Abstract
Flowering plants display a remarkable range of inflorescence architecture, and pedicel characteristics are one of the key contributors to this diversity. However, very little is known about the genes or the pathways that regulate pedicel development. The brevipedicellus (bp) mutant of Arabidopsis thaliana displays a unique phenotype with defects in pedicel development causing downward-pointing flowers and a compact inflorescence architecture. Cloning and molecular analysis of two independent mutant alleles revealed that BP encodes the homeodomain protein KNAT1, a member of the KNOX family. bp-1 is a null allele with deletion of the entire locus, whereas bp-2 has a point mutation that is predicted to result in a truncated protein. In both bp alleles, the pedicels and internodes were compact because of fewer cell divisions; in addition, defects in epidermal and cortical cell differentiation and elongation were found in the affected regions. The downward-pointing pedicels were produced by an asymmetric effect of the bp mutation on the abaxial vs. adaxial sides. Cell differentiation, elongation, and growth were affected more severely on the abaxial than adaxial side, causing the change in the pedicel growth angle. In addition, bp plants displayed defects in cell differentiation and radial growth of the style. Our results show that BP plays a key regulatory role in defining important aspects of the growth and cell differentiation of the inflorescence stem, pedicel, and style in Arabidopsis.
The building blocks of the plant architecture (body plan) are composed of reiterative units referred to as phytomers and are elaborated during different phases of development (1). In Arabidopsis thaliana, three types of phytomers have been described (2). The number of units and their size variations among these three main types of phytomers in different plant species contribute to the tremendous architectural diversity observed in flowering plants (3). The activity of the shoot apical meristem (SAM), together with additional meristems, regulates the growth and development of all three types of phytomers (3–5). The SAM contains three major domains defined by cytoplasmic densities and cell division rates: the central zone, which is responsible for maintaining the pluripotent stem cells; the peripheral zone, which is involved in the production of lateral organs; and the rib zone, from which the bulk of the stem is derived (6). Recent studies in Arabidopsis have shown that several genes including SHOOTMERISTEMLESS (STM), WUSCHEL, and CLAVATA-family receptor kinases and their putative ligands define key functions in the SAM (7–10).
In Arabidopsis the inflorescence constitutes the major part of the shoot and thus contributes significantly to the overall shoot architecture. Several genes have been identified in Arabidopsis that play key roles in defining the architecture of the shoot/inflorescence. For example, dwarf plants with uniform effects on all phytomers have been associated with altered levels of or defects in the signaling pathways of certain plant hormones (gibberellins or brassinosteroids; refs. 11 and 12 and references therein). The supershoot (13) and altered meristem program (14) mutants display abnormally high levels of cytokinins and produce extensive branching and altered shoot and inflorescence architecture. Auxin polar transport mutants such as pinformed (15) and pinoid (16) form inflorescences that are reduced to pin-like structures that do not produce any lateral organs or meristems. A compact inflorescence is caused by the erecta mutation, which involves a putative receptor kinase (17). An even stronger effect on inflorescence architecture is conferred in a Landsberg erecta (Ler) background by the brevipedicellus (bp) mutation, which is defined by a recessive mutant with compact internodes and short, downward-pointing pedicels (18).
The role of homeobox genes in defining body plan and their evolutionary relationships in animals is well documented (19, 20). More recently, several plant knotted-like homeobox (KNOX) genes have been identified, which form two classes based on sequence similarities and expression domains (21–23). In Arabidopsis, there are four different class I KNOX genes, STM, KNAT1, KNAT2, and KNAT6 (8, 24, 25). STM is expressed in the central zone (8), whereas KNAT1 and KNAT2 are expressed in the peripheral zone of the SAM (24). KNAT1 is expressed also in the cortical cell layers of the inflorescence stem (peduncle) and pedicel (24). The expression of STM, KNAT1, and KNAT2 is down-regulated in the leaf primordia and developing leaves (24) by ASYMMETRIC LEAVES 1 and 2 (26, 27). Ectopic expression of KNAT1 and KNAT2 in leaves induces altered symmetry and cell fate and ectopic meristem/shoot formation from the adaxial surface (28). Recessive mutations in the prototypical class I KNOX gene, maize knotted1, produce defective meristems, whereas gain-of-function mutations resulting in ectopic expression of maize KNOX genes disrupt normal leaf development (22, 29). In addition, loss-of-function mutations in rice OSH15 affect the shoot architecture (30). In Arabidopsis, loss-of-function mutations in class I KNOX genes are known only for STM, suggesting a critical role in SAM maintenance and function (8). Significantly, however, no such mutations have been described previously for KNAT1, hampering study of the role of this homeobox gene in plant development.
In this report we provide a molecular definition of the BP locus and show that it encodes the homeodomain (HD) protein KNAT1. Anatomical and developmental studies provided further insights into its function in effecting the development and growth of the inflorescence internodes, pedicels, and carpels.
Materials and Methods
Plant Material and Genetic Analysis.
Plants were grown at 22°C (90% relative humidity) under fluorescent and incandescent light at ≈60 μE/m2/s (E = 1 mol of photons) with 16-h days. bp seeds were obtained from the Arabidopsis Biological Resources Center (Ohio State University, Columbus, OH, stock no. CS30; ref. 18). We designated this allele bp-1. A second allele (bp-2) was isolated from promoter-tagged Arabidopsis lines in an RLD background, introgressed into Ler, and backcrossed five times with wild type (wt). bp-2 was introduced into a Columbia (Col) wt background from Ler and backcrossed thrice.
Histology.
Plant samples were fixed for 24 h at room temperature in formalin/acetic acid/alcohol and paraffin embedded as described (31). Serial sections were taken at 8 μm on a rotary microtome, attached to glass slides with Mayer's egg albumin (Sigma) solution, and dried on a warming tray (42°C). The sections were stained in toluidine blue O after removal of the embedding medium and observed under a Leitz microscope. Images were captured by using an Optronics International (Chelmsford, MA) DEI 750 digital microscope camera.
Scanning Electron Microscopy (SEM).
For SEM the samples were fixed in 3% glutaraldehyde and processed as described (32). Samples were mounted on aluminum stubs and coated with gold in an Edwards S150B sputter coater. Observations were made with a Phillips SEM 505 at 30 kV and recorded by using Polaroid type 665 P/N. Images were scanned and enhanced by using Adobe PHOTOSHOP 4.0.
Suppression Subtractive Hybridization.
Total RNA was harvested from stem/pedicel and leaf tissues of Col wt by using Trizol Reagent (Life Technologies, Rockville, MD). Poly(A)+ RNA was isolated by using mRNA spin columns (CLONTECH). cDNA synthesis was carried out by using a cDNA synthesis kit (Life Technologies). A total of 2 μg each of leaf cDNA (driver) and stem/pedicel cDNA (tester) was digested with HaeIII (New England Biolabs) and used for suppression subtractive hybridization as described (33). The subtracted mix was 32P-labeled by using a RediPrime kit (AP Biotech, Piscataway, NJ) and used to screen bacterial artificial chromosome (BAC) DNA preparations as described below.
Southern Blot.
BAC clones from chromosome 4 were obtained from the Arabidopsis Biological Resources Center. DNA was prepared from 10-ml cultures of BACs T17A2, T13D4, F9M13, T12G3, T28D5, T15F16, T3F12, T32A17, T3H13, F23J3, T8A17, T30A10, T15G18, T25P22, and T24H23 by using an alkaline lysis miniprep method (34). BAC DNA was digested with BamHI, EcoRI, or HindIII (Life Technologies), fractionated on a 0.8% agarose gel, and then blotted to a Zeta Probe membrane (BioRad) using standard procedures (34). The blot was probed with the pooled subtracted mix representing cDNAs expressed in stem/pedicel tissue, prepared as described above. Genomic DNA (5 μg) isolated from leaves (35) of wt and bp plants was digested by using 30 units of BamHI or EcoRI (Life Technologies) at 37°C for 8 h, processed as described above, and probed with the 32P-labeled KNAT1 reverse transcription (RT)–PCR product from Col wt. Hybridization proceeded for 3 h (BAC screen) or overnight (genomic Southern blot) at 65°C in QuickHyb hybridization solution (Stratagene); the most stringent wash was in 0.1× SSC (0.15 M sodium chloride/0.015 M sodium citrate, pH 7)/0.1% SDS at 65°C. The blots were exposed to X-Omat AR film (Kodak) overnight at −70°C.
PCR, RT-PCR, and DNA Sequencing.
See Table 1, which is published as supporting information on the PNAS web site, www.pnas.org, for details of primer sequences and amplification conditions. RT was carried out by using 3–5 μg of total RNA from stem tissue of wt (Col, Ler, and RLD) and bp plants and Superscript II RT (Life Technologies). To amplify the KNAT1 ORF, 1 μl of cDNA was used for PCR with primers 954 and 955 and Pfu polymerase (1 unit). Amplification of the cytosolic glyceraldehyde-3-phosphate dehydrogenase cDNA (36) from the same cDNA pools was performed under the same conditions. KNAT1-encoding PCR products were cloned and sequenced by primer walking using an ABI 377 DNA sequencer.
BP Complementation Constructs.
Two different complementation constructs were prepared. Plasmid pRD400-951/955 consisted of the 1.5-kb predicted BP/KNAT1 promoter (Col wt) linked to the KNAT1 ORF (Col wt) and Nos terminator in pRD400 (37). A second complementation vector, pRD400-951/956, consisted of the BP/KNAT1 promoter fused to the KNAT1-encoding genomic fragment (Col wt). Agrobacterium tumefaciens GV3101 containing these recombinant constructs was used to transform bp-2 (Ler) plants by vacuum infiltration (38).
Results
Phenotypic Characteristics of bp Plants.
The bp mutant was reported first in Ler (bp-1; ref. 18) and has been used since as a genetic marker for chromosome 4. No other bp alleles have been reported nor have developmental studies on bp appeared in the literature. While screening Arabidopsis promoter-tagged lines (RLD background), we identified a line that showed a bp-like phenotype. Subsequent genetic analysis revealed that this mutation (bp-2) was allelic to bp-1, but bp-2 was not linked to a transferred DNA insertion (data not shown). Because bp-1 (in Ler) showed a stronger phenotype than bp-2 (in RLD), we introduced bp-2 into Ler and studied the developmental defects. Unless stated otherwise, the phenotypic and developmental characteristics of bp-1 and bp-2 are designated as bp where they were similar; where differences were observed, the bp-1 and bp-2 alleles are distinguished.
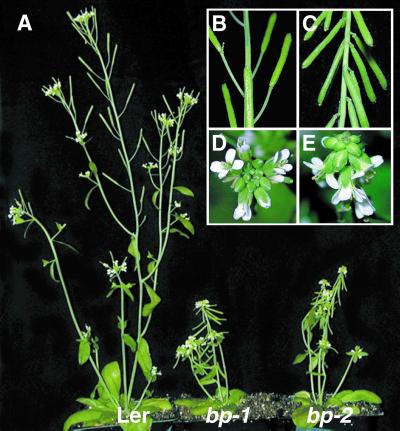
In bp plants the earliest signs of abnormalities were evident at the time of bolting, with more compactly arranged floral buds at the apex; the effects were more pronounced when the first few coflorescence internodes from the rosette leaves started elongating (Fig. 1 A, D, and E). At maturity, bp plants displayed a marked reduction in overall height, primarily as a result of shortened internodes (Fig. 1 A–C); moreover, the floral internodes were affected to a greater extent than the coflorescence internodes (Fig. 2). Additionally, bending at nodes was observed, and this phenotype was more severe in bp-1 than in bp-2 plants. bp-2 in RLD (the original isolate) and Col backgrounds showed similar patterns of defects, although the reduction in internodal lengths was less severe than observed in the Ler background (see Fig. 8, which is published as supporting information on the PNAS web site).
Figure 1.
Phenotypes of 6-week-old Ler wt, bp-1 Ler, and bp-2 Ler plants. (A) Whole plant. Close-ups of floral nodes with siliques of Ler wt (B) and bp-1 Ler (C) and close-ups of the inflorescence apex in Ler wt (D) and bp-1 Ler (E) are shown.
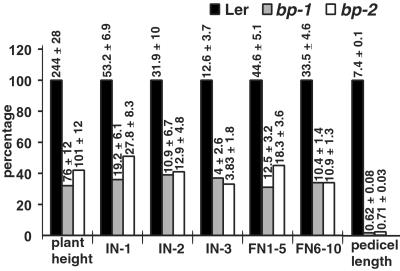
Figure 2.
Comparison of internode and pedicel lengths between Ler wt, bp-1 Ler, and bp-2 Ler. The histograms represent the percentage reduction for bp-1 and bp-2; the actual measurements in mm (mean values ± SD of 30 data points) are shown above the corresponding bars. The average pedicel lengths represent the values for the floral nodes 1–5. IN-1, IN-2, and IN-3, coflorescence nodes 1, 2, and 3, respectively; FN1–5, floral nodes 1–5; FN6–10, floral nodes 6–10.
bp Affects Cell Division and Cell Differentiation in the Internodes of the Inflorescence.
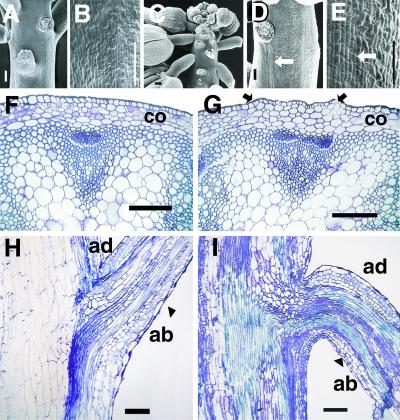
SEM analysis of bp plants showed that the floral buds began pointing downward quite early in their development, and the internodal elongation was reduced significantly (Fig. 3 A and C). The peduncle surface showed stripes consisting of cell files (≈15 cells in width) with defects in epidermal cell differentiation [defined by alterations in mutant lines in cell size, shape, and/or cell type (stomata) in relation to similar regions in wt] associated with regions below the nodes (Fig. 3 B, D, and E). Cross sections through internodes in bp indicate that the overall radial pattern, in terms of tissue types, was very similar to the wt (Fig. 3 F and G). However, small sectors with defects in epidermal cell differentiation were observed (Fig. 3G) and corresponded to the stripes of differentiation-defective cells observed by SEM (Fig. 3 D and E). Furthermore, the cortical cells below these sectors were defective in differentiation (indicated by a lack of chloroplasts), and the cells were relatively larger with less intercellular space (Fig. 3G and data not shown). Longitudinal sections through the nodes showed sectors of epidermal and subepidermal defects (Fig. 3 H and I). Because the cell number per unit area along the main axis of the peduncle in bp was comparable to that in wt, the reduced internodal length was interpreted to be a result of fewer cell divisions.
Figure 3.
SEM micrographs of inflorescences from Ler (A and B) and bp-1 Ler (C–E). (A) Ler wt floral nodes. (B) Ler wt peduncle internode magnified to show differentiated epidermal cells. (C) bp-1 floral nodes. (D and E) bp-1 peduncle internode showing stripes of less differentiated epidermal cells (arrows) that originate below the node. The anatomy of the peduncle of Ler wt (F and H) and bp-1 (G and I) is shown. (F and G) Cross sections through the internodal region of the peduncle of Ler wt and bp-1, respectively. (H and I) Longitudinal sections through the nodal region of Ler wt and bp-1, respectively. The arrows in G demarcate a band of less differentiated cells that originate below the node. co, cortical cell layer; ad, adaxial; ab, abaxial. (Bars: A, B, D, and E–I, 0.1 mm; C, 1 mm.)
bp Causes Defects in Pedicel Development.
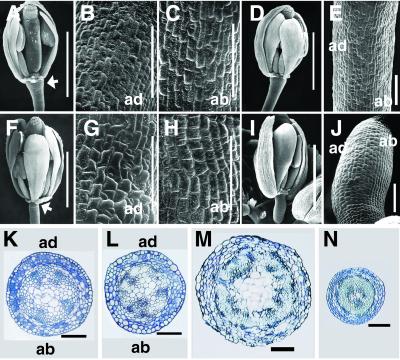
Pedicels in bp plants at all the floral nodes showed a drastic reduction in length compared with wt (Fig. 2) in addition to downward-pointing siliques (Fig. 1 A–C). The severity of the latter phenotype conferred by bp-2 varied in different backgrounds from downward-pointing (Ler) to less acute and variable bending in RLD and Col backgrounds (see Fig. 8). Because very little is known about pedicel development in any plant species including Arabidopsis, we determined the comparative ontogeny in Ler wt and bp. Pedicel initiation was observed first around stage 3 flowers, followed by elaboration of the pedicel with coordinated development on both the abaxial and adaxial sides and along the proximo-distal axis (Fig. 9, which is published as supporting information on the PNAS web site). The first signs of epidermal differentiation (defined by characteristic changes in cell shape and the appearance of stomata) were observed on the abaxial side at stage 9 and was followed closely by differentiation on the adaxial side in subsequent stages (Fig. 9). By stage 12, epidermal cell differentiation was completed with no apparent differences observed between the abaxial and adaxial sides in the wt (Fig. 4 A–C). In bp, no detectable differences from wt were observed up to stage 3 (Fig. 9). However, the pedicel differentiation and elaboration processes lagged behind the wt, and the first sign of epidermal cell differentiation was observed only at stage 12, restricted to the adaxial surface; no corresponding differentiation was observed on the abaxial side even by the mature stage (Fig. 4 F–J). Anatomical analysis showed that although the major part of the pedicel in bp contained defects in the differentiation of abaxial-side epidermal cells and cortical cells (Fig. 4 K and L), the distal region including the receptacle was affected more strongly with a significantly reduced pith region, cell size and differentiation, and radial growth (Fig. 4 M and N). Longitudinal sections through the pedicels also showed that the cells in the epidermal layer and cortical tissues on the abaxial side were less elongated (Fig. 3 H and I). Furthermore, there were fewer cells in the proximo-distal axis of the pedicel, indicative of fewer cell divisions. Although there were no apparent defects observed in the sepals, petals, and stamens, the carpels showed detectable differences in bp. Notably, there was reduced radial growth of the style (Fig. 5 A, C, D, and F). The epidermal and cortical cells of the style, especially in the lateral axis, were defective in differentiation and elongation, and as a consequence the arrangement of stigmatic papillae was altered significantly (Fig. 5 A, B, D, and E). These observations suggest that BP also plays a role in maintaining the normal radial growth of the style.
Figure 4.
Pedicel development in Ler wt (A–E, K, and L) and bp-1 Ler (F–J, M, and N). (A–C) SEM of pedicel of stage 12 flower of Ler wt (A) showing complete epidermal differentiation on both the adaxial (B) and abaxial (C) sides. (F–H) Pedicel of stage 12 flower of bp-1 (F) with narrow distal end (arrow), differentiated adaxial (G), and less differentiated abaxial (H) sides. (D and E) SEM of stage 13 flower of Ler wt and its pedicel, respectively. Stage 13 flower of bp-1 (I) and its pedicel (J) show a less differentiated abaxial side. (K–N) Cross section through the midregion of the pedicel of Ler wt (K) and bp-1 (L) and the distal end of the pedicel of Ler wt (M) and bp-1 (N). ad, adaxial; ab, abaxial. (Bars: A, D, F, and I, 1 mm; B, C, E, G, H, J–N, 0.1 mm.)
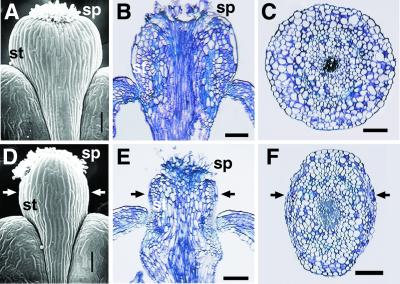
Figure 5.
SEM of the style of a stage 17 flower of Ler wt (A) and bp-1 Ler (D) and longitudinal sections through the style of Ler (B) and bp-1 (E). Cross sections through the style of Ler wt (C) and bp-1 (F) are shown. The arrows in D–F indicate the lateral axis. sp, stigmatic papillae; st, style. (Bar, 0.1 mm.)
BP Encodes KNAT1.
The developmental and anatomical studies suggested that the defects in bp were associated only with the peduncle and certain parts of the flower but not with the leaves. We therefore assumed that BP may be expressed predominantly in stem and pedicel tissues in wt plants and used this expression pattern in combination with genetic data to isolate BP. The approach we developed is based on the availability of overlapping genomic clones spanning the predicted chromosomal region and requires the production of a pool of differentially expressed cDNAs representing the assumed expression pattern of the target gene. Potential targets may be identified by hybridizing these probes to BACs that span the predicted genomic target region. To clone BP, we first narrowed the search to a region between DET1 and the centromere on chromosome 4 based on genetic maps compiled from several data sets (www.arabidopsis.org; ref. 39). To produce probes reflecting the anticipated expression pattern of BP, we isolated poly(A)+ RNA from both stem/pedicel and leaf tissues in Col wt plants and performed a subtraction using leaf cDNA as driver. The pooled subtracted products then were used as a probe in a Southern blot with 15 BACs as targets spanning a region of ≈1.5 Mb on chromosome 4 between DET1 and the centromere. A BamHI fragment of ≈20 kb from BAC F9M13 was the only band that showed any hybridization to the subtracted probe (Fig. 10, which is published as supporting information on the PNAS web site). BAC F9M13 (GenBank accession no. AC006267) contains a single gene on this 20-kb BamHI fragment within a region rich in repeats. Subsequent fingerprinting of F9M13 with this probe (data not shown) confirmed that the probe detected the previously reported homeobox gene KNAT1 (24). Thus, KNAT1 was the only gene identified in the target area of the genome that fit the expression profile expected of BP. Southern blot analysis of the KNAT1 locus in wt and bp revealed that bp-1 (Ler) appeared to lack KNAT1 entirely, suggesting a deletion of the gene. In contrast, bp-2 (RLD) appeared to contain an intact KNAT1 locus (Fig. 6A). Furthermore, bp-1 (Ler) produced no detectable KNAT1 transcripts, whereas bp-2 in all backgrounds produced an apparently full-length transcript (Fig. 6B). Sequences of the KNAT1 cDNAs were determined for wt (Ler and RLD) and bp-2. Ler and RLD wt KNAT1 ORFs each encoded predicted proteins of 400 aa (Fig. 7) compared with 398 aa for Col wt (24). Minor sequence dissimilarity among the three wt KNAT1 cDNAs was detected (Fig. 7). The sequences of KNAT1 from bp-2 in all backgrounds were identical, revealing that no recombination had occurred at this locus during introgression of bp-2, and contained polymorphisms compared with the wt (Fig. 7). Notably, bp-2 contained a C → T transition corresponding to position 535 of the Col wt ORF, changing codon 179 from CAG to TAG and thereby introducing a stop codon and resulting in a predicted protein that was truncated upstream of both the ELK region and HD. Considering that bp-1 correlated with the absence of the KNAT1 gene and its expression, and bp-2 contained a truncated predicted KNAT1 protein, we hypothesized that bp corresponded to KNAT1.
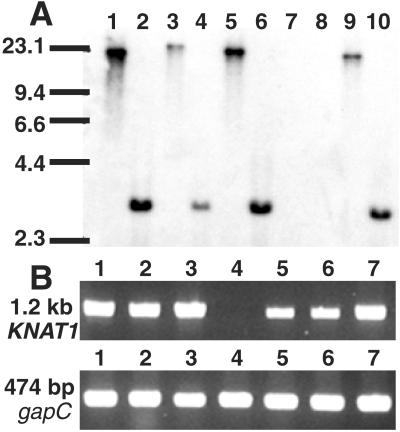
Figure 6.
Southern blot and RT-PCR of KNAT1. (A) Southern blot. Genomic DNA from Col wt (lanes 1 and 2), Ler wt (lanes 3 and 4), RLD wt (lanes 5 and 6), bp-1 Ler (lanes 7 and 8), and bp-2 RLD (lanes 9 and 10) was digested with BamHI (lanes 1, 3, 5, 7, and 9) or EcoRI (lanes 2, 4, 6, 8, and 10) and probed with the KNAT1 cDNA. Sizes of the molecular weight standards (kb) are indicated. (B) RT-PCR using KNAT1 primers 954 and 955. Lane 1, Col wt; lane 2, RLD wt; lane 3, Ler wt; lane 4, bp-1 Ler; lane 5, bp-2 Ler; lane 6, bp-2 RLD; lane 7, bp-2 Col. The same cDNA pools were amplified with primers specific for glyceraldehyde-3-phosphate dehydrogenase (gapC; ref. 36).
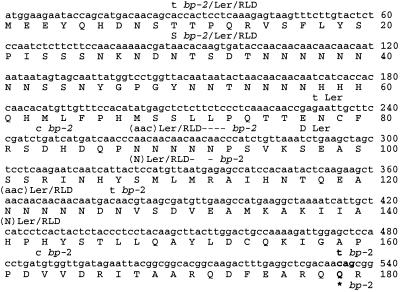
Figure 7.
Sequences of the polymorphic regions of the BP-encoding cDNAs from Col wt, RLD wt, Ler wt, and bp-2. Numbering is shown for the Col wt sequence (GenBank accession no. U14174). Stop codons are indicated by an asterisk (*), nucleotide and amino acid deletions relative to Col wt are indicated by a dash (−), and nucleotide and amino acid insertions relative to Col wt are indicated in parentheses ( ). The C → T transition that causes a stop codon at position 535 in bp-2 is shown in bold. Nucleotides downstream of position 540 were identical among all the BP-encoding genes analyzed and are not shown.
KNAT1 Complements the bp Mutant Phenotype.
Transformation of bp-2 (Ler) with the genomic clone of KNAT1 resulted in 20 transformants; four were rescued completely to wt, whereas the others were rescued partially. Southern analysis confirmed that these complemented lines contained the KNAT1 wt transgene (data not shown). Further analysis of two single transgene copy lines showed a 3:1 (wt/bp) segregation pattern in the T2 generation, providing genetic confirmation of complementation. Additional experiments with the KNAT1 ORF also produced two fully complemented lines, suggesting that the KNAT1 coding region is sufficient for rescuing bp. Therefore, KNAT1 corresponds to the BP locus, and we propose that the latter name be retained in view of its earlier designation (18).
BP Expression Is Consistent with a Role in the Normal Development of the Pedicel, Peduncle, and Style.
Having established that KNAT1 and BP are the same gene, we sought to determine whether the previous studies on KNAT1 transcript localization in situ (24) and the expression pattern of the BP∷β-glucuronidase (GUS) construct in Arabidopsis (Fig. 11, which is published as supporting information on the PNAS web site) could provide clues to the function of BP in regulating inflorescence architecture. The earliest GUS expression was found in embryonic stages in the SAM region (Fig. 11 A and B). In young seedlings, however, expression was observed in the SAM and also in the hypocotyl (Fig. 11 C and D). During the reproductive phase, GUS was expressed in the inflorescence apex, floral primordia, peduncle, pedicels, and styles (Fig. 11 E–G). Analysis of longitudinal and transverse sections through the peduncle and pedicels revealed that expression was localized to the cortical cells and was not found in the epidermal and pith tissues (Fig. 11 G–I). Taken together, the BP∷GUS expression pattern in the peduncle, pedicel, and style was consistent with the KNAT1 in situ results (24) and also correlated well with the phenotypic alterations in bp.
Discussion
Although bp is a classical mutant of Arabidopsis (18) and presents a unique architectural phenotype, its molecular identity had not been determined thus far. Similarly, the in vivo function of KNAT1 has been elusive, because there has been no loss-of-function mutant described. This study establishes that BP encodes KNAT1 (hereafter BP). BP is the product of a single-copy gene and shares significant homology with a number of other HD proteins, in particular the class I KNOX proteins from Arabidopsis: STM (41%), KNAT2 (40%), and KNAT6 (39%). In addition, BP is similar to maize RS1 and rice OSH15 (30, 40) both in predicted amino acid sequence (53 and 52%, respectively) and expression pattern; the latter two genes likely are orthologs (22). Furthermore, osh15 loss-of-function mutants share some phenotypic similarities with bp, although the rice panicle and pedicels are unaffected (30). It is possible that BP, OSH15, and RS1 form an orthologous group, but the acquisition of similar functions by convergence cannot be ruled out. Although loss of function is a definite consequence of deleting the entire protein coding region in bp-1, the similarity of the developmental defects in bp-2 suggests that the highly conserved HD and/or the flanking ELK domain is essential for its in vivo function. These regions are essential also in other HD proteins from plants and animals (19, 41). Recently, BP has been shown to form functional heterodimers with BEL1 in yeast two-hybrid assays (42), suggesting that the various phenotypic consequences of bp might be caused by perturbations of interactions of BP with other HD proteins.
The effects of bp indicate that its function is critical for normal pedicel and inflorescence development. BP is unique among the homeobox genes in Arabidopsis, because it seems to play diverse roles in defining the inflorescence architecture (8, 9). The regions affected by bp (peduncle, pedicel, and style) share radial structure, suggesting a role for BP in mediating coordinated radial growth in these organs. The effects in these regions include defective cell division, cell differentiation, and cell elongation, suggesting that BP may operate through these cellular processes. The shortened floral internodes and pedicels were caused primarily by decreased cell division, a key cellular process that is connected directly to plant growth and development (43). Recent studies raise the possibility that BP may operate through cytokinin-mediated signaling to regulate pedicel and internodal growth. For example, misexpression of BP in leaves implicates cytokinin signaling in at least some of the gain-of-function phenotypes (28). In addition, expression of cytokinin oxidase in transgenic tobacco plants results in reduced cytokinin levels and dwarf plants with shortened internodes caused by decreased cell divisions (44), which is reminiscent of bp plants. The reduction in cell divisions in bp did not show any polarity in either the internode or the pedicel, but the defects in cell differentiation and elongation were not as uniform. Instead, regions of the peduncle below the nodes and the abaxial side of the pedicel were affected more strongly; the lateral axis of the style was also affected (Figs. 3–5). Furthermore, the defects in the epidermal layer were always associated with similar defects in the flanking cortical cells. However, in situ (24) and BP∷GUS expression analyses (Fig. 11) show that BP transcripts are found in the cortical tissue of both affected and unaffected regions but not in the epidermis. This observation suggests that the proper differentiation of the epidermal layer, including the appearance of stomata in the affected regions, may use positional cues from the neighboring cortical cells. Alternatively, transport of BP transcript or protein, as observed for other HD proteins (45, 46), may be involved in regulating the localized cell differentiation in the peduncle, pedicel, and style. These data collectively suggest that BP acts differentially in different parts of the plant. It differs in this respect from OSH15, in which loss-of-function in rice causes uniform defects in both epidermal and hypodermal cells (30). Although KNAT2 shares overlapping expression domains with BP (24, 26), its functions are likely different in regulating inflorescence architecture; otherwise the bp phenotype would have been either suppressed or less severe. Once loss-of-function mutants are available for KNAT2, this possibility can be addressed directly.
As noted from the above, the effects of bp are distinct from that of the mutations that cause overall dwarfism even though both include shortened internodes. The downward-pointing silique phenotype was not a result of a stunted shoot or inflorescence. For instance, bp-2 in Col and RLD backgrounds did not reduce the pedicel length as much as it did in Ler, but all three had downward-pointing siliques (Figs. 1 A–C and 8). Pedicel ontogeny in wt plants revealed that around stage 9 the abaxial side was developmentally more advanced than the adaxial side, although at maturity this asymmetry was less evident (Figs. 4 A–E and 9). In bp, however, the abaxial side cells did not progress through their normal program of differentiation as observed in wt, whereas the elongation and differentiation of the adaxial side cells was consistently ahead (Figs. 4 F–J and 9). Coupled with the effects of fewer cell divisions along both sides, this strong asymmetry resulted in the downward-pointing, shortened pedicels observed in the mutant. These asymmetric effects suggest that the downstream effectors of differentiation in the abaxial side may be particularly critical and/or sensitive to BP function (24). Alternatively, BP may suppress a default developmental program, giving rise to polarity in lateral organs such as the pedicel; in bp mutants, the default program would be active, resulting in a strongly asymmetrical organ. Further experiments are required to resolve these and other possibilities.
Although BP expression was found in embryonic and vegetative meristems (Fig. 11 A–D), bp mutants did not have an obvious phenotype in this regard, suggesting that other genes such as STM may provide overlapping functions in early stages of development. The effects of bp therefore may be restricted to those domains in which no overlapping functions are provided by other genes. The results presented here provide functional insights into how BP regulates inflorescence architecture in Arabidopsis. The results also have broader implications for understanding the diversity of pedicel architecture observed in flowering plants.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resources Center for bp-1 seeds and BAC clones, S. Hake for providing KNAT1∷GUS lines, G. Haughn and M. Byrne for suggestions and for sharing unpublished results, A. Cutler, M. Wilkinson, and J. Zou for critical comments on the manuscript, two anonymous reviewers and the editor for helpful comments and suggestions, and M. Martin for technical assistance. This is publication 43808 from the National Research Council of Canada.
Abbreviations
- SAM
shoot apical meristem
- Ler
Landsberg erecta
- bp
brevipedicellus
- HD
homeodomain
- wt
wild type
- Col
Columbia
- SEM
scanning electron microscopy
- BAC
bacterial artificial chromosome
- RT
reverse transcription
- GUS
β-glucuronidase
Footnotes
References
- 1.Sussex I M, Kerk N M. Curr Opin Plant Biol. 2001;4:33–37. doi: 10.1016/s1369-5266(00)00132-1. [DOI] [PubMed] [Google Scholar]
- 2.Schultz E A, Haughn G W. Plant Cell. 1991;3:771–781. doi: 10.1105/tpc.3.8.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steeves T A, Sussex I M. Patterns in Plant Development. Cambridge, U.K.: Cambridge Univ. Press; 1989. [Google Scholar]
- 4.Medford J I, Behringer F J, Callos J D, Feldmann K A. Plant Cell. 1992;4:631–643. doi: 10.1105/tpc.4.6.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simon R. Semin Cell Dev Biol. 2001;12:357–362. doi: 10.1006/scdb.2001.0264. [DOI] [PubMed] [Google Scholar]
- 6.Bowman J L, Eshed Y. Trends Plant Sci. 2000;5:110–115. doi: 10.1016/s1360-1385(00)01569-7. [DOI] [PubMed] [Google Scholar]
- 7.Brand U, Hobe M, Simon R. BioEssays. 2001;23:134–141. doi: 10.1002/1521-1878(200102)23:2<134::AID-BIES1020>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 8.Long J A, Moan E I, Medford J I, Barton M K. Nature (London) 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- 9.Mayer K F, Schoof H, Haecker A, Lenhard A, Jurgens G, Laux T. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 10.Clark S E. Nat Rev Mol Cell Biol. 2001;2:276–284. doi: 10.1038/35067079. [DOI] [PubMed] [Google Scholar]
- 11.Hedden N P, Kamiya Y. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- 12.Richards D E, King K E, Ait-ali T, Harberd N P. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:67–88. doi: 10.1146/annurev.arplant.52.1.67. [DOI] [PubMed] [Google Scholar]
- 13.Tantikanjana T, Yong J W, Letham D S, Griffith M, Hussain M, Ljung K, Sandberg G, Sundaresan V. Genes Dev. 2001;15:1577–1588. doi: 10.1101/gad.887301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaudhury A M, Letham S, Craig S, Dennis E S. Plant J. 1993;4:907–916. [Google Scholar]
- 15.Okada K, Ueda J, Komaki M K, Bell C J, Shimura Y. Plant Cell. 1991;3:677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett S R M, Alvarez J, Bossinger G, Smyth D R. Plant J. 1995;8:505–520. [Google Scholar]
- 17.Torii K U, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier R F, Komeda Y. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koornneef M, Eden J v, Hanhart C J, Stam P, Braaksma F J, Feenstra W J. J Hered. 1983;74:265–272. [Google Scholar]
- 19.Gehring W J, Affolter M, Burglin T. Annu Rev Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- 20.Kappen C. Proc Natl Acad Sci USA. 2000;97:4481–4486. doi: 10.1073/pnas.97.9.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bharathan G, Janssen B, Kellogg E, Sinha N. Mol Biol Evol. 1999;16:553–563. doi: 10.1093/oxfordjournals.molbev.a026136. [DOI] [PubMed] [Google Scholar]
- 22.Reiser L, Sanchez B P, Hake S. Plant Mol Biol. 2000;42:151–166. [PubMed] [Google Scholar]
- 23.Serikawa K A, Martinez-Laborda A, Zambryski P. Plant Mol Biol. 1996;32:673–693. doi: 10.1007/BF00020208. [DOI] [PubMed] [Google Scholar]
- 24.Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. Plant Cell. 1994;6:1859–1876. doi: 10.1105/tpc.6.12.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y. Development (Cambridge, UK) 2001;128:1771–1783. doi: 10.1242/dev.128.10.1771. [DOI] [PubMed] [Google Scholar]
- 26.Ori N, Eshed Y, Chuck G, Bowman J L, Hake S. Development (Cambridge, UK) 2000;127:5523–5532. doi: 10.1242/dev.127.24.5523. [DOI] [PubMed] [Google Scholar]
- 27.Byrne M, Barley R, Curtis M, Arroyo J, Dunham M, Hudson A, Martienssen R. Nature (London) 2000;408:967–971. doi: 10.1038/35050091. [DOI] [PubMed] [Google Scholar]
- 28.Chuck G, Lincoln C, Hake S. Plant Cell. 1996;8:1277–1289. doi: 10.1105/tpc.8.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vollbrecht E, Reiser L, Hake S. Development (Cambridge, UK) 2000;127:3161–3172. doi: 10.1242/dev.127.14.3161. [DOI] [PubMed] [Google Scholar]
- 30.Sato Y, Sentoku N, Miura Y, Hirochika H, Kitano H, Matsuoka M. EMBO J. 1999;18:992–1002. doi: 10.1093/emboj/18.4.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansen D A. Plant Microtechnique. New York: McGraw–Hill; 1940. [Google Scholar]
- 32.Venglat S P, Sawhney V K. Planta. 1996;1968:480–487. doi: 10.1007/BF00620066. [DOI] [PubMed] [Google Scholar]
- 33.Diatchenko L, Lau Y-F C, Campbell A P, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov E D, Siebert P D. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smit J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- 35.Dellaporta S. In: The Maize Handbook. Freeling M, Walbot V, editors. New York: Springer; 1994. pp. 522–525. [Google Scholar]
- 36.Shih M-C, Heinrich P C, Goodman H M. Gene. 1991;104:133–138. doi: 10.1016/0378-1119(91)90242-4. [DOI] [PubMed] [Google Scholar]
- 37.Datla R S S, Hammerlindl J K, Panchuk B, Pelcher L E, Keller W. Gene. 1992;122:383–384. doi: 10.1016/0378-1119(92)90232-e. [DOI] [PubMed] [Google Scholar]
- 38.Bechtold N, Ellis J, Pelletier G. C R Acad Sci Ser III. 1993;316:1194–1199. [Google Scholar]
- 39.Pepper A, Delaney T, Washburn T, Poole D, Chory J. Cell. 1994;78:109–116. doi: 10.1016/0092-8674(94)90577-0. [DOI] [PubMed] [Google Scholar]
- 40.Schneeberger R G, Becraft P W, Hake S, Freeling M. Genes Dev. 1995;9:2292–2304. doi: 10.1101/gad.9.18.2292. [DOI] [PubMed] [Google Scholar]
- 41.Williams R W. BioEssays. 1998;20:280–282. doi: 10.1002/(SICI)1521-1878(199804)20:4<280::AID-BIES2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 42.Bellaoui M, Pidkowich M S, Samach A, Kushalappa K, Kohalmi S E, Modrusan Z, Crosby W L, Haughn G W. Plant Cell. 2001;13:2455–2470. doi: 10.1105/tpc.010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyerowitz E M. Cell. 1997;88:299–308. doi: 10.1016/s0092-8674(00)81868-1. [DOI] [PubMed] [Google Scholar]
- 44.Werner T, Motyaka V, Strand M, Schmulling T. Proc Natl Acad Sci USA. 2001;98:10487–10492. doi: 10.1073/pnas.171304098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lucas W J, Bouché-Pillon S, Jackson D P, Nguyen L, Baker L, Ding B, Hake S. Science. 1995;270:1980–1983. doi: 10.1126/science.270.5244.1980. [DOI] [PubMed] [Google Scholar]
- 46.Kim M, Canio W, Kessler S, Sinha N. Science. 2001;293:287–289. doi: 10.1126/science.1059805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.