Abstract
Background: Rapid genetic screening for cytochrome P450 (CYP) 2C9 variants may play a role in improving the efficacy and safety of warfarin in individuals with CYP2C9 variants. The feasibility of prospective CYP2C9 model-based warfarin dosing has not yet been assessed.
Objectives: To evaluate the feasibility of applying a CYP2C9 gene-based warfarin dosing model in clinical practice.
Design: Prospective, randomized, single-blinded clinical pilot trial.
Setting: Large multispecialty group practice.
Patients: Candidates were recruited from a list of clinic patients eligible for warfarin initiation. This included patients with newly diagnosed thromboembolic disease or atrial arrhythmia, as well as patients anticipating elective valvuloplasty or arthroplasty. Patients who previously received warfarin were excluded.
Interventions: Subjects were randomized to receive either 1) a standard initiation dose of 5 mg warfarin/day, or 2) rapid CYP2C9 genotyping and an initiation dose determined using parameters estimated from a previously published multivariate model [including age, body size, co-morbidity (e.g., diabetes), clinical indication (e.g., valvuloplasty) and CYP2C9 genotype].
Measurements: Primary outcome measurements were patient willingness to participate, physician willingness to refer, sample processing time, ability to administer calculated dosage and adequacy of follow-up.
Limitations: This pilot trial was designed to assess the feasibility of model-based warfarin dosing. Power was insufficient for statistical comparison of adverse event rates.
Results: Forty-three of 117 patients had no prior warfarin treatment and were eligible. Five declined to participate. Twenty patients were randomized to a standard initiation dose of 5 mg daily. Eighteen patients were randomized to model-based dosing. All but one participant received the assigned initiation dose. Blood draw to dosage calculation time (including genotyping) required approximately 4 hours. Six adverse events occurred within the standard dosing group, and two adverse events occurred within the model-based dosing group.
Conclusions: Prospective application of a multivariate CYP2C9 gene-based warfarin dosing model is feasible.
Keywords: Adverse events, Anticoagulation, Cytochrome P450, CYP2C9, Warfarin, Multivariate dosing models, Genetic screening
Retrospective clinical studies have demonstrated that individuals with mutant allelic variants of the cytochrome P450 (CYP) 2C9 genotype slowly metabolize warfarin and may be at increased risk of hemorrhage during initiation of therapy.1–6 A variety of other factors also lead to variable patient responses to warfarin treatment. We previously developed a multivariate warfarin dosing model that incorporates age, body size, co-morbidity (diabetes), clinical indication (valve replacement) and CYP2C9 genotype.7 This multivariate model appears to explain 33.7% of the overall variability in warfarin maintenance dose.7
While rapid genetic screening for CYP2C9 variants is now possible, questions remain regarding its application feasibility in patients scheduled for the initiation of anticoagulant therapy. Before gene-based warfarin dosing models are uniformly applied, the following fundamental issues need to be addressed: 1) whether patients will be receptive to genetic testing before treatment, 2) whether the consent, sample collection, sample transfer, genotyping and model-based dose initiation can be conducted efficiently and 3) whether the results can be made available to a clinician prior to the time when warfarin would be typically administered.
Therefore, we performed a prospective clinical pilot trial to assess the feasibility of CYP2C9 model based warfarin dosing at the time of diagnosis. A thorough evaluation of the acceptance and feasibility of this approach is necessary before large-scale prospective randomized trials can be conducted in an efficient and cost-effective manner.
Methods
The trial protocol received prior approval by the Marshfield Clinic Research Foundation Institutional Review Board.
Enrollment
The sole inclusion criterion for this trial was eligibility for warfarin therapy based on diagnosis. Exclusion criteria included antiphospholipid antibodies, contraindications for warfarin, previous exposure to warfarin, liver disease, renal disease and non-Caucasian race, since ethnicity is known to influence allele frequency and this dosing model was derived in a Caucasian population.8–10 Patients under the age of 40 were also excluded because data from the original model did not include information on patients below this age.7
Eligible patients were identified for recruitment into this trial from six points of contact: vascular laboratory (ultrasound screening for thromboembolic disease), electrocardiography laboratory (screening for patients with new atrial arrhythmias), department of orthopedics (postoperative hip and knee arthroplasty patients), department of cardiology (elective valvuloplasty patients), and outpatient and inpatient pharmacies (screening for low molecular weight heparin therapy).
The clinical community at Marshfield Clinic was informed of the trial and enrollment was opened for a 1-month period. This pilot trial was then closed for a 30-day period, by design, to allow review of the enrollment process. This interim period was performed in order to evaluate patient enrollment and trial efficiency and to institute strategies to improve the study design. We then tested the modified study design for an additional month and reassessed the efficiency of conducting the study. Since the primary aim of the trial was to establish feasibility (rather than statistically meaningful differences in outcome), trial enrollment was time-limited (i.e., 2 months), rather than limited by a power-driven estimate of adequate sample size.
Participation
Patient receptiveness to phlebotomy and genotyping was quantified and recorded as the percentage of subjects who agreed to participate, in relation to all those who were identified as potentially eligible and therefore offered the opportunity. Upon identification, each patient who agreed to participate in the trial met with a research coordinator. Informed consent was obtained and all relevant clinical information was entered into an interactive, multi-functional database. This database required double entry confirmation of all data, and it contained a concealed randomization table that automatically randomized each patient to either standard dosing or multivariate model-based dosing. The clinical parameters incorporated in the model included genotype, age, body surface area, the presence/absence of a valve replacement, diabetes mellitus as a comorbidity and relevant interaction terms (see table 5 ▶ in reference 7). The INR target was not found to be predictive of dose after adjustment for these parameters and, therefore, was not included in the model. However, there was not much variability in the INR targets in this pilot trial (79% with midpoint 2.5, range 1.9 to 3.0).
Table 5.
Outcome measures by dosing group.
| Standard dosing | Model-based dosing | |
| Warfarin dosing (mg/week) | Mean (S.D./range) | Mean (S.D./range) |
| Initial dose | 35.0 (0.0/35.0–35.0) | 32.2 (9.1/17.5–59.5) |
| Stable dose | 32.6 (16.8/7.0–85.0) | 28.9 (11.5/12.0–50) |
| Mean (S.D.) | Mean (S.D.) | |
| Percent time INR in range | 41.5 (24.9) | 41.7 (25.4) |
| n(%) | n(%) | |
| Maximum INR >4.0 (number of patients) | 6 (30.0) | 6 (33.3) |
| Relative dosing success* | Mean (S.D.) | Mean (S.D.) |
| Absolute deviation (initial vs. stable) | 10.9 (10.3) | 10.3 (9.3) |
| n(%) | n(%) | |
| Superior regimen (retrospective)† | 13 (35.1) | 20 (54.1) |
*For each dosing scheme, relative dosing success is based on all 37 patients who remained on warfarin since the initial dose (whether actually assigned or hypothetical) could be computed under each scheme for all patients.
†The two dosing schemes (standard vs. model-based) were compared, retrospectively, for each individual patient to determine which scheme better matched the final stable dose.
Specimen processing
Immediately upon enrollment and phlebotomy, the blood sample was transported to the laboratory for DNA extraction. On-call laboratory personnel entered patient identifiers in the database and received the randomization status (for immediate or delayed genotyping). Patients randomized to model-based dosing were genotyped immediately (i.e., prospectively) for CYP2C9 with a goal of completing the genetic assay within 4 to 6 hours. Patients in the standard dosing group were genotyped retrospectively (i.e., following completion of the 28-day warfarin initiation phase) to allow genotypic information to be included in all subsequent analyses. Doses for both groups were communicated to each respective referring physician (i.e., the attending physician responsible for the patient’s care) within 4 to 6 hours after blood draw.
Genotyping
Blood samples were encrypted prior to genotype determination. After a phlebotomist drew 10 ml of blood into a tube containing sodium citrate, the sample was stored at 4°C until DNA extraction. Genomic DNA was extracted from whole blood using the Gentra Capture Column Kit (Gentra Systems, Inc., Minneapolis, Minnesota). Genotypes CYP2C9 *1/*1, *1/*2, *2/*2, *1/*3, *2/*3 and *3/*3 were determined using the CYP2C9 Mutation Detection Kit and LightCycler (Roche Diagnostics Corp., Idaho Technology Inc., Idaho Falls, Idaho). Two point mutations (C/T at base 430 for CYP2C9 *2 and A/C at base 1075 for CYP2C9 *3) were simultaneously detected by dual color emission and melting curve analysis. Briefly, 100 ng of purified DNA was combined with unlabeled primer, hybridization probes specific for bases 430 and 1075, FastStart Taq DNA polymerase (Roche Diagnostics Corp.) reaction buffer, dNTPs and FastStart Taq DNA polymerase in a pre-cooled LightCycler capillary tube. A positive, heterozygous control (supplied from Roche Laboratories) and a negative control (sterile, ultrapure dH2O) were prepared and analyzed with each patient DNA sample. DNA samples were denatured at 95°C for 10 minutes followed by 45 cycles each of 95°C x 10 seconds, 55°C x 10 seconds and 72°C x 10 seconds. Fluorescence was acquired once per cycle at the end of the annealing phase. Amplicon melting curves were analyzed by slow, gradual heating from 40°C to 80°C at a rate of 0.1°C/second. Fluorescent emissions at 640 nM and 705 nM were acquired continuously throughout the melting phase. Color compensation using a LightCycler Color Compensation Set (Roche Diagnostics Corp.) was performed after melting curve analysis.
The melting peaks for the 2C9 *2 variant occur at 50°C and at 61°C for the wild type allele (2C9 *1). The melting peaks for the 2C9 *3 variant occur at 52°C and at 60°C for the wild type allele. Genotypes were determined by review of the melting curves for individual DNA samples noting the relationship with controls. The sensitivity and accuracy of this assay have previously been shown to be 100% by comparison with sequencing data.7
Flow of information
Clinical and genetic data were entered into the interactive database as soon as they were available, and the database calculated the trial dose based upon randomization. The calculated dosage was then automatically forwarded through electronic mail to the nursing staff of the clinical anticoagulation service. Upon receipt, the initial warfarin dosing recommendation was communicated to the attending physician (or his/her designee) by either a trial physician or a certified registered nurse from the anticoagulation service. Coordination of nursing staff was required to facilitate proper dosing and follow-up. In all cases, the trial coordinator confirmed correct administration of the intended dose and, in all cases, the process was directly supervised by one of three trial physicians. In one case, the trial physician adjusted the recommended dose (from 5.5 mg to 6.0 mg) for the first day, but all subsequent doses were according to protocol.
All patients randomized to the standard dosing arm received an initial daily warfarin dose of 5 mg.11–13 Initial doses of other than 5 mg/day would likely have implied to the attending physician that the patient was randomized to the model-based dosing arm. This study is therefore single-blinded by definition. Attending physicians were not otherwise informed of the treatment arm.
Clinical follow-up
Inpatients received daily assessment of their prothrombin time expressed as an international normalized ratio (INR). Outpatient INR values were checked at a frequency determined by the anticoagulation service. Dosing adjustments were made according to a standardized, validated algorithm used routinely by the anticoagulation service.14 This algorithm includes consideration of differing target INR values based on the clinical indication for anticoagulation, and the current study protocol allowed an option for dosing modification according to physician discretion.
All patients were followed on protocol for 4 weeks (28 days) after initiation of warfarin therapy. If chronic anticoagulation was indicated beyond that point, the responsibility for assessing each patient’s INR value and adjusting warfarin dose was returned to the primary physician who may or may not have opted to utilize continued warfarin management by the anticoagulation service.
Data analysis
Data are presented with standard descriptive summaries. Although we report demographic and outcome data by randomized group, the study was not powered with the intent of testing hypotheses related to differences in outcomes between study groups. As such, there is potential for extraneous group differences, and all observed outcomes should be considered descriptive.
Results
Logistical assessment
Of 117 patients screened, 74 were excluded, in most cases due to prior treatment with warfarin (see Methods section for detailed discussion of exclusion criteria). Of the 43 eligible for trial participation, 5 patients declined to participate. The 38 (88%) patients who elected to participate were randomized to either standard (n=20) or model-based dosing (n=18) groups (table 1 ▶).
Table 1.
Enrollment efficiency.
| First month of pilot | Last month of pilot | |
| Patient eligibility | 18 eligible/48 screened (38%) | 25 eligible/69 screened (36%) |
| Patient receptivity | 15 enrolled/18 eligible (83%) | 23 enrolled/25 eligible (92%) |
| Prospective sample genotyping success | 15/15 (100%) | 23/23 (100%) |
| Dosing calculator application | 15/15 (100%) | 23/23 (100%) |
All patients in the standard dosing arm received 5 mg of oral warfarin daily upon initiation. Of the 18 patients in the model-based dosing group, 17 received the initiation dose calculated from the multivariate model. Based upon physician discretion, one patient received 6 mg of warfarin instead of the calculated dose of 5.5 mg. No patients in either group were lost to follow-up.
Collectively, the average time from blood draw to entry of the genotype into the dose calculator was 249 minutes (approximately 4 hours). In no case did the period between blood draw and completion of the genotyping process exceed 420 minutes (7 hours; table 2 ▶). No differences in sample processing time were seen between the first and last months of the pilot trial.
Table 2.
Genotyping timeframe for patients receiving model-based warfarin dose.
| Times (minutes) | ||
| Process steps | Median | Range |
| Blood draw to arrival in lab | 20 | 5–86 |
| Arrival to completion of extraction | 93 | 25–315 |
| Extraction to start of LightCycler assay | 15 | 0–94 |
| Assay beginning to assay end | 60 | 60–70 |
| End assay to dose calculation | 10 | 0–30 |
| Total time blood draw to dose | 225 | 110–432 |
During the “closed” 30-day period between the first and second enrollment months, two changes in the trial were made. First, early in the trial it was noted that frequent INR determinations were not being obtained on study patients who had entered nursing homes following hospital discharge. Therefore, an institutional review board-approved protocol amendment was performed mid-trial to allow the trial coordinator to intervene and assure that the nursing home physicians ordered an INR determination within the first 7 days of follow-up.
Second, it also became apparent that although the initial follow-up process was successful, it was not sustainable by any one individual physician. Consequently, a centralized dosing mechanism was implemented to facilitate communication of recommended doses, and to ensure appropriate follow-up. All corresponding orders were supervised and co-signed by the coordinating trial physician while the participant was an inpatient. All patients were then enrolled in the outpatient anticoagulation clinic after discharge, where subsequent dosing recommendations were coordinated through nursing staff and co-signed by each patient’s primary care physician.
Patient demographics
The demographic characteristics of the standard and model-based dosing groups were similar (table 3 ▶). However, a difference in the clinical indications for anticoagulation was observed, with more atrial fibrillation/flutter patients randomized to the standard dosing group and more deep vein thrombosis/pulmonary embolism patients randomized to the model-based dosing group. Corresponding recruitment strategies were not different between these two groups.
Table 3.
Patient demographics.
| Standard dosingn= 20 Mean (S.D.) | Model-based dosingn= 18 Mean (S.D.) | |
| Age (years) | 70.5 (13.3) | 68.8 (11.3) |
| Height (cm) | 167 (8.9) | 168 (10.0) |
| Weight (kg) | 91 (30.4) | 96 (32.0) |
| Body surface area (m2) | 2.0 (0.3) | 2.0 (0.3) |
| n(%) | n(%) | |
| Gender | ||
| Male | 9 (45%) | 8 (44%) |
| Female | 11 (55%) | 10 (56%) |
| Diabetes | ||
| Yes | 6 (30%) | 5 (28%) |
| No | 14 (70%) | 13 (72%) |
| Current smoker | ||
| Yes | 3 (15%) | 3 (17%) |
| No | 17 (85%) | 15 (83%) |
| Concomitant CYP2C9 medications | ||
| Inducer | 0 | 0 |
| Inhibitor | 7 (35%) | 6 (33%) |
| Substrate | 10 (50%) | 12 (67%) |
| Treatment indication | ||
| Atrial fibrillation/flutter | 9 (45%) | 3 (17%) |
| Prosthetic valve | 4 (20%) | 4 (22%) |
| Prosthetic joint | 3 (15%) | 3 (17%) |
| Deep venous thrombosis/pulmonary embolism | 3 (15%) | 6 (33%) |
| Other | 1 (5%) | 2 (11%) |
Genotype distribution
The CYP2C9 genotype distribution was quite similar within the two groups. Patients with a wild type homozygous CYP2C9 genotype (*1/*1) were observed at a frequency of 65% in the standard dosing group and 61% in the model-based dosing group (table 4 ▶).
Table 4.
Patient genotype by dosing group.
| *1/*1 | *1/*2 | *1/*3 | *2/*2 | *2/*3 | *3/*3 | Total | ||
| Standard | n | 13 | 3 | 2 | 0 | 1 | 1 | 20 |
| % | 65.0 | 15.0 | 10.0 | 0.0 | 5.0 | 5.0 | ||
| Model-based | n | 11 | 4 | 0 | 2 | 1 | 0 | 18 |
| % | 61.1 | 22.2 | 0.0 | 11.1 | 5.6 | 0.0 | ||
| Total | 24 | 7 | 2 | 2 | 2 | 1 | 38 |
Preliminary outcomes analyses
Although the primary aim of this trial was to assess the feasibility rather than the efficacy of using a dosing model, we also summarized clinically important short-term outcomes. Outcomes with respect to INR and warfarin dosing are presented in table 5 ▶. The groups were similar with respect to the percentage of time with INR in-range (about 42% for each) and with respect to the percentage of patients where the INR exceeded 4 at some point (30% for standard and 33% for model-based dosing).
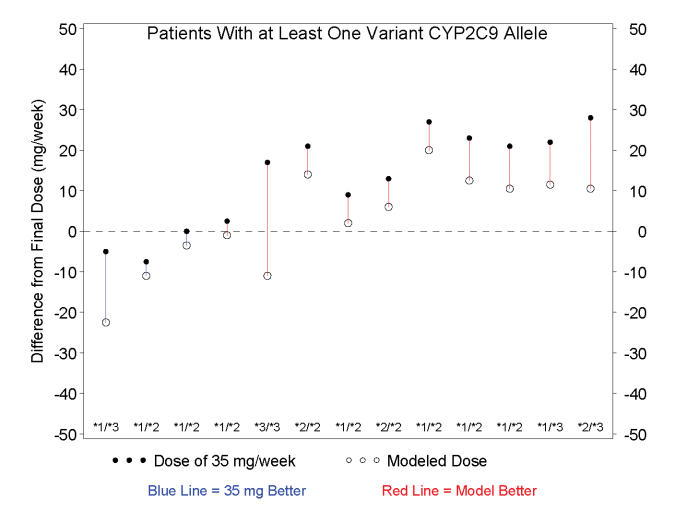
A major clinical challenge in warfarin therapy is the selection of a correct initiation dose (i.e., estimating the maintenance dose that will eventually be needed for each individual patient). We therefore also attempted to quantify how closely the assigned initial dose approximated the observed final (stable) dose for each patient in this trial. We report comparisons across two study groups of interest: 1) the total trial population and 2) the patients with at least one abnormal CYP2C9 allele. In the total trial population, the model-based dose was a better predictor of stable dose in 54% of subjects (20 of the 37 subjects; table 5 ▶). However, in those patients with at least one abnormal CYP2C9 allele, the model-based dose was a better predictor of stable dose in 77% of subjects (10 of 13 subjects; figure 1 ▶). After accounting for all parameters, the model calculated weekly dose estimates for each of the following genotypes: *1/*1, 35.3 ± 11.0 mg/wk (n=24); *1/*2, 28.5 ± 3.1 mg/wk (n=7); *1/*3, 21.0 ± 4.9 mg/wk (n=2); *2/*2, 28.0 ± 0.0 mg/wk (n=2); *2/*3, 17.5 ± 0.0 mg/wk (n=2) and *3/*3, 7.0 mg/wk (n=1). These dosing requirements are consistent with previously published observations.1,7
Figure 1.
Difference between the actual final maintenance dose and the dose predicted by the multivariate dosing calculator in only those patients with at least one variant CYP2C9 allele. Genotype for each individual is shown. In 10 of 13 patients with at least one variant CYP2C9 allele, the dose predicted by the model (open circles) was closer to the individual’s actual final maintenance dose than the dose assumed to be standard (closed circles).
Adverse events are listed in table 6 ▶. Six warfarin-related events (in five patients) occurred among the standard dosing group, while only two (in two patients) occurred in the model-based dosing group.
Table 6.
Adverse events per patient group.
| Standard dosing | Model-based dosing | |
| Warfarin-related | n = 5 patients | n = 2 patients |
| Hemorrhagic, gastrointestinal bleed | 1 | 2 |
| Hemorrhagic, hematuria | 1 | 0 |
| Hemorrhagic, epistaxis | 2 | 0 |
| Thromboembolic, deep venous thrombosis | 1 | 0 |
| Thromboembolic, pulmonary embolism | 1 | 0 |
| Warfarin-unrelated | n = 3 patients | n = 3 patients |
| Back pain | 0 | 1 |
| Dizziness | 2 | 0 |
| Hyperglycemia | 1 | 0 |
| Incision infection | 0 | 1 |
| Rash (trunk) | 0 | 1 |
| Tympanic membrane perforation | 1 | 0 |
| Urinary tract infection | 0 | 1 |
| Vaginal yeast infection | 0 | 1 |
| Bridging therapy | ||
| Subcutaneous low molecular weight heparin | 0 | 0 |
| Intravenous heparin | 0 | 0 |
| Reversal of anticoagulation | ||
| Subcutaneous vitamin K | 2 | 0 |
| Intravenous fresh frozen plasma | 0 | 0 |
Discussion
Clinical utility
This pilot trial has demonstrated that the application of a CYP2C9 gene-based multivariate warfarin dosage calculator is feasible from the standpoint of both health care infrastructure and patient willingness to participate. CYP2C9 genotyping supplies and instrumentation are readily available through commercial sources so that the technology is not a limiting factor. It remains to be determined whether larger prospective studies will provide enough clinical benefit to 1) justify multivariate model-based dosing and 2) justify the inclusion of genetic information in these models. Additional outcome-based pilot trials are needed prior to implementing a larger study.
Interestingly, there were six warfarin-related adverse events (e.g., gastrointestinal bleed, hemorrhagic stroke) among the standard dosing group and only two among the model-based dosing group. Whether this difference can be replicated in studies involving larger numbers of patients remains to be determined. Such studies are currently being organized.
The data in figure 1 ▶ show that patients with a variant CYP2C9 genotype tend to receive a more appropriate initial warfarin dose if a model-based dosing approach is applied prospectively. Since variant genotypes for CYP2C9 are found in a large percent of the general population,7 and since a higher rate of adverse events occur early in warfarin treatment,14 a multivariate warfarin dosing model that includes CYP2C9 genotype may potentially have significant clinical benefit. However, the economic implications of these findings remain undetermined. Given what is known regarding the prevalence of CYP2C9 genetic variation, a high percentage of patients with variant alleles are likely to be revealed through repeated INR determinations, even without genetic tests. Thus, benefits of gene-based dosing (potentially fewer adverse events in both outpatient and inpatient settings, reduced length of hospital stays and reduced patient morbidity and mortality) may need to be realized early in therapy, perhaps even within the first 14 to 28 days of warfarin initiation, to be meaningful from a pharmacoeconomic standpoint.
Recent preliminary calculations of cost-effectiveness have suggested that as few as 44 patients need to be screened for CYP2C9 genotype before benefits in excess of costs begin to accrue by the avoidance of one adverse event.15 The present study included slightly fewer total subjects (38 patients), but benefits were suggested by the avoidance of an additional four adverse events within the model-based dosing group. An additional advantage of the model used in the present study is that clinical data were added to genetic data. These data elements are typically maintained by integrated health care delivery systems and may be efficiently acquired from electronic medical records. As such, these data elements would not be expected to add significantly to the cost of developing and/or optimizing a gene-based dosing intervention.
Conclusion
Based on the results of this pilot trial, it may be concluded that prospective application of CYP2C9 gene-based multivariate warfarin dosing calculators are both technically feasible and acceptable to patients and providers. Those patients with a variant CYP2C9 genotype appear likely to benefit from the use of gene-based models to dose the drug, warfarin.
Acknowledgments
We would like to acknowledge the timely and efficient DNA testing conducted by Sherry Salzman, Kai Qi Zhang and Steven Offer. We would also like to acknowledge the following research coordinators for their assistance in the recruitment of patients: Dawn David, Sandy Freeman, Kathy Mancl and Lori Scheller. We would also like to thank Jaime Steward for her work with the trial databases and quality assurance, Melissa Mikelson and Marshfield Clinic Care Management nursing staff for providing guidance on the protocol-based daily dosing and Marshfield Clinic Research Foundation for providing assistance in the preparation of this manuscript through the services of Linda Weis, Alice Stargardt and Graig Eldred.
Grant Support: Marshfield Clinic, Research Foundation to M.A.H. and R.A.W.
References
- 1.Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM, Rettie AE. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA 2002;287:1690–1698. [DOI] [PubMed] [Google Scholar]
- 2.Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet 1999;353:717–719. [DOI] [PubMed] [Google Scholar]
- 3.Loebstein R, Yonath H, Peleg D, Almog S, Rotenberg M, Lubetsky A, Roitelman J, Harats D, Halkin H, Ezra D. Interindividual variability in sensitivity to warfarin – Nature or nurture? Clin Pharmacol Ther 2001;70:159–164. [DOI] [PubMed] [Google Scholar]
- 4.Taube J, Halsall D, Baglin T. Influence of cytochrome P-450 CYP2C9 polymorphisms on warfarin sensitivity and risk of over-anticoagulation in patients on long-term treatment. Blood 2000;96:1816–1819. [PubMed] [Google Scholar]
- 5.Scordo MG, Pengo V, Spina E, Dahl ML, Gusella M, Padrini R. Influence of CYP2C9 and CYP2C19 genetic polymorphisms on warfarin maintenance dose and metabolic clearance. Clin Pharmacol Ther 2002;72:702–710. [DOI] [PubMed] [Google Scholar]
- 6.Voora D, McLeod HL, Eby C, Gage BF. Use of pharmacogenetics to guide warfarin therapy. Drugs Today (Barc) 2004;40:247–257. [DOI] [PubMed] [Google Scholar]
- 7.Hillman MA, Wilke RA, Caldwell MD, Berg RL, Glurich I, Burmester JK. Relative impact of covariates in prescribing warfarin according to CYP2C9 genotype. Pharmacogenetics 2004;14:539–547. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan-Klose TH, Ghanayem BI, Bell DA, Zhang ZY, Kaminsky LS, Shenfield GM, Miners JO, Birkett DJ, Goldstein JA. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics 1996;6:341–349. [DOI] [PubMed] [Google Scholar]
- 9.Leung AY, Chow HC, Kwong YL, Lie AK, Fung AT, Chow WH, Yip AS, Liang R. Genetic polymorphism in exon 4 of cytochrome P450 CYP2C9 may be associated with warfarin sensitivity in Chinese patients. Blood 2001;98:2584–2587. [DOI] [PubMed] [Google Scholar]
- 10.Xie HG, Prasad HC, Kim RB, Stein CM. CYP2C9 allelic variants: ethnic distribution and functional significance. Adv Drug Deliv Rev 2002;54:1257–1270. [DOI] [PubMed] [Google Scholar]
- 11.Harrison L, Johnston M, Massicotte MP, Crowther M, Moffat K, Hirsh J. Comparison of 5-mg and 10-mg loading doses in initiation of warfarin therapy. Ann Intern Med 1997;126:133–136. [DOI] [PubMed] [Google Scholar]
- 12.Ansell J, Hirsh J, Dalen J, Bussey H, Anderson D, Poller L, Jacobson A, Deykin D, Matchar D. Managing oral anticoagulant therapy. Chest. 2001;119:22S–38S. [DOI] [PubMed] [Google Scholar]
- 13.Hirsh J, Fuster V, Ansell J, Halperin JL; American Heart Association; American College of Cardiology Foundation. American Heart Association/American College of Cardiology Foundation guide to warfarin therapy. Circulation 2003;107:1692–1711. [DOI] [PubMed] [Google Scholar]
- 14.Marshfield Medical Research and Education Foundation. Assessing impact of organizational interventions – Marshfield Clinic’s Coumadin Clinic Evaluation: Final Report. Submitted to Agency for Healthcare Research and Quality (AHRQ), 13 May, 2001.
- 15.Higashi MK, Veenstra DL. Managed care in the genomics era: assessing the cost effectiveness of genetic tests. Am J Manag Care 2003;9:493–500. [PubMed] [Google Scholar]