Abstract
Cyanobacteria are the only prokaryotes known thus far possessing regulation of physiological functions with approximate daily periodicity, or circadian rhythms, that are controlled by a cluster of three genes, kaiA, kaiB, and kaiC. Here we demonstrate considerably higher genetic polymorphism and extremely rapid evolution of the kaiABC gene family in a filamentous cyanobacterium, Nostoc linckia, permanently exposed to the acute natural environmental stress in the two microsite evolutionary models known as “Evolution Canyons,” I (Mount Carmel) and II (Upper Galilee) in Israel. The family consists of five distinct subfamilies (kaiI–kaiV) comprising at least 20 functional genes and pseudogenes. The obtained data suggest that the duplications of kai genes have adaptive significance, and some of them are evolutionarily quite recent (≈80,000 years ago). The observed patterns of within- and between-subfamily polymorphisms indicate that positive diversifying, balancing, and purifying selections are the principal driving forces of the kai gene family's evolution.
The problem of the relative importance of various driving forces in genome evolution requires a comprehensive critical testable approach. The challenging question is that of how selective or nonselective factors influence the evolution of organisms living under contrasting ecological conditions.
Lower Nahal Oren, Mount Carmel, and Lower Nahal Keziv, western Upper Galilee, Israel, known as “Evolution Canyons” (ECs) I and II, respectively (1–3), represent microsites with highly contrasting environmental conditions on opposite slopes separated by only 300–400 m at the top and 50–100 m at the bottom. They were eroded in the Plio-Pleistocene period. The “African” south-facing slopes (SFSs) receive up to 600% more solar radiation and are warmer, drier, and spatiotemporally more heterogeneous and fluctuating than “European” north-facing slopes (NFSs). Such ecological contrasts on a microscale provide an excellent opportunity to test various hypotheses regarding the influence of natural environmental stress on patterns of genome evolution and shaping genetic variability of populations (1), adaptive radiation, and incipient speciation across life (2, 3).
Cyanobacteria (blue-green algae, Cyanoprokaryota, Cyanophyta) can occupy various ecotopes and have high ecological adaptability (4) and thus are suitable model organisms for testing various predictions about probable evolutionary forces affecting patterns of genetic polymorphism under contrasting ecological conditions. Thus far, extensive studies have been done to ascertain molecular and genetic mechanisms of cyanobacterial responses to various stresses (see refs. 5–9).
However, despite the extensive studies of cyanobacteria, data about the effect of long-time natural environmental stress on the level and patterns of nucleotide polymorphism in these prokaryotes are lacking. Such data can highlight the evolutionary forces contributing to adaptive genetic radiation and speciation. This issue is of particular interest, because it pertains to processes shaping genetic variation at intra- and interpopulation levels under sharp environmental gradients at microscales, where microevolution can be tracked effectively (3).
According to the niche-width variation hypothesis (10) and the environmental theory of genetic diversity (11–13), populations from spatiotemporally more variable and/or ecologically stressful habitats should display higher morphological and genetic polymorphism. Filamentous cyanobacterium Nostoc linckia is a ubiquitous species inhabiting both slopes of the two ECs (14). Because of its sessile character, the cyanobacterium is exposed permanently to fluctuating complex environmental stress (light, temperature, desiccation, etc.) and thus is expected to follow the above predictions. Our recent study showed significantly higher levels of intra- and interpopulation amplified fragment length (15) and HIP1 (16) polymorphisms of Nostoc on the SFS compared with the NFS.
On the other hand, according to the neutral theory, nucleotide polymorphism in a population is determined by effective population size and the mutation rate, θ = 4Neμ (17). It suggests that a large effective population size and high mutation rate should increase the level of nucleotide variation. In the ECs, the much higher level of UV radiation received by the SFS may result in a higher mutation rate and, along with the abundance of the cyanobacterium, a higher nucleotide polymorphism when compared with the NFS.
Among environmental factors affecting biota at the ECs, the level of solar and UV radiation seems to be the most stressful and contrasting for the slopes. We therefore hypothesized that genes related to light-dependent physiological and biochemical processes should be suitable to test the above hypotheses and thus chose genes regulating circadian rhythms in N. linckia.
Regulation of physiological functions with approximate daily periodicity, or circadian rhythms, is a common feature of eukaryotes. Among prokaryotes, only cyanobacteria show this rhythmicity, which is regulated by a cluster of three genes, kaiA, kaiB, and kaiC (18, 19). The kai genes have been studied most extensively in the unicellular cyanobacterium Synechococcus. They were shown to operate as a single unit and to have a complicated mode of regulation; kaiA positively regulates kaiBC promoter, whereas overexpression of kaiC represses it (18, 20). Recently kaiC was reported to have two kaiA-binding domains (21) and to be a crucial component of clock precession in cyanobacteria (22). Circadian pacemakers in cyanobacteria were shown to influence the expression of a wide variety of genes including those regulating nitrogen fixation (23), cell division (24), and other metabolic processes (25, 26). Furthermore, close matching between the endogenous clock and the temporal environmental cycle improves the reproductive fitness of cyanobacteria (27). In all cyanobacteria studied thus far, the kaiABC cluster was reported as a single copy (18, 19).
In the present study we address several problems. First, what are the level and patterns of sequence polymorphism in kai genes of N. linckia from the ECs microsites? Second, what evolutionary factors contribute most significantly to the observed level of the polymorphism? Third, is there any adaptive significance to the observed polymorphism?
Materials and Methods
Cyanobacterial Strains.
Five cyanobacterial strains from each of 12 sampling stations (three stations on each slope of both ECs) were analyzed. The strains were maintained on BG-11-0 media (Sigma) with 1.5% agar. For DNA isolation, the cultures were grown on 50 ml of BG-11-0 liquid media for 1 month to be prepared for the subsequent DNA isolation.
DNA Isolation, Amplification, and Sequencing.
Total genomic DNA from the cyanobacterial lines was isolated by the miniprep procedure using CTAB (28) with some minor modifications. PCR primers were designed based on the published partial sequences of the kaiBC region in Cylindrospermum PCC 7417 (GenBank accession no. AF222605) and Nodularia PCC 73104 (AF222602). The amplifications were performed with forward primer kF37 (5′-AGC CGA AGA AGA TAA AAT-3′) and reverse primer kR910 (5′-GAC GTT CTC CTT CTA AAA-3′). The names of the primers refer to the respective nucleotide position in the Nodularia PCC 73104 kaiBC sequence. PCR was made in a total of 20 μl of reaction mix containing 1 unit of REDTaq DNA polymerase (Sigma), 10 ng of template, 1× REDTaq PCR buffer, 1.5 mM MgCl2, 250 μM dNTPs, and 0.25 μM of each primer. The PCR program consisted of one cycle at 94°C for 3 min followed by 45 cycles of 1 min at 94°C, 50 sec at 50°C, and 50 sec at 72°C and by final extension at 72°C for 10 min. The PCR product then was purified from 1% agarose gel with the QIAquick gel-extraction kit (Qiagen) and either used for direct sequencing (BigDye sequencing kit, Perkin–Elmer Applied Biosystems) or cloned in pGEM-T Easy vector (Promega). The cloned fragments were then isolated with QIAprep Spin miniprep kit (Qiagen) and sequenced with M13 primers. The fragments, differing by only a few singletons, were sequenced repeatedly after each additional PCR to avoid confusing with PCR errors. The newly determined kaiBC sequences of N. linckia were deposited in the GenBank, DDBJ, and EMBL databases (Table 1).
Table 1.
List of the kai gene family members obtained from the analyzed cyanobacterial strains in the ECs
| Location | Station | Haplotype | Allele | Total length (intergenic spacer) of the sequenced fragment, bp | GenBank accession no. |
|---|---|---|---|---|---|
| Lower Nahal | SFS1 | A | kaiIa* | 1039 (274) | AY051296 |
| Oren (EC I) | D | kaiIId1 | 1264 (499) | AY051303 | |
| kaiIId2 | 1264 (499) | AY051304 | |||
| kaiIIId1 | 871 (106) | AY051308 | |||
| kaiIIId2 | 878 (113) | AY051309 | |||
| E | kaiIIe | 1208 (443) | AY051301 | ||
| kaiIe | 1039 (274) | AY051297 | |||
| ψkaiIe | 1038 (274) | AY051298 | |||
| kaiIIIe | 878 (113) | AY051310 | |||
| F | kaiIIf | 1264 (499) | AY051302 | ||
| kaiIa | 1039 (274) | AY051296 | |||
| kaiIIIf1 | 871 (106) | AY051306 | |||
| kaiIIIf2 | 871 (106) | AY051307 | |||
| B | kaiIVb | 841 (76) | AY051312 | ||
| SFS2 | C | kaiIVc | 841 (76) | AY051313 | |
| Lower Nahal | SFS1 | G | kaiVg | 843 (78) | AY051317 |
| Keziv (EC II) | kaiIIIg | 848 (83) | AY051311 | ||
| ψkaiIg | 1037 (274) | AY051300 | |||
| ψkaiIIg | 1228 (464) | AY051305 | |||
| SFS2 | H | kaiVh | 843 (78) | AY051316 | |
| kaiIh | 1039 (274) | AY051299 |
Ubiquitous in all the locations unless specified, also occurs in haplotype F among the other kai gene family members. Roman numbers indicate gene subfamilies.
Data Analyses.
The sequences were aligned with CLUSTALW (29). Synonymous (Ks) and nonsynonymous (Ka) nucleotide substitutions were calculated by using the modified Nei–Gojobori method (30) with Jukes–Cantor correction and the transition/transversions ratio equal to 3. We also estimated a codon usage bias measured by the codon bias index (CBI; ref. 31). It may range from 0 (no codon bias) to 1 (maximum codon bias). Respective computations were performed by using DNASP, version 3.53 (32). The Poisson random field model with maximum likelihood estimation (33) was applied to test whether selection operates on shaping the observed nucleotide polymorphism. Phylogenetic inferences for the obtained genes were made by using the maximum likelihood approach (34) realized in DAMBE, version 4.0.57 (35). Published kai sequences of Nodularia PCC 73104 (AF222602), Cylindrospermum PCC 7417 (AF222605), and Nostoc punctiforme (preliminary sequence data were obtained from the Department of Energy Joint Genome Institute at www.jgi.doe.gov/JGI_microbial/html/index.html) were used as an outgroup. The statistical significance of the constructed trees was evaluated by bootstrapping with 1,000 replications.
Results
The amplified segment of the kaiABC cluster in N. linckia included parts of the kaiB and kaiC genes and an intergenic spacer, ≈1 kb length in total. In the 60 strains analyzed (30 from each EC), we found at least eight haplotypes, which differed by either nucleotide sequence or the number of gene copies presented. In some strains from the most stressful upper SFS1 and SFS2 stations of both canyons, the kai cluster was represented by multiple copies, at least two or four (Table 1). The strains from all other sampling stations of both canyons had the same single completely monomorphic copy of the cluster, kaiIa. Multibanded patterns of the amplicons observed on agarose gel referred to the indel polymorphism in the kaiBC intergenic spacer.
Phylogeny of kai Multigene Family and the Time of the Duplications.
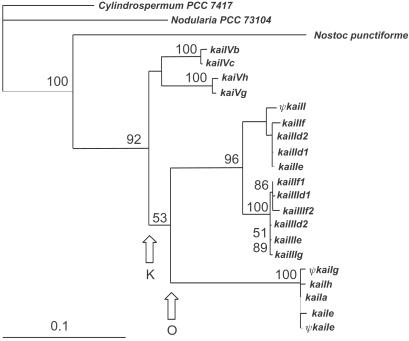
The constructed tree clearly indicates that the kai gene family is monophyletic and results from multiple duplication events (Fig. 1). All members of the family are subdivided into five groups, or subfamilies, by their nucleotide homology. Some cyanobacterial haplotypes may vary in the number of genes of different subfamilies (Table 1). The topology of the obtained phylogenetic tree suggests different times for the first duplication events occurring in the ECs (Fig. 1). A few duplications (for example, between kaiIIId1 and kaiIIId2) were quite recent.
Figure 1.
Maximum likelihood phylogenetic tree of the kai multigene family. The first duplication events in EC I and II are depicted as O and K, respectively. Bootstrap values (1,000 replications) are indicated at nodes. The bootstrap values <50% are not shown.
The molecular clock hypothesis was tested by the maximum likelihood test and was rejected if the genes of all the species (i.e., including those from the closely related Nostocaceae) were included in the test because of the faster rate of substitution in N. linckia kai gene family. However, the hypothesis was confirmed if this family was excluded. Having paleontological data about the origin of family Nostocaceae (≈1.6 billion years ago; refs. 36 and 37) and the average pairwise number of synonymous substitutions per synonymous site (30) between Nodularia, Cylindrospermum, and N. punctiforme kai genes (Ks = 0.706), we thus could determine the neutral substitution rate in non-linckian lineages. It appeared to be 2.21 × 10−10 substitutions per site per year. The approximate time of the duplications was computed based on the data of geological history of the canyons (1, 38) and pairwise Ks values of the respective gene descendants. For example, having the first duplication event in Nahal Oren (Fig. 1 O) ≈5 million years ago and Ks between kaiIe and kaiIIe equal to 0.420, we obtain the rate of 4.20 × 10−8 substitutions per site per year or ≈190 times higher than in the non-linckian lineages. Furthermore, taking Ks between kaiIIId1 and kaiIIId2 as 0.007, we finally can set the time of duplication to be ≈80,000 years ago.
Nucleotide Polymorphism and Diversity Among Members of the kai Multigene Family.
Translated amino acid sequences of the fragments showed high similarity to the respective cyanobacterial sequences reported previously (19). Some of the obtained nucleotide sequences differed either by a few synonymous substitutions or by silent substitutions in intergenic spacers, and therefore their amino acid sequences were identical. A number of the kaiBC sequences proved to be pseudogenes caused by frameshift mutations. For example, ψkaiIg resulted from the deletion of a dinucleotide at position 75 of the kaiB gene, ψkaiIIg (from the deletion of a nucleotide at position 635 of kaiC).
Overall, the deduced kai amino acid sequences in N. linckia shared 90.9% identity in the 254-residue region amplified. Walker's motif As (P-loop 1, residues 91–98), reported to be important in ATP/GTP binding and circadian rhythm regulation (18, 39), is not conserved; in the copy kaiIIId1, occurring on the stressful SFS in EC I, polar amino acid serine was replaced by nonpolar proline. Another motif, DXXG (where X is any residue), which is essential also for circadian oscillation (39), occurs twice in the studied region (residues 113–116 and 156–159) and is not conserved in the first case (either P115S or P115A). The most variable region within residues 51–61 is likely related to desiccation stress response; substitutions there are mostly in favor of hydrophobic amino acids.
The sequence alignment of 1,268 bp showed a total of 241 segregating sites (including those in indels) or 19%, of which 119 (15.6%) were in coding regions, and 122 (24.3%) were in the intergenic spacer. A large proportion of polymorphism in the spacer referred to indels that results in variation of its length from 76 (kaiIV) to 499 bp (kaiII).
Data on nucleotide polymorphism within paralogous subfamilies of kai multigene family are presented in Table 2. They suggest that although the total level of within-subfamily polymorphism essentially is the same, its patterns may vary significantly between different parts of the studied region. For example, the rate of synonymous substitutions in the kaiIII subfamily (Ks = 0.0037) is about six times lower as that in the kaiII subfamily (Ks = 0.0227), whereas in the intergenic spacer we observe an opposing picture (0.0149 and 0.0032, respectively). Members of the kaiIV subfamily show no silent substitutions in the kaiB gene and the intergenic spacer but indicate the highest Ks level in the kaiC gene (0.0347). In all the subfamilies (except kaiIII), the level of silent substitutions in the intergenic spacer is considerably lower than in the coding regions of the kai cluster. This result suggests that both coding regions and the spacer are under different selective constraints. The same is referred to the different subfamilies; presumably oldest, kaiV has a Ks value virtually the same as the most recent kaiII (0.0224 and 0.0227, respectively).
Table 2.
Data on synonymous and nonsynonymous substitutions and nucleotide diversity in the different kai domains within the subfamilies of the kai gene family in N. linckia
| Kai subfamily | No. of the member genes determined |
kaiB (135 bp)
|
Intergenic spacer (79–502 bp)
|
kaiC (627 bp)
|
Total in coding regions
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ks | Ka | π, SE | π | Ks | Ka | π, SE | Ks | Ka | π, SE | ||
| I | 5 | 0.0131 | 0.0041 | 0.0061 ± 0.0023 | 0.0029 | 0.0192 | 0.0042 | 0.0077 ± 0.0017 | 0.0181 | 0.0042 | 0.0074 ± 0.0018 |
| II | 5 | 0.0000 | 0.0079 | 0.0060 ± 0.0022 | 0.0032 | 0.0278 | 0.0034 | 0.0090 ± 0.0026 | 0.0227 | 0.0042 | 0.0085 ± 0.0025 |
| III | 6 | 0.0000 | 0.0131 | 0.0100 ± 0.0022 | 0.0149 | 0.0045 | 0.0021 | 0.0027 ± 0.0007 | 0.0037 | 0.0040 | 0.0040 ± 0.0008 |
| IV | 2 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0347 | 0.0021 | 0.0096 ± 0.0048 | 0.0284 | 0.0017 | 0.0079 ± 0.0039 |
| V | 2 | 0.0316 | 0.0000 | 0.0074 ± 0.0037 | 0.0000 | 0.0204 | 0.0000 | 0.0048 ± 0.0024 | 0.0224 | 0.0000 | 0.0053 ± 0.0026 |
| Average | — | 0.2480 | 0.0073 | 0.0523 ± 0.0061 | 0.0941 | 0.2226 | 0.0098 | 0.0522 ± 0.0062 | 0.2264 | 0.0093 | 0.0522 ± 0.0061 |
Values of nucleotide diversity (π) were obtained according to Nei (62) with Jukes–Cantor correction for multiple substitutions; in the intergenic spacer segregating sites in indels were considered.
A distinctive feature of polymorphism within kai gene subfamilies is that only singletons represent it (of course, here we do not consider the subfamilies kaiIV and kaiV made up of only two members). Moreover, there are only two mutations shared between the subfamilies (Table 3). Overall, between-subfamily variability accounts for 93% of the total variability observed in the kai family. These facts provide proof for the recent duplication of the genes and their primarily functional divergence.
Table 3.
Variation within and between subfamilies of the kai gene family in N. linckia
| Subfamilies | kaiI | kaiII | kaiIII | kaiIV | kaiV |
|---|---|---|---|---|---|
| kaiI | 0.006 ± 0.002 | 130/0 | 137/0 | 131/1 | 147/0 |
| kaiII | 0.161 ± 0.014 | 0.007 ± 0.001 | 28/0 | 72/1 | 72/0 |
| kaiIII | 0.115 ± 0.012 | 0.036 ± 0.005 | 0.006 ± 0.001 | 72/0 | 71/0 |
| kaiIV | 0.094 ± 0.011 | 0.093 ± 0.010 | 0.088 ± 0.010 | 0.007 ± 0.003 | 61/0 |
| kaiV | 0.114 ± 0.013 | 0.092 ± 0.010 | 0.086 ± 0.010 | 0.078 ± 0.010 | 0.005 ± 0.003 |
Between-subfamilies diversity and its standard error are shown below the diagonal. The pairwise numbers of fixed/shared mutations are indicated above the diagonal. The diagonal shows within-subfamily diversity. The Tamura–Nei model of substitutions (63) was used for the calculations.
Codon Usage Bias and G+C Content.
The studied region in all kai subfamilies, as well as in the kaiBC region from the other Nostocaceae, appears to have strong codon usage bias (Table 4). In N. linckia the mean value of CBI was 0.473. The analysis of relative codon usage tables (data not shown) indicates that the bias is in favor of NNT and NNA codons over NNC and NNG codons, which is expressed in the lower G+C content (0.359) in the kaiB gene compared with kaiC (0.423), especially at third codon positions (0.236 and 0.350, respectively). Estimating codon usage in complete kaiB and kaiC sequences of the closest relative N. punctiforme results in a CBI of 0.545 and 0.487, respectively. Based on the partial sequences of kaiB and kaiC from N. linckia and N. punctiforme and their respective values of CBI (data not shown), we could suppose rather firmly that the values of CBI for complete sequences of these genes in N. linckia are similar to those in N. punctiforme. Significantly different CBI values between kaiB and kaiC is a very interesting fact, taking into account that both these genes are regulated by a single promoter (18). In the most recent subfamilies kaiII and kaiIII, this bias is lower (P < 0.001) than in the other evolutionarily older subfamilies but still higher than in kaiC.
Table 4.
Comparative data on codon usage and G+C content in the studied region of the kai gene family and some other genes of N. linckia and other cyanobacteria
| Gene (accession number) | Promoter/ intergenic spacer*
|
Coding region
|
|||
|---|---|---|---|---|---|
| G+C | CBI | G+C2 | G+C3s | G+C | |
| kai | |||||
| Nodularia PCC 73104 | 0.286 | 0.425 | 0.368 | 0.336 | 0.419 |
| Cylindrospermum PCC 7417 | 0.289 | 0.398 | 0.375 | 0.353 | 0.423 |
| N. punctiforme | 0.246 | 0.538 | 0.370 | 0.273 | 0.400 |
| kaiI, N. linckia | 0.350 | 0.452 | 0.373 | 0.323 | 0.413 |
| kaiII, N. linckia | 0.405 | 0.482 | 0.378 | 0.334 | 0.414 |
| kaiIII, N. linckia | 0.314 | 0.490 | 0.375 | 0.332 | 0.412 |
| kaiIV, N. linckia | 0.226 | 0.470 | 0.366 | 0.342 | 0.415 |
| kaiV, N. linckia | 0.321 | 0.458 | 0.374 | 0.320 | 0.411 |
| Average in kai of N. linckia | 0.368 | 0.473 | 0.374 | 0.330 | 0.413 |
| Acetyl-CoA synthase, N. linckia (AY037787) | — | 0.712 | 0.463 | 0.918 | 0.661 |
| dnaK, N. linckia (AF388880) | 0.374 | 0.409 | 0.439 | 0.421 | 0.490 |
| sodF, N. linckia (AY055243) | 0.320 | 0.252 | 0.370 | 0.464 | 0.455 |
| Ribosomal protein L12, Synechocystis (X53178)† | 0.368 | 0.693 | 0.398 | 0.408 | 0.484 |
| trpA, N. punctiforme (AF288131) | 0.478 | 0.405 | 0.439 | 0.409 | 0.487 |
G + C2 − GC content at the second codon positions; G + C3s − GC content at third (synonymous) codon positions; G + C − GC content in the genomic region.
In kai gene.
Ref. 64.
Discussion
The phenomenon of the kai multigene family is quite unique for prokaryotes. In fact, this family has resulted from duplications of a whole cluster of three genes (≈3 kb) rather than of a single gene. We have not found data in the available literature about duplication of such a long functional region of genomic DNA in prokaryotes.
The kai family contains at least five distinct subfamilies. Two of them, kaiIV and kaiV, occur only in EC I and EC II, respectively. Interestingly, genes of the kaiIV subfamily were found only in single-copy haplotypes, whereas genes of the kaiV subfamily were observed in multicopy haplotypes (Table 1).
Rapid Evolution of the Kai Gene Family Under the Local Environmental Stress.
The kai multigene family is relatively large and evolutionarily recent. In N. linckia, we found at least 20 different functional genes and pseudogenes, and further sequencing, most likely, will reveal additional members. The absence of respective homologs in many other species of cyanobacteria, and even closely related Nostocaceae (e.g., Nodularia, Cylindrospermum, and N. punctiforme), indicates that origin and radiation of the family occurred after the speciation of N. linckia. A milestone question is, did the duplications occur before or after the canyons evolved? The high number of singletons, low within-subfamily and high between-subfamily diversity, the occurrence of the multiple-copy haplotypes only in the stressful SFS stations of both canyons, and low nucleotide polymorphism in the pseudogenes support the latter assumption; this gene family originated and has evolved along with the geological evolution of the microsites EC I and EC II. There are a lot of data about numerous gene families in prokaryotes (i.e., refs. 40–43) that suggest that the number of universally conserved gene families is small and thus assumes horizontal gene transfer and genome fusion as major forces in the evolution of prokaryotes (44). In the case of the kai gene family, we do not have evidence for the latter; instead, a stressful environment and high UV radiation probably give rise to genome instability and more replication errors, which may lead to frequent duplications.
The general areas of the canyons initially were occupied most likely by strains with a single ancestral copy of the kai cluster. Deepening the relief of the canyons by increasing erosion during the uplift of Mt. Carmel resulted in sharpening the interslope environmental conditions at a microscale and, consequently, in further single-copy kai cluster evolution. At some period, different patterns of geological development of the canyons brought about unequal rates of molecular evolution in the kai subfamilies and, consequently, different times for the first cluster's duplications in the canyons (Fig. 1). Despite the potential for a high migration rate between the slopes and the canyons (which is confirmed by the common occurrence of haplotypes with the invariable copy kaiIa), genetic diversity within the gene family is determined largely by the local contrasting microclimatic factors in the microsites. This is confirmed by the fact that all the haplotypes are unique to the canyons in which they occur. Furthermore, they are not observed even in the lowest stations, SFS3, of the same African slopes of the canyons. The unique nonsense mutations in all observed pseudogenes also support the hypothesis that they evolved independently. Additionally, if one supposes that the multiple-copy haplotypes were introduced into the canyons and then evolved further, one should expect to find such haplotypes on the two opposite slopes, but this was not the case.
Driving Forces of Kai Gene Family Evolution: Selection vs. Neutrality.
According to the theory elaborated thus far, there are three potential ways of the further evolution of duplicated genes: (i) one of the copies maintains the original function, whereas another may accumulate deleterious mutations and become nonfunctional (nonfunctionalization), (ii) one copy acquires a novel, advantageous function and becomes rapidly fixed in a population, and another copy still keeps the original function (neofunctionalization), and (iii) duplication may lead to the partitioning of original functions such that both new copies have reduced efficiency compared with the ancestral gene (subfunctionalization; refs. 45–49).
In the origin and evolution of the kai multigene family, one can clearly determine the case of nonfunctionalization (pseudogenes ψkaiIe, ψkaiIg, and ψkaiIIg). On the other hand, neofunctionalization and subfunctionalization seem not to have occurred during this evolution. The former demands sufficiently large divergence between the copies to attain a new function by one of them. For the kaiII and kaiIII subfamilies, it is hardly probable because of the short period elapsed since the duplication. In turn, subfunctionalization implies lower fitness of each newly arising gene, but it is not the case for the kai subfamilies, because almost all multiple-copy haplotypes (except haplotype D) contain virtually the same kaiIa allele, which occurs in the single-copy strains ubiquitous in all locations and, consequently, have at least the same or similar fitness. Therefore, there is no evidence for the decreased fitness of the newly duplicated genes. Instead, our data suggest that these duplications may have adaptive significance and most likely increase fitness. A similar assumption, that gene duplications (at least in enteric bacteria) are quite rapid and perhaps the first response to strong selection, was made in the early Escherichia coli literature (50). We propose to consider such a way of evolution of duplicated genes as superfunctionalization (reinforcement).
Indeed, evolution generally should increase fitness of organisms to ensure their survival. In the ECs, rise in the daily abundance of UV and visible radiation on the SFS, compared with the NFS, should favor temporal intracellular adaptation to the local day/night conditions to optimize programming for the cellular photochemically dependent processes. This process led to either the adaptive evolution of a single copy of the kai cluster or the appearance of multiple copies. Recently Haack and Roth (51) showed that in populations of enteric bacteria, spontaneous tandem chromosomal duplications themselves are fairly common; most of them are unstable and lost, but some may confer a selective advantage under specific conditions. Our data on patterns of synonymous and nonsynonymous substitutions in the different domains of the studied region of the kai cluster (Table 1) provide further evidence for the predominance of selective factors operating to shape the observed polymorphism. The most recently duplicated subfamilies kaiII and kaiIII have only replacement substitutions fixed in the kaiB gene, providing clear evidence for positive selection. Nonsynonymous mutations arising de novo in this domain of the subfamilies after the duplication conferred new functional features to the gene apparently had adaptive significance and thus became maintained in the population. For instance, replacing polar amino acid serine by nonpolar proline in ATP/GTP binding site of kaiIIId1 likely is adaptive to desiccation, because proline was shown to be related to drought-stress response (52–54). The substitutions in the 51–61 amino acid region probably have similar adaptive significance, because they are generally in favor of hydrophobic amino acids. The strains with the multiple gene copies seem to have a selective advantage under harsh conditions merely because nonsense mutations in one or even two copies of the cluster (as in haplotypes E and G, Fig. 1) do not result in the death of the mutant. By contrast, polymorphism of kaiI, kaiIV, and kaiV subfamilies has already acquired adaptive significance to the specific local environmental conditions, such that most of the replacements become deleterious and, therefore are eliminated by purifying selection. Thus, the low within-subfamily nucleotide polymorphism is governed by purifying selection, whereas the high haplotype diversity is maintained by diversifying and stabilizing selections.
Comparison of codon usage in kaiBC genes to that in some highly and moderately expressed genes (Table 4) suggests that the kai genes are highly expressed. Of the two kai genes, kaiB has a stronger codon usage bias and probably higher expressivity than kaiC. Codon usage patterns usually are well conserved during evolution and confer selective constraints to the rate of synonymous substitutions (55). In the kai gene family, despite the high between-subfamily rate of synonymous substitutions, the within-subfamily rate is low (Table 2), and both are constrained to maintain the low GC content. The observed codon bias in the kai genes in favor of A/T-ended codons may be related to an overall high A+T content of the genome.
The maximum likelihood estimates using the Poisson random field model (33) give further strong support for selection as a principle driving force of kai gene family evolution (Table 5). They suggest that selection operates against disfavored amino acids (γ = −3.76) and in favor of synonymous substitutions (γ = 2.84), which confirms the general prevalence of purifying selection in the whole studied kaiBC region. However, in the particular kai regions and subfamilies, different types of selection may be predominant as is, for example, a case of positive selection described above for kaiB gene of kaiII and kaiIII subfamilies.
Table 5.
Maximum likelihood estimates of selection intensity and mutation rate in the kai gene family of N. linckia based on the Poisson random field model
| Sites | No. of sites | u | μg | Ne* | γ | LLexp | LLobs | P, one d.f. |
|---|---|---|---|---|---|---|---|---|
| Synonymous | 178 sites | 5.84 ± 1.44 | 3.28 × 10−2 ± 0.81 × 10−2 | 7.81 × 106 ± 1.93 × 106 | 2.84 ± 3.84 | −38.799 | −35.342 | 0.0085 |
| Nonsynonymous | 254 codons | 9.43 ± 3.56 | n.a. | n.a. | −3.76 ± 2.46 | −19.984 | −13.672 | 0.0004 |
u, total mutation rate (scaled to Ne); μg, mutation rate per site per generation (scaled to Ne); γ, selection coefficient (scaled to Ne); LLexp, expected log likelihood (provided γ = 0); LLobs, observed log likelihood (for the obtained γ); n.a., not applied; d.f., degree of freedom.
Estimated with μg = 4.20 × 10−9 (assuming per site per year μ = 4.20 × 10−8 and 10 generations per year on average).
The scaled per site per generation mutation rate obtained by using θ is consistent with that computed with maximum likelihood estimates (μg, Table 5). Actually, given synonymous θ = 0.162 and, on average, 10 generations of Nostoc per year (56) yields μg = 0.81 × 10−2, which is only four times less than the value obtained with maximum likelihood estimates. However, under conditions on the SFS of both canyons, the life cycle of cyanobacteria may increase considerably in length. The longer generation may take place on the one hand because of generally favorable conditions in the rainy season (October–May) when vegetative stages of Nostoc prevail, and on the other hand, during the dry season (May–October) because of longer intervals between the spores' production and germination (56). Therefore, Nostoc on the SFS of both canyons most likely has 4–5 generations per year. In such a case, values of μg calculated with the two mentioned methods become virtually the same. Such consistency of the results obtained by different approaches strongly confirms our assumptions about the patterns of the kai gene family evolution.
Do neutral factors operate to shape polymorphism in the kai genes? According to the neutral theory, neutral genetic diversity (θ) in a haploid population is governed by the effective population size (Ne) and mutation rate (μ), θ = 2Neμ (17). Thus, large population size and high mutation rate should increase the level of polymorphism. As stated above, the neutral mutation rate in the kai genes from N. linckia in the stressful SFS stations of the ECs was determined to be 4.20 × 10−8 substitutions per site per year, which is almost 200 times higher than in the kai genes of other cyanobacteria. It also is almost 10–20 times higher than the rate of neutral substitutions in endosymbiotic E. coli and Buchnera (57). The effective population size of N. linckia from those slopes is estimated to be 7.81 × 106 ± 1.93 × 106 (Table 5). It is almost 2 orders of magnitude lower than the estimated Ne for E. coli (33) and, most likely, significantly lower than the Ne for N. linckia from the NFS of the canyons. The smaller effective population size thus may promote faster fixation of neutral and slightly advantageous substitutions. But this factor apparently plays a minor role in the evolution of the kai gene family.
An intriguing question concerns the level of polymorphism in the kaiBC intergenic spacer. In all the subfamilies except kaiIII, it indicates significantly lower nucleotide diversity than in the coding regions (Table 2), although it was expected to be at least as high as the level of synonymous substitutions. There are numerous data that suggest that intergenic spacers and 5′-flanking sequences may play an important role in the regulation of transcription (58–60). Therefore, although the intergenic spacer was reported to have no functional significance in the kai cluster (18), our data on its nucleotide polymorphism may suggest differently, at least in some kai subfamilies.
The long-term study of the evolutionary patterns under an acute microclimatic stress in EC I revealed that, across phylogeny, permanent ecological stress usually results in a higher mutation rate and an increase in genetic polymorphism (2, 3, 61). This higher polymorphism in the cyanobacteria may arise, in addition to other factors, from the multiple gene duplication events, which probably are one of the ways that cyanobacteria adapt to the extreme and fluctuating environment. Under these conditions, evolution of functionally important genes such as those regulating circadian rhythms is governed primarily by various types of selection rather than by purely neutral factors. The described example of extremely rapid evolution of the clock gene family controlling circadian rhythmicity, one of the fundamental features in most organisms, helps to better understand the processes underlying adaptation by natural selection to environmental stress.
Acknowledgments
We thank Mrs. T. Krugman and Dr. N. Satish for help in collecting cyanobacterial strains and technical assistance in DNA isolation. We also thank Prof. O. Savolainen (University of Oulu), Dr. C. Vogl (University of Munich), and Prof. D. Hartl (Harvard University) for valuable comments on the manuscript. This work was supported by the Israeli National Science Foundation Grant 98-04-4963, the Israeli Discount Bank Chair of Evolutionary Biology, and the Ancell-Teicher Research Foundation for Genetics and Molecular Evolution.
Abbreviations
- EC
Evolution Canyon
- SFS
south-facing slope
- NFS
north-facing slope
- CBI
codon bias index
References
- 1.Nevo E. Proc R Soc London B. 1995;262:149–155. [Google Scholar]
- 2.Nevo E. Theor Popul Biol. 1997;52:231–243. doi: 10.1006/tpbi.1997.1330. [DOI] [PubMed] [Google Scholar]
- 3.Nevo E. Proc Natl Acad Sci USA. 2001;98:6233–6240. doi: 10.1073/pnas.101109298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitton B A. In: Survival and Dormancy of Microorganisms. Hennis Y, editor. New York: Wiley; 1987. pp. 109–167. [Google Scholar]
- 5.Clarke A K, Soitamo A, Gustafsson P, Oquist G. Proc Natl Acad Sci USA. 1993;90:9973–9977. doi: 10.1073/pnas.90.21.9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kulkarni R D, Golden S S. J Bacteriol. 1994;176:959–965. doi: 10.1128/jb.176.4.959-965.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erdmann N, Effmert U, Fulda S, Oheim S. Curr Microbiol. 1997;35:348–355. doi: 10.1007/s002849900267. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka N, Nakamoto H. FEBS Lett. 1999;458:117–123. doi: 10.1016/s0014-5793(99)01134-5. [DOI] [PubMed] [Google Scholar]
- 9.Vass I, Kirilovsky D, Perewoska I, Mate Z, Nagy F, Etienne A L. Eur J Biochem. 2000;267:2640–2648. doi: 10.1046/j.1432-1327.2000.01274.x. [DOI] [PubMed] [Google Scholar]
- 10.Van Valen L. Am Nat. 1965;99:377–390. [Google Scholar]
- 11.Soulé M, Stewart B R. Am Nat. 1970;104:85–97. [Google Scholar]
- 12.Nevo E. Evol Biol. 1988;23:217–246. [Google Scholar]
- 13.Nevo E. J Exp Zool. 1998;282:95–119. [Google Scholar]
- 14.Vinogradova O N, Kovalenko O V, Wasser S P, Nevo E, Kislova O A, Belikova O A. Algologia. 2000;5:46–55. [Google Scholar]
- 15.Satish N, Krugman T, Vinogradova O N, Nevo E, Kashi Y. Microb Ecol. 2001;42:306–316. doi: 10.1007/s00248-001-0013-0. [DOI] [PubMed] [Google Scholar]
- 16.Krugman T, Satish N, Vinogradova O N, Beharav A, Kashi Y, Nevo E. Evol Ecol Res. 2001;3:899–915. doi: 10.1007/s00248-001-0013-0. [DOI] [PubMed] [Google Scholar]
- 17.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, U.K.: Cambridge Univ. Press; 1983. [Google Scholar]
- 18.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson C R, Tanabe A, Golden S S, Johnson C H, Kondo T. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 19.Lorne J, Scheffer J, Lee A, Painter M, Miao V P. FEMS Microbiol Lett. 2000;189:129–133. doi: 10.1111/j.1574-6968.2000.tb09218.x. [DOI] [PubMed] [Google Scholar]
- 20.Johnson C H, Golden S S, Kondo T. Trends Microbiol. 1998;6:407–410. doi: 10.1016/s0966-842x(98)01356-0. [DOI] [PubMed] [Google Scholar]
- 21.Taniguchi Y, Yamaguchi A, Hijikata A, Iwasaki H, Kamagata K, Ishiura M, Go M, Kondo T. FEBS Lett. 2001;496:86–90. doi: 10.1016/s0014-5793(01)02408-5. [DOI] [PubMed] [Google Scholar]
- 22.Xu Y, Mori T, Johnson C H. EMBO J. 2000;19:3349–3357. doi: 10.1093/emboj/19.13.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y B, Dominic B, Mellon M T, Zehr J P. J Bacteriol. 1998;180:3598–3605. doi: 10.1128/jb.180.14.3598-3605.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori T, Binder B, Johnson C H. Proc Natl Acad Sci USA. 1996;93:10183–10188. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang T-C, Chen H-M, Pen S-Y, Chen T H. Planta. 1994;193:131–136. [Google Scholar]
- 26.Johnson C H, Golden S S. Annu Rev Microbiol. 1999;53:389–409. doi: 10.1146/annurev.micro.53.1.389. [DOI] [PubMed] [Google Scholar]
- 27.Ouyang Y, Andersson C R, Kondo T, Golden S S, Johnson C H. Proc Natl Acad Sci USA. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golden S S, Brusslan J, Haselkorn R. Methods Enzymol. 1987;153:215–231. doi: 10.1016/0076-6879(87)53055-5. [DOI] [PubMed] [Google Scholar]
- 29.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nei M, Gojobori T. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 31.Morton B R. J Mol Evol. 1993;37:273–280. doi: 10.1007/BF00175504. [DOI] [PubMed] [Google Scholar]
- 32.Rozas J, Rozas R. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- 33.Hartl D L, Moriyama E N, Sawyer S A. Genetics. 1994;138:227–234. doi: 10.1093/genetics/138.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Felsenstein J. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 35.Xia X. Data Analysis in Molecular Biology and Evolution. Boston: Kluwer; 2000. [Google Scholar]
- 36.Tyler S A, Barghoorn E S. Science. 1954;119:606–608. doi: 10.1126/science.119.3096.606. [DOI] [PubMed] [Google Scholar]
- 37.Cloud P E. Science. 1965;148:27–35. doi: 10.1126/science.148.3666.27. [DOI] [PubMed] [Google Scholar]
- 38.Karcz Y. Bull Res Counc Isr G. 1959;8:119–130. [Google Scholar]
- 39.Nishiwaki T, Iwasaki H, Ishiura M, Kondo T. Proc Natl Acad Sci USA. 2000;97:495–499. doi: 10.1073/pnas.97.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hill K E, Marchesi J R, Weightman A J. J Bacteriol. 1999;181:2535–2547. doi: 10.1128/jb.181.8.2535-2547.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makarova K S, Aravind L, Galperin M Y, Grishin N V, Tatusov R L, Wolf Y I, Koonin E V. Genome Res. 1999;9:608–628. [PubMed] [Google Scholar]
- 42.Yanai I, Camacho C J, DeLisi C. Phys Rev Lett. 2000;85:2641–2644. doi: 10.1103/PhysRevLett.85.2641. [DOI] [PubMed] [Google Scholar]
- 43.Carlyon J F, Roberts D M, Marconi R T. Microb Pathog. 2000;28:89–105. doi: 10.1006/mpat.1999.0326. [DOI] [PubMed] [Google Scholar]
- 44.Koonin E V, Galperin M Y. Curr Opin Genet Dev. 1997;7:757–763. doi: 10.1016/s0959-437x(97)80037-8. [DOI] [PubMed] [Google Scholar]
- 45.Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970. [Google Scholar]
- 46.Walsh J B. Genetics. 1985;110:345–364. doi: 10.1093/genetics/110.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clark A G. Proc Natl Acad Sci USA. 1994;91:2950–2954. doi: 10.1073/pnas.91.8.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sidow A. Curr Opin Genet Dev. 1996;6:715–722. doi: 10.1016/s0959-437x(96)80026-8. [DOI] [PubMed] [Google Scholar]
- 49.Force A, Lynch M, Pickett F B, Amores A, Yan Y L, Postlethwait J. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langridge J. Mol Gen Genet. 1969;105:74–83. doi: 10.1007/BF00750315. [DOI] [PubMed] [Google Scholar]
- 51.Haack K R, Roth J R. Genetics. 1995;141:1245–1252. doi: 10.1093/genetics/141.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshiba Y, Kiyosue T, Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. Plant Cell Physiol. 1997;38:1095–1102. doi: 10.1093/oxfordjournals.pcp.a029093. [DOI] [PubMed] [Google Scholar]
- 53.Harrak H, Chamberland H, Plante M, Bellemare G, Lafontaine J G, Tabaeizadeh Z. Plant Physiol. 1999;121:557–564. doi: 10.1104/pp.121.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Menke U, Renault N, Mueller-Roeber B. Plant Physiol. 2000;122:677–686. doi: 10.1104/pp.122.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ikemura T. Mol Biol Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 56.Kondratieva N V, Posudin, Yu I, Belikova O A. Ukr Bot J. 1987;44:21–25. [Google Scholar]
- 57.Ochman H, Elwyn S, Moran N A. Proc Natl Acad Sci USA. 1999;96:12638–12643. doi: 10.1073/pnas.96.22.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Busby S J, Reeder R H. Cell. 1983;34:989–996. doi: 10.1016/0092-8674(83)90556-1. [DOI] [PubMed] [Google Scholar]
- 59.Flavell R B, O'Dell M, Sharp P, Nevo E, Beiles A. Mol Biol Evol. 1986;3:547–558. [Google Scholar]
- 60.Lang W H, Morrow B E, Ju Q, Warner J R, Reeder R H. Cell. 1994;79:527–534. doi: 10.1016/0092-8674(94)90261-5. [DOI] [PubMed] [Google Scholar]
- 61.Lamb B C, Saleem M, Scott W, Thapa N, Nevo E. Genetics. 1998;149:87–99. doi: 10.1093/genetics/149.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ. Press; 1987. [Google Scholar]
- 63.Tamura K, Nei M. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 64.Sibold C, Subramanian A R. Biochim Biophys Acta. 1990;1050:61–68. doi: 10.1016/0167-4781(90)90142-o. [DOI] [PubMed] [Google Scholar]