the C-terminal amino acid in each fragment was predicted from the N-terminal cleavage site, the predicted molecular mass of each fragment in SDS/PAGE and the consensus amino acids of C-terminal cleavage sites [C-terminal sides of arginine or lysine for trypsin, of glutamate (or aspartate) for endoproteinase Glu-C, and of lysine for endoproteinase Lys-C]. A summary of the results is schematically represented in Figure 3(B). It seems highly possible that all of the C-terminal cleavage sites were located in subregion V (aa 451–515). These observations suggest low protease accessibility and tight folding of the core stress-sensing region.
Figure 3. Partial proteolysis of the yeast Ire1 luminal domain.
(A) As described in the Experimental section, His8–rP[I-V] was incubated with trypsin, or endoproteinase Glu-C or Lys-C, and fractionated by SDS/12% PAGE. The gel was then stained with Coomassie Blue and photographed. All lanes are from the same gel. Positions of undigested His8–rP[I-V] and unidentified proteins are respectively marked by white arrowheads and asterisks (*). The major fragments that appear in Table 1 are marked by black arrows and are labelled alphabetically. Lane 1, molecular-mass markers [unstained Precision Plus Protein Standards (Bio-Rad)] (sizes are indicated in kDa); lane 2, undigested; lane 3, trypsin (15 ng); lane 4, trypsin (30 ng); lane 5, Glu-C (240 ng); lane 6, Lys-C (120 ng). (B) Proteolytic cleavage sites in His8–rP[I-V] are represented schematically by arrows for trypsin (○), Glu-C (●) and Lys-C (*) respectively.
Table 1. Partial-proteolysis fragments of (His)8-rP[I-V].
The protein bands (partial-proteolysis fragments) labelled alphabetically in Figure 3(A) were subjected to N-terminal sequencing, the results of which were used to decide their N-terminal positions. The molecular masses were calculated from the migration on SDS/PAGE, which was performed several times to yield errors less than 0.5 kDa. These data were used for prediction of the C-terminal positions. When the fragments seemed to be produced by cleavage at their N-terminal ends only, the ‘position of C-terminal aa’ is indicated as 520.
| Fragment | Protease | N-terminal sequence | Position of N-teminal aa | Predicted molecular mass (kDa) | Predicted position of C-teminal aa |
|---|---|---|---|---|---|
| a | Trypsin | TTEGL | 60 | 52 | 520 |
| b | Trypsin | LSSYP | 69 | 50 | 515 |
| c | Trypsin | RANKK | 85 | 47 | 501 or 502 or 503 |
| d | Trypsin | RAANS | 92 | 46 | 501 or 502 or 503 |
| e | Glu-C | VASTK | 35 | 55 | 520 |
| f | Glu-C | VASTK | 35 | 52 | 495 or 498 |
| g | Glu-C | GLPNM | 63 | 49 | 484 |
| h | Glu-C | VASTK | 35 | 47 | 451 or 454 |
| i | Glu-C | GLPNM | 63 | 44 | 451 or 454 |
| j | Glu-C | GLPNM | 63 | 43 | 451 or 454 |
| k | Lys-C | NINSP | 50 | 53 | 520 |
| l | Lys-C | LSSYP | 69 | 50 | 520 |
| m | Lys-C | NINSP | 50 | 46 | 464 |
| n | Lys-C | LSSYP | 69 | 43 | 455 |
| o | Lys-C | GRRAA | 90 | 41 | 455 |
Dimer formation of the core stress-sensing region and its abolishment by an internal deletion of subregion IV
To examine which subregion in Ire1 contributes to dimer formation in vivo, we constructed various FLAG- or HA-tagged wild-type and mutant version of Ire1s as shown in Figure 4(A). Cells that co-expressed C-terminal FLAG-tagged wild-type Ire1 (Ire1–FLAG:WT) and C-terminal truncated HA-tagged Ire1 [Ire1(ΔC)–HA:WT] were subjected to anti-HA immunoprecipitation. In response to ER stress, the co-immunoprecipitated FLAG-tagged wild-type Ire1 was detected by anti-FLAG Western blotting, suggesting that Ire1 formed dimers under the ER stress (Figure 4B, lanes 1 and 2). We then generated internal deletion mutant versions of Ire1–FLAG and Ire1(ΔC)–HA. One mutation, named Δ145, was a 10-aa deletion in subregion II, and another mutation, named Δ327, was also a 10-aa deletion, which was positioned in subregion IV (see Figure 4A and [20] for exact deletion positions). A similar result was obtained when both FLAG-tagged subregion IIΔ145 mutant (Ire1–FLAG:Δ145) and HA-tagged subregion IIΔ145 mutant [Ire1(ΔC)–HA:Δ145] were co-expressed in the cells (Figure 4B, lanes 3 and 4). Conversely, when both FLAG-tagged and HA-tagged subregion IVΔ327 mutants [Ire1–FLAG:Δ327 and Ire1(ΔC)–HA:Δ327 respectively] were co-expressed with or without tunicamycin, there was no band blotted with anti-FLAG antibody after anti-HA immunoprecipitation (Figure 4B, lanes 5 and 6). These results clearly indicate that subregion IV is required for dimer formation in vivo, and that both FLAG-tagged and HA-tagged subregion IVΔ327 mutants exist as a monomer.
We next checked the in vitro oligomeric status of the luminal domain of yeast Ire1, using native PAGE analysis. Since on native PAGE, His8–rP[I-V] migrated too broadly to be detected as a protein band, we used another recombinant protein in which large parts of subregions I and V were truncated. This protein was double-tagged with MBP and His8 residues, and was named MBP–rP[II-IV]–His8 (see Figure 1B). We also generated internal deletion mutant versions of MBP–rP[II-IV]–His8, named Δ145 and Δ327, which were 10-aa deletions in subregion II and subregion IV respectively (Figure 1B). BiP was used as a standard marker protein, since purified BiP forms a homodimer as described previously [26]. Under the reducing conditions of SDS/PAGE, wild-type and internal deletion mutant proteins migrated as single bands corresponding to their molecular masses, and BiP migrated at 70 kDa (Figure 5A, lanes 7–10). Conversely, native PAGE analysis showed that BiP migrated as two bands, indicating a monomer and dimer (Figure 5A, lane 5), as reported previously [26]. Since the Δ327 mutant existed as a monomer in the in vivo experiment described above, the band position of Δ327 MBP–rP[II-IV]–His8 should be a monomer (Figure 5A, lane 3). On the other hand, wild-type and Δ145 mutant MBP–rP[II-IV]–His8 proteins migrated slowly compared with Δ327 mutant protein, indicating that these proteins exist as a dimer (Figure 5A, lanes 2–4). When an excess of wild-type sample was loaded on to the gel, in addition to the major original band, a faint faster migrating band appeared, whose position corresponded to that of Δ327 mutant protein (results not shown). This result also supports the suggestion that MBP–rP[II-IV]–His8 exists mostly as a dimer.
The in vitro self-binding ability of the core stress-sensing region and its mutants was checked further by pull-down assays to exhibit binding of these proteins with the differently tagged versions of the same proteins (Figure 5B). MBP–rP[II-IV]–HA and its Δ145 and Δ327 deletion mutants were the modified versions of MBP–rP[II-IV]–His8 and its deletion mutants, in which the C-terminal tag was changed from the His8 tag to an HA epitope (see Figure 1B). The HA-tagged proteins were bound to anti-HA antibody–agarose beads, which were incubated further with the His8-tagged versions of the same proteins, washed, and subjected to SDS/PAGE to detect the bound proteins. As shown in Figure 5(B), the anti-HA antibody–agarose beads trapped MBP–rP[II-IV]–HA and its deletion mutants with almost equal efficiency (Figure 5B, middle panel, black arrowheads). MBP–rP[II-IV]–His8 was trapped by MBP–rP[II-IV]–HA with a detectable efficiency, and was diminished not by the Δ145 mutation but, by the Δ327 mutation (Figure 5B, middle panel, white arrowheads). The protein bands marked by white arrowheads correspond not to degradation products of the HA-tagged proteins, but exactly to the His8-tagged proteins, since those bands were also detected by anti-His-tag Western blotting. The right-hand panel in Figure 5(B) presents a negative-control experiment showing that the His8-tagged proteins were not non-specifically trapped by anti-HA antibody–agarose beads. In the middle panel of Figure 5(B), it should be noted that, even for the wild-type version, the protein bands corresponding to the His8-tagged proteins were fainter than those corresponding to the HA-tagged proteins. This is probably because dimerization of two proteins carrying the same tag occurred competitively. Based on these observations, we have concluded that MBP–rP[II-IV]–His8 possesses dimer-forming ability, which is significantly weakened not by the Δ145 mutation, but by the Δ327 mutation.
Subregion IV possesses self-binding ability
Since these findings suggest the contribution of subregion IV to dimer formation of the core stress-sensing region in vitro and in vivo, we next asked if subregion IV possesses self-binding ability. We first tried native PAGE analysis of some recombinant fragments corresponding to subregion IV only, but these fragments were so aggregative that they stacked at the start point of electrophoresis. Then, as shown in Figure 6, binding between two subregion-IV recombinant fragments carrying different tags was detected by in vitro pull-down assays. MBP–rP[IV]–His8 was a subregion-IV recombinant fragment double-tagged with MBP and His8 residues, while, to generate GST–rP[IV]–His8, the MBP tag of MBP–rP[IV]–His8 was replaced by a GST tag (see Figure 1B). Furthermore, MBP–rP[IV]–His8, MBP–rP[III]–His8 and MBP–rP[V]–His8 (see Figure 1B) were subjected to the same assay. Because of rather low yield of these proteins, which were expressed in yeast cytosol, protein bands on SDS/PAGE were visualized not by Coomassie Blue staining, but by silver staining or Western blotting. In the pull-down assay, GST–rP[IV]–His8 was bound to glutathione–Sepharose beads, which were incubated further with the MBP-tagged proteins, washed and subjected to SDS/PAGE. Figure 6(A) shows the binding of GST–rP[IV]–His8 to glutathione–Sepharose beads. In the middle panel of Figure 6(B), pull-down of MBP–rP[IV]–His8 by GST–rP[IV]–His8 was clearly observed, while that of MBP–rP[III]–His8 or MBP–rP[V]–His8 was hardly detectable. The right panel of Figure 6(B) presents a negative control experiment that shows the MBP-tagged proteins were not non-specifically trapped by glutathione–Sepharose beads. Therefore we have concluded that subregion IV possesses self-binding ability.
Entire deletion of subregion IV abolished the activity of Ire1
In our previous study, subregion IV, together with subregion II, was defined as a region in which internal 10-aa deletions impair the ability of Ire1 to up-regulate the UPR pathway. In order to confirm that subregion IV is indispensable for Ire1 activity, we checked the in vivo phenotypes of Ire1 mutants in which the entire sequence of subregion IV was deleted. The ΔIV and ΔIV–V mutant versions (see Figure 1B for deletion positions) of the IRE1 gene were introduced into a Δire1 strain, and their ability to up-regulate the UPR pathway in response to ER stress was monitored by the UPRE–lacZ reporter assay. As shown in Figure 2, this assay indicated that neither of the mutants retained the activity of Ire1.
DISCUSSION
The first finding in the present study is that the CS and NQ mutants of yeast Ire1 (Figure 1B) retained the ability to respond to ER stress (Figure 2). Similar observations with human IRE1α were reported by Liu et al. [15,16]. Therefore we have concluded that disulphide bond formation and N-glycosylation of the Ire1 luminal domain are dispensable for regulation of the overall Ire1 protein, and that bacterial and eukaryotic cytosolically synthesized recombinant fragments of the Ire1 luminal domain should function as well as those that had matured under native conditions in the ER. Furthermore, this insight indicates that the possibility that ER stress directly up-regulates Ire1 by inhibiting these post-translational modifications of Ire1 itself is unlikely.
The main part of the present study reports the in vitro characterization of recombinant fragments of the Ire1 luminal domain, which is likely to support the structure–function relationship predicted from our previous in vivo mutation study. At first, partial proteolysis His8–rP[I-V] (Figure 3) suggested tight folding of subregions II–IV (namely the core stress-sensing region). This insight is consistent with our previous observation that Ire1 lost activity when internal 10-aa deletions in either subregion II or subregion IV were introduced [20]. Moreover, high protease accessibility of subregion V supports our idea that BiP targets this subregion (Figure 3 and Table 1) [20].
Liu et al. [17] suggested that one important role of the Ire1 luminal domain is to contribute to homodimer formation of the overall Ire1 protein, and, in their later reports [15,16], the luminal domain of human IRE1α was indeed shown to possess dimer-forming ability. Consistent with this, we demonstrated in the present study that the core stress-sensing region of yeast Ire1 exists as a dimer in vitro (Figure 5). This dimer formation was abolished not by the Δ145 mutation, but by the Δ327 mutation (Figure 5). Furthermore, in vivo dimerization of Ire1, which occurred clearly in response to ER stress, was also impaired not by the Δ145 mutation, but by the Δ327 mutation (Figure 4, and [20]). Therefore we believe that the core stress-sensing region dimerized in vitro in the same way as it naturally did in vivo.
As described in the Introduction, the significance of subregion IV has been obscure. However, the contribution of subregion IV to the dimerization of the overall Ire1 protein is strongly suggested by the above-mentioned in vitro and in vivo phenotypes of the Δ327 mutation, which is located in subregion IV. The finding that subregion IV possessed self-binding ability (Figure 6) also supports this idea. The indispensability of subregion IV was also indicated by our finding that the ΔIV and ΔIV–V mutations (Figure 1B) abolished Ire1 activity completely (Figure 2).
We speculate that a 10-aa deletion causes perturbation of the folding status in only subregion domains. For instance, in the present study, a 10-aa deletion mutant in subregion IV (Δ327) lost the ability of dimer formation, indicating that one of the functions of subregion IV is dimerization of Ire1. All serial 10-aa deletion mutants in subregion IV, including Δ327, still have the ability to dissociate BiP in response to ER stress [20]. Therefore a 10-aa deletion in subregion IV did not disturb the function of subregion II, which could contribute to the dissociation of BiP from Ire1. In contrast, the Δ145 mutant in subregion II has lost the ability to dissociate BiP from Ire1 in response to ER stress [20], but had the ability of dimer formation (Figures 4 and 5).
There remain some problems that we could not address in the present study. First, our preliminary study suggested that, in addition to subregion IV, subregion II has self-binding ability (results not shown). It remains to be determined how the distinct self-binding abilities of subregions II and IV contribute to dimer formation of the overall luminal domain. Secondly, we failed to generate an in vitro experimental system in which the Ire1 luminal domain or its fragment switches between monomeric and dimeric forms. We believe that, as a next step in our work, such an experimental system will provide the definitive answer to the question of how ER stress up-regulates Ire1.
Acknowledgments
We thank Dr Kazutoshi Mori (Kyoto University, Kyoto, Japan) for yeast strains and plasmids, Junko Tsukamoto for peptide sequencing and Miki Matsumura for technical assistance. This work was supported by Grants-in-Aids for Scientific Research on Priority Areas (14037240 to K.K., 15030232 to Y.K.) and for 21st Century COE Research from MEXT, and JSPS.KAKENHI (15570160 to Y.K.). M.T. was supported by the grant 14037240.
References
- 1.Mori K., Ma W., Gething M. J., Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 2.Cox J. S., Shamu C. E., Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 3.Shamu C. E., Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 4.Bertolotti A., Zhang Y., Hendershot L. M., Harding H. P., Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 5.Sidrauski C., Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 6.Cox J. S., Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 7.Mori K., Kawahara T., Yoshida H., Yanagi H., Yura T. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- 8.Mori K., Ogawa N., Kawahara T., Yanagi H., Yura T. mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4660–4665. doi: 10.1073/pnas.050010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruegsegger U., Leber J. H., Walter P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell. 2001;107:103–114. doi: 10.1016/s0092-8674(01)00505-0. [DOI] [PubMed] [Google Scholar]
- 10.Tirasophon W., Welihinda A. A., Kaufman R. J. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X. Z., Harding H. P., Zhang Y., Jolicoeur E. M., Kuroda M., Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwawaki T., Hosoda A., Okuda T., Kamigori Y., Nomura-Furuwatari C., Kimata Y., Tsuru A., Kohno K. Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat. Cell Biol. 2001;3:158–164. doi: 10.1038/35055065. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 14.Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., Clark S. G., Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature (London) 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 15.Liu C. Y., Wong H. N., Schauerte J. A., Kaufman R. J. The protein kinase/endoribonuclease IRE1 that signals the unfolded protein response has a luminal N-terminal ligand-independent dimerization domain. J. Biol. Chem. 2002;277:18346–18356. doi: 10.1074/jbc.M112454200. [DOI] [PubMed] [Google Scholar]
- 16.Liu C. Y., Xu Z., Kaufman R. J. Structure and intermolecular interactions of the luminal dimerization domain of human IRE1α. J. Biol. Chem. 2003;278:17680–17687. doi: 10.1074/jbc.M300418200. [DOI] [PubMed] [Google Scholar]
- 17.Liu C. Y., Schroder M., Kaufman R. J. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J. Biol. Chem. 2000;275:24881–24885. doi: 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- 18.Kimata Y., Kimata Y. I., Shimizu Y., Abe H., Farcasanu I. C., Takeuchi M., Rose M. D., Kohno K. Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol. Biol. Cell. 2003;14:2559–2569. doi: 10.1091/mbc.E02-11-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamura K., Kimata Y., Higashio H., Tsuru A., Kohno K. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem. Biophys. Res. Commun. 2000;279:445–450. doi: 10.1006/bbrc.2000.3987. [DOI] [PubMed] [Google Scholar]
- 20.Kimata Y., Oikawa D., Shimizu Y., Kimata Y. I., Kohno K. A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1. J. Cell Biol. 2004;167:445–456. doi: 10.1083/jcb.200405153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser C., Michaelis S., Mitchell A. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1994. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. [Google Scholar]
- 22.Muhlrad D., Hunter R., Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J., Fritsch E. F., Maniatis T. 2nd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 24.Mori K., Sant A., Kohno K., Normington K., Gething M. J., Sambrook J. F. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohno K., Normington K., Sambrook J., Gething M. J., Mori K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 1993;13:877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tokunaga M., Kawamura A., Kohno K. Purification and characterization of BiP/Kar2 protein from Saccharomyces cerevisiae. J. Biol. Chem. 1992;267:17553–17559. [PubMed] [Google Scholar]