Abstract
Oral infectious diseases, including dental caries, various forms of periodontitis and oral malodor, are not caused by a single pathogen.The etiology of these diseases is known to be associated with bacterial accumulation and plaque composition on the hard and soft tissues of the oral cavity. Therefore, the quantitative, as well as qualitative, analysis of the microorganisms present in oral biofilms, namely dental plaque, subgingival plaque and tongue debris, is important for diagnosis and rational treatment decisions.The quantitative microbial analysis of oral multi-species biofilms also provides useful information for establishing the etiology of oral infectious diseases. Recently, a 5′ fluorogenic, nuclease-based, real-time polymerase chain reaction (PCR) technique has been increasingly employed for the quantitative microbial assessment of the human oral cavity. We review the development and use of TaqMan real-time PCR for quantifying oral bacteria, its role in the diagnosis of oral infectious diseases and their microbial etiology.
Keywords: TaqMan real-time PCR, Quantification, Oral bacteria, Biofilm
The human oral cavity contains more than 500 bacterial species that interact with each other and with their host tissues.1,2 These complex interactions result in the formation of microbial biofilms as dental plaque, subgingival plaque and tongue surface debris leading to dental caries, periodontal disease and oral malodor. Recent studies have suggested that several oral bacteria are opportunistic pathogens that cause systemic disease, such as bacterial endocarditis,3–5 aspiration pneumonia,6 pre-term low birth weight7 and cardiovascular disease.8 Consequently, it is important to know the microbial composition of the oral cavity for the diagnosis and rational treatment of these diseases. Among the various procedures used to detect oral bacteria, such as microbial culture, immunological assays, enzymatic methods and molecular biology, a polymerase chain reaction (PCR)-based diagnostic technique has become a powerful and increasingly popular tool due to its rapidity, sensitivity and specificity.9 However, it is difficult to quantify accurately the number of bacteria using conventional PCR because the reactions are evaluated after gene amplification is completed.10 In addition, the quantification of PCR products can be affected by contamination, interfering substances and unequal amounts of collected samples.11 PCR is difficult to use for routine diagnosis due to the time required for sample handling and post-PCR analysis.
Recently, a real-time PCR assay using a TaqMan probe (a fluorescent DNA probe based on the 5′ to 3′ exonuclease activity of Taq polymerase) has been developed for quantitative DNA analysis.12 The oligonucleotide probe, with a reporter fluorescent dye attached to its 5′ end and a quencher dye attached to its 3′ end, hybridizes to the target gene. During PCR amplification, the quencher dye is cleaved by the 5′ nuclease activity of Taq polymerase resulting in the accumulation of reporter fluorescence. The release of the fluorescent dye during amplification allows for rapid detection and quantification of DNA.13 Compared with other methods for quantifying oral pathogens, including flow cytometry14 and DNA-DNA hybridization,15 the TaqMan real-time PCR assay has advantages in terms of sensitivity and rapidity. Furthermore, by using universal primers and probes, the total number of bacteria and the proportion of target microorganisms in single clinical specimens can be calculated. The SYBR Green I assay can also be used with another type of real-time PCR. SYBR Green I dye binds specifically to double-stranded DNA and enables the detection of products accumulating during the PCR.16 This assay has the advantage of being simple and less expensive but does not use a probe specific for the nucleotide sequence of the target microorganism. On the other hand, the high specificity of the primers and probes in TaqMan PCR enable it to distinguish target pathogens from the numerous closely related species in the oral cavity. Quantitative analysis is essential for monitoring therapeutic efficacy in clinical trials, gaining insight into oral biofilms and diagnosis of oral infectious diseases. This review describes the development and application of TaqMan PCR for determining the quantitative microbial composition of multi-species biofilms in the human oral cavity.
Development of TaqMan Real-Time PCR for the Detection of Oral Bacteria
The use of TaqMan technology for the quantitative analysis of oral bacteria is a recent phenomenon; its development is outlined in table 1 ▶. Many oligonucleotide primers and probes for target microorganisms are derived from 16S ribosomal RNA (rRNA) gene sequences, although some are based on species-specific gene sequences associated with virulence factors.17–19 As each bacterium may contain a variable number of 16S rRNA sequences, 16S rRNA-derived primers and probes may overestimate the number of target microorganisms. Primers and probes based on a single copy gene sequence would provide a more accurate measure of the absolute number of bacteria. The amplification and detection are generally carried out with the ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Foster City, CA). The oligonucleotide primers and probe sequences designed for this system can be used with other systems, such as the LightCycler (Roche Diagnostics GmbH, Penzberg, Germany) (see Authors’ Reply),20 but a new standard curve should be generated from the amplification plot for each system employed for diagnosis and monitoring.
Table 1.
Development of TaqMan real-time PCR for the detection of human oral bacteria.
| Bacterial strain | Sequence of primer or probe (5′–3′) | Target genes | Clinical specimens | Equipment | Reference |
| Organism associated with periodontitis, oral malodor, advanced caries, and pulp inflammation | |||||
| Actinobacillus actinomycetemcomitans | Forward, CAG CAT CTG CGA TCC CTG TA Reverse, TCA GCC CTT TGT CTT TCC TAG GT Probe, TCG AGT ATT CCT CAA GCA TTC TCG CAC G |
lktA | Subgingival plaque Saliva | ABI PRISM 7700 Sequence Detection Systema | 17 |
| Forward, CCC ATC GCT GGT TGG TTA Reverse, GGC ACG TAG GCG GAC C Probe, CCT CTG TAT ACG CCA TTG TAG CAC GTG TGT |
16S rRNA gene | Subgingival plaque Tongue surface debris |
LightCycler systemb | 24 | |
| Forward, ACG CAG ACG ATT GAC TGA ATT TAA Reverse, GAT CTT CAC AGC TAT ATG GCA GCT A Probe, TCA CCC TTC TAC CGT TGC CAT GGG |
lktC | Subgingival plaque | ABI PRISM 7700 Sequence Detection System | 19 | |
| Porphyromonas gingivalis | Initial forward, GGA TTA GAT ACC CTG GTA GTC Initial reverse, GGA GTA TTT AGC CTT Second forward, CAT CGG TAG TTG CTA ACA GTT TTC G Second reverse, CCG ACC TCT ACA TTA TCA G Probe, ATG ACG TCA AAT CAG CAC GGC CCT TAC AT |
16S rRNA gene and 23S rRNA gene | Subgingival plaque | ABI PRISM 7700 Sequence Detection System | 30 |
| Forward, TCG GTA AGT CAG CGG TGA AAC Reverse, GCA AGC TGC CTT CGC AAT Probe, CTC AAC GTT CAG CCT GCC GTT GAA A |
16S rRNA gene | Carious dentine | ABI PRISM 7700 Sequence Detection System | 23 | |
| Forward, TAC CCA TCG TCG CCT TGG T Reverse, CGG ACT AAA ACC GCA TAC ACT TG Probe, GCT AAT GGG ACG CAT GCC TAT CTT ACA GCT |
16S rRNA gene | Subgingival plaque Saliva |
ABI PRISM 7700 Sequence Detection System | 17 | |
| Forward, ACC TTA CCC GGG ATT GAA ATG Reverse, CAA CCA TGC AGC ACC TAC ATA GAA Probe, ATG ACT GAT GGT GAA AAC CGT CTT CCC TTC |
16S rRNA gene | Subgingival plaque Tongue surface debris |
LightCycler system | 24 | |
| Forward, CCT ACG TGT ACG GAC AGA GCT ATA Reverse, AGG ATC GCT CAG CGT AGC ATT Probe, TCG CCC GGG AAG AAC TTG TCT TCA |
Arg- gingipain gene | Subgingival plaque | ABI PRISM 7700 Sequence Detection System | 19 | |
| Porphyromonas endodontalis | Forward, GCT GCA GTC CAA CTG TAG TCT TG Reverse, TCA GTG TCA GAC GGA GCC TAG TAC Probe, CAT TCC GCA TAC CTT CGG TCT CCT CTA GC |
16S rRNA gene | Carious dentine | ABI PRISM 7700 Sequence Detection System | 23 |
| Micromonas micros | Forward, AGT GGG ATA GCC GTT GGA AA Reverse, GAC GCG AGC CCT TCT TAC AC Probe, ACC GCA TGA GAC CAC AGA ATC GCA |
16S rRNA gene | Carious dentine | ABI PRISM 7700 Sequence Detection System | 23 |
| Treponema denticola | Forward, TAA TAC CGA ATG TGC TCA TTT ACA T Reverse, CTG CCA TAT CTC TAT GTC ATT GCT CTT Probe, GAC GGG GGC CCG CAC AAG CGG |
16S rRNA gene | Subgingival plaque | iCycler iQ detection systemc | 39 |
| Forward, AGA GCA AGC TCT CCC TTA CCG T Reverse, TAA GGG CGG CTT GAA ATA ATG A Probe, CAG CGT TCG TTC TGA GCC AGG ATC A |
16S rRNA gene | Subgingival plaque Saliva |
ABI PRISM 7700 Sequence Detection System | 36 | |
| Forward, CCG AAT GTG CTC ATT TAC ATA AAG GT Reverse, GAT ACC CAT CGT TGC CTT GGT Probe, ATG GGC CCG CGT CCC ATT AGC |
16S rRNA gene | Subgingival plaque Tongue surface debris |
LightCycler system | 24 | |
| Treponema vincentii | Forward, GTC TCA ATG GTT CAT AAG AA Reverse, CAA GCC TTA TCT CTA AGA CT Probe, GAC GGG GGC CCG CAC AAG CGG |
16S rRNA gene | Subgingival plaque | iCycler iQ detection system | 39 |
| Treponema medium | Forward, CAC TCA GTG CTT CAT AAG Reverse, CCG GCC TTA TCT CTA AGA CC Probe, GAC GGG GGC CCG CAC AAG CGG GG |
16S rRNA gene | Subgingival plaque | iCycler iQ detection system | 39 |
| Tannerella forsythia | Forward, GGG TGA GTA ACG CGT ATG TAA CCT Reverse, ACC CAT CCG CAA CCA ATA AA Probe, CCC GCA ACA GAG GGA TAA CCC GG |
16S rRNA gene | Subgingival plaque | ABI PRISM 7700 Sequence Detection System | 21 |
| Forward, ATC CTG GCT CAG GAT GAA CG Reverse, TAC GCA TAC CCA TCC GCA A Probe, ATG TAA CCT GCC CGC AAC AGA GGG ATA AC |
16S rRNA gene | Subgingival plaque | ABI PRISM 7700 Sequence Detection System | 22 | |
| Forward, AGC GAT GGT AGC AAT ACC TGT C Reverse, TTC GCC GGG TTA TCC CTC Probe, TGA GTA ACG CGT ATG TAA CCT GCC CGC |
16S rRNA gene | Subgingival plaque Tongue surface debris |
LightCycler system | 24 | |
| Forward, TCC CAA AGA CGC GGA TAT CA Reverse, ACG GTC GCG ATG TCA TTG T Probe, CCG CGA CGT GAA ATG GTA TTC CTC |
BspA antigen gene | Subgingival plaque | ABI PRISM 7700 Sequence Detection System | 19 | |
| Fusobacterium spp. | Forward, CGC AGA AGG TGA AAG TCC TGT AT Reverse, TGG TCC TCA CTG ATT CAC ACA GA Probe, ACT TTG CTC CCA AGT AAC ATG GAA CAC GAG |
16S rRNA gene | Subgingival plaque | ABI PRISM 7700 Sequence Detection System | 22 |
| Forward, AAG CGC GTC TAG GTG GTT ATG T Reverse, TGT AGT TCC GCT TAC CTC TCC AG Probe, CAA CGC AAT ACA GAG TTG AGC CCT GCA TT |
16S rRNA gene | Carious dentine | ABI PRISM 7700 Sequence Detection System | 23 | |
| Prevotella spp. | Forward, CCA GCC AAG TAG CGT GCA Reverse, TGG ACC TTC CGT ATT ACC GC Probe, AAT AAG GAC CGG CTA ATT CCG TGC CAG |
16S rRNA gene | Carious dentine | ABI PRISM 7700 Sequence Detection System | 23 |
| Prevotella melaninogenica | Forward, GTG GGA TAA CCT GCC GAA AG Reverse, CCC ATC CAT TAC CGA TAA ATC TTT A Probe, CAA ATC TGA TGC CGT CAT CGA AGA CTA TGC |
16S rRNA gene | Carious dentine | ABI PRISM 7700 Sequence Detection System | 23 |
| Prevotella intermedia | Forward, TCC ACC GAT GAA TCT TTG GTC Reverse, ATC CAA CCT TCC CTC CAC TC Probe, CGT CAG ATG CCA TAT GTG GAC AAC ATC G |
16S rRNA gene | Subgingival plaque Tongue surface debris |
LightCycler system | 24 |
| Prevotella nigrescens | Forward, CCG TTG AAA GAC GGC CTA A Reverse, CCC ATC CCT TAC CGG RA Probe, CCC GAT GTG TTT CAT TGA CGG CAT C |
16S rRNA gene | Subgingival plaque Tongue surface debris |
LightCycler system | 24 |
| Cariogenic pathogen | |||||
| Streptococcus mutans | Forward, GCC TAC AGC TCA GAG ATG CTA TTC T Reverse, GCC ATA CAC CAC TCA TGA ATT GA Probe, TGG AAA TGA CGG TCG CCG TTA TGA A |
gtfB | Dental plaque Saliva |
ABI PRISM 7700 Sequence Detection System | 18 |
| Streptococcus sobrinus | Forward, TTC AAA GCC AAG ACC AAG CTA GT Reverse, CCA GCC TGA GAT TCA GCT TGT Probe, CCT GCT CCA GCG ACA AAG GCA GC |
gtfT | Dental plaque Saliva |
ABI PRISM 7700 Sequence Detection System | 18 |
| Early microbial colonizer in human dental biofilm | |||||
| Streptococcus gordonii | Forward, GGT GTT GTT TGA CCC GTT CAG Reverse, AGT CCA TCC CAC GAG CAC AG Probe, AAC CTT GAC CCG CTC ATT ACC AGC TAG TAT G |
The gene encoding putative type I site- specific DNase | Dental plaque | ABI PRISM 7700 Sequence Detection System | 58 |
| Streptococcus mitis | Forward, GAG TCC TGC ATC AGC CAA GAG Reverse, GGA TCC ACC TTT TCT GCT TGA C Probe, TGT TCC CAA GTG GAG CCA ACC AAA CT |
The gene encoding putative Glycine-rich protein | Dental plaque | ABI PRISM 7700 Sequence Detection System | 58 |
| Actinomyces naeslundii | Forward, TCG AAA CTC AGC AAG TAG CCG Reverse, AGA GGA GGG CCA CAA AAG AAA Probe, GGG TAC TCT AGT CCA AAC TGG CGG ATA GCG |
The gene encoding unknown protein | Dental plaque | ABI PRISM 7700 Sequence Detection System | 58 |
| Actinomyces viscosus | Forward, ATG TGG GTC TGA CCT GCT GC Reverse, CAA AGT CGA TCA CGC TCC G Probe, ACG GAG GTC GGG AAC GGT GGA AG |
The gene encoding unknown protein | Dental plaque | ABI PRISM 7700 Sequence Detection System | 58 |
| Universal | |||||
| Forward, GGA TTA GAT ACC CTG GTA GTC Initial reverse, GGA GTA TTT AGC CTT Second reverse, TAC CTT GTT ACG ACT T Probe, TGA CGG GCG GTG TGT ACA AGG C |
16S rRNA gene and 23S rRNA gene | ABI PRISM 7700 Sequence Detection System | 30 | ||
| Forward, CCA TGA AGT CGG AAT CGC TAG Reverse, GCT TGA CGG GCG GTG T Probe, TAC AAG GCC CGG GAA CGT ATT CAC CG |
16S rRNA gene | ABI PRISM 7700 Sequence Detection System | 21 | ||
| Forward, TCC TAC GGG AGG CAG CAG T Reverse, GGA CTA CCA GGG TAT CTA ATC CTG TT Probe, CGT ATT ACC GCG GCT GCT GGC AC |
16S rRNA gene | ABI PRISM 7700 Sequence Detection System | 49 | ||
| Forward, CGC TAG TAA TCG TGG ATC AGA ATG Reverse, TGT GAC GGG CGG TGT GTA Probe, CAC GGT GAA TAC GTT CCC GGG C |
16S rRNA gene | ABI PRISM 7700 Sequence Detection System | 17 | ||
a Applied Biosystems, Foster City, CA
b Roche Diagnostics GmbH, Penzberg, Germany
c Bio-Rad Laboratories Inc., Hercules, CA
Specimens are taken from the regions associated with infectious disease: supragingival dental plaque with dental caries,18 subgingival plaque with periodontitis,17,19,21,22 carious dentine with infected pulp or chronic caries23 and tongue surface debris with oral malodor.24,25 In combination with such specimens, we normally use saliva as a PCR template that reflects the overall condition of the oral cavity. In fact, bacteria causing dental caries and periodontitis are more frequently detected in saliva than in the dental plaque.17,18,26
Detection of the Relative Amount of the Target DNA in Clinical Specimens
Porphyromonas gingivalis, a black-pigmented, gram-negative, anaerobic rod, is considered to be the most important pathogen in adult-onset periodontitis27,28 and oral malodor.29 Lyons et al.30 developed a nested, two-step, real-time PCR for comparing the number of P. gingivalis cells with that of the total bacterial cells in subgingival dental plaque. The first amplification targeted conserved sequences in the bacterial 16S and 23S rRNA genes. In the second stage, nested PCRs were used to quantify the number of P. gingivalis cells or the total number of bacterial cells. With previous methods it has been especially difficult to determine the total number of bacteria. This study showed that real-time PCR offers the prospect of determining the number of P. gingivalis and the total number of bacteria in plaque samples directly without culturing. To measure the relative amount of P. gingivalis and Actinobacillus actinomycetemcomitans in subgingival dental plaque and saliva samples, we normalized the number of target DNA copies to that of the 16S rRNA gene using a simplification of the comparative threshold cycle (ΔΔCt) method that was provided in User Bulletin #2 of ABI PRISM 7700 Sequence Detection System by Applied Biosystems.17 The critical threshold cycle (Ct) is defined as the first cycle in which fluorescence is detectable above the background and is inversely proportional to the logarithm of the initial number of template molecules. The fold-difference (N) in the number of the target organism-specific gene copies relative to the number of 16S rRNA gene copies was determined as follows:
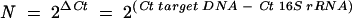 |
The ΔCt values for the sample and the 16S rRNA were determined by subtracting the average Ct value for the target gene from that of the 16S rRNA gene. The simplified ΔΔCt method provides the required simplicity and rapidity for clinical diagnosis.
Using TaqMan real-time PCR and the simplified ΔΔCt method, we performed quantitative analysis of P. gingivalis to clarify the relationship between the bacterial numbers and the periodontal status.31 Before periodontal treatment, a significant positive correlation between the log-translated number of P. gingivalis and pocket depth was found (y=1.14x − 2.01, r2=0.37, p<0.0001). This indicated that the number of P. gingivalis cells increased approximately 10-fold for each 1 mm increase in pocket depth. Simple regression analysis of the proportion of P. gingivalis bacteria in relation to pocket depth also showed a positive correlation (y=4.69x − 13.4, r2=0.40, p<0.0001), which indicated that the percentage of P. gingivalis cells to the total bacterial cells in subgingival plaque specimens increased 4.69% for each 1 mm increase in pocket depth. Subsequently, changes in pocket depth and the number of P. gingivalis bacteria before and after treatment were monitored. There were significant reductions in both the absolute number (Wilcoxon’s signed-rank test, p<0.01) and the percentage (p<0.001) of P. gingivalis cells before and after periodontal treatment. As there was a significant difference (paired t-test, p<0.0001) in pocket depth before and after treatment, these results suggest that the absolute and relative numbers of P. gingivalis bacteria are closely related to periodontal status and that quantitative analysis of this bacterium is important for evaluating periodontal therapy.
Development of TaqMan Assay for Quantifying Oral Treponemes
In addition to P. gingivalis, oral treponemes have been implicated in periodontitis32,33 and oral malodor.34 These helically shaped spirochetes are present in significantly elevated numbers in plaque samples from deep-pocket sites of patients with severe periodontitis.35 Most oral spirochetes either cannot yet be grown or are extremely difficult to grow in culture because they have lost the ability to synthesize many essential molecules that they normally obtain from their host.2 Real-time PCR is a valuable tool for detection of such organisms that are difficult to grow in culture. This assay is able to detect and quantify the target organisms directly in oral specimens.
Using TaqMan real-time PCR, we evaluated the absolute and relative numbers of Treponema denticola bacteria in 53 subgingival dental plaque samples from nine patients prior to periodontal treatment.36 Although T. denticola is the most-studied spirochete due to its virulence in periodontal disease and is one of the predominant oral treponemes found in the gingival crevice, it is difficult to grow in culture.37,38 Simple regression analysis showed a significant positive correlation between the log-transformed number of T. denticola cells and pocket depth (y=0.61x + 1.51, r2=0.23, p<0.0001). A significant positive correlation was also observed between the percentage of T. denticola bacteria and pocket depth (y=4.90x − 12.60, r2=0.28, p<0.0001). Asai et al.39 developed a TaqMan real-time PCR assay to detect the absolute number of three oral treponemes, T. denticola, Treponema vincentii and Treponema medium in subgingival plaque. They found that T. denticola and T. medium were detected in patients with serious periodontal diseases (pocket depths 4 mm to 8 mm), whereas T. vincentii was detected mainly in shallow pockets of 4 mm to 5 mm depth.
Previously, dark-field microscopy was used to establish the relationship between number of subgingival spirochetes and severity of periodontitis.40 Dark-field microscopy is suitable for determining the overall number of spirochetes but is unsuitable for identifying specific spirochete species in clinical specimens due to similarities in shape. These reports demonstrate that TaqMan real-time PCR is suitable for detection and quantification of oral treponemes in cases where identification using culture and visual observation is difficult. In addition, TaqMan PCR is able to differentiate between spirochete species and, therefore, may be useful in studying the etiological role of different oral spirochetes at each stage of periodontitis.
Quantitative Studies Showing the Relationship Between Periodontal Bacteria
Periodontitis is thought to arise from complex microflora consisting of putative periodontopathic bacteria, such as P. gingivalis, T. denticola, Tannerella forsythia (formerly Bacteroides forsythus or Tannerella forsythensis)41 and Fusobacterium spp.42,43 These bacteria have been co-isolated from affected periodontal sites,44,45 and it has been suggested that periodontitis is initiated by cooperation between the different species. We established a TaqMan real-time PCR assay to quantify the absolute and relative numbers of T. forsythia and Fusobacterium spp. in clinical specimens and to demonstrate their co-localization in periodontal pockets.22 Spearman’s rank correlation showed a significant positive correlation between the percentage of each bacterium and the pocket depth of each sample (r=0.48 for T. forsythia, and r=0.50 for Fusobacterium spp., p<0.001). There was a significant correlation between the proportions of both bacteria in each pocket (r=0.75, p<0.001). Between-subject analysis was performed to avoid cross-contamination of sites; the correlation between the mean proportions of these two bacteria for each subject was likewise very strong (r=0.88, p<0.001). The significant relationship between the two bacteria and pocket depth suggests that both species grow well in deeper periodontal pockets. Furthermore, when Fusobacterium spp. and T. forsythia were quantified in samples from periodontal pockets of different depths, both bacteria were detected more frequently in samples from periodontal pockets ≥4 mm (χ2 test, p<0.001). T. forsythia was not detected in subgingival plaque, from where Fusobacterium spp. were absent (Fisher’s exact probability test, p<0.001) or in samples from shallow pockets ≤3 mm depth. In fact, T. forsythia was not detected at any Fusobacterium-negative site (p=0.03) suggesting co-localization between T. forsythia and Fusobacterium spp. in deep periodontal pockets. These preliminary tests show that real-time PCR may be used to clarify the relationship between oral periodontopathic bacteria in periodontal pockets.
TaqMan Assay for Quantification of Periodontal Bacteria Associated With Oral Malodor
Oral malodor is mostly caused by oral pathology, mainly from malodorous compounds produced by oral bacteria. Major compounds responsible for oral malodor are volatile sulfur compounds (VSCs), including hydrogen sulfide (H2S), methyl mercaptan (CH3SH) and dimethyl sulfide (CH3SCH3).46 About 90% of the VSCs in the oral cavity are H2S and CH3SH24 and some periodontal pathogens such as P. gingivalis, Prevotella intermedia, T. denticola and Fusobacterium nucleatum generate significant amounts of H2S and CH3SH from L-cysteine and L-methionine.47 Consequently, there has been renewed interest in the relationship between oral malodor and periodontal disease. On the other hand, it has been reported that the main source of odor is usually the tongue microbiota.48 Tanaka et al.25 employed TaqMan real-time PCR to measure the number of anaerobes in tongue biofilm samples and examined the association between oral malodor and tongue periodontal pathogens including P. gingivalis, P. intermedia, Prevotella nigrescens, T. denticola and T. forsythia. Species-specific primer and probe sets were designed from the variable regions of the 16S rRNA gene sequences24 and a universal primer and probe set was used for quantifying the total proportion of bacterial cells in specimens.49 Among the VSCs, H2S concentration was strongly correlated with the amount of P. intermedia and P. nigrescens bacteria, whereas CH3SH concentration was strongly correlated with P. gingivalis and P. nigrescens (Spearman’s rank correlation coefficient, p<0.0001). The total proportion of these five anaerobes on the tongue dorsa was strongly correlated with total VSC level (r=0.88), but only weakly correlated with the degree of organolepsis (r=0.29) suggesting that these periodontal pathogens on the tongue dorsa contribute greatly to VSC production. Furthermore, the proportions of P. gingivalis, P. nigrescens and T. forsythia and the total of the five anaerobes on the tongue dorsa were higher in patients with periodontitis than in those without periodontitis. Recently, the importance of oral malodor has been recognized as it carries considerable social stigma in our modern society. Quantification of VSC-producing bacteria is important for diagnosis and therapeutic assessment of oral malodor. Conventional tests, gas chromatography, organoleptic tests and portable sulfide monitors are essential tools for evaluating oral malodor, however, these tests cannot measure any direct correlation between the disease and pathogenic bacteria. In addition to these conventional tools, real-time PCR will support diagnosis of oral malodor pathogens and contribute to control of VSC production.
Quantification of Cariogenic Pathogens and Early Microbial Colonizers in Dental Biofilms
Streptococcus mutans (serotype c, e and f mutans streptococci) and Streptococcus sobrinus (serotype d and g mutans streptococci) are directly involved in formation of human dental caries.50,51 The water-insoluble forms of glucan produced from dietary sucrose by S. mutans and S. sobrinus are closely associated with pathogenicity. These glucose polymers provide a structure for aggregation of mutans and other oral streptococci and modify dental biofilm porosity, thus increasing availability of nutrients for continued bacterial metabolism. Consequently, the levels of these cariogenic pathogens in the human oral cavity are predictive of caries activity and the transmission risk of mutans streptococci. Although various methods have been developed to identify mutans streptococci, a quantitative assay as an index of caries activity has not yet been established. We have described a TaqMan real-time PCR assay for quantitative detection of S. mutans and S. sobrinus in saliva and dental plaque.18 The S. mutans- and S. sobrinus-specific primers and probes were derived from the gtfB52 and gtfT53 genes, which encode glucosyltransferases involved in the synthesis of insoluble forms of glucan. The limits of detection and quantification by real-time PCR were linear over the range 1.7× 101 to 1.7×107 colony forming units (CFU) for S. mutans and 1.1× 101 to 1.1× 106 CFU per reaction mixture for S. sobrinus. Using this assay, the numbers of S. mutans and S. sobrinus in saliva and dental plaque were examined in 10 patients. The absolute and relative numbers of these bacteria have been shown to vary between saliva and dental plaque. Both bacteria were detected more frequently in saliva: 10 in saliva and 8 in dental plaque for S. mutans and 7 in saliva and 5 in dental plaque for S. sobrinus. Our results suggest that saliva may be suitable for estimating individual caries risk, but the localized caries risk may be closely dependent on the microbial composition of dental plaque. Furthermore, based on the notion that dental plaque is a complex microbial community growing as a biofilm on enamel surfaces, the most recent evidence suggests that dental caries have a multi-bacterial etiology and, therefore, it is important to gain insight into the total bacterial composition of dental plaque.
During dental biofilm formation, Actinomyces spp. and several Streptococcus spp., including Streptococcus mitis, Streptococcus oralis, Streptococcus sanguinis and Streptococcus gordonii, adhere to the acquired pellicle on the tooth surface and are the predominant colonizers in the very early stages.54 Different specific intra- and inter-generic co-aggregations between these primary colonizers and secondary colonizers have been demonstrated,55–57 and it appears that early colonizers of dental plaque play an important role in progressive biofilm formation and the characterization of mature dental plaque. However, there have been no reports of any specific relationship between mutans streptococci and other oral bacteria. Quantitative analysis is required not only for accurate diagnosis of caries risk but also for studying biofilm formation and etiology. We developed a real-time PCR technique for quantitative detection of four early colonizer microorganisms in dental biofilms: Actinomyces naeslundii, Actinomyces viscosus, S. mitis and S. gordonii.58 Using TaqMan PCR, we calculated the numbers of these four early colonizers and two cariogenic bacteria, S. mutans and S. sobrinus, in dental plaque. Specimens were obtained from the buccal side of the upper first molar in 5 individuals who had refrained from brushing for 12 hours. Large numbers (≥103 copies) of S. mitis were detected in all samples, while S. gordonii and A. viscosus were minor species in terms of prevalence (2 for S. gordonii and 1 for A. viscosus) and number (theoretical data below the detection limits except for 1 specimen). The frequency and number of S. mutans, S. sobrinus and A. naeslundii differed among individuals. As TaqMan PCR can monitor bacterial numbers in biofilms, it can be used to clarify the behavior of oral bacteria in dental biofilm formation. Thus, it may play a role in future biofilm research.
Designing Species-Specific Primers and TaqMan Probes Using Genomic Subtractive Hybridization
The key to the success of the TaqMan method is designing specific primers and probes based on the nucleotide sequence of the target microorganism. Genes encoding species-specific proteins or 16S rRNA are generally used as targets for real-time PCR.18,23 However, it is difficult to isolate specific nucleotide sequences from the genes of closely related species. The classification of the viridans group streptococci, including S. mitis, S. oralis and S. gordonii, has long been considered difficult. Indeed, on the basis of 16S rRNA sequence, S. mitis and S. oralis share >99% sequence identity with each other and ~97% sequence identity with S. gordonii.59 Although the DNA–DNA hybridization between strains belonging to the mitis group showed that the DNA–DNA similarity values for the entire chromosome are estimated to be <60%,59 it is difficult to identify species-specific DNA sequences because much of the genome project for these bacteria has not yet been finished. Actinomyces spp. also shows extensive phenotypic and serologic variations.60 Most of the former A. viscosus have been classified as A. naeslundii genospecies 2.61 We employed genomic subtractive hybridization to identify genomic differences between the two closely related strains of Streptococcus or Actinomyces spp. and designed species-specific primers and probes for TaqMan real-time PCR.58 Subtractive hybridization was initially developed to identify differences in cDNA pools,62 but it has also been successfully used to identify genomic differences between closely related strains of Helicobacter pylori,63 Mycobacterium tuberculosis64 and Neisseria meningitidis.65 Chromosomal DNA of S. oralis ATCC 10557 was used as driver DNA for S. gordonii DL1 and S. mitis 903, and chromosomal DNA of A. naeslundii ATCC 51655 and A. viscosus ATCC 43146 were used as driver DNAs for each other. The oligonucleotide adaptors used in the first subtractive hybridization were 5′-GATCCTCGGTGA-3′ (RBam12) and 5′-AGCACTCTCCAGCCTCTCACCGAG-3′ (RBam24), and the second adaptors were 5′-GATCCGTTCATG-3′ (JBam12) and 5′-ACCGACGTCGACTATCCATGAACG-3′(JBam24). The amplification products of the second-round PCR, which appeared as a smear of products <700 bp upon electrophoresis with ethidium bromide, were digested with Sau3AI cloned into BamHI-digested pBluescript II SK+ (Stratagene, La Jolla, CA) and used to transform Escherichia coli DH5α (TAKARA Bio Company, Shiga, Japan). Eight colonies were selected randomly from each cDNA bank containing 200 colonies and the nucleotide sequences were determined using an ABI PRISM 310 genomic analyzer (Applied Biosystems, Foster City, CA). A total of 32 fragments of 150 bp to 550 bp contained diverse sequences. Sequence homology between the subtracted fragments and those in the GenBank database was determined using BLAST66 by means of the WWW server at the National Center for Biotechnology Information (Bethesda, MD; http://www.ncbi.nlm.nih.gov/BLAST). A nucleotide database search (blastn) revealed that 21 insertions have no significant sequence homology to any known DNA sequences in GenBank, and the nucleotide sequences of 7 fragments were detected in the driver bacterial DNA. Three insertions are homologous to 16S rRNA genes of oral bacteria and another 8 insertions share homology with genes encoding unknown proteins. The S. gordonii-, S. mitis-, A. naeslundii-and A. viscosus-specific primers and TaqMan probes derived from the subtractive DNA fragments are listed in table 1 ▶. The blastx similarities of the protein-encoding region in the fragments showed expected values of >0.1, except for the S. gordonii sequence (E=7e−37). This technique is useful for designing species-specific primers and probes for real-time PCR, especially for organisms for which whole genome sequence information is not yet available.
Conclusion
TaqMan real-time PCR has the advantage of providing rapid, accurate, sensitive and quantitative detection. It allows the continuous monitoring of DNA amplification eliminating the need for gel electrophoresis. Its rapidity and ease of handling are very important for the diagnosis of infections requiring timely identification of bacterial pathogens, such as bacterial meningitis, septicemia and communicable respiratory diseases.67,68 Oral infectious diseases are not caused by a single critical pathogen. The etiologies of dental caries, periodontal diseases and oral malodor are associated with bacterial accumulation and plaque composition. Therefore, the key for diagnosis of oral infections is to establish the microbial composition and the number of opportunistic pathogens present in oral biofilms rather than the rapid identification of the microorganisms concerned. TaqMan real-time PCR is an excellent method for the quantitative detection of oral bacteria and for the determination of relative numbers of different bacteria. Using genomic subtractive hybridization, it is easy to design primers and TaqMan probes for target microorganisms by directly subtracting the chromosomal DNA of a related strain from that of the target bacterium.
The human oral cavity contains more than 500 species of bacteria that colonize host tissues and co-aggregate with one another and other oral microorganisms.1 Following the proposal that the specific adhesion between different bacterial species contributes to the development of dental plaque,69 the microbial composition of dental plaque and the co-aggregation of the dominant organisms in biofilm formation have been extensively studied.2,57,70–73 Various procedures have been used to study these bacterial interactions, including laser confocal microscopy, spectrophotometry, co-aggregation assays, bacterial culture, molecular biology and microcalorimetry.57,71,74–76 These in vitro studies have shed light on many aspects of specific interactions between oral bacteria in dental biofilm. TaqMan real-time PCR can be used for direct target quantification without culturing and is useful for detecting species that are extremely difficult to grow in culture. This technique allows the visualization of the microbial ecosystem in the oral cavity and will likely be increasingly employed in future biofilm research.
Grant Support: Research fellowships from the Japan Society for the Promotion of Science for Young Scientists (N.S.), a research grant from the Fukushima Society for the Promotion of Medicine (N.S.), a research grant from the Nakatomi Foundation (A.Y.), and a research grant from the Clinical Research Foundation (A.Y.).
References
- 1.Kroes I, Lepp PW, Relman DA Bacterial diversity within the human subgingival crevice. Proc Natl Acad Sci USA 1999;96:14547–14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. J Bacteriol 2001;183:3770–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Douglas CW, Heath J, Hampton KK, Preston FE. Identity of viridans streptococci isolated from cases of infective endocarditis. J Med Microbiol 1993;39:179–182. [DOI] [PubMed] [Google Scholar]
- 4.Tao L, Herzberg MC. Identifying in vivo expressed streptococcal genes in endocarditis. Methods Enzymol 1999;310:109–116. [DOI] [PubMed] [Google Scholar]
- 5.Berbari EF, Cockerill FR 3rd, Steckelberg JM. Infective endocarditis due to unusual or fastidious microorganisms. Mayo Clin Proc 1997;72:532–542. [DOI] [PubMed] [Google Scholar]
- 6.Scannapieco FA. Role of oral bacteria in respiratory infection. J Periodontol 1999;70:793–802. [DOI] [PubMed] [Google Scholar]
- 7.Offenbacher S, Jared HL, O’Reilly PG, Wells SR, Salvi GE, Lawrence HP, Socransky SS, Beck JD. Potential pathogenic mechanisms of periodontitis associated pregnancy complications. Ann Periodontol 1998;3:233–250. [DOI] [PubMed] [Google Scholar]
- 8.Genco R, Offenbacher S, Beck J. Periodontal disease and cardiovascular disease: epidemiology and possible mechanisms. J Am Dent Assoc 2002;133(Suppl):14S–22S. [DOI] [PubMed] [Google Scholar]
- 9.Rodu B. The polymerase chain reaction: the revolution within. Am J Med Sci 1990;299:210–216. [DOI] [PubMed] [Google Scholar]
- 10.Kainz P. The PCR plateau phase - towards an understanding of its limitations. Biochim Biophys Acta 2000;1494:23–27. [DOI] [PubMed] [Google Scholar]
- 11.Wilson IG. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol 1997;63:3741–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA 1991;88:7276–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res 1996;6:986–994. [DOI] [PubMed] [Google Scholar]
- 14.Veal DA, Deere D, Ferrari B, Piper J, Attfield PV. Fluorescence staining and flow cytometry for monitoring microbial cells. J Immunol Methods 2000;243:191–210. [DOI] [PubMed] [Google Scholar]
- 15.Socransky SS, Smith C, Martin L, Paster BJ, Dewhirst FE, Levin AE. “Checkerboard” DNA-DNA hybridization. Biotechniques 1994;17:788–792. [PubMed] [Google Scholar]
- 16.Morrison TB, Ma Y, Weis JH, Weis JJ. Rapid and sensitive quantification of Borrelia burgdorferi-infected mouse tissues by continuous fluorescent monitoring of PCR. J Clin Microbiol 1999;37:987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida A, Suzuki N, Nakano Y, Oho T, Kawada M, Koga T. Development of a 5′ fluorogenic nuclease-based real-time PCR assay for quantitative detection of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. J Clin Microbiol 2003;41:863–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshida A, Suzuki N, Nakano Y, Kawada M, Oho T, Koga T. Development of a 5′ nuclease-based real-time PCR assay for quantitative detection of cariogenic dental pathogens Streptococcus mutans and Streptococcus sobrinus. J Clin Microbiol 2003;41:4438–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morillo JM, Lau L, Sanz M, Herrera D, Martin C, Silva A. Quantitative real-time polymerase chain reaction based on single copy gene sequence for detection of periodontal pathogens. J Clin Periodontol 2004;31:1054–1060. [DOI] [PubMed] [Google Scholar]
- 20.Boutaga K, van Winkelhoff AJ, Vandenbroucke-Grauls CM, Savelkoul PH. Considerations in evaluating the applicability of universal detection of oral pathogens. J Clin Microbiol 2004;42:4414; author reply 4414–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shelburne CE, Prabhu A, Gleason RM, Mullally BH, Coulter WA. Quantitation of Bacteroides forsythus in subgingival plaque comparison of immunoassay and quantitative polymerase chain reaction. J Microbiol Methods 2000;39:97–107. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki N, Yoshida A, Saito T, Kawada M, Nakano Y. Quantitative microbiological study of subgingival plaque by real-time PCR shows correlation between levels of Tannerella forsythensis and Fusobacterium spp. J Clin Microbiol 2004;42:2255–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin FE, Nadkarni MA, Jacques NA, Hunter N. Quantitative microbiological study of human carious dentine by culture and real-time PCR: association of anaerobes with histopathological changes in chronic pulpitis. J Clin Microbiol 2002;40:1698–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuboniwa M, Amano A, Kimura KR, Sekine S, Kato S, Yamamoto Y, Okahashi N, Iida T, Shizukuishi S. Quantitative detection of periodontal pathogens using real-time polymerase chain reaction with TaqMan probes. Oral Microbiol Immunol 2004;19:168–176. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka M, Yamamoto Y, Kuboniwa M, Nonaka A, Nishida N, Maeda K, Kataoka K, Nagata H, Shizukuishi S. Contribution of periodontal pathogens on tongue dorsa analyzed with real-time PCR to oral malodor. Microbes Infect 2004;6:1078–1083. [DOI] [PubMed] [Google Scholar]
- 26.Sakamoto M, Takeuchi Y, Umeda M, Ishikawa I, Benno Y. Rapid detection and quantification of five periodontopathic bacteria by real-time PCR. Microbiol Immunol 2001;45:39–44. [DOI] [PubMed] [Google Scholar]
- 27.Zambon JJ, Reynolds HS, Slots J. Black-pigmented Bacteroides spp. in the human oral cavity. Infect Immun 1981;32:198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slots J, Genco RJ. Black-pigmented Bacteroides species, Capnocytophaga species, and Actinobacillus actinomycetemcomitans in human periodontal disease: virulence factors in colonization, survival, and tissue destruction. J Dent Res 1984;63:412–421. [DOI] [PubMed] [Google Scholar]
- 29.Nakano Y, Yoshimura M, Koga T. Correlation between oral malodor and periodontal bacteria. Microbes Infect 2002;4:679–683. [DOI] [PubMed] [Google Scholar]
- 30.Lyons SR, Griffen AL, Leys EJ. Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J Clin Microbiol 2000;38:2362–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawada M, Yoshida A, Suzuki N, Nakano Y, Saito T, Oho T, Koga T. Prevalence of Porphyromonas gingivalis in relation to periodontal status assessed by real-time PCR. Oral Microbiol Immunol 2004;19:289–292. [DOI] [PubMed] [Google Scholar]
- 32.Riviere GR, Smith KS, Carranza N Jr, Tzagaroulaki E, Kay SL, Dock. Subgingival distribution of Treponema denticola, Treponema socranskii, and pathogen-related oral spirochetes: prevalence and relationship to periodontal status of sampled sites. J Periodontol 1995;66:829–837. [DOI] [PubMed] [Google Scholar]
- 33.Takeuchi Y, Umeda M, Sakamoto M, Benno Y, Huang Y, Ishikawa I. Treponema socranskii, Treponema denticola, and Porphyromonas gingivalis are associated with severity of periodontal tissue destruction. J Periodontol 2001;72:1354–1363. [DOI] [PubMed] [Google Scholar]
- 34.Kazor CE, Mitchell PM, Lee AM, Stokes LN, Loesche WJ, Dewhirst FE, Paster BJ. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J Clin Microbiol 2003;41:558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simonson LG, Goodman CH, Bial JJ, Morton HE. Quantitative relationship of Treponema denticola to severity of periodontal disease. Infect Immun 1988;56:726–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshida A, Kawada M, Suzuki N, Nakano Y, Oho T, Saito T, Yamashita Y. TaqMan real-time polymerase chain reaction assay for the correlation of Treponema denticola numbers with the severity of periodontal disease. Oral Microbiol Immunol 2004;19:196–200. [DOI] [PubMed] [Google Scholar]
- 37.Choi BK, Lee HJ, Kang JH, Jeong GJ, Min CK, Yoo YJ. Induction of osteoclastogenesis and matrix metalloproteinase expression by the lipooligosaccharide of Treponema denticola. Infect Immun 2003;71:226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishihara K, Kuramitsu HK, Okuda K. A 43-kDa protein of Treponema denticola is essential for dentilisin activity. FEMS Microbiol Lett 2004;232:181–188. [DOI] [PubMed] [Google Scholar]
- 39.Asai Y, Jinno T, Igarashi H, Ohyama Y, Ogawa T. Detection and quantification of oral treponemes in subgingival plaque by real-time PCR. J Clin Microbiol 2002;40:3334–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Armitage GC, Dickinson WR, Jenderseck RS, Levine SM, Chambers DW. Relationship between the percentage of subgingival spirochetes and the severity of periodontal disease. J Periodontol 1982;53:550–556. [DOI] [PubMed] [Google Scholar]
- 41.Maiden MF, Cohee P, Tanner AC. Proposal to conserve the adjectival form of the specific epithet in the reclassification of Bacteroides forsythus Tanner et al. 1986 to the genus Tannerella Sakamoto et al. 2002 as Tannerella forsythia corrig., gen. nov., comb. nov. Request for an Opinion. Int J Syst Evol Microbiol 2003;53:2111–2112. [DOI] [PubMed] [Google Scholar]
- 42.Takamatsu N, Yano K, He T, Umeda M, Ishikawa I. Effect of initial periodontal therapy on the frequency of detecting Bacteroides forsythus, Porphyromonas gingivalis, and Actinobacillus actinomycetemcomitans. J Periodontol 1999;70:574–580. [DOI] [PubMed] [Google Scholar]
- 43.Haffajee AD, Socransky SS. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000 1994;5:78–111. [DOI] [PubMed] [Google Scholar]
- 44.Simonson LG, McMahon KT, Childers DW, Morton HE. Bacterial synergy of Treponema denticola and Porphyromonas gingivalis in a multinational population. Oral Microbiol Immunol 1992;7:111–112. [DOI] [PubMed] [Google Scholar]
- 45.Socransky SS, Haffajee AD, Dzink JL, Hillman JD. Associations between microbial species in subgingival plaque samples. Oral Microbiol Immunol 1988;3:1–7. [DOI] [PubMed] [Google Scholar]
- 46.Kleinberg I, Westbay G. Oral malodor. Crit Rev Oral Biol Med 1990;1:247–259. [DOI] [PubMed] [Google Scholar]
- 47.Persson S, Edlund MB, Claesson R, Carlsson J. The formation of hydrogen sulfide and methyl mercaptan by oral bacteria. Oral Microbiol Immunol 1990;5:195–201. [DOI] [PubMed] [Google Scholar]
- 48.Rosenberg M. Clinical assessment of bad breath: current concepts. J Am Dent Assoc 1996;127:475–482. [DOI] [PubMed] [Google Scholar]
- 49.Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 2002;148:257–266. [DOI] [PubMed] [Google Scholar]
- 50.Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev 1986;50:353–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koga T, Oho T, Shimazaki Y, Nakano Y. Immunization against dental caries. Vaccine 2002;20:2027–2044. [DOI] [PubMed] [Google Scholar]
- 52.Shiroza T, Ueda S, Kuramitsu HK. Sequence analysis of the gtfB gene from Streptococcus mutans. J Bacteriol 1987;169:4263–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hanada N, Yamashita Y, Shibata Y, Sato S, Katayama T, Takehara T, Inoue M. Cloning of a Streptococcus sobrinus gtf gene that encodes a glucosyltransferase which produces a high-molecular-weight water-soluble glucan. Infect Immun 1991;59:3434–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li J, Helmerhorst EJ, Leone CW, Troxler RF, Yaskell T, Haffajee AD, Socransky SS, Oppenheim FG. Identification of early microbial colonizers in human dental biofilm. J Appl Microbiol 2004;97:1311–1318. [DOI] [PubMed] [Google Scholar]
- 55.Kolenbrander PE, Andersen RN. Characterization of Streptococcus gordonii (S. sanguis) PK488 adhesin-mediated coaggregation with Actinomyces naeslundii PK606. Infect Immun 1990;58:3064–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamont RJ, El-Sabaeny A, Park Y, Cook GS, Costerton JW, Demuth DR. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 2002;148:1627–1636. [DOI] [PubMed] [Google Scholar]
- 57.Maeda K, Nagata H, Yamamoto Y, Tanaka M, Tanaka J, Minamino N, Shizukuishi S. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus oralis functions as a coadhesin for Porphyromonas gingivalis major fimbriae. Infect Immun 2004;72:1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suzuki N, Nakano Y, Yoshida A, Yamashita Y, Kiyoura Y. Real-time TaqMan PCR for quantifying oral bacteria during biofilm formation. J Clin Microbiol 2004;42:3827–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawamura Y, Hou XG, Sultana F, Miura H, Ezaki T. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol 1995;45:406–408. [DOI] [PubMed] [Google Scholar]
- 60.Johnson JL, Moore LV, Kaneko B, Moore WE. Actinomyces georgiae sp. nov., Actinomyces gerencseriae sp. nov., designation of two genospecies of Actinomyces naeslundii, and inclusion of A. naeslundii serotypes II and III and Actinomyces viscosus serotype II in A. naeslundii genospecies 2. Int J Syst Bacteriol 1990;40:273–286. [DOI] [PubMed] [Google Scholar]
- 61.Putnins EE, Bowden GH. Antigenic relationships among oral Actinomyces isolates, Actinomyces naeslundii genospecies 1 and 2, Actinomyces howellii, Actinomyces denticolens, and Actinomyces slackii. J Dent Res 1993;72:1374–1385. [DOI] [PubMed] [Google Scholar]
- 62.Lisitsyn N, Lisitsyn N, Wigler M. Cloning the differences between two complex genomes. Science 1993;259:946–951. [DOI] [PubMed] [Google Scholar]
- 63.Akopyants NS, Fradkov A, Diatchenko L, Hill JE, Siebert PD, Lukyanov SA, Sverdlov ED, Berg DE. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc Natl Acad Sci USA 1998;95:13108–13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mahairas GG, Sabo PJ, Hickey MJ, Singh DC, Stover CK. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J Bacteriol 1996;178:1274–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perrin A, Nassif X, Tinsley C. Identification of regions of the chromosome of Neisseria meningitidis and Neisseria gonorrhoeae which are specific to the pathogenic Neisseria species. Infect Immun 1999;67:6119–6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997;25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Corless CE, Guiver M, Borrow R, Edwards-Jones V, Fox AJ, Kaczmarski EB. Simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J Clin Microbiol 2001;39:1553–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Elden LJ, Nijhuis M, Schipper P, Schuurman R, van Loon AM. Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J Clin Microbiol 2001;39:196–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gibbons RJ, van Houte J. On the formation of dental plaques. J Periodontol 1973;44:347–360. [DOI] [PubMed] [Google Scholar]
- 70.Cisar JO, Kolenbrander PE, McIntire FC. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect Immun 1979;24:742–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kolenbrander PE, Andersen RN, Moore LV. Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect Immun 1989;57:3194–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Becker MR, Paster BJ, Leys EJ, Moeschberger ML, Kenyon SG, Galvin JL, Boches SK, Dewhirst FE, Griffen AL. Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol 2002;40:1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol 2003;30:644–654. [DOI] [PubMed] [Google Scholar]
- 74.Grenier D, Mayrand D. Nutritional relationships between oral bacteria. Infect Immun 1986;53:616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Postollec F, Norde W, van der Mei HC, Busscher HJ. Enthalpy of interaction between coaggregating and non-coaggregating oral bacterial pairs—a microcalorimetric study. J Microbiol Methods 2003;55:241–247. [DOI] [PubMed] [Google Scholar]
- 76.Palmer RJ Jr, Kazmerzak K, Hansen MC, Kolenbrander PE. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect Immun 2001;69:5794–5804. [DOI] [PMC free article] [PubMed] [Google Scholar]


