Abstract
Cancer remains the second leading cause of mortality globally, necessitating the development of novel therapeutic agents. In this work, we synthesized 34 derivatives of nitrated N-substituted-4-hydroxy-2-quinolone-3-carboxamides, which were spectroscopically analyzed using FT-IR, NMR (1H and 13C), and elemental analysis. Derivatives tailored with m-CF3 (10), m-OCH3 (13), m-Cl (16), and m-F (20) benzyl moiety exhibited distinctive cytotoxicity against human colon cancer (HCT-116) cells with IC50s of 23.41, 27.14, 28.43, and 22.95 µM. Analogue 11 showed 100% inhibitory activity against ovarian cancer (NCI/ADR-RES), colon cancer (COLO 205), CNS cancer (SF-295), and melanoma (SK-MEL-2) cells. Cheminformatics analysis further revealed insights into the physicochemical and drug-like properties of these analogues, highlighting their potential to bind PI3Kα through alignment with key pharmacophoric features required for effective enzyme interaction. Molecular docking studies against both wild-type and mutant PI3Kα elucidated binding interactions, suggesting that specific substituents enhance selectivity and potency. This study highlights the therapeutic potential of quinolone derivatives in targeting cancer-related pathways and contributes valuable data to the ongoing search for more effective anticancer therapies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13065-025-01616-w.
Keywords: Anticancer agents, Cheminformatics, HCT-116, PC-3, Pathway analysis, Nitrated N-benzyl-4-hydroxy-2-quinolone-3-carboxamides
Introduction
Cancer remains a major challenge for the medical community, being the second leading cause of death worldwide, with approximately 19.3 million new cases and 10 million deaths reported in 2020 alone [1–4]. Projections indicate around 27 million cases by 2050, resulting in 17.5 million deaths annually [5]. Various risk factors, including cigarette smoking [6], alcohol consumption [7], obesity [8], psychological stress [9], exposure to hazardous environmental agents [9], and UV radiation [10], significantly contribute to cancer incidence. Additionally, infectious diseases such as Helicobacter pylori [11], human papillomavirus (HPV) [12], and hepatitis B virus (HBV) [13] play a role in cancer prognosis. Recent research suggests that changes in the microbiome may also influence disease pathways including cancer pathways, highlighting an emerging area of interest in cancer research [14, 15].
Furthermore, cancer is characterized by abnormal and uncontrolled cellular responses and pathways that disrupt essential processes like proliferation and apoptosis [16, 17]. Malignant cells can invade and damage surrounding tissues, leading to metastasis [18, 19]. Such behaviors stem from mutations that aberrantly activate critical signaling pathways involved in cellular proliferation and survival [20, 21]. Notable pathways implicated in various cancers include MAPK/ERK [21, 22], PI3K/PKB (AKT)/mTOR [23–26], GSK3β/β-catenin/Wnt [27, 28], EGFR/RAS/RAF [29], and JAK/STAT [30].
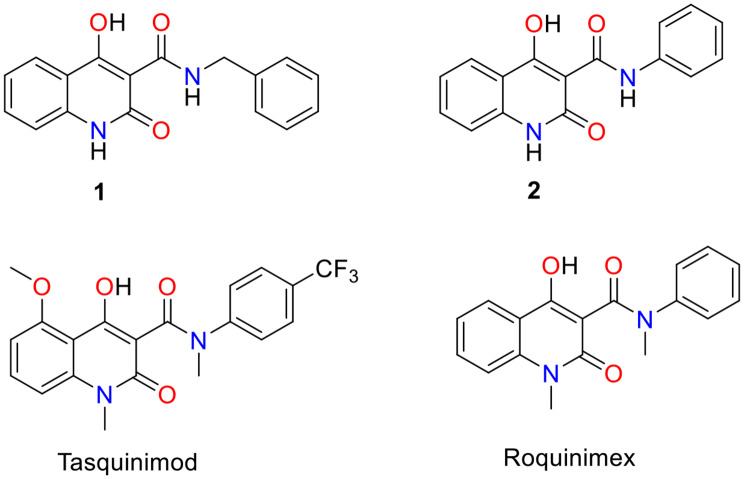
Numerous studies have highlighted the importance of the quinolone scaffold as an effective class of anticancer agents. This distinctive structure suppresses tumor growth through various mechanisms, including modulation of nuclear receptor activity, disruption of cell migration, induction of cell cycle arrest, promotion of apoptosis, and inhibition of angiogenesis [31]. Notably, quinolone derivatives have demonstrated anticancer efficacy against several types of cancer, including colorectal, breast, renal, and lung cancers [32–34]. Our research team has focused extensively on the quinolone core, identifying N-benzyl-4-hydroxy-2-quinolone-3-carboxamide (1) through pharmacophore modeling and pharmacophore-based virtual screening [35]. Further optimization led to the discovery of N-phenyl-4-hydroxy-2-quinolone-3-carboxamide (2) (Fig. 1) (Supplementary Figure S1) [36].
Fig. 1.
The structures of 1, 2, Tasquinimod, and Roquinimex
Several quinolone-based derivatives have since been synthesized and biologically evaluated for their anticancer properties. Fortunately, these derivatives have shown significant activity against human colon carcinoma (HCT-116) cells [37, 38], along with notable selectivity for the mutant H1047R PI3Kα over the wild-type PI3Kα [35, 36]. Furthermore, two quinolones are undergoing clinical trials as anticancer agents (angiogenesis inhibitors); Tasquinimod and Roquinimex (Linomide®) (Fig. 1) [39, 40].
The fruitful outcome of our ongoing research has encouraged us to tailor the quinolone core structure with a nitro group on two positions. Studies showed that NO reacts directly with DNA, causing base deamination, nitration, oxidation, and inhibition of DNA repair [41]. Numerous studies showed that compounds containing NO functionalities exert cytotoxicity [42–45]. In this framework, a series of nitrated N-substituted-4-hydroxy-2-quinolone-3-carboxamides has been synthesized to probe the significance of incorporating a nitro moiety on the anticancer activity and to elaborate the structure-activity relationship (SAR) of the analogs. Biological examinations have been probed against HCT-116 and human prostate cancer (PC-3) cell lines.
Furthermore, we undertook a computational approach to assess perturbed cancer pathways in the selected model cell lines and conducted molecular descriptor analysis to compare the molecular diversity in the multidimensional physicochemical space [46–48]. It is known that computational chemical biology studies further elucidate the relationship between molecular structure and activity as has been reported in the scientific literature. Docking studies allow scientists to gain further insights into the binding interactions between small molecules and their target proteins (or some of their target proteins), providing detailed information about the orientation and affinity of ligands within the binding site when they are compared with the crystalized molecule that shares a similar core structure. Ultimately, our research contributes to the broader goal of developing novel targeted therapies for cancer, addressing an urgent medical need.
Materials and methods
Chemistry
All the chemicals and solvents used in this project have been purchased from the corresponding companies (SD Fine-Chem Limited (SDFCL), Acros Organics, Sigma-Aldrich, Fluka, Sharlau, Tedia, Gainland Chemical Company (GCC), and BBC Chemicals). n-hexane (95%), anhydrous tetrahydrofuran (THF), N,N-dimethylformamide (DMF), dimethyl sulfoxide (DMSO), and petroleum ether supplied (M-Tedia). Ethanol (C2H5OH) and chloroform (CHCl3) (Emsure Company). Methanol (CH3OH) and sodium bicarbonate (NaHCO3) (Sigma-Aldrich). Ethyl acetate (C4H8O2) and anhydrous sodium sulfate (Na2SO4) (Fisher Scientific). Sulfuric acid (H2SO4) and sodium ethoxide (C2H5ONa) (Honeywell/ Fluka). Acetone 99.8% (LABCHEM), hydrochloric acid (HCl) 35.4% (Alpha-Chemika), silicone liquid, and calcium chloride (BBC Chemicals). 2-Amino-4-nitrobenzoic acid, 2-amino-5-nitrobenzoic acid (95%). Benzyl amine, 3-trifluromethyl benzyl amine, 4-trifluromethyl benzyl amine, 2-methoxy benzyl amine, 3-methoxy benzyl amine, 4-methoxy benzyl amine, 2-chloro benzyl amine, 3-chloro benzyl amine, 4-chloro benzyl amine, 2-fluro benzyl amine, 3-fluro benzyl amine, 4-fluro benzyl, 3-methyl benzyl amine, 4-methyl benzyl amine, 4-hydroxy benzyl amine, aniline, 2-fluro aniline, 3-fluro aniline, 4-fluro aniline, p-aniside, 4-aminophenol, 2-amino-5-methyl benzoic acid, 2-amino-4-chloro benzoic acid, phenyl hydrazine. All the previously mentioned chemicals were of analytical grade and highly purified, hence they were utilized without further purification. Thin-layer chromatography (TLC) was performed on 20 × 20 cm and 0.20 mm thickness of pre-coated aluminum sheets with fluorescent silica gel (Macherey-Nagel, Germany) and visualized using UV light (254/366nm). Evaporation of ordinary solvents was carried out using a Rota vapor model R-215 (Buchi, Switzerland) linked to a vacuum pump v-700 and heating water bath (Hei-VAP value digital, Heidolph). The melting point was measured using Gallenkamp melting point apparatus. Hot plates and magnetic stirrers were obtained from Thermo Scientific Cimarec. Shimadzu IR Affinity FTIR spectrophotometer was used to record Infrared (IR) spectra; samples were mixed with potassium bromide (Sigma-Aldrich) and pressed into a disc. 1H and 13C Nuclear Magnetic Resonance (NMR) spectra were analyzed by Bruker NanoBay 400 MHz spectrophotometer (the Hashemite University), chemical shifts are expressed in δ (ppm) and (J) coupling constant values are presented in Hz (Hertz) using TMS internal reference; DMSO and/or NaOD were/was used to dissolve the samples. Elemental analyses (the Hashemite University) were conducted using a Euro Vector (Italy) elemental analyzer, model EUROEA3000 A.
Synthesis of targeted compounds
Ethyl 2-amino-4-nitrobenzoate (5a)
Amount of 2-amino-4-nitrobenzoic acid (3a) (5.0 g, 27.4 mmol) was mixed with an excess amount of absolute ethanol(4) (450 ml). The mixture was placed in an ice bath for 10 min followed by adding H2SO4 (7.5 ml) dropwise. The reaction was refluxed for 3 days and the progress of the reaction was monitored using TLC. After cooling, the excess ethanol was removed by evaporation under reduced pressure. To get rid of the residual acid, extraction using (5%) NaHCO3 solution and CHCl3 was applied three times, followed by twice extraction with CHCl3 and water (25 ml) to remove any salt formed by acid and bicarbonate reaction. Anhydrous Na2SO4 was used to dry the organic layer, then the product was obtained by evaporating under reduced pressure. Orange powder (5.4 g) with yield 96%, Rf = 0.67 (n-Hexane: Ethylacetate) (8:2); m.p: 89–90 °C; 1H-NMR (400 MHz, DMSO-d6): δ = 1.33 (t, J = 7.0 Hz, 3 H, - CH3), 4.30 (q, J = 7.2 Hz, 2 H, -OCH2), 7.13 (s, 2 H, NH2), 7.25 (d, J = 7.2 Hz, 1H, Ar-H), 7.67 (s, 1H, Ar-H), 7.93 (d, J = 9.2 Hz, 1H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 14.5 (1 C), 61.2 (1 C), 108.5 (1 C), 111.3 (1 C), 113.8 (1 C), 133.2 (1 C), 151.2 (1 C), 152.7 (1 C), 166.6 (1 C) ppm; IR (KBr disc): 3487, 3373, 2989, 1743,1699, 1637, 1589, 1517, 1444, 1392, 1350, 1301 cm− 1; Elemental Anal. Calcd. for C12H10N2O6: C, 51.81; H, 3.62; N, 10.07. Found: C, 51.21; H, 3.60; N, 10.13.
Ethyl 2-amino-5-nitrobenzoate (5b)
Amount of 2-amino-5-nitrobenzoic acid (3b) (5.0 g, 27.4mmol) was mixed with an excess amount of absolute ethanol(4) (450 ml). The mixture was placed in an ice bath for 10 min followed by adding H2SO4 (7.5 ml) dropwise. The reaction was refluxed for 3 days and the progress of the reaction was monitored using TLC. After cooling, the excess ethanol was removed by evaporation under reduced pressure. To get rid of the residual acid, extraction using (5%) NaHCO3 solution and CHCl3 was applied three times, followed by twice extraction with CHCl3 and water (25 ml) to remove any salt formed by acid and bicarbonate reaction. Anhydrous Na2SO4 was used to dry the organic layer, then the product was obtained by evaporating under reduced pressure. A yellow powder (6.44 g, 93%); Rf = 0.47 (CHCl3: MeOH) (10 ml:3drops); m.p 144–146 °C; 1H-NMR (500 Hz, DMSO-d6): δ = 1.33 (t, J = 7.1 Hz, 3 H, CH3), 4.31 (q, J = 7.1 Hz, 2 H, OCH2), 6.88 (d, J = 9.3 Hz, 1H, Ar-H), 7.81 (s, 2 H, NH2), 8.06 (d, J = 9.3 Hz, 1H, Ar-H), 8.55 (s, 1H, Ar-H) ppm; 13C-NMR (100 Hz, DMSO-d6): δ = 14.5 (1 C), 61.2 (1 C), 108.2 (1 C), 117.1 (1 C), 128.6 (1 C), 129.1 (1 C), 135.5 (1 C), 156.2 (1 C), 166.4 (1 C) ppm. Elemental Anal. Calcd. for C12H10N2O6: C, 51.81; H, 3.62; N, 10.07. Found: C, 51.45; H, 3.79; N, 9.72.
Ethyl 4-hydroxy-7-nitro-2-quinolone-3-Carboxylate (7a)
A mixture of ethyl 2-amino-4-nitrobenzoate (5a) (5.0 g, 23.8 mmol), an excess amount of diethylmalonate (6) (38.1 ml, 249.1 mmol), and sodium ethoxide (4.9 g, 71.4 mmol) in DMSO (25 ml) were prepared and refluxed for 3 days. After cooling, the resulted solution was acidified with 0.5 M HCl to reach a pH value of 4.5-5.0, the formed solid was filtered and washed with water then CH3OH (10 ml), and dried in a vacuum oven at 70 °C for 1 hr. Yellow powder (6 g) with yield 91%, Rf = 0.58 (n-C6H14: EtOAc) (2:8); m.p: 268–270 ºC; 1H-NMR (400 MHz, DMSO-d6): δ = 1.04 (t, J = 7.2 Hz, 3 H, - CH3), 4.30 (q, J = 7.2 Hz, 2 H, -OCH2), 6.99 (s, 1H, Ar-H), 7.36 (d, J = 6.4 Hz, 1H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 18.8 (1 C), 56.5 (1 C), 108.5 (1 C), 114.5 (1 C), 117.4 (1 C), 126.2 (1 C), 129.0 (1 C), 147.8 (1 C), 150.6 (1 C), 172.1 (1 C), 172.8 (1 C), 173.1 (1 C) ppm; IR (KBr disc): 3080, 2981, 2937, 2843, 2808, 1741, 1681, 1635, 1566, 1531, 1506, 1473, 1415, 1350, 1327 cm− 1; Elemental Anal. Calcd. for C17H13N3O5: C, 60.18; H, 3.86; N, 12.38. Found: C, 60.51; H, 3.43; N, 12.34.
Ethyl 4-hydroxy-6-nitro-2-quinolone-3-carboxylate (7b)
A mixture of ethyl 2-amino-5-nitrobenzoate (5b) (5.0 g, 23.8 mmol), an excess amount of diethylmalonate (6) (38.1 ml, 249.1 mmol), and sodium ethoxide (4.9 g, 71.4 mmol) in DMSO (25 ml) were prepared and refluxed for 3 days. After cooling, the resulted solution was acidified with 0.5 M HCl to reach a pH value of 4.5-5.0, the formed solid was filtered and washed with water then CH3OH (10 ml), and dried in a vacuum oven at 70 °C for 1 hr. A beige powder (1.75 g, 66%); Rf = 0.48 (CHCl3: MeOH) (8 ml:2 ml); decomposed at 280 °C; 1H-NMR (500 Hz, DMSO-d6): δ = 12.18 (s, 1H, NH) 1.30 (t, J = 7.1 Hz, 3 H, CH3), 4.32 (q, J = 7.1 Hz, 2 H, OCH2), 7.50 (d, J = 9.1 Hz, 1H, Ar-H), 8.40 (dd, J = 9.1 Hz, 1H, Ar-H), 8.78 (s, 1H, Ar-H) ppm; 13C-NMR (100 Hz, DMSO-d6): δ = 14.7 (1 C), 58.7 (1 C), 122.5 (2 C), 102.2 (1 C), 123.1 (1 C), 123.4 (1 C), 135.4 (1 C), 156.8 (1 C), 172.0 (1 C), 174.2 (1 C),174.6 (1 C) ppm. Elemental Anal. Calcd. for C17H13N3O5: C, 60.18; H, 3.86; N, 12.38. Found: C, 59.63; H, 4.28; N, 12.04.
N-Benzyl 4-hydroxy-7-nitro-2-quinolone-3-carboxamide (9)
A mixture of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and benzylamine (8i) (1.2 g, 11.2 mmol) was prepared in THF (25 ml). Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated using methanol and the precipitate was collected by suction filtration at RT. Finally, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Mustard powder (0.36 g) with yield 26%, Rf = 0.61 (n-C6H14: ETOAc) (6:4); m.p: decomposed at 320 °C; 1H-NMR (400 MHz, DMSO-d6 + NaOD): δ = 4.45 (s, 2 H, - CH2), 7.20–7.55 (m, 6 H, Ar-H), 7.74 (m, 1H, Ar-H), 7.95 (s, 1H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6 + NaOD): δ = 42.5 (1 C), 102.8 (1 C), 108.7 (1 C), 117.2 (1 C), 126.8 (1 C), 127.5 (1 C), 127.6 (1 C), 127.7 (1 C), 127.8 (1 C), 128.5 (1 C), 129.5 (1 C), 141.2 (1 C), 148.2 (1 C), 149.9 (1 C), 171.5 (1 C), 174.5 (1 C), 175.3 (1 C) ppm; IR (KBr disc): 3228, 3209, 3178, 3068, 3030, 2935, 2864, 1647, 1556, 1450, 1423, 1348 cm− 1; Elemental Anal. Calcd. for C17H13N3O5: C, 60.18; H, 3.86; N, 12.38. Found: C, 60.51; H, 3.93; N, 11.84.
N-(3-Triflourobenzyl)-4-hydroxy-7-nitro-2-quinolone-3-carboxamide (10)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and 3-trifluorobenzylamine (8ii) (1.9 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated using methanol and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Yellow powder (0.60 g) with yield 41%, Rf = 0.57 (n-C6H14: EtOAc) (6:4); m.p: 236–239 °C; 1H-NMR (400 MHz, DMSO-d6): δ = 4.67 (s, 2 H, -CH2), 7.57–7.73 (m, 4 H, Ar-H ), 7.94 (d, J = 7.6 Hz, 1H, Ar-H), 8.08 (d, J = 8.8 Hz, 2 H, Ar-H), 10.60 (s, 1H, -NH amide ring), 12.20 (s, 1H, -OH) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 42.3 (1 C), 98.5 (1 C), 111.4 (1 C), 120.6 (1 C), 124.4 (1 C), 124.6 (2 C), 126.0 (1 C), 126.6 (1 C), 129.2 (2 C), 129.8 (1 C), 130.0 (1 C), 150.3 (1 C), 163.1 (1 C), 170.8 (1 C), 176.7 (1 C) ppm; IR (KBr disc): 3277, 3163, 3082, 3001, 2941, 2866, 1662, 1604, 1560, 1535, 1490, 1429, 1350, 1330 cm− 1; Elemental Anal. Calcd. for C18H12F3N3O5: C, 53.08; H, 2.97; N, 10.32. Found: C, 53.12; H, 3.02; N, 9.88.
N-(4-Triflourobenzyl)-4-hydroxy-7-nitro-2-quinolone-3-carboxamide (11)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and 4-trifluorobenzylamine (8iii) (1.9 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated using methanol and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Yellow powder (0.42 g) with yield 43%, Rf = 0.46 (n-C6H14: ETOAc) (6:4); m.p: 200–204 °C; 1H-NMR (400 MHz, DMSO-d6): δ = 4.70 (s, 2 H, - CH2), 7.40–7.47 (m, 2 H, Ar-H ), 7.50 (d, J = 8.4 Hz, 1H, Ar-H), 7.60(d, J = 8.0 Hz, 1H, Ar-H), 7.59–7.70 (m, 1H, Ar-H ), 7.72–7.81 (m, 1H, Ar-H ), 7.83–7.85 (m, 1H, Ar-H ), 11.9 (s, 1H, -NH amide), 12.00 (s, 1H, -NH amide ring), 12.20 (s, 1H, -OH) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 42.4 (1 C), 98.5 (1 C), 110.8 (1 C), 115.3 (1 C), 125.8 (1 C), 126.2 (1 C), 127.6 (3 C), 127.9 (3 C), 138.6 (1 C), 145.1 (1 C), 149.4 (1 C), 166.5 (1 C), 170.0 (1 C), 176.2 (1 C) ppm; IR (KBr disc): 3277, 3163, 3082, 3001, 2941, 2866, 1662, 1604, 1560, 1535, 1490, 1429, 1350, 1330 cm-1; Elemental Anal. Calcd. for C18H12F3N3O5: C, 53.08; H, 2.97; N, 10.32. Found: C, 49.88; H, 3.02; N, 8.93.
N-(2-Methoxybenzyl)-4-hydroxy-7-nitro-2-quinolone-3-carboxamide (12)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and 2-methoxybenzylamine (8iv) (1.5 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated using methanol and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Yellow powder (0.50 g) with yield 38%, Rf = 0.48 (n-C6H14: EtOAc) (6:4); m.p: 230–234 °C; 1H-NMR (400 MHz, DMSO-d6): δ = 3.91 (s, 3 H, - OCH3), 4.50 (s, 2 H, - CH2), 6.95–7.03 (m, 2 H, Ar-H ), 7.05–7.30 (m, 2 H, Ar-H), 7.97 (d, J = 8.4 Hz, 1H, Ar-H), 8.10 (d, J = 10.8 Hz, 2 H, Ar-H), 10.57 (s, 1H, -NH amide), 11.30 (s, 1H, -NH amide ring), 12.15 (s, 1H, -OH) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 42.0 (1 C), 55.9 (1 C), 111.3 (2 C), 116.5 (1 C), 120.8 (2 C), 125.6 (1 C), 126.7 (1 C), 129.1 (2 C), 129.4 (1 C), 139.1 (1 C), 150.4 (1 C), 157.5 (1 C), 163.1 (1 C), 170.4 (1 C), 171.7 (1 C) ppm; IR (KBr disc): 3284, 3147, 3082, 2947, 2841, 1668, 1604, 1560, 1492, 1462, 1429, 1348, 1330 cm− 1; Elemental Anal. Calcd. for C18H15N3O6: C, 58.54; H, 4.09; N, 11.38. Found: C, 59.01; H, 4.53; N, 10.88.
N-(3-Methoxybenzyl)-4-hydroxy-7-nitro-2-quinolone-3-carboxamide (13)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and 3-methoxybenzylamine (8v) (1.5 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated using methanol and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Yellow powder (0.50 g) with yield 39%, Rf = 0.43 (n-C6H14: EtOAc) (6:4); m.p: 210–213 °C; 1H-NMR (400 MHz, DMSO-d6): δ = 3.74 (s, 3 H, - OCH3), 4.55 (s, 2 H, - CH2), 6.93–6.85 (m, 3 H, Ar-H ), 7.27 (s, 1H, Ar-H ), 7.93 (s, 1H, Ar-H), 7.95–8.09 (m, 2 H, Ar-H ), 10.50 (s, 1H, -NH amide ring), 12.15 (s, 1H, -OH) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 42.7 (1 C), 55.5 (1 C), 98.5 (1 C), 111.4 (1 C), 113.0 (1 C), 113.8 (1 C), 116.5 (1 C), 120.1 (2 C), 126.6 (1 C), 130.1 (1 C), 139.1 (1 C), 139.9 (1 C), 150.3 (1 C), 159.9 (1 C), 162.9 (1 C), 170.6 (1 C), 171.7 (1 C) ppm; IR (KBr disc): 3265, 3082, 2937, 2868, 2839, 1668, 1608, 1558, 1489, 1452, 1435, 1417, 1350, 1334 cm− 1; Elemental Anal. Calcd. for C18H15N3O6: C, 58.54; H, 4.09; N, 11.38. Found: C, 59.06; H, 3.94; N, 11.19.
N-(4-Methoxybenzyl)-4-hydroxy-7-nitro-2-quinolone-3 Carboxamide (14)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and 4-methoxybenzylamine (8vi) (1.5 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC the product was precipitated using methanol and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Mustard powder (0.45 g) with yield 35%, Rf = 0.53 (n-C6H14: EtOAc) (6:4); m.p: decomposed at 228 °C; 1H-NMR (400 MHz, DMSO-d6): δ = 3.74 (s, 3 H, - OCH3), 4.47 (s, 2 H, - CH2), 6.89–7.17 (m, 3 H, Ar-H), 7.18–7.27 (m, 3 H, Ar-H), 7.23 (s, 1H, Ar-H), 10.52 (s, 1H, -NH amide), 11.32 (s, 1H, -NH amide ring), 11.88 (s, 1H, -OH) ppm;13C-NMR (100 MHz, DMSO-d6 + NaOD): δ = 13.2 (1 C), 42.2 (1 C), 55.5 (1 C), 113.8 (1 C), 114.0 (1 C), 114.4 (1 C), 115.0 (1 C), 127.9 (1 C), 129.5 (1 C), 129.7 (2 C), 123.3 (1 C), 135.9 (1 C), 139.0 (1 C), 139.8 (1 C), 159.0 (1 C), 166.8 (1 C), 169.5 (1 C), 171.8 (1 C) ppm; IR (KBr disc): 3263, 3165, 3080, 2999, 2935, 2839, 1664, 1610, 1560, 1512, 1456, 1427, 1348, 1300 cm− 1; Elemental Anal. Calcd. for C18H15N3O6: C, 58.54; H, 4.09; N, 11.38. Found: C, 58.65; H, 4.29; N, 11.04.
N-(2-Chlorobenzyl)-4-hydroxy-7-nitro-2-quinolone-3-carboxamide (15)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and 2-chlorobenzylamine (8vii) (1.6 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated using methanol and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Yellow powder (0.70 g) with yield 52%, Rf = 0.54 (n-C6H14: EtOAc) (2:8); m.p: 248–250 °C; 1H-NMR (400 MHz, DMSO-d6 + NaOD): δ = 4.55 (s, 2 H, - CH2), 7.17–7.52 (m, 4 H, Ar-H) 7.63 (d, J = 8.4 Hz, 1H, Ar-H), 7.77 (d, J = 6.0 Hz, 1H, Ar-H), 8.04 (t, J = 8.4 Hz, 1H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6 + NaOD): δ = 42.7 (1 C), 102.2 (1 C), 109.1 (1 C), 113.3 (1 C), 117.4 (1 C), 126.8 (1 C), 127.5 (2 C), 128.5 (1 C), 129.2 (1 C), 129.3 (1 C), 132.4 (1 C), 138.6 (1 C), 148.3 (1 C), 150.5 (1 C), 171.7 (1 C), 175.7 (1 C). ppm; IR (KBr disc): 3398, 3367, 3336, 3267, 3184, 3070, 2997, 2945, 2864, 1658, 1554, 1423, 1346 cm− 1; Elemental Anal. Calcd. for C17H12ClN3O5: C, 54.63; H, 3.24; N, 11.24. Found: C, 54.86; H, 3.35; N, 11.24.
N-(3-Chlorobenzyl)-4-hydroxy-7-nitro-2-quinolone-3-carboxamide (16)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and 3-chlorobenzylamine (8viii) (1.6 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated using methanol and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Yellow powder (0.63 g) with yield 47%, Rf = 0.5 (n-C6H14: EtOAc) (6:4); m.p: 202–203 °C; 1H-NMR (400 MHz, DMSO-d6): δ = 4.55 (s, 2 H, - CH2), 7.31–7.40 (m, 3 H, Ar-H), 7.57 (s, 1H, Ar-H), 7.91 (d, J = 8.8 Hz, 1H, Ar-H), 8.07 (s, 1H, Ar-H), 8.12 (d, J = 8.8 Hz, 1H, Ar-H), 10.70 (s, 1H, -NH amide), 11.84 (s, 1H, -NH amide ring), 12.09 (s, 1H, -OH) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 42.6 (1 C), 98.3 (1 C), 112.1 (1 C), 115.7 (1 C), 127.4 (1 C), 127.7 (1 C), 127.9 (1 C), 128.5 (1 C), 129.0 (1 C), 130.1 (1 C), 133.5 (1 C), 138.2 (1 C), 141.9 (1 C), 149.9 (1 C), 164.3 (1 C), 170.5 (1 C), 172.8 (1 C) ppm; IR (KBr disc): 3263, 3143, 3068, 2987, 2935, 2860, 1666, 1602, 1571, 1558, 1523, 1429, 1346 cm− 1; Elemental Anal. Calcd. for C17H12ClN3O5: C, 54.63; H, 3.24; N, 11.24. Found: C, 54.78; H, 3.15; N, 11.01.
N-(4-Chlorobenzyl)-4-hydroxy-7-nitro-2-quinolone-3-carboxamide (17)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and 4-chlorobenzylamine (8ix) (1.6 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated using methanol and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. White powder (0.63 g) with yield 47%, Rf = 0.5 (n-C6H14: EtOAc) (2:8); m.p: 232–234 °C; 1H-NMR (400 MHz, DMSO-d6): δ = 4.57 (s, 2 H, - CH2), 6.35 (d, J = 10.0 Hz, 1H, Ar-H), 6.53 (d, J = 6.8 Hz, 1H, Ar-H), 7.35 (s, 1H, Ar-H), 7.47 (d, J = 7.2 Hz, 1H, Ar-H), 7.59 (d, J = 3.2 Hz, 1H, Ar-H), 7.67 (d, J = 8.4 Hz, 1H, Ar-H), 7.99 (d, J = 8.4 Hz, 1H, Ar-H), 10.60 (s, 1H, -NH amide), 11.35 (s, 1H, -NH amide ring), 12.23 (s, 1H, -OH) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 42.0 (1 C), 92.4 (1 C), 96.0 (1 C), 104.1 (1 C), 112.0 (1 C), 116.6 (1 C), 128.9 (1 C), 129.8 (1 C), 131.6 (1 C), 135.3 (1 C), 138.0 (1 C), 150.5 (1 C), 154.8 (1 C), 163.7 (1 C), 171.6 (1 C), 172.3 (1 C), 192.6 (1 C) ppm; IR (KBr disc): 3230, 3172, 3086, 3030, 2937, 2868, 1662, 1643, 1560, 1490, 1427, 1348 cm-1; Elemental Anal. Calcd. for C17H12ClN3O5: C, 54.63; H, 3.24; N, 11.24. Found: C, 54.87; H, 3.65; N, 10.90.
N-(Pyridin-4-ylmethyl)-4-hydroxy-7-nitro-2-quinolone-3-carboxamide (18)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and 4-(aminomethyl)pyridine (8x) (1.2 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated using methanol and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Yellow powder (0.48 g) with yield 39%, Rf = 0.5 (n-C6H14: EtOAc) (2:8); m.p: decomposed at 280 °C; 1H-NMR (400 MHz, DMSO-d6): δ = 4.63 (s, 2 H, - CH2), 6.36 (d, J = 8.0 Hz, 2 H, Ar-H), 6.52 (d, J = 6.8 Hz, 2 H, Ar-H), 7.30 (d, J = 8.0 Hz, 2 H, Ar-H), 7.60 (d, J = 8.8 Hz, 1H, Ar-H), 8.52 (s, 1H, Ar-H), 10.66 (s, 1H, -NH amide ring), 11.38 (s, 1H, -OH) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 41.4(1 C), 92.4 (1 C), 96.1 (1 C), 104.1 (1 C), 112.0 (1 C), 122.6 (1 C), 125.9 (1 C), 141.9 (1 C), 148.4 (1 C), 149.8 (1 C), 150.1 (1 C), 150.8 (1 C), 154.9 (1 C), 163.7 (1 C), 171.9 (1 C), 172.2 (1 C) ppm; IR (KBr disc): 3442, 3342, 3221, 3084, 2941, 1635, 1612, 1550, 1421, 1352, 1219, 1130 cm− 1; Elemental Anal. Calcd. for C16H12N4O5: C, 56.47; H, 3.55; N, 16.46. Found: C, 56.24; H, 3.30; N, 16.06.
N-(2-Flurobenzyl)-4-hydroxy-7-nitro-2-quinolone-3-carboxamide (19)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and 2-flurobenzylamine (8xi) (1.4 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated using methanol and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Pale brown powder (0.40 g) with yield 32%, Rf = 0.55 (n-C6H14: EtOAc) (6:4); m.p: 242–245 °C; 1H-NMR (400 MHz, DMSO-d6 + NaOD): δ = 4.52 (s, 2 H, - CH2), 7.14 (t, J = 7.6 Hz, 1H, Ar-H), 7.24–7.32 (m, 3 H, Ar-H), 7.42 (t, J = 7.4 Hz, 1H, Ar-H), 7.75 (s, 1H, Ar-H), 7.95–8.10 (m, 1H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6 + NaOD): δ = 42.6 (1 C), 103.2 (1 C), 115.1 (1 C), 115.4 (1 C), 117.4 (1 C), 124.7 (1 C), 126.8 (1 C), 128.2 (1 C), 128.8 (1 C), 129.0 (1 C), 148.2 (1 C), 150.4 (1 C), 159.3 (1 C), 161.8 (1 C), 171.6 (1 C), 174.8 (1 C), 175.6 (1 C) ppm; IR (KBr disc): 3257, 3161, 3082, 2962, 2931, 2852, 1668, 1625, 1558, 1537, 1490, 1409, 1350, 1332 cm− 1; Elemental Anal. Calcd. for C17H12FN3O5: C, 57.15; H, 3.39; N, 11.76. Found: C, 57.40; H, 3.46; N, 11.52.
N-(3-Flurobenzyl)-4-hydroxy-7-nitro-2-quinolone-3-carboxamide (20)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and 3-flurobenzylamine (8xii) (1.4 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated using methanol and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Yellow powder (0.6 g) with yield 47%, Rf = 0.52 (n-C6H14: EtOAc) (6:4); m.p: 215–218 °C; 1H-NMR (400 MHz, DMSO-d6 + NaOD): δ = 4.50 (s, 2 H, - CH2), 7.00 (t, J = 7.6 Hz, 1H, Ar-H), 7.12 (d, J = 8.0 Hz, 1H, Ar-H) 7.18 (d, J = 8.0 Hz, 1H, Ar-H), 7.30–7.36 (m, 2 H, Ar-H), 7.76 (s, 1H, Ar-H), 7.95–8.10 (m, 1H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6 + NaOD): δ = 41.5 (1 C), 102.4 (1 C), 108.5 (1 C), 113.5 (1 C), 114.2 (1 C), 117.3 (1 C), 123.5 (1 C), 126.8 (1 C), 128.4 (1 C), 130.6 (1 C), 144.7 (1 C), 150.1 (1 C), 161.3 (1 C), 163.9 (1 C), 171.6 (1 C), 174.6 (1 C), 175.5 (1 C) ppm; IR (KBr disc): 3265, 3217, 3161, 3088, 2962, 2862, 1940, 1664, 1558, 1531, 1487, 1429, 1402, 1350 cm− 1; Elemental Anal. Calcd. for C17H12FN3O5: C, 57.15; H, 3.39; N, 11.76. Found: C, 57.23; H, 3.55; N, 11.50.
N-(4-Flurobenzyl)-4-hydroxy-7-nitro-2-quinolone-3-carboxamide (21)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and 4-flurobenzylamine (8xiii) (1.4 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated using methanol and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Pale yellow powder (0.7 g) with yield 55%, Rf = 0.48 (n-C6H14: EtOAc) (6:4); m.p: 233–235 °C; 1H-NMR (400 MHz, DMSO-d6 + NaOD): δ = 4.48 (s, 2 H, - CH2), 7.10 (d, J = 8.4 Hz, 2 H, Ar-H), 7.12–7.38 (m, 3 H, Ar-H), 7.88 (s, 1H, Ar-H), 8.02 (d, J = 8.4 Hz, 1H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6 + NaOD): δ = 41.3 (1 C), 101.9 (1 C), 109.1 (1 C), 115.1 (1 C), 115.3 (1 C), 116.8 (1 C), 126.9 (1 C), 128.5 (1 C), 129.6 (1 C), 137.7 (1 C), 146.1 (1 C), 148.3 (1 C), 160.2 (1 C), 162.6 (1 C), 171.2 (1 C), 173.7 (1 C), 175.6 (`1 C) ppm; IR (KBr disc): 3273, 3161, 3080, 2958, 2927, 2860, 1944, 1662, 1604, 1556, 1508, 1433, 1350 cm-1; Elemental Anal. Calcd. for C17H12FN3O5: C, 57.15; H, 3.39; N, 11.76. Found: C, 57.44; H, 3.53; N, 11.74.
N-(3-Methylbenzyl)-4-hydroxy-7-nitro-2-quinolone-3 Carboxamide (22)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and 3-methylbenzylamine (8xiv) (1.4 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated using methanol and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Yellow-orange powder (0.94 g) with yield 75%, Rf = 0.62 (n-C6H14: EtOAc) (6:4); m.p: decomposed at 256 °C; 1H-NMR (400 MHz, DMSO-d6 + NaOD): δ = 2.30 (s, 3 H, - CH3), 4.48 (s, 2 H, - CH2), 6.44 (s, 1H, Ar-H ), 7.01–7.18 (m, 4 H, Ar-H ), 7.78 (s, 1H, Ar-H), 8.01 (d, J = 7.6 Hz, 1H, Ar-H) 10.49 (s, 1H, -NH amide ring), 12.18 (s, 1H, -OH) ppm; 13C-NMR (100 MHz, DMSO-d6 + NaOD): δ = 21.4 (1 C), 42.7 (1 C), 98.4 (1 C), 111.4 (1 C), 116.5 (1 C), 119.0 (1 C), 125.1 (1 C), 126.5 (1 C), 128.4 (1 C), 128.6 (2 C), 129.0 (1 C), 138.2 (1 C), 139.0 (1 C), 150.3 (1 C), 162.9 (1 C), 170.5 (1 C), 171.7 (1 C) ppm; IR (KBr disc): 3257, 3163, 3080, 2989, 2924, 2854, 1670, 1602, 1556, 1529, 1487, 1442, 1415, 1344 cm− 1; Elemental Anal. Calcd. for C18H15N3O5: C, 61.19; H, 4.28; N, 11.89. Found: C, 61.57; H, 4.34; N, 11.76.
N-(4-Methylbenzyl)-4-hydroxy-7-nitro-2-quinolone-3 Carboxamide (23)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and 4-methylbenzylamine (8xv) (1.4 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated using methanol and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Yellow powder (0.63 g) with yield 50%, Rf = 0.62 (n-C6H14: EtOAc) (6:4); m.p: 225–228 °C; 1H-NMR (400 MHz, DMSO-d6): δ = 3.37 (s, 3 H, - CH3), 4.54 (s, 2 H, - CH2), 6.82 (d, J = 8.0 Hz, 1H, Ar-H ), 6.87 (d, J = 7.6 Hz, 1H, Ar-H ), 7.42 (d, J = 7.4 Hz, 2 H, Ar-H), 7.33 (d, J = 8.8 Hz, 1H, Ar-H), 7.85 (d, J = 8.8 Hz, 1H, Ar-H), 7.99 (s, 1H, Ar-H), 10.52 (s, 1H, -NH amide), 11.03 (s, 1H, -NH amide ring), 11.80 (s, 1H, -OH) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 21.1 (1 C), 42.6 (1 C), 98.8 (1 C), 110.7 (1 C), 114.9 (1 C), 126.3 (1 C), 128.0 (1 C), 129.0 (1 C), 129.5 (1 C), 129.7 (1 C), 131.4 (1 C), 137.2 (1 C), 138.7 (1 C), 139.1 (1 C), 149.2 (1 C), 166.7 (1 C), 169.6 (1 C), 175.8 (1 C) ppm; IR (KBr disc): 3267, 3080, 2999, 2926, 2864, 1666, 1610, 1558, 1452, 1427, 1348 cm− 1; Elemental Anal. Calcd. for C18H15N3O5: C, 61.19; H, 4.28; N, 11.89. Found: C, 60.64; H, 4.48; N, 11.17.
N-(4-Hydroxy benzyl)-4-hydroxy-7-nitro-2-quinolone-3 Carboxamide (24)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and 4-hydroxybenzylamine (8xvi) (1.4 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated using methanol and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Yellow powder (0.4 g) with yield 66%, Rf = 0.33 (n-C6H14: EtOAc) (6:4); m.p: 238–240 °C; 1H-NMR (400 MHz, DMSO-d6 + NaOD): δ = 4.50 (s, 2 H, - CH2), 6.74 (d, J = 8.4 Hz, 1H, Ar-H ), 7.50 (d, J = 8.6 Hz, 2 H, Ar-H), 7.95 (d, J = 8.8 Hz, 1H, Ar-H), 8.00-8.20 (m, 3 H, Ar-H ), 9.41 (s, 1H, -NH amid), 10.47 (s, 1H, -NH amide ring), 12.17 (s, 1H, -OH) ppm; 13C-NMR (100 MHz, DMSO-d6 + NaOD): δ = 42.4 (1 C), 98.4 (1 C), 111.5 (1 C), 115.8 (2 C), 116.5 (1 C), 119.1 (1 C), 126.6 (1 C), 128.4 (1 C), 129.5 (2 C), 139.1 (1 C), 150.4 (1 C), 157.2 (1 C), 163.0 (1 C), 170.3 (1 C), 171.8 (1 C) ppm; IR (KBr disc): 3257, 3163, 3080, 2924, 2854, 1670, 1602, 1556, 1529, 1487, 1442, 1415, 1344 cm− 1; Elemental Anal. Calcd. for C17H13N3O6: C, 57.47; H, 3.69; N, 11.83. Found: C, 57.08; H, 3.42; N, 11.45.
N-(Phenyl)-4-hydroxy-7-nitro-2-quinolone-3-carboxamide (25)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and aniline (8 xvii) (1.0 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The product was precipitated on the inner flask and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Yellow powder (0.7 g) with yield 60%, Rf = 0.48 (n-C6H14: EtOAc) (2:8); m.p: decomposed at 340 °C; 1H-NMR (400 MHz, DMSO-d6 + NaOD): δ = 6.90 (t, J = 7.2 Hz, 1H, Ar-H), 7.22 (t, J = 7.6 Hz, 2 H, Ar-H), 7.38 (d, J = 2.4 Hz, 1H, Ar-H), 7.71 (d, J = 8.0 Hz, 2 H, Ar-H), 7.81 (d, J = 2.0 Hz, 1H, Ar-H), 8.08 (s, 1H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6 + NaOD): δ = 67.5 (1 C), 102.6 (1 C), 108.6 (1 C), 117.6 (1 C), 119.9 (2 C), 121.5 (1 C), 126.9 (1 C), 128.3 (1 C), 128.9 (1 C), 141.6 (1 C), 148.4 (1 C), 150.4 (1 C), 169.7 (1 C), 174.8 (1 C), 176.0 (1 C) ppm; IR (KBr disc): 3034, 2989, 2931, 2864, 1668, 1606, 1550, 1531, 1489, 1413, 1369, 1344 cm− 1; Elemental Anal. Calcd. for C16H11N3O5: C, 59.08; H, 3.41; N, 12.92. Found: C, 58.79; H, 3.80; N, 12.49.
N-(2-Flurophenyl)-4-hydroxy-7-nitro-2-quinolone-3-carboxamide (26)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and 2-fluroaniline (8 xviii) (1.1 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated on the inner flask and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Yellow powder (0.8 g) with yield 65%, Rf = 0.4 (n-C6H14: EtOAc) (5:5); m.p: decomposed at 345 °C; 1H-NMR (400 MHz, DMSO-d6 + NaOD): δ = 6.86–6.93 (m, 1H, Ar-H), 7.00 (t, J = 7.6 Hz, 1H, Ar-H), 7.16 (t, J = 9.2 Hz, 1H, Ar-H), 7.73 (d, J = 8.4 Hz, 1H, Ar-H), 7.81 (s, 1H, Ar-H), 8.07 (d, J = 8.4 Hz, 1H, Ar-H), 8.63–8.67 (m, 1H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6 + NaOD): δ = 102.5 (1 C), 108.8 (1 C), 114.8 (1 C), 117.5 (1 C), 121.7 (1 C), 122.2 (1 C), 124.4 (1 C), 127.0 (1 C), 128.1 (1 C), 148.5 (1 C), 150.5 (1 C), 151.4 (1 C), 153.8 (1 C), 169.9 (1 C), 176.3 (1 C), 174.6 (1 C) ppm; IR (KBr disc): 3165, 3088, 2993, 2939, 2862, 1664, 1604, 1550, 1539, 1489, 1463, 1409, 1348 cm-1; Elemental Anal. Calcd. for C16H10FN3O5: C, 55.98; H, 2.94; N, 12.24. Found: C, 56.02; H, 2.85; N, 12.21.
N-(3-Flurophenyl)-4-hydroxy-7-nitro-2-quinolone-3-carboxamide (27)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and 3-fluroaniline (8xix) (1.1 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated on the inner flask and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Yellow powder (0.4 g) with yield 33%, Rf = 0.45 (n-C6H14: EtOAc) (2:8); m.p: decomposed at 350 °C; 1H-NMR (400 MHz, DMSO-d6 + NaOD): δ = 6.69 (t, J = 6.0 Hz, 1H, Ar-H), 7.17–7.26 (m, 2 H, Ar-H), 7.35–7.38 (m, 1H, Ar-H), 7.94 (d, J = 12.4 Hz, 1H, Ar-H), 7.75–7.89 (m, 1H, Ar-H), 8.04 (d, J = 8.8 Hz, 1H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6 + NaOD): δ = 102.3 (1 C), 106.6 (1 C), 107.5 (1 C), 108.6 (1 C), 115.3 (1 C), 117.6 (1 C), 126.9 (1 C), 128.2 (1 C), 130.1 (1 C), 148.5 (1 C), 150.5 (1 C), 161.7 (1 C), 164.1 (1 C), 169.8 (1 C), 174.6 (1 C), 176.2 (1 C) ppm; IR (KBr disc): 3174, 3070, 2987, 2937, 2868, 2376, 2314, 1668, 1608, 1531, 1489, 1411, 1342 cm− 1; Elemental Anal. Calcd. for C16H10FN3O5: C, 55.98; H, 2.94; N, 12.24. Found: C, 55.94; H, 2.84; N, 12.02.
N-(4-Flurophenyl)-4-hydroxy-7-nitro-2-quinolone-3-carboxamide (28)
A combination of ethyl 4-hydroxy-7-nitro-2-quinolone-3-carboxylate (7a) (1.0 g, 3.6 mmol) and 4-fluroaniline (8xx) (1.1 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated on the inner flask and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Yellow powder (0.5 g) with yield 41%, Rf = 0.45 (n-C6H14: EtOAc) (2:8); m.p: decomposed at 320 °C; 1H-NMR (400 MHz, DMSO-d6 + NaOD): δ = 6.99–7.03 (m, 2 H, Ar-H), 7.35–7.38 (m, 1H, Ar-H), 7.70–7.73 (m, 2 H, Ar-H), 7.80 (s, 1H, Ar-H), 8.04 (d, J = 8.8 Hz, 1H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6 + NaOD): δ = 102.4 (1 C), 108.7 (1 C), 115.2 (1 C), 115.4 (1 C), 117.6 (1 C), 121.2 (1 C), 126.9 (1 C), 128.2 (1 C), 137.9 (1 C), 148.4 (1 C), 150.4 (1 C), 156.1 (1 C), 169.5 (1 C), 174.6 (1 C), 175.9 (1 C), 174.6 (1 C) ppm; IR (KBr disc): 3055, 2991, 2937, 2866, 2376, 2310, 1674, 1622, 1550, 1537, 1508, 1492, 1427, 1411, 1346 cm− 1; Elemental Anal. Calcd. for C16H10FN3O5: C, 55.98; H, 2.94; N, 12.24. Found: C, 55.78; H, 3.02; N, 11.95.
N-(2-Trifluromethyl phenyl)-4-hydroxy-6-nitro-2-quinolone-3-carboxamide (29)
A combination of ethyl 4-hydroxy-6-nitro-2-quinolone-3-carboxylate (7b) (1.0 g, 3.6 mmol) and 2- trifluroaniline (8xxi) (1.1 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated on the inner flask and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. White powder (g) with yield of 57% (0.8gm); Rf = 0.79 (CHCl3: MeOH) (9.7 ml:0.3 ml); m.p: decomposed at 345 °C; 1H-NMR (400 MHz, DMSO-d6 + NaOD): δ = 7.41 (m, 4 H, Ar-H), 7.70–7.73 (m, 2 H, Ar-H), 7.88 (d, J = 4.0 Hz, 1H, Ar-H), 9.94 (s, 1H, -NH amide ring), 11.89 (s, 1H, -NH amide), 12.10 (s, 1H, -OH) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 96.7 (1 C), 115.9 (3 C), 118.6 (2 C), 123.2 (2 C), 127.1 (3 C), 134.1 (1 C), 137.8 (1 C), 162.1(1 C), 167.8 (2 C), 171.2 (1 C) ppm; IR (KBr disc): 1300, 1375, 1417, 1456, 1529, 1556, 1598, 1629, 1658, 1911, 2308, 2883.58, 2976, 3051, 3089, 3151 cm− 1. Elemental Anal. Calcd. for C17H10F3N3O5: C, 51.92; H, 2.56; N, 10.68. Found: C, 52.70; H, 2.94; N, 10.11.
N-(3-Trifluromethyl phenyl)-4-hydroxy-6-nitro-2-quinolone-3-carboxamide (30)
A combination of ethyl 4-hydroxy-6-nitro-2-quinolone-3-carboxylate (7b) (1.0 g, 3.6 mmol) and 3- trifluroaniline (8xxii) (1.1 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated on the inner flask and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. White powder (5.4 g) with yield 29% (0.42 gm); Rf = 0.81 (CHCl3: MeOH) (9.7 ml:0.3 ml); m.p: decomposed at 355 °C; 1H-NMR (400 MHz, DMSO-d6 + NaOD): δ = 7.33 (m, 3 H, Ar-H), 7.95 (m, 3 H, Ar-H), 8.21 (s, 1H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 67.5 (1 C), 100.9 (1 C), 101.1 (1 C), 119.1 (1 C), 119.9 (1 C), 122.5 (1 C), 122.6 (1 C), 122.9 (1 C), 123.1 (1 C), 124.5 (1 C), 124.7 (1 C),137.5 (1 C), 138.1 (1 C), 170.6 (1 C), 170.9 (1 C), 174.4 (1 C), 174.3 (1 C) ppm; IR (KBr disc): 1338, 1377, 1421, 1481, 1498, 1525, 1616, 1940, 2862, 2920, 3005, 3089, 3163, 3442, 3532 cm− 1. Elemental Anal. Calcd. for C17H10F3N3O5: C, 51.92; H, 2.56; N, 10.68. Found: C, 52.65; H, 3.20; N, 11.35.
N-(4-Trifluromethyl phenyl)-4-hydroxy-6-nitro-2-quinolone-3-carboxamide (31)
A combination of ethyl 4-hydroxy-6-nitro-2-quinolone-3-carboxylate (7b) (1.0 g, 3.6 mmol) and 4- trifluroaniline (8 xxiii) (1.1 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated on the inner flask and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Coffee brown powder with yield 39% (0.5 gm); Rf = 0.79 (CHCl3: MeOH) (9.7 ml:0.3 ml); m.p: decomposed at 265 °C; 1H-NMR (400 MHz, DMSO-d6 + NaOD): δ = 7.39 (d, J = 8.1 Hz, 2 H, Ar-H), 7.88 (m, 3 H, Ar-H), 8.09 (d, J = 7.5 Hz, 1H, Ar-H); 8.29 (s, 1H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 58.9 (1 C), 100.7 (2 C), 118.2 (1 C), 122.5 (3 C), 122.8 (3 C), 124.8 (2 C), 138.8 (1 C), 149.1 (1 C), 167.9 (1 C), 170.3 (1 C), 173.8 (1 C) ppm; IR (KBr disc): 1338, 1377, 1421, 1481, 1498, 1523, 1616, 1674, 1940, 2860, 2918, 3005, 3088, 3442, 3523 cm− 1. Elemental Anal. Calcd. for C17H10F3N3O5: C, 51.92; H, 2.56; N, 10.68. Found: C, 51.36; H, 3.04; N, 10.02.
N-phenyl-4-hydroxy-6-nitro-2-quinolone-3-carboxamide (32)
A combination of ethyl 4-hydroxy-6-nitro-2-quinolone-3-carboxylate (7b) (1.0 g, 3.6 mmol) and aniline (8 xxiv) (1.1 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated on the inner flask and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 h. An off-white powder (0.70 g, 60%); Rf = 0.7 (CHCl3: MeOH) (9 ml:1 ml); decomposed at 300 °C; 1 H-NMR (500 Hz DMSO-d6): δ = 12.59 (s, 1 H, OH), 12.04 (s, 1 H, NH), 11.80 (s, 1 H, NH), 8.33–8.70 (m, 3 H, Ar-H), 7.21–7.64 (m, 5 H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 176.95 (1 C), 175.0 (1 C), 169.35 (1 C), 155.72 (1 C), 141.06 (1 C), 136.38 (1 C), 128.96 (2 C), 124.07(1 C), 123.59 (1 C), 122.82 (1 C), 121.87 (1 C), 119.91 (2 C), 119.91 (1 C), 101.70 (1 C) ppm. Elemental Anal. Calcd. For C16H11N3O5: C, 59.08; H, 3.41; N, 12.92. Found: C, 59.0; H, 3.24; N, 12.50.
N-(2-Carboxy phenyl)-4-hydroxy-6-nitro-2-quinolone-3-carboxamide (33)
A combination of ethyl 4-hydroxy-6-nitro-2-quinolone-3-carboxylate (7b) (1.0 g, 3.6 mmol) and 2- amino benzoic acid (8xxv) (1.1 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated on the inner flask and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Yellow powder with yield of 23% (0.3gm); Rf = 0.3 (CHCl3: MeOH) (9.7 ml:0.3 ml); m.p: 140–142 °C; 1H-NMR (400 MHz, DMSO-d6 + NaOD): δ = 7.69 (m, 4 H, Ar-H), 8.06 (s, 1H, Ar-H), 8.21 (m, 2 H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 108.1 (1 C), 114.8 (3 C), 119.3 (1 C), 126.6 (2 C), 128.3 (1 C), 128.6 (1 C), 129.4 (3 C), 134.9 (2 C), 156.7 (1 C), 166.6 (1 C), 170.3 (1 C) ppm; IR (KBr disc): 1441, 1479, 1571, 1629, 1691, 2441, 2983, 3090, 3334, 3446 cm− 1. Elemental Anal. Calcd. for C17H11N3O7: C, 55.29; H, 3.00; N, 11.38. Found: C, 54.95; H, 3.24; N, 11.01.
N-(3-Carboxy phenyl)-4-hydroxy-6-nitro-2-quinolone-3-carboxamide (34)
A combination of ethyl 4-hydroxy-6-nitro-2-quinolone-3-carboxylate (7b) (1.0 g, 3.6 mmol) and 3- amino benzoic acid (8xxvi) (1.1 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated on the inner flask and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. 1H-NMR (400 MHz, DMSO-d6 + NaOD): δ = 6.94 (d, J = 8 Hz, 1 H, Ar-H), 7.17–7.21 (m, 2 H, Ar-H), 7.49–7.57 (m, 2 H, Ar-H), 7.81 (d, J = 8 Hz, 1 H, Ar-H), ), 7.89–7.91 (m, 1H, Ar-H), 8.15 (s, 1H, -NH amide), 8.79 (s, 1H, -OH) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 101.01 (1 C), 120.61 (1 C), 121.30 (1 C), 122.27 (1 C), 122.84 (1 C), 123.68 (1 C), 124.06 (1 C), 127.64 (1 C), 128.27 (1 C), 135.95 (1 C),136.24 (1 C), 141.35 (1 C), 155.81 (1 C), 169.07 (1 C), 171.33 (1 C), 175.67 (1 C), 176.97 (1 C) ppm; IR (KBr disc): 1267.23, 1309.67, 1334.74, 1342.46, 1382.96, 1425.40, 1479.40, 1483.26, 1535.34, 1573.91, 1604.77, 1658.78, 1693.50, 1703.14, 1720.50, 2555.68, 2920.23, 2985.81, 2014.74, 3059.10, 3433.29, 3522.02 cm− 1. Elemental Anal. Calcd. for C17H11N3O7: C, 55.29; H, 3.00; N, 11.38. Found: C, 54.95; H, 3.24; N, 11.01.
N-(4-Carboxy phenyl)-4-hydroxy-6-nitro-2-quinolone-3-carboxamide (35)
A combination of ethyl 4-hydroxy-6-nitro-2-quinolone-3-carboxylate (7b) (1.0 g, 3.6 mmol) and 4- amino benzoic acid (8xxvii) (1.1 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated on the inner flask and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. Coffee brown powder with a yield 47% (0.5 gm); Rf = (CHCl3: MeOH) (9.7 ml:0.3 ml); m.p: decomposed at 300 °C; 1H-NMR (400 MHz, DMSO-d6 + NaOD): δ = 7.92 (m, 3 H, Ar-H), 8.19 (m, 3 H, Ar-H), 8.25 (s, 1H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 67.5 (1 C), 101.0 (1 C), 118.2 (2 C), 122.1 (1 C), 122.6 (1 C), 123.6 (1 C), 124.1 (1 C), 130.4 (2 C), 132.8 (1 C), 136.6 (1 C), 142.8 (1 C),155.8 (1 C), 169.2 (1 C), 171.6 (1 C), 177.1 (1 C) ppm; IR (KBr disc): 1377, 1427, 1479, 1498, 1531, 1598, 1620, 1670, 2858, 2920, 3007, 3086, 3460, 3523 cm-1. Elemental Anal. Calcd. for C17H11N3O7: C, 55.29; H, 3.00; N, 11.38. Found: C, 55.85; H, 2.74; N, 11.86.
N-(4-Methoxyphenyl)-4-hydroxy-6-nitro-2-quinolone-3-carboxamide (36)
A combination of ethyl 4-hydroxy-6-nitro-2-quinolone-3-carboxylate (7b) (1.0 g, 3.6 mmol) and 4-methoxy aniline (8xxviii) (1.1 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated on the inner flask and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. An off-white powder with a yield of 63% (0.8gm); Rf = 0.61 (CHCl3: MeOH) (9.7 ml:0.3 ml); decomposed at 305 °C; 1H-NMR (500 Hz, DMSO-d6): δ = 3.65 (s, 3 H, CH3), 6.75 (d, J = 8.2 Hz, 2 H, Ar-H), 6.83 (d, J = 8.6 Hz, 1H, Ar-H), 7.55 (d, J = 8.2 Hz, 2 H, Ar-H), 7.81 (d, J = 8.2 Hz, 1H, Ar-H), 8.71 (s, 1H, Ar-H), 12.11 (s, 1H, NH), 12.53 (s, 1H, NH ppm; 13C-NMR (100 Hz, DMSO-d6): δ = 55.3 (1 C), 100.9 (1 C), 113.9 (2 C), 121.0 (2 C), 121.8 (1 C), 122.6 (1 C), 123.0 (1 C), 123.7(1 C), 134.4 (1 C), 135.9 (1 C), 154.1 (1 C), 155.9 (1 C), 168.7 (1 C), 175.6 (1 C), 176.6 (1 C) ppm. IR (KBr disc): 1319, 1338, 1379, 1436, 1483, 1568, 1595, 1622, 1693, 2441, 2991, 3093, 3344, 3446, 3456 cm− 1. Elemental Anal. Calcd. for C17H13N3O6: C, 57.47; H, 3.69; N, 11.83. Found: C, 57.65; H, 3.77; N, 11.74.
N-(4-Hydroxyphenyl)-4-hydroxy-6-nitro-2-quinolone-3-carboxamide (37)
A combination of ethyl 4-hydroxy-6-nitro-2-quinolone-3-carboxylate (7b) (1.0 g, 3.6 mmol) and 4- aminophenol (8xxix) (1.1 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated on the inner flask and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. A yellow powder (0.9 g, 73%); Rf = 0.5 (CHCl3: MeOH) (9 ml:1 ml); decomposed at 315 °C; 1H-NMR (500 Hz DMSO-d6): δ = 17.06 (s, 1H, OH), 12.45 (s, 1H, NH), 12.05 (s, 1H, NH), 8.72 (s, 1H, Ar-H), 7.82 (d, J = 8.8 Hz, 1H, Ar-H), 7.09 (d, J = 7.3 Hz, 2 H, Ar-H), 6.88 (d, J = 8.85 Hz, 1H, Ar-H), 6.27 (d, J = 7.85 Hz, 2 H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 176.21 (1 C), 175.94 (1 C), 168.34 (1 C), 165.44 (1 C), 155.67 (1 C), 136.31 (1 C), 125.24 (1 C), 123.81 (1 C), 123.59 (1 C), 122.78 (1 C), 122.51 (2 C), 122.09 (1 C), 118.55 (2 C), 101.82 (1 C) ppm. Elemental Anal. Calcd. for C16H11N3O6: C, 56.31; H, 3.25; N, 12.31. Found: C, 56.0; H, 3.20; N, 12.20.
N-(5-Methylbenzoic acid)-4-hydroxy-6-nitro-2-quinolone-3-carboxamide (38)
A combination of ethyl 4-hydroxy-6-nitro-2-quinolone-3-carboxylate (7b) (1.0 g, 3.6 mmol) and 2-amino-5- methylbenzoic acid (8xxx) (1.1 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated on the inner flask and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. A beige powder (0.85 g, 62%); Rf = 0.42 (CHCl3: MeOH) (1 ml:9 ml); Mp 294–296 °C; 1H-NMR (500 Hz DMSO-d6): δ = 17.0 (s, 1H, OH), 12.39 (s, 1H, NH), 12.36 (s, 1H, NH), 8.64 (s, 1H, Ar-H), 8.42 (d, J = 7.2 Hz, 1H, Ar-H), 8.12 (d, J = 6.96 Hz, 1H, Ar-H), 7.75 (s, 1H, Ar-H), 7.46 (d, J = 8.24 Hz, 1H, Ar-H), 7.4 (d, 1H, Ar-H), 2.33 (s,3 H, CH3) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 176.0 (1 C), 175.53 (1 C), 174.0 (1 C), 170.0 (1 C), 156.28 (1 C), 137.23 (1 C), 135.98 (1 C), 130.42 (1 C), 129.60 (1 C), 129.05 (2 C), 123.75 (1 C), 123.75 (2 C), 122.70 (2 C), 121.84 (1 C), 103.03 (1 C), 20.84 (1 C) ppm. Elemental Anal. Calcd. for C18H13N3O7: C, 56.40; H, 3.42; N, 10.96. Found: C, 56.20; H, 3.30; N, 10.75.
N-4-Chlorobenzoic acid-4-hydroxy-6-nitro-2-quinolone-3-carboxamide (39)
A combination of ethyl 4-hydroxy-6-nitro-2-quinolone-3-carboxylate (7b) (1.0 g, 3.6 mmol) and 2-amino-4- chlorobenzoic acid (8xxxi) (1.1 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated on the inner flask and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. A beige powder (0.73 g, 50%); Rf = 0.45 (CHCl3: MeOH) (4 drops:10 ml); Mp 279–281 °C; 1H-NMR (500 Hz DMSO-d6): δ = 16.31 (s, 1H, OH), 13.67 (s, 1H, OH), 13.16 (s, 1H, NH), 12.37 (s, 1H, NH), 8.58 (s, 1H, Ar-H), 8.4 (d, 1H, Ar-H), 7.90 (d, J = 8.44 Hz, 2 H, Ar-H), 7.42 (d, J = 9.04 Hz, 1H, Ar-H), 7.29 (d, J = 8.28 1H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 176.57 (1 C), 175.40 (1 C), 171.48 (1 C), 169.68 (1 C), 156.42 (1 C), 141.17 (1 C), 136.14 (1 C), 133.26 (1 C), 131.68 (1 C), 127.89 (1 C), 123.97 (1 C), 123.86 (1 C), 122.75 (2 C), 120.62 (1 C), 120.13 (1 C), 102.71 (1 C) ppm. Elemental Anal. Calcd. for C16H10ClN3O5: C, 53.42; H, 2.80; Cl, 9.85; N, 11.68. Found: C, 53.30; H, 2.70; N, 11.60.
N-4-Pyridin-4-hydroxy-6-nitro-2-quinolone-3-carboxamide (40)
A combination of ethyl 4-hydroxy-6-nitro-2-quinolone-3-carboxylate (7b) (1.0 g, 3.6 mmol) and 4-Amino Pyridin (8xxxii) (1.1 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated on the inner flask and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. 1H-NMR (400 MHz, DMSO-d6): δ = 11.38 (s, 1H, -OH), 10.66 (s, 1H, -NH amide ring), 8.52 (s, 1H, Ar-H), 7.60 (d, J = 8.8 Hz, 1H, Ar-H), 7.30 (d, J = 8.0 Hz, 2 H, Ar-H), 6.52 (d, J = 6.8 Hz, 2 H, Ar-H), 6.36 (d, J = 8.0 Hz, 2 H, Ar-H) ppm; 13C-NMR (100 MHz, DMSO-d6): δ = 172.2 (1 C), 171.9 (1 C), 163.7 (1 C), 154.9 (1 C), 150.8 (1 C), 150.1 (1 C), 149.8 (1 C), 148.4 (1 C), 141.9 (1 C), 125.9 (1 C), 122.6 (1 C), 112.0 (1 C), 104.1 (1 C), 96.1 (1 C), 92.4 (1 C) ppm; IR (KBr disc): 3442, 3342, 3221, 3084, 2941, 1635, 1612, 1550, 1421, 1352, 1219, 1130 cm− 1. Elemental Anal. Calcd. for C15H10N4O5: C, 55.22; H, 3.09; N, 17.17. Found: C, 54.50; H, 3.01; N, 17.0.
N-Anilino-4-hydroxy-6-nitro-2-quinolone-3-carboxamide (41)
A combination of ethyl 4-hydroxy-6-nitro-2-quinolone-3-carboxylate (7b) (1.0 g, 3.6 mmol) and phenylhydrazine (8xxxiii) (1.1 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated on the inner flask and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 hr. 1H-NMR (400 MHz, DMSO-d6 + NaOD): δ = 6.89–7.02 (m, 2 H, Ar-H), 7.21–7.25 (m, 2 H, Ar-H), 7.36 (s, 1H, Ar-H), 7.71 (d, J = 8 Hz, 1 H, Ar-H), 7.89–7.90 (m, 1H, Ar-H), 8.77 (s, 1H, -NH amide ring), ), 8.78 (s, 1H, -NH amide) 8.80 (s, 1H, -OH) ppm. 13C-NMR (100 MHz, DMSO-d6): δ = 101.07 (1 C), 119.80 (1 C), 121.60 (1 C), 122.92 (1 C), 123.72 (1 C), 124.01 (1 C), 128.68 (1 C), 128.96(2 C), 129.56 (1 C), 135.97 (1 C), 141.48 (1 C), 156.41 (1 C), 169.32 (1 C), 176.06 (1 C), 177.12 (1 C) ppm. IR (KBr disc): 692.44, 754.17, 806.25, 827.46, 871.82, 1076.28, 1157.29, 1267.23, 1340.53, 1379.10, 1429.25, 1475.54, 1496.76, 1556.55, 1616.35, 1660.71, 2910.58, 2978.09, 3053.32, 3361.93 cm− 1. Elemental Anal. Calcd. for C16H12N4O5: C, 56.47; H, 3.55; N, 16.46. Found: C, 63.83; H, 4.08; N, 13.18.
N-Benzyl-4-hydroxy-6-nitro-2-quinolone-3-carboxamide (42)
A combination of ethyl 4-hydroxy-6-nitro-2-quinolone-3-carboxylate (7b) (1.0 g, 3.6 mmol) and Benzylamine (8xxxiv) (1.1 g, 11.2 mmol) was placed together in 25 ml of THF. Few drops of DMF were added and the solution was refluxed for 72 h. The reaction progress and the formation of amide were monitored using TLC. The product was precipitated on the inner flask and the precipitate was collected by suction filtration at RT. At last, the product was washed with H2O, CH3OH, and THF then dried in a vacuum oven at 70 °C for 1 h. A pale yellow powder (0.60 g, 49%); Rf = 0.7 (n-Hexane: Ethyl acetate: MeOH) (1 ml:8.3 ml:0.7 ml); Mp 266–268 °C; 1 H-NMR (500 Hz DMSO-d6): δ = 17.42 (s, 1 H, OH), 12.27 (s, 1 H, NH), 10.34 (s, 1 H, NH), 8.56 (s, 1 H, Ar-H), 8.36 (d, J = 8.9 Hz, 1 H, Ar-H), 7.40 (d, J = 8.8 Hz, 1 H, Ar-H), 7.24–7.32 (m, 5 H, Ar-H), 4.55 (s, 2 H, CH2) ppm; 13C-NMR (125 MHz, DMSO-d6): δ = 176.62 (1 C), 175.77 (1 C), 171.18 (1 C), 155.91 (1 C), 141.04 (1 C), 136.18 (1 C), 128.69 (2 C), 127.64 (2 C), 126.86 (1 C), 123.89 (1 C), 123.58 (1 C), 122.69 (1 C), 122.11 (1 C), 101.15 (1 C), 42.07 (1 C) ppm. Elemental Anal. Calcd. for C17H13N3O5: C, 60.18; H, 3.86; N, 12.38. Found: C, 60.0; H, 3.76; N, 12.18.
Biological studies: reagents and dyes
The Dulbecco’s modified Eagle’s medium (DMEM) was used to culture all the cancer cells grown in the lab. DMEM was purchased from GE Healthcare Life Sciences, HyClone Laboratories (Logan, UT, USA). 0.25% trypsin was used to detach cells and was purchased from Corning Life Sciences (VWR International, LLC, Radnor, PA, USA) including 2.2 mM EDTA lysis buffer. Phosphate buffer saline (PBS) was purchased from Media Tech, Inc. (Manassas, VA, USA). Dimethylthiazol-2-yl-2, 5-diphenyltetrazolium bromide (MTT) was purchased from Calbiochem EMD Millipore (Billerica, MA, USA) [49].
Cell line and culture conditions
Prostate cancer (PC-3) and colon cancer (HCT-116) cells were a gift from late Dr. Gary Kruh, University of Illinois at Chicago. The cells were grown as an adherent monolayer in a cell culture flask in culture medium containing DMEM, supplemented with 4.5 g of glucose, 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin). The cells were maintained in an incubator, at 37 °C with 5% CO2 and a relative humidity of 95% [49, 50]. Other Growth conditions and reagents were prescribed in our previous work [38, 51, 52].
Cytotoxicity assay
The dimethylthiazol-2-yl-2,5-diphenyltetrazolium bromide (MTT) cytotoxicity assay was performed to identify the hits at various drug concentrations (0 and 10 µM). Hit screening was performed in HCT-116 and PC-3 cells. Briefly, 3000 cells/ well was seeded in a 96 well plate and allowed to attach overnight. The following day, the cells were treated with the test compounds (0 and 10 µM ). Hits were identified as compounds with an IC50 of less than 10 µM. The identified hits were further tested in both HCT-116 and PC-3 cells at various concentrations between 0 and 10 µM.
Computational methods
Molecular structures
All molecular structures were sketched using ChemDraw and saved in a sdf file format which was used as an input for ChemAxon Standardizer [53] to prepare the molecules for descriptor generation by adding hydrogens and aromatize all aromatic rings in the input structures. Details about structure preparation and molecular diversity assessments are reported elsewhere [54].
Molecular descriptors
Two dimensional (2D) alvaDesc [55–58] molecular descriptors to compare the calculated physicochemical properties of the synthesized compounds. We calculated 3874 2D molecular descriptors out of 5305 available descriptors after excluding all 3D descriptors. Next, 1849 2D descriptors were used for the principal component analysis (PCA). We also generated extended connectivity fingerprints (ECFP4) to assess molecular diversity in terms of chemical fragments. ECFP4, were used for representing the structures of our synthesized chemical compounds before performing similarity assessments. ECFPs are circular topological fingerprints that can be calculated easily and rapidly; they are not predefined and can represent an essentially infinite number of different molecular features (including stereochemical information) [59].
Docking and scoring
Molecular docking is a computational technique that simulates ligand binding to its molecular target by predicting the preferred orientation and conformation of the ligand within the binding site(s) of the molecular target. Scoring is the process of evaluating and quantifying the strength of the ligand-target interactions based on the predicted binding pose. Different scoring functions can be used to calculate docking scores as surrogate to estimating the binding affinities or free energies of the ligand-protein interaction. Higher negative docking scores indicate better binding. In this study we relied on induced-fit docking (IFD) software from Maestro and IFD scores for evaluating the binding potential of a group of synthesized carboxamide derivatives to their putative protein targets identified herein.
Ligand preparation for docking
All small-molecule chemical structures were prepared as the following: (1) generate 3D coordinates of all potential ligands based on the template of the co-crystallized ligand (X6K) in 4L23 using “Build” wizard in Maestro [60], (2) the 3D ligand coordinates were energetically treated using the “ligprep” script in Maestro [60]. LigPrep probed stereoisomerism, tautomerism, ring conformations, and ionization state. LigPrep generated diverse chemical and structural features from a single structure.
Protein preparation
The x-ray structures of human WT PI3Kα (PDB ID: 4L23) [61] and MUT (H1047R) PI3Kα (PDB ID: 3HHM) [62] were obtained from the Protein Data Bank (PDB) repository. Protein preparation module in Maestro [60] was employed to fill up the missing sequences, cap the N-and C-termini, minimize the hydrogen atoms, and optimize protein’s H-bond organization. Next, the proteins’ sidechains were further energetically minimized to reduce steric clashes.
Induced-fit docking
Induced-Fit Docking (IFD) was used to explore the affinities of our synthesized compounds to proteins included in an in-house drug target database. The co-ligands X6K/4L23 were marked as centroids in the binding clefts. The Vander Waals scaling factors for receptors and ligands were calibrated to 0.5 to provide adequate plasticity for the best docked ligand poses. For other parameters, the default values were used. The ligand conformation with the highest XP Glide binding score was reported. Docking scores were represented in term of Kcal/mol and the more the negative docking score infers the better the binder.
Results and discussion
Chemistry
Target compounds (9–42) were synthesized to investigate the effect of presenting different functionalities at the carboxamide side chain on the bioactivity of nitrated 4-hydroxy-2-quinolone scaffold. Ethyl anthranilate was produced by reacting anthranilic acid with an excess of ethanol in acidic media under reflux. Compounds 5 and 6 were refluxed in a basic medium to produce the target scaffold (7) with a yield of about 91% as shown in Fig. 2. Thin layer chromatography (TLC) was used to observe the progress of the reaction. Compound (7) was collected after ethyl anthranilate spot was no longer appeared on TLC. Regard compounds (9–42); they were synthesized by reacting (7) with excess of the corresponding amines (R-NH2) using THF and DMF under reflux, as stated in Table 1. Characterization of the chemical structures of the targeted compounds was carried out using NMR, FTIR, and elemental analysis The obtained data are shown in the experimental section along with the target structures.
Fig. 2.
Conditions: (a): (i) H2SO4, Reflux, 72 h. (b): (ii) NaOC2H5, DMSO, Reflux, 72 h. (c): (iii) DMF, THF Reflux, 48 h
Table 1.
The chemical structures of 4-hydroxy-7-nitro-2-quinolone 3-carboxamides (9–28) and 4-hydroxy-6-nitro-2-quinolone 3-carboxamides (29–42)

Biological evaluation of the synthesized compounds
To assess the anti-cancer activity of compounds (7 (a, b), 9–42), we probed their anti-proliferative activity in human colon cancer (HCT-116), colorectal adenocarcinoma (Caco-2), and prostate cancer cell (PC-3) cell lines. Two parallel biological investigations were accomplished in Toledo University in USA against PC-3 for analogues (7a, 9–28) and in the University of Jordan against HCT-116 and Caco-2 for derivatives (7a, 9–28, 32, 34, 36–42).
The malignant human colon carcinoma cell line (HCT-116) harbors both wild-type (WT) and mutant (MUT) (H1047R) PI3Kα and it was produced from a primary tumor following a specific tissue culture protocol [63]. HCT-116 cells express estrogen receptor β (ERβ), prostaglandin E2 (PGE2) receptors, matrix metalloproteinase 9 (MMP-9), and AMP-activated protein kinase (AMPK) [64]. HCT-116 expresses cell death regulator protein (B-cell lymphoma 2 (Bcl-2) and Fas receptor (tumor necrosis factor receptor superfamily 6 (TNFRSF6) or CD95) [65, 66]. HCT-116 cells encode PI3K, peroxisome proliferator-activated receptor gamma (PPAR-γ), and nonsteroidal anti-inflammatory drug-activated gene-1 (NAG-1) [67]. HCT-116 cells express epidermal growth factor (EGF) [68, 69], adenosine receptor (AR) [70], protein phosphatase 2 A (PP2A) [71], chemokines [72], caspases (3, 8, and 9) [73], death receptors (DR4 and DR5) [73], tumor suppressor gene (p53) [74], fibronectin receptor (α5β1 integrin) [75], proliferating cell nuclear antigen (PCNA) (lectin-like transcript 1 (LLT1)) [76], carcinoembryonic antigen (CEA) [77, 78], and ROS-mediated apoptotic pathways [79]. The extracellular-signal-regulated kinase (1/2) (ERK) (1/2)/MAPK [80, 81], PI3K/AKT [81–83], stromal cell-derived factor-1 (SDF-1)/chemokine receptor type 4 (CXCR4) [84, 85], and JAK/STAT [86, 87] pathways are essential for HCT-116 function. Histone deacetylase (HDAC-1, HDAC-2, HDAC-3) [88] and DNA methyltransferases (DNMTs) are expressed in HCT-116 cells [89].
The human colorectal adenocarcinoma (Caco-2) cell line encodes EGF, EGFR, retinoic acid binding protein I, and retinol binding protein II [90, 91]. The proliferated Caco-2 expresses MAPK, ERK1/2, JNK, protein kinase C (PKC-α), and MMP-9 [92, 93]. The PI3K/AKT pathway and histone deacetylase (HDAC-1, HDAC-2, and HDAC-3) controls Caco-2 growth [94].
The malignant human prostate cancer cell line (PC-3) expresses androgen receptor [95], PPAR-γ [96], human leukocyte antigen (HLA) (HLA1 and HLA9) [97], transforming growth factor-β (TGFβ) [98], tyrosine protein kinase (c-Met) [99], HDACs (HDAC-1, HDAC-2, and HDAC-3) [100], and EGFR [101]. The PI3K/AKT [102], nuclear factor NF-Kappa B (NF-κB) [103], c-Met/AKT/mTOR [104], and JAK/STAT [105] pathways are crucial for PC-3 function. Biological data revealed that the synthesized analogues exerted an inhibitory activity against PC-3 cells (Fig. 3).
Fig. 3.
A comparison of the % cell survival of synthesized compounds against PC-3. The compound IDs are on the x-axis and the % cell survival values are on the y-axis
Biological data against HCT-116 and Caco-2 cells showed that the antiproliferative activities of benzyl-bearing derivatives (9–24) surpass those of analogues with aniline moiety (25–28, 37, 41–42) indicating that the flexibility of the side chain mediated by benzyl group assists in the accommodation of ligands in the binding pocket (Table 2). Flexible side chains could play important roles in orientating the ligands deeply in protein binding sites [106]. Biological results demonstrated that compound bearing unsubstituted benzyl moiety (9) induces the antiproliferative activity in HCT-116 and Caco-2 cells, whereas, the substituted benzyl analogues provoke potent activity interrogating the significance of tailored benzyl motif (10–11, 13–14, 16, 19–24). Selective inhibitory activity against HCT-116 has been noticed for compounds (9–28, 36–37, 41–42) (Table 2).
Table 2.
The inhibitory activity (IC50 uM) of compounds (7 (a, b), 9–42) and selectivity-fold against HCT-116, SD never exceeded 5%, (n = 9), and treatment for 48 h
| Compound | IC50 uM | Selectivity Fold | Compound | IC50 uM | Selectivity Fold | ||
|---|---|---|---|---|---|---|---|
| HCT-116 | CaCo-2 | HCT-116 | CaCo-2 | ||||
| 7a | 74.98 | 913.8 | 12 | 26 | 139.3 | 469.3 | 3.4 |
| 7b | 101.0 | 178.7 | 1.8 | 27 | 332.2 | 479.0 | 1.4 |
| 9 | 60.39 | 95.3 | 1.6 | 28 | 119.7 | 205.1 | 1.7 |
| 10 | 23.41 | 58.08 | 2.5 | 29 | 11.9 | 76.0 | 6 |
| 11 | 30.82 | 69.94 | 2.3 | 30 | 15.6 | 47.5 | 3 |
| 12 | 83.22 | 128.77 | 1.6 | 31 | 51.6 | 59.8 | 1 |
| 13 | 27.14 | 77.18 | 2.8 | 32 | 170.6 | NOT ACTIVE | - |
| 14 | 56.99 | 116.7 | 2.0 | 33 | 38.7 | 67.3 | 1.7 |
| 15 | 101.1 | 171.0 | 1.7 | 34 | 121.5 | 200 | |
| 16 | 28.43 | 70.52 | 2.5 | 35 | 37.8 | 32.9 | 0.9 |
| 17 | 127.5 | 194.4 | 1.5 | 36 | 76.86 | 504 | 6.6 |
| 18 | 207.7 | 652.4 | 3.0 | 37 | 447.1 | 756 | 1.7 |
| 19 | 39.08 | 94.12 | 2.4 | 38 | 476.7 | NOT ACTIVE | - |
| 20 | 22.95 | 84.01 | 3.7 | 39 | 2304 | NOT ACTIVE | - |
| 21 | 43.68 | 235.77 | 5.4 | 40 | 65.4 | 61.9 | 0.9 |
| 22 | 37.42 | 98.26 | 2.6 | 41 | 167.1 | 325.6 | 1.9 |
| 23 | 39.11 | 60.57 | 1.5 | 42 | 181 | 346 | 1.9 |
| 24 | 51.39 | 199.26 | 3.9 | LY294002 | 7.4 | ||
| 25 | 229.8 | 385.7 | 1.7 | ||||
Concerning the 4-hydroxy-7-nitro-2-quinolone 3-carboxamides (9–28), the biological studies illustrated that the prospective compounds exhibited distinct inhibitory activity in Caco-2 and HCT-116 cells (Table 2). Compound 20 appears to be the most potent against HCT-166 cells while compound 10 exerts the most anti-proliferative activity against Caco-2 cells. The inhibitory activity in HCT-116 cells for compounds (9–14, and 22–25) reveals that tailoring the benzyl motif with m-CF3 (10), m-OCH3 (13), and m-CH3 (22) induces the activity implying that hydrophobic force drives ligand/receptor interaction. Additionally, incorporating p-CF3 (11) or p-CH3 (23) potentiates the activity interrogating that hydrophobic interaction mediates ligand/receptor complex formation. However, o- OCH3 (12) and p-OCH3 (14) attenuate the activity suggesting that the O-atom impedes the proper orientation of CH3 moiety. Indeed, the activity of (9) emphasizes the significance of tailoring the benzyl motif. Interestingly, p-OCH3 (14) and p-OH (24) exhibit similar activity interrogating that O atom shields the orientation of -CH3 in the binding site. The activity of m-Cl (16) agrees with that of (10) and confirms that hydrophobic interaction guides ligand/receptor interaction. Contrary, the activity of p-Cl (17) suggests that the electronegativity of –Cl might prohibit its accommodation in the binding site; though, -CH3, -CF3, and –Cl are isostere. And, the activity of o-Cl (15) might infer that o-substitution is not favored. Comparing the activity of 9 and 18 implies that H-bond and/or polarity weaken(s) the activity and thus in turn underlines that hydrophobicity guides ligand/receptor binding. The inhibitory activity in Caco-2 cells for compounds (9–14, and 24) proclaims that m-CF3 (10) enhances the activity indicating that hydrophobic and/ or H-bond direct(s) ligand/PI3Kα interaction. However, the activity of m-OCH3 (13) anticipates that O atom hinders the proper pose in the binding cleft. Comparing the activity of m-CH3 (22) to those of m-CF3 (10) and m-OCH3 (13) predicts that the electronegativity and hydrophobic features proceed ligand/receptor interaction. The activity of p-OH (24) infers that H-bond interaction is not involved in ligand/receptor complex formation.
Tailoring the benzyl ring with p-CF3 (11) or p-CH3 (23) enhances the activity supposing that hydrophobic interaction drives ligand/receptor complex formation. In contrast, p-OCH3 (14) weakens the activity suggesting that the O-atom hinders the proper accommodation of CH3 moiety in the binding cleft. Verily, the activity of 9 highlights the importance of substituting the benzyl motif. Moreover, the activity of m-Cl (16) accords with that of (10) and approves that hydrophobic interaction guides ligand/receptor binding. The activity of o-Cl (15) and p-Cl (17) implies that o-and p-Cl are not favored. Eventually, the activity of (18) interrogates that H-bond and/or polarity decrease(s) the activity pinpointing the significance of hydrophobicity. Interestingly, the similar profile of the prospective compounds in both cell lines motivates us to identify potential receptor(s) and explore its/their signaling pathway(s). It’s worth noting that compounds bearing fluorinated benzyl motif (19, 20, and 21) exert higher activity in HCT-116 and Caco-2 compared to those of fluorinated phenyl derivatives (26, 27, and 28) implying that elongation of carboxamide side chain by one carbon incites the activity. Additionally, o-F (26) and p-F (28) exhibit better activity in HCT-116 interrogating that H-bond acceptor is preferred on o-and p-position. The activity of 25 indicates that tailoring the phenyl motif is required to induce the activity. Eventually, the activity of 7a in HCT-116 and Caco-2 hypothesizes that –NO2 moiety incites the toxicity in HCT-116.
Concerning the 4-hydroxy-6-nitro-2-quinolone 3-carboxamides (29–42), the biological studies revealed that phenyl analogue (32) exerts better activity than its peer analogue (25), whereas, the benzyl derivative (9) exerts potent activity than that of its counterpart compound (42). Such results infer that phenyl bearing derivative (32) potentiates the activity for 4-hydroxy-6-nitro-2-quinolone 3-carboxamides, whereas benzyl bearing derivative (9) incites the activity for 4-hydroxy-7-nitro-2-quinolone 3-carboxamides. Tailoring the phenyl moiety with p-OCH3 analogue (36) exhibits potent and selective activity against HCT-116, whereas, attaching p-OCH3 on the benzyl moiety (14) potentiates the selectivity against HCT-116 and activity against HCT-116 and Caco-2 cells.
Interestingly, compounds (30 and 31) exert comparable cytotoxicity to those of 10 and 11 interrogating the significance of –CF3 motif on m- and p- positions. Indeed, the activity of o-CF3 (29) aligns with those of 30 and 31. Furthermore, the activity of o- and p- COOH (33 and 35) provides a further proof to the significance of the electronegativity and/or H-bond inferring the importance of water solubility that assists in their distribution.
In addition, Compound 11 was screened against a panel of cancer cells by the National Cancer Institute (NCI) biological laboratories facilities in USA [107] (Figs. 4 and 5) (Supplementary Table S1). Interestingly, 11 exerted potent inhibitory activity against leukemia (CCRF-CEM, HL-60(TB), K-S62, and RPMI-8226, SR), melanoma (LOXIMVI, SK-MEL-5, and UACC), non-small cell lung cancer (NSCLC) (EKVX, NCI-H460, and NCI-H522), colon (HCT-15), CNS (SF-295), ovarian (NCI/ADR-RES), breast (MDA-MB-468), prostate, and renal cancer (ACHN, CAKI-1, and U—31) cells and particularly against leukemia (HL-60 (TB)) (≈ 95%), melanoma (SK-MEL-5) (≈ 94%), and breast cancer (MDA-MB-468) (≈ 92%). Astonishingly, 100% inhibitory activity was shown against ovarian cancer (NCI/ADR-RES), colon cancer (COLO 205), CNC cancer (SF-295), and melanoma (SK-MEL-2) cells.
Fig. 4.
The growth % of analogue 11 against a panel of cancer cells
Fig. 5.
The mean growth % of analogue 11 against a panel of cancer cells
In order to evaluate the toxicity of analogues against human normal cells and to infer their selectivity against cancer cells, compound 11 was screened against an adult primary dermal fibroblast (HDFa) (PCS-201-012). Interestingly, 11 exerted 50-fold selective toxicity against HCT-116 and 22-fold against Caco-2 (Table 3). Such finding implies the safety profile of the verified analogues against normal cell and paves the way for in vivo studies.
Table 3.
The (IC50 uM) against primary dermal fibroblast PCS-201-012
| Compound | IC50 uM | Selectivity Fold against HCT-116 | Selectivity Fold against Caco-2 |
|---|---|---|---|
| 11 | 1559 | 50.6 | 22.3 |
We hypothesized that the observed activities can be attributed to the strategic incorporation of the nitro moiety, which is known to enhance cytotoxic effects through mechanisms such as DNA damage and disruption of repair pathways [108, 109]. The structural diversity of our derivatives (which is discussed in the computational section of the results) allowed us to explore how different functional groups influence biological activity. Notably, compounds with flexible side chains, particularly those bearing benzyl moieties, exhibited superior activity compared to those with more rigid structures. This flexibility may facilitate better binding interactions with target proteins, enhancing their efficacy as inhibitors.
Computational chemical biology analysis
Analyzing cancer pathways in HCT-116 and PC-3
Two cancer cell lines known to overexpress PI3K, colorectal HCT-116 and prostate PC-3, were used to study the anticancer effects of our compounds in vitro. In order to assess the similarities between these two cell lines in terms of perturbed cancer pathways, we compared the number of perturbed genes across 10 cancer pathways monitored by cBioPortal project [110] including cell cycle, HIPPO, MYC, NOTCH, NRF2, PI3K, RTK-RAS, TGF-Beta, TP53 and WNT. The results of this analysis are shown in Fig. 6. We found that both HCT-116 and PC-3 have more than one perturbed gene affecting seven of these pathways (HIPPO, MYC, NOTCH, PI3K, RTK-RAS, TP53 and WNT). However, perturbations in cell cycle and TGF-Beta were associated with HCT-116. NRF2 pathway was not perturbed in either cell line. These results confirm that both cell lines are suitable for studying the effects of PI3K inhibition on cancer pathways.
Fig. 6.

Comparison between two studied cancer cell lines HCT-116 and PC-3. The affected cancer pathways are plotted on the x-axis excluding NRF2 pathway since it isn’t perturbed in either cell line. The number of perturbed genes in each tracked pathway is plotted on the y-axis This plot was generated based on data mined from cBioPortal [110]
Cheminformatics evaluation
The cheminformatics analysis of the 34 synthesized quinolone derivatives provided valuable insights into their molecular characteristics and the structural diversity within their physicochemical space. The molecular structures were saved in SDF format and standardized using ChemAxon’s Standardizer [53]. These standardized 2D structures were then used as input for alvaDesc to compute thousands of molecular descriptors [55–57]. Figure 7 shows the results of principal component analysis (PCA) based on 1,857 descriptors listed in Supplementary Table S2. Clustering of compounds in PCA space suggests structural similarities that may underlie shared biological activities or mechanisms of action, offering a foundation for further drug optimization.
Fig. 7.
Graphical representation of the principal component analysis (PCA) of calculated 2D alvaDesc molecular descriptors for the synthesized active compounds. PC1 explaining 50.79% of the variation in descriptor values is on the x-axis, and PC2 explaining 16.61% of the variation in descriptor values is on the y-axis
To evaluate drug likeness, ten drug likeness descriptors were calculated using alvaDesc [58], as listed in Table 4. Figure 8 presents radar plots summarizing eight key indices: DLS_01, DLS_02, DLS_03, DLS_06, DLS_07, DLS_cons, Ro5, and cRo5. Most compounds exhibit balanced drug-likeness descriptor profiles, indicating favorable physicochemical properties for drug development.
Table 4.
Drug likeness scores used to assess synthesized compounds
| Score Index | Description |
|---|---|
| DLS_01 | Modified Lipinski’s Rule-based score [6] (4 rules). nRules: number of unsatisfied Ro5 rules. |
| DLS_02 | Modified drug-like score based on 6 rules [7]: nHDon ≤ 5, (N + O) 1–8, MW 200–450, MLOGP − 2.0 to 4.5, RBN 1–9, nCIC ≤ 5. |
| DLS_03 | Modified drug-like score based on 6 rules [8]: nHDon ≤ 5, (N + O) ≤ 10, MW 200–500, MLOGP − 5.0 to 5.0, RBN ≤ 8, charge − 2 to 2. |
| DLS_04 | Modified drug-like score based on 7 rules [9] including nHDon ≤ 5, (N + O) 2–10, MW 78–500, MLOGP − 0.5 to 5.0, C3p 0.15–0.8, h-p 0.6–1.6, Unsat-p 0.10–0.45. |
| DLS_05 | Modified drug-like score based on 2 rules [10]: NO_C3 0.10–1.80, Unsat-p ≤ 0.43. |
| DLS_06 | Modified drug-like score based on 6 rules from Rishton [11]: nHDon ≤ 5, (N + O) ≤ 10, MW ≤ 500, MLOGP ≤ 5, RBN ≤ 10, TPSA ≤ 140. |
| DLS_07 | Modified drug-like score based on 2 rules [12]: RBN ≤ 10, TPSA ≤ 140 or (nHDon + nHAcc) ≤ 12. |
| DLS_cons | Consensus drug-like score: Average of all DLS indices. |
| Ro5 | Lipinski Alert Index: Based on the number of Lipinski’s Rule of Five violations (MW > 500, LogP > 5, nHDon > 5, nHBAcc > 10). Compounds with zero or one violation are considered drug-like. |
| cRo5 | Complementary Lipinski Alert Index: Designed to evaluate compounds that fall outside the traditional Ro5 space, such as natural products or macrocycles. It considers flexibility, polarity, and solubility to assess non-Ro5 compliance drug-likeness. |
nHDon: number of hydrogen bond donors, nHBAcc: Number of hydrogen bond acceptors, MLOGP: Moriguchi’s logP, MW: molecular weight, RBN: rotatable bond number, nCIC: number of rings, C3p: ratio of sp³-hybridized carbon atoms (Csp³) to the total number of non-halogen atoms in the molecule, h-p represents the ratio of hydrogen atoms to the total number of non-halogen atoms in a molecule, Unsat-p: the degree of unsaturation in a molecule, normalized by the number of atoms that are not directly bonded to hydrogen or halogens, nX: number of halogens, TPSA: total polar surface area, Ro5: Lipinski’s role of five, cRo5: complementary Lipinski alert index, LogP: octanol-water partition coefficient
Fig. 8.
Radar plots for 34 compounds showing drug-like indices
However, compounds bearing OH, COOH, or hydrazine substituents, such as compounds 9, 18, and 24, show compressed or asymmetrical radar areas, with notably lower scores in descriptors such as DLS_03 (which incorporates polarity and hydrogen bonding alerts) and DLS_05 (which evaluates lead-likeness based on unsaturation and N/O content). These deviations suggest that such polar or reactive groups may negatively affect drug-likeness by impairing solubility, permeability, or metabolic stability. In contrast, derivatives lacking these functionalities generally exhibit more symmetrical and extended radar plots, especially in Ro5 and DLS_cons, reflecting greater compliance with Lipinski’s Rule of Five and stronger consensus across drug likeness models. These findings support a structure–property relationship and provide preliminary QSAR based justification for prioritizing specific scaffolds in anticancer drug development.
Overlaying synthesized analogues with pharmacophoric features of PI3Kα
To further expand upon the cheminformatics analysis and investigate the binding potential of the synthesized analogues (compounds 9–42) molecular modeling was performed by mapping the 3D coordinates of synthesized compounds against our pharmacophore model for active PI3Kα inhibitors. This pharmacophore model shown in Fig. 9, which encapsulates the key molecular features required for PI3Kα binding, served as a template to assess the fit of our compounds to these essential binding groups. Modeling results showed that the analogues indeed align with the pharmacophoric features characteristic of known active PI3Kα inhibitors (Fig. 9). This alignment suggests that the synthesized compounds possess the critical pharmacophoric features necessary for effective binding to the PI3Kα target, providing a rational explanation for their observed cytotoxicity. Moreover, these findings support the hypothesis that the anticancer activity of the analogues may be attributed to the inhibition of PI3Kα, highlighting the importance of these molecular interactions in their mechanism of action.
Fig. 9.
The pharmacophore model of PI3Kα inhibitors with 36 (pink color) and 37 (yellow color). The pharmacophoric groups are demonstrated as F1: aromatic ring; F2: aromatic ring or H-bond acceptor; F3: aromatic ring or hydrophobic motif; F4 and F5: H-bond acceptor. Picture visualized by MOE [111]
These insights not only confirm the potential of the analogues as PI3Kα inhibitors but also pave the way for further exploration of their inhibitory mechanisms. Understanding how these compounds interact with the PI3Kα binding site will be crucial in optimizing their efficacy and designing more potent derivatives in the future.
Docking and scoring against wild-type and mutant PI3Kα
The synthesized quinolone derivatives (7a–7b and 9–33), designed based on selective PI3Kα inhibitors previously reported by our group [36], were evaluated for their binding profiles against both wild-type (WT, PDB ID: 4L23) [112] and mutant PI3Kα H1047R (MUT, PDB ID: 3HHM) [62]. Structure-guided induced-fit docking (IFD) [60] was performed to accommodate side-chain flexibility and capture conformational changes in the kinase domain during ligand binding. Compounds exhibiting anticancer activity in HCT-116 and Caco-2 cells were prioritized for docking.
IFD results showed that compounds 7a–7b and 9–33 effectively occupy the PI3Kα active site. Notably, compound 10 closely mimicked the binding pose of the co-crystallized ligands X6K and KWT in WT and MUT structures, respectively, confirming its favorable orientation and interaction (Fig. 10A–F). Key binding residues identified across complexes included S774, W780, K802, D810, Y836, E849, V851, N853, S854, Q859, S919, and D933 (Table 5), many of which are known to contribute to kinase–ligand recognition [38, 113].
Fig. 10.
Binding poses and structural superpositions of selected quinolone analogues docked into PI3Kα kinase domains. (A) IF docked poses in 4L23 (WT PI3Kα). (B) Overlay of compound 10 (yellow) with native ligand X6K (green) in 4L23. (C) IF docked poses in 3HHM (H1047R mutant). (D) Overlay of compound 10 (yellow) with native ligand KWT (green) in 3HHM. (E) Binding of compound 21 (green) in the kinase domain of wild-type PI3Kα (PDB ID: 4L23). (F) Binding of compound 21 (green) in the kinase domain of mutant PI3Kα H1047R (PDB ID: 3HHM). Binding residues and ligand backbones are shown as stick models. H atoms are omitted for clarity. Carbon atoms are shown in yellow or green (ligands) and grey (residues); nitrogen is blue, oxygen is red. Visualizations were rendered in PYMOL [60]
Table 5.
The IFD scores (Kcal/mol) and H-bonding against native (PDB ID: 4L23) and mutant PI3Kα (PDB ID: 3HHM)
| CPD ID | NATIVE PI3Kα (PDB ID: 4L23) | MUTANT PI3Kα (PDB ID: 3HHM) | ||
|---|---|---|---|---|
| Docking Score |
H-Bond | Docking Score | H-Bond | |
| 7a | -8.26 | Y836, D933 | -8.03 | V851 |
| 7b | -8.26 | V851, S854 | -7.76 | E849, V851 |
| 9 | -8.59 | K802, Y836, V851, D933 | -7.43 | V851, N853, S854 |
| 10 | -8.42 | V851 | -8.84 | V851, N853, S854 |
| 11 | -8.15 | Y836, V851 | -7.78 | V851, S854 |
| 12 | -8.21 | Y836, V851 | -8.94 | E849, V851, N853, S854 |
| 13 | -8.40 | K802,N853, V851, | -8.38 | V851, N853, S854 |
| 14 | -8.06 | K802, Y836, V851 | -8.44 | V851 |
| 15 | -7.44 | K802, Y836, V851 | -7.69 | V851, Q859 |
| 16 | -8.33 | Y836, V851 | -7.88 | V851 |
| 17 | -8.60 | K802, Y836, V851 | -6.73 | S774, V851, D933 |
| 18 | -8.50 | Y836, V851 | -7.98 | V851 |
| 19 | -8.89 | K802, V851 | -7.41 | V851, Q859 |
| 20 | -8.43 | W780, V851, D933 | -8.12 | V851 |
| 21 | -7.95 | E849, V851, S854 | -7.92 | V851, N853, S854 |
| 22 | -8.22 | V851, D933 | -8.75 | E849, V851, N853, S854 |
| 23 | -10.21 | K802, V851, Q859, D933 | -8.31 | V851, N853, S854, S919 |
| 24 | -8.92 | N853, V851, S854 | -7.85 | V851, N853, S854 |
| 25 | -8.82 | V851 | -8.71 | V851, N853, S854 |
| 26 | -9.37 | N853, V851, S854 | -8.19 | V851, N853, S854 |
| 27 | -8.92 | V851 | -7.88 | V851, N853, S854 |
| 28 | -8.68 | V851 | -8.84 | V851, Q859 |
| 29 | -9.25 | V851, Q859 | -8.58 | E849, V851 |
| 30 | -9.16 | V851, Q859 | -8.67 | V851 |
| 31 | -10.54 | K802, D810, Y836, V851, Q859 | -8.79 | V851, S854, Q859 |
| 32 | -9.29 | E849, V851 | -8.38 | V851, Q859, S919 |
| 33 | -8.55 | V851, Q859, D933 | -7.98 | E849, V851 |
Molecular dynamics (MD) simulations previously conducted by our group [114] further supported the docking results. The H1047R mutation, known to enhance PI3Kα activity through oncogenic signaling [115], disrupted key hydrogen bonding interactions and increased conformational flexibility, especially within the L1 and L2 loop regions, which adopted a hook-like shape that promotes membrane association. This mutation also increased the distance between the catalytic and regulatory subunits and resulted in a more compact binding cleft; structural changes commonly associated with dysregulated kinase activity [116].
Despite these conformational differences, most compounds exhibited similar docking scores for both WT and MUT forms, suggesting robust backbone affinity. However, variation in substituent positioning and electronic properties influenced binding interactions and scores, especially with residues such as S774, S854, and Q859, indicating potential for fine-tuning selectivity. Overall, docking and MD results provided computational validation on the ability of these quinolone scaffolds to engage both PI3Kα isoforms and offer a foundation for further rational design of selective inhibitors in cancer therapy.
In summary, there is a clear and moderate relationship between the docking scores for wild type PI3Kα and the experimental IC₅₀ values. Compounds that docked more strongly, reflected by more negative docking scores, generally showed better anticancer activity. This suggests that the docking model is useful for predicting activity against the wild type enzyme. However, the weaker correlation between docking scores and IC₅₀ values for the mutant PI3Kα (H1047R) indicates that other factors, such as changes in protein shape, binding flexibility, or how the compound behaves in cells, may also play important roles in determining how well these compounds work against the mutant target.
Conclusions
We have successfully synthesized and evaluated a series of nitrated N-substituted 4-hydroxy-2-quinolone-3-carboxamides, highlighting their potential as anticancer agents. These derivatives demonstrated potent inhibitory activities against HCT-116, PC-3, and Caco-2 cell lines, with IC50 values indicating substantial antiproliferative effects. Clearly, compounds with flexible benzyl substituents were associated with enhanced activities. These results align with existing literature on the anticancer potential of quinolone-based compounds, which are known to modulate key cellular processes involved in tumor growth and metastasis. Docking studies reinforced the hypothesis of selective inhibition of PI3Kα, especially in its mutant form, suggesting avenues for future therapeutic development. Cheminformatics analysis and pharmacophore mapping results provided a more comprehensive evaluation of the therapeutic potential of synthesized analogues which could aid future developmental efforts of more potent and drug-like anticancer agents.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors acknowledge the generous support from the Deanship of Scientific Research at Al-Zaytoonah University of Jordan for the professional chemical laboratories facilities and the computational resources and databases (Grant numbers: 2023-2022/17/50 and 2025-2024/06/29), Deanship of Scientific Research, the University of Jordan, and the Hashemite University, the Chemistry Departments, for spectroscopic facilities. We thank also the College of Pharmacy, the University of Jordan, for offering cell culture laboratory and equipment. We would like to thank the Department of Medicinal and Biological Chemistry/College of Pharmacy and Pharmaceutical Sciences at the University of Toledo for their support to accomplish the anticancer screening. We thank the National Cancer Institute (NCI) in USA for biological laboratories facilities for further screening of the topmost active analogue against a panel of 60 cancer cells.
Author contributions
Conceptualization: DAS, RH, and SKB; Data curation: RAI, NSH, DAS, RH, SKB, KS, and RAK; Formal analysis: DAS, RH, GAS, KS, RAK and SKB; Funding acquisition DAS, RH, and SKB; Investigation RAI, NSH, DAS, RH, SKB, and KS; Methodology: RAI, NSH, DAS, RH, SKB, SB, AKT, and AMAZ; Project administration: DAS; Resources: DAS, RH, SKB, KS, and RAK; Software: DAS and RH; Supervision: DAS; Validation: DAS, RH, and SKB; Writing - review & editing: RAI, DAS, RH, SKB, and KS. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the funding received from the Deanship of Scientific Research and Graduate Studies at Al-Zaytoonah University of Jordan (Grant numbers: 2023 − 2022/17/50 and 2025 − 2024/06/29).
Data availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files). The NCI calculated IC50 for compound **11** is embedded in the Supplementary Table S1 (Excel Spreadsheet) and the 2D calculated descriptors for the synthesized analogues are shown in Supplementary Table S2 (Excel Spreadsheet).
Declarations
Ethics approval and consent to participate
The research was conducted at Al-Zaytoonah University of Jordan, the University of Jordan, Toledo University, and the National Cancer Institute (NCI). No human participants, identifiable human data, or animal subjects were involved. As the study did not involve human subjects, it is exempt from the requirement for ethics committee or Institutional Review Board (IRB) approval. Accordingly, ethics approval and informed consent were not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Baghban R, Afarid M, Soleymani J, Rahimi M. Were magnetic materials useful in cancer therapy? Biomed Pharmacother. 2021;144:112321. 10.1016/j.biopha.2021.112321. [DOI] [PubMed] [Google Scholar]
- 2.Tran KB, Lang JJ, Compton K, Xu R, Acheson AR, Henrikson HJ, et al. The global burden of cancer attributable to risk factors, 2010–19: a systematic analysis for the global burden of disease study 2019. Lancet. 2022;400(10352):563–91. 10.1016/S0140-6736(22)01438-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Osman S, Raza A, Al-Zaidan L, Inchakalody VP, Merhi M, Prabhu KS, et al. Anti-cancer effects of tranilast: an update. Biomed Pharmacother. 2021;141:111844. 10.1016/j.biopha.2021.111844. [DOI] [PubMed] [Google Scholar]
- 5.Anjum J, Mitra S, Das R, Alam R, Mojumder A, Emran TB, et al. A renewed concept on the MAPK signaling pathway in cancers: polyphenols as a choice of therapeutics. Pharmacol Res. 2022;184:106398. 10.1016/j.phrs.2022.106398. [DOI] [PubMed] [Google Scholar]
- 6.Hecht SS, Hatsukami DK. Smokeless tobacco and cigarette smoking: chemical mechanisms and cancer prevention. Nat Rev Cancer. 2022;22(3):143–55. 10.1038/s41568-021-00423-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi Y-J, Myung S-K, Lee J-H. Light alcohol drinking and risk of cancer: a meta-analysis of cohort studies. Cancer Res Treat. 2018;50(2):474–87. 10.4143/crt.2017.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez-Gutierrez L, Motiño O, Barriuso D, de la Puente-Aldea J, Alvarez-Frutos L, Kroemer G, et al. Obesity-Associated colorectal cancer. Int J Mol Sci. 2024;25(16). 10.3390/ijms25168836. [DOI] [PMC free article] [PubMed]
- 9.Liang H, Zhou X, Zhu Y, Li D, Jing D, Su X, et al. Association of outdoor air pollution, lifestyle, genetic factors with the risk of lung cancer: A prospective cohort study. Environ Res. 2022;114996. 10.1016/j.envres.2022.114996. [DOI] [PubMed]
- 10.Zeinali M, Abbaspour-Ravasjani S, Soltanfam T, Paiva-Santos AC, Babaei H, Veiga F, et al. Prevention of UV-induced skin cancer in mice by gamma oryzanol-loaded nanoethosomes. Life Sci. 2021;283:119759. 10.1016/j.lfs.2021.119759. [DOI] [PubMed] [Google Scholar]
- 11.Duan Y, Xu Y, Dou Y, Xu D. Helicobacter pylori and gastric cancer: mechanisms and new perspectives. J Hematol Oncol. 2025;18(1):10. 10.1186/s13045-024-01654-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan CK, Aimagambetova G, Ukybassova T, Kongrtay K, Azizan A. Human papillomavirus infection and cervical cancer: epidemiology, screening, and vaccination—review of current perspectives. J Oncol. 2019;2019:3257939. 10.1155/2019/3257939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mani SKK, Andrisani O. Hepatitis B virus-associated hepatocellular carcinoma and hepatic cancer stem cells. Genes. 2018;9(3):137. 10.3390/genes9030137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hajjo R, Sabbah DA, Al Bawab AQ. Unlocking the potential of the human Microbiome for identifying disease diagnostic biomarkers. Diagnostics. 2022;12(7):1742. 10.3390/diagnostics12071742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al Bataineh MT, Alzaatreh A, Hajjo R, Banimfreg BH, Dash NR. Compositional changes in human gut microbiota reveal a putative role of intestinal mycobiota in metabolic and biological decline during aging. J Nutr Health Aging. 2021;6:269–83. 10.3233/NHA-210130. [Google Scholar]
- 16.Ikhmais BA, Hammad AM, Abusara OH, Hamadneh L, Abumansour H, Abdallah QM, et al. Investigating carvedilol’s repurposing for the treatment of Non-Small cell lung cancer via aldehyde dehydrogenase activity modulation in the presence of β-Adrenergic agonists. Curr Issues Mol Biol. 2023;45(10):7996–8012. 10.3390/cimb45100505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abusara OH, Ibrahim AI, Issa H, Hammad AM, Ismail WH. In vitro evaluation of ALDH1A3-Affinic compounds on breast and prostate cancer cell lines as single treatments and in combination with doxorubicin. Curr Issues Mol Biol. 2023;45(3):2170–81. 10.3390/cimb45030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Mansoori L, Elsinga P, Goda SK. Bio-vehicles of cytotoxic drugs for delivery to tumor specific targets for cancer precision therapy. Biomed Pharmacother. 2021;144:112260. 10.1016/j.biopha.2021.112260. [DOI] [PubMed] [Google Scholar]
- 19.Nurmik M, Ullmann P, Rodriguez F, Haan S, Letellier E. In search of definitions: Cancer-associated fibroblasts and their markers. Int J Cancer. 2020;146(4):895–905. 10.1002/ijc.32193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Che N, Zhao X, Zhao N, Zhang Y, Ni C, Zhang D, et al. The role of different PI3K protein subtypes in the metastasis, angiogenesis and clinical prognosis of hepatocellular carcinoma. Ann Diagn Pathol. 2021;53:151755. 10.1016/j.anndiagpath.2021.151755. [DOI] [PubMed] [Google Scholar]
- 21.Lau A, Le N, Nguyen C, Kandpal RP. Signals transduced by Eph receptors and Ephrin ligands converge on MAP kinase and AKT pathways in human cancers. Cell Signal. 2022;104:110579. 10.1016/j.cellsig.2022.110579. [DOI] [PubMed] [Google Scholar]
- 22.Asl ER, Amini M, Najafi S, Mansoori B, Mokhtarzadeh A, Mohammadi A, et al. Interplay between MAPK/ERK signaling pathway and micrornas: A crucial mechanism regulating cancer cell metabolism and tumor progression. Life Sci. 2021;278:119499. 10.1016/j.lfs.2021.119499. [DOI] [PubMed] [Google Scholar]
- 23.Sabbah DA, Hajjo R, Bardaweel SK, Zhong HA. Targeting the PI3K/AKT signaling pathway in anticancer research: a recent update on inhibitor design and clinical trials (2020–2023). Expert Opin Ther Pat. 2024;34(3):141–58. 10.1080/13543776.2024.2338100. [DOI] [PubMed] [Google Scholar]
- 24.Sabbah DA, Hajjo R, Bardaweel SK, Zhong HA. Phosphatidylinositol 3-kinase (PI3K) inhibitors: a recent update on inhibitor design and clinical trials (2016–2020). Expert Opin Ther Pat. 2021;31(10):877–92. 10.1080/13543776.2021.1924150. [DOI] [PubMed] [Google Scholar]
- 25.Iksen PS, Pongrakhananon V. Targeting the PI3K/AKT/mTOR signaling pathway in lung cancer: an update regarding potential drugs and natural products. Molecules. 2021;26(13). 10.3390/molecules26134100. [DOI] [PMC free article] [PubMed]
- 26.Occhiuzzi MA, Lico G, Ioele G, De Luca M, Garofalo A, Grande F. Recent advances in PI3K/PKB/mTOR inhibitors as new anticancer agents. Eur J Med Chem. 2022;114971. 10.1016/j.ejmech.2022.114971. [DOI] [PubMed]
- 27.Lin J, Song T, Li C, Mao W. GSK-3β in DNA repair, apoptosis, and resistance of chemotherapy, radiotherapy of cancer. Biochim Biophys Acta Mol Cell Res. 2020;1867(5):118659. 10.1016/j.bbamcr.2020.118659. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Zhu Y, Sun C, Wang T, Shen Y, Cai W, et al. Feedback activation of basic fibroblast growth factor signaling via the Wnt/β-Catenin pathway in skin fibroblasts. Front Pharmacol. 2017;8:32. 10.3389/fphar.2017.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hajjo R, Sabbah DA, Bardaweel SK, Zhong HA. Targeting the EGFR/RAS/RAF signaling pathway in anticancer research: a recent update on inhibitor design and clinical trials (2020–2023). Expert Opin Ther Pat. 2024;34(1–2):51–69. 10.1080/13543776.2024.2327307. [DOI] [PubMed] [Google Scholar]
- 30.Gutiérrez-Hoya A, Soto-Cruz I. Role of the JAK/STAT pathway in cervical cancer: its relationship with HPV E6/E7 oncoproteins. Cells. 2020;9(10). 10.3390/cells9102297. [DOI] [PMC free article] [PubMed]
- 31.Azzman N, Anwar S, Syazani Mohamed WA, Ahemad N. Quinolone derivatives as anticancer agents: importance in medicinal chemistry. Curr Top Med Chem. 2024;24(13):1134–57. 10.2174/0115680266300736240403075307. [DOI] [PubMed] [Google Scholar]
- 32.Singh Y, Bhatia N, Biharee A, Kulkarni S, Thareja S, Monga V. Developing our knowledge of the quinolone scaffold and its value to anticancer drug design. Expert Opin Drug Discov. 2023;18(10):1151–67. 10.1080/17460441.2023.2246366. [DOI] [PubMed] [Google Scholar]
- 33.Sharma V, Das R, Mehta DK, Sharma D, Sahu RK. Exploring quinolone scaffold: unravelling the chemistry of anticancer drug design. Mini Rev Med Chem. 2022;22(1):69–88. 10.2174/1389557521666210112142136. [DOI] [PubMed] [Google Scholar]
- 34.Ilakiyalakshmi M, Arumugam Napoleon A. Review on recent development of Quinoline for anticancer activities. Arab J Chem. 2022;15(11):104168. 10.1016/j.arabjc.2022.104168. [Google Scholar]
- 35.Sabbah DA, Simms NA, Brattain MG, Vennerstrom JL, Zhong H. Biological evaluation and Docking studies of recently identified inhibitors of phosphoinositide-3-kinases. Bioorg Med Chem Lett. 2012;22(2):876–80. 10.1016/j.bmcl.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sabbah DA, Simms NA, Wang W, Dong Y, Ezell EL, Brattain MG, et al. N-Phenyl-4-hydroxy-2-quinolone-3-carboxamides as selective inhibitors of mutant H1047R phosphoinositide-3-kinase (PI3Kα). Bioorg Med Chem. 2012;20(24):7175–83. 10.1016/j.bmc.2012.09.059. [DOI] [PubMed] [Google Scholar]
- 37.Sabbah DA, Hishmah B, Sweidan K, Bardaweel S, AlDamen M, Zhong HA, et al. Structure-based design: synthesis, X-ray crystallography, and biological evaluation of N-substituted-4-hydroxy-2-quinolone-3-carboxamides as potential cytotoxic agents. Anticancer Agents Med Chem. 2018;18(2):263–76. 10.2174/1871520617666170911171152. [DOI] [PubMed] [Google Scholar]
- 38.Sabbah DA, Hasan SE, Abu Khalaf R, Bardaweel SK, Hajjo R, Alqaisi KM, et al. Molecular modeling, synthesis and biological evaluation of N-phenyl-4-hydroxy-6-methyl-2-quinolone-3-carboxamides as anticancer agents. Molecules. 2020;25(22):5348. 10.3390/molecules25225348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin Y, Liu YQ, Zhu KA, Hu MQ, Li Z, Min XJ. Tasquinimod enhances the sensitivity of ovarian cancer cells to cisplatin by regulating the Nur77-Bcl-2 apoptotic pathway. Adv Clin Exp Med. 2024;33(2):151–61. 10.17219/acem/166044. [DOI] [PubMed] [Google Scholar]
- 40.Fan R, Satilmis H, Vandewalle N, Verheye E, Vlummens P, Maes A, et al. Tasquinimod suppresses tumor cell growth and bone resorption by targeting immunosuppressive myeloid cells and inhibiting c-MYC expression in multiple myeloma. J Immunother Cancer. 2023;11(1):e005319. 10.1136/jitc-2022-005319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mintz J, Vedenko A, Rosete O, Shah K, Goldstein G, Hare JM, et al. Current advances of nitric oxide in cancer and anticancer therapeutics. Vaccines (Basel). 2021;9(2). 10.3390/vaccines9020094. [DOI] [PMC free article] [PubMed]
- 42.Güngör T, Tokay E, Güven Gülhan Ü, Hacıoğlu N, Çelik A, Köçkar F, et al. Prodrugs for nitroreductase based cancer therapy-4: towards prostate cancer targeting: synthesis of N-heterocyclic nitro prodrugs, Ssap-NtrB enzymatic activation and anticancer evaluation. Bioorg Chem. 2020;105:104450. 10.1016/j.bioorg.2020.104450. [DOI] [PubMed] [Google Scholar]
- 43.Farag AB, Othman AH, El-Ashrey MK, Abbas SE, Elwaie TA. New 6-nitro-4-substituted Quinazoline derivatives targeting epidermal growth factor receptor: design, synthesis and in vitro anticancer studies. Future Med Chem. 2024;16(19):2025–41. 10.1080/17568919.2024.2389772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spasova M, Stoyanova N, Nachev N, Ignatova M, Manolova N, Rashkov I, et al. Innovative fibrous materials loaded with 5-Nitro-8-hydroxyquinoline via electrospinning/electrospraying demonstrate antioxidant, antimicrobial and anticancer activities. Antioxid (Basel). 2023;12(6). 10.3390/antiox12061243. [DOI] [PMC free article] [PubMed]
- 45.Ayad Mohamed Rasheed H, Al-Majidi SMH. 5-nitro Isatin containing heterocyclics derivatives: synthesis, antioxidant activity, anticancer activity and molecular Docking. Nat Prod Res. 2025;39(1):56–65. 10.1080/14786419.2023.2250898. [DOI] [PubMed] [Google Scholar]
- 46.Hajjo R, Sabbah DA, Bardaweel SK. Chemocentric informatics analysis: dexamethasone versus combination therapy for COVID-19. ACS Omega. 2020;5(46):29765–79. 10.1021/acsomega.0c03597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bardaweel SK, Al-salamat H, Hajjo R, Sabbah D, Almutairi S. Unveiling the intricacies of monoamine Oxidase-A (MAO-A) Inhibition in colorectal cancer: computational systems biology, expression patterns, and the anticancer therapeutic potential. ACS Omega. 2024;9(33):35703–17. 10.1021/acsomega.4c04100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bardaweel SK, AlOmari R, Hajjo R. Integrating computational and experimental chemical biology revealed variable anticancer activities of phosphodiesterase isoenzyme 5 inhibitors (PDE5i) in lung cancer. RSC Med Chem. 2024;15(8):2882–99. 10.1039/D4MD00364K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balaji S, Neupane R, Malla S, Khupse R, Amawi H, Kumari S, et al. IND-2, a Quinoline derivative, inhibits the proliferation of prostate cancer cells by inducing oxidative stress, apoptosis and inhibiting topoisomerase II. Life. 2022;12(11):1879. 10.3390/life12111879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neupane R, Malla S, Abou-Dahech MS, Balaji S, Kumari S, Waiker DK, et al. Antiproliferative efficacy of N-(3-chloro-4-fluorophenyl)-6, 7-dimethoxyquinazolin-4-amine, DW-8, in colon cancer cells is mediated by intrinsic apoptosis. Molecules. 2021;26(15):4417. 10.3390/molecules26154417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sabbah DA, Haroon RA, Bardaweel SK, Hajjo R, Sweidan K. N-Phenyl-6-Chloro-4-Hydroxy-2-Quinolone-3-CarboxAmides: molecular docking, synthesis, and biological investigation as anticancer agents. Molecules. 2021;26:73. 10.3390/molecules26010073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sabbah DA, Samarat HH, Al-Shalabi E, Bardaweel SK, Hajjo R, Sweidan K, et al. Design, synthesis, and biological examination of N‐Phenyl‐6‐fluoro‐4‐hydroxy‐2‐quinolone‐3‐carboxamides as anticancer agents. ChemistrySelect. 2022;7(19):e202200662. 10.1002/slct.202200662. [Google Scholar]
- 53.ChemAxon. Cheminformatics software for the next generation of scientists; available from: https://chemaxon.com/
- 54.Hajjo R, Grulke CM, Golbraikh A, Setola V, Huang XP, Roth BL, et al. Development, validation, and use of quantitative structure-activity relationship models of 5-hydroxytryptamine (2B) receptor ligands to identify novel receptor binders and putative valvulopathic compounds among common drugs. J Med Chem. 2010;53(21):7573–86. 10.1021/jm100600y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mauri A, Bertola M, AlvaBuilder:. A software for de Novo molecular design. J Chem Inf Model. 2024;64(7):2136–42. 10.1021/acs.jcim.3c00610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mauri A, Bertola M, Alvascience. A new software suite for the QSAR workflow applied to the Blood–Brain barrier permeability. Int J Mol Sci. 2022;23(21):12882. 10.3390/ijms232112882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mauri A, alvaDesc. A tool to calculate and analyze molecular descriptors and fingerprints. In: Roy K, editor. Ecotoxicological QSARs. New York, NY: Springer US; 2020. pp. 801–20. [Google Scholar]
- 58.alvaDesc. 2.0. https://chm.kode-solutions.net/pf/alvadesc-2-0/ Accessed 28 January 2025.
- 59.Boldini D, Ballabio D, Consonni V, Todeschini R, Grisoni F, Sieber SA. Effectiveness of molecular fingerprints for exploring the chemical space of natural products. J Cheminform. 2024;16(1):35. 10.1186/s13321-024-00830-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schrödinger. Protein Preparation wizard, maestro, macromodel, QPLD-dock, and Pymol. Portland, OR, U.S.A.: Schrödinger, LLC; 2022. p. 97204. [Google Scholar]
- 61.Zhao Y, Zhang X, Chen Y, Lu S, Peng Y, Wang X, et al. Crystal structures of PI3Kalpha complexed with PI103 and its derivatives: new directions for inhibitors design. ACS Med Chem Lett. 2013;5(2):138–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mandelker D, Gabelli SB, Schmidt-Kittler O, Zhu J, Cheong I, Huang C-H, et al. A frequent kinase domain mutation that changes the interaction between PI3K alpha and the membrane. Proc Natl Acad Sci USA. 2009;106(40):16996–7001. 10.1073/pnas.0908444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brattain MG, Levine AE, Chakrabarty S, Yeoman LC, Willson JKV, Long B. Heterogeneity of human colon carcinoma. Cancer Metastasis Rev. 1984;3(3):177–91. 10.1007/bf00048384. [DOI] [PubMed] [Google Scholar]
- 64.Aladelokun O, Lu L, Zheng J, Yan H, Jain A, Gibson J, et al. Growth characteristics of HCT116 xenografts lacking asparagine synthetase vary according to sex. Hum Genomics. 2024;18(1):67. 10.1186/s40246-024-00635-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fries BD, Hummon AB. FAS inhibited proteomics and phosphoproteomics profiling of colorectal cancer spheroids shows activation of ferroptotic death mechanism. J Proteome Res. 2024;23(9):3904–16. 10.1021/acs.jproteome.4c00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang D, Li X, Song D, Chen S, Zhang Z, Cao S, et al. Atractylenolide III induces apoptosis by regulating the Bax/Bcl-2 signaling pathway in human colorectal cancer HCT-116 cells in vitro and in vivo. Anticancer Drugs. 2022;33(1):30–47. 10.1097/cad.0000000000001136. [DOI] [PubMed] [Google Scholar]
- 67.Kolawole OR, Kashfi K. NSAIDs and cancer resolution: new paradigms beyond cyclooxygenase. Int J Mol Sci. 2022;23(3):1432. 10.3390/ijms23031432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pisheh L, Matis S, Taglieri M, Di Gregorio L, Benelli R, Poggi A. EGFR-Targeted Antibody-Drug conjugate to different aminobisphosphonates: direct and indirect antitumor effects on colorectal carcinoma cells. Cancers (Basel). 2024;16(7). 10.3390/cancers16071256. [DOI] [PMC free article] [PubMed]
- 69.Unson S, Chang TC, Yang YN, Wang SH, Huang CH, Crawford DR, et al. Heteronemin and tetrac induce Anti-Proliferation by blocking EGFR-Mediated signaling in colorectal cancer cells. Mar Drugs. 2022;20(8):482. 10.3390/md20080482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khan S, Mentrup HL, Novak EA, Siow VS, Wang Q, Crawford EC, et al. Cyclic GMP-AMP synthase contributes to epithelial homeostasis in intestinal inflammation via Beclin-1-mediated autophagy. FASEB J. 2022;36(5):e22282. 10.1096/fj.202200138R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sakowicz-Burkiewicz M, Kitowska A, Grden M, Maciejewska I, Szutowicz A, Pawelczyk T. Differential effect of adenosine receptors on growth of human colon cancer HCT 116 and HT-29 cell lines. Arch Biochem Biophys. 2013;533(1–2):47–54. 10.1016/j.abb.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 72.Elemam NM, Al-Jaderi Z, Hachim MY, Maghazachi AA. HCT-116 colorectal cancer cells secrete chemokines which induce chemoattraction and intracellular calcium mobilization in NK92 cells. Cancer Immunol Immunother. 2019;68(6):883–95. 10.1007/s00262-019-02319-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao X, Feng X, Wang C, Peng D, Zhu K, Song JL. Anticancer activity of Nelumbo nucifera stamen extract in human colon cancer HCT-116 cells in vitro. Oncol Lett. 2017;13(3):1470–8. 10.3892/ol.2016.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kappel S, Ross-Kaschitza D, Hauert B, Rother K, Peinelt C. p53 alters intracellular Ca(2+) signaling through regulation of TRPM4. Cell Calcium. 2022;104:102591. 10.1016/j.ceca.2022.102591. [DOI] [PubMed] [Google Scholar]
- 75.Pelillo C, Mollica H, Eble JA, Grosche J, Herzog L, Codan B, et al. Inhibition of adhesion, migration and of α5β1 integrin in the HCT-116 colorectal cancer cells treated with the ruthenium drug NAMI-A. J Inorg Biochem. 2016;160:225–35. 10.1016/j.jinorgbio.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 76.Malaer JD, Mathew PA. Role of LLT1 and PCNA as natural killer cell immune evasion strategies of HCT 116 cells. Anticancer Res. 2020;40(12):6613–21. 10.21873/anticanres.14686. [DOI] [PubMed] [Google Scholar]
- 77.He S, Li S, Guo J, Zeng X, Liang D, Zhu Y, et al. CD166-specific CAR-T cells potently target colorectal cancer cells. Transl Oncol. 2023;27:101575. 10.1016/j.tranon.2022.101575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li M, Li S, Zhao R, Lv J, Zheng D, Qin L, et al. CD318 is a target of chimeric antigen receptor T cells for the treatment of colorectal cancer. Clin Exp Med. 2023;23(6):2409–19. 10.1007/s10238-022-00967-1. [DOI] [PubMed] [Google Scholar]
- 79.Jin H, Kim HS, Yu ST, Shin SR, Lee SH, Seo GS. Synergistic anticancer effect of docosahexaenoic acid and Isoliquiritigenin on human colorectal cancer cells through ROS-mediated regulation of the JNK and cytochrome c release. Mol Biol Rep. 2021;48(2):1171–80. 10.1007/s11033-021-06159-6. [DOI] [PubMed] [Google Scholar]
- 80.Huang E-Y, Chang J-C, Chen H-H, Hsu C-Y, Hsu H-C, Wu K-L. Carcinoembryonic antigen as a marker of radioresistance in colorectal cancer: a potential role of macrophages. BMC Cancer. 2018;18(1):321. 10.1186/s12885-018-4254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Phang CW, Karsani SA, Sethi G, Abd Malek SN, Flavokawain C. Inhibits cell cycle and promotes apoptosis, associated with Endoplasmic reticulum stress and regulation of MAPKs and Akt signaling pathways in HCT 116 human colon carcinoma cells. PLoS ONE. 2016;11(2):e0148775. 10.1371/journal.pone.0148775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang WS, Hsieh MC, Huang CY, Kuo YH, Tung SY, Shen CH, et al. The association of CXC receptor 4 mediated signaling pathway with Oxaliplatin-Resistant human colorectal cancer cells. PLoS ONE. 2016;11(9):e0159927. 10.1371/journal.pone.0159927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hamadneh L, Abuarqoub R, Alhusban A, Bahader M. Upregulation of PI3K/AKT/PTEN pathway is correlated with glucose and glutamine metabolic dysfunction during Tamoxifen resistance development in MCF-7 cells. Sci Rep. 2020;10(1):21933. 10.1038/s41598-020-78833-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen CY, Yang SH, Chang PY, Chen SF, Nieh S, Huang WY, et al. Cancer-Associated-Fibroblast-Mediated paracrine and autocrine SDF-1/CXCR4 signaling promotes stemness and aggressiveness of colorectal cancers. Cells. 2024;13(16):1334. 10.3390/cells13161334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sarkhosh-Inanlou R, Imani M, Sam MR. The response of PIK3CA/KRAS-mutant colorectal cancer stem-like cells to RGD-peptide frac produced by the strawberry anemone: A promising water-soluble peptide-based inhibitor of metastasis-driver gene CXCR4, stem cell regulatory genes and self-renewal. Biomed Pharmacother. 2020;132:110807. 10.1016/j.biopha.2020.110807. [DOI] [PubMed] [Google Scholar]
- 86.Dariya B, Muppala S, Srivani G, Momin S, Alam A, Saddala MS. Targeting STAT proteins via computational analysis in colorectal cancer. Mol Cell Biochem. 2021;476(1):165–74. 10.1007/s11010-020-03893-6. [DOI] [PubMed] [Google Scholar]
- 87.Hu Y, Shen Y, Zhao Y, Tang Y, Liu C, Gu Y, et al. Genomic distribution of signal transducer and activator of transcription (STAT) family in colorectal cancer. Hum Cell. 2023;36(1):286–95. 10.1007/s13577-022-00815-0. [DOI] [PubMed] [Google Scholar]
- 88.Sanaei M, Kavoosi F. Effect of sodium butyrate on p16INK4a, p14ARF, p15INK4b, class I HDACs (HDACs 1, 2, 3) class II HDACs (HDACs 4, 5, 6), cell growth Inhibition and apoptosis induction in pancreatic cancer AsPC-1 and colon cancer HCT-116 cell lines. Asian Pac J Cancer Prev. 2022;23(3):795–802. 10.31557/apjcp.2022.23.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Laranjeira ABA, Nguyen D, Pelosof LC, Doroshow JH, Yang SX. Upregulation of TET2 and resistance to DNA methyltransferase (DNMT) inhibitors in DNMT1-Deleted cancer cells. Diseases. 2024;12(7). 10.3390/diseases12070163. [DOI] [PMC free article] [PubMed]
- 90.Pisheh L, Matis S, Taglieri M, Di Gregorio L, Benelli R, Poggi A. EGFR-Targeted Antibody–Drug conjugate to different aminobisphosphonates: direct and indirect antitumor effects on colorectal carcinoma cells. Cancers. 2024;16(7):1256. 10.3390/cancers16071256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Abdelaal MR, Ibrahim E, Elnagar MR, Soror SH, Haffez H. Augmented therapeutic potential of EC-Synthetic retinoids in Caco-2 cancer cells using an in vitro approach. Int J Mol Sci. 2022;23(16):9442. 10.3390/ijms23169442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chu C, Ru H, Chen Y, Xu J, Wang C, Jin Y. Gallic acid attenuates LPS-induced inflammation in Caco-2 cells by suppressing the activation of the NF-κB/MAPK signaling pathway. Acta Biochim Biophys Sin (Shanghai). 2024;56(6):905–15. 10.3724/abbs.2024008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kaur H, Moreau R. mTORC1 Silencing during intestinal epithelial Caco-2 cell differentiation is mediated by the activation of the AMPK/TSC2 pathway. Biochem Biophys Res Commun. 2021;545:183–8. 10.1016/j.bbrc.2021.01.070. [DOI] [PubMed] [Google Scholar]
- 94.Korsten S, Vromans H, Garssen J, Willemsen LEM. Butyrate protects barrier integrity and suppresses immune activation in a Caco-2/PBMC Co-Culture model while HDAC Inhibition mimics butyrate in restoring Cytokine-Induced barrier disruption. Nutrients. 2023;15(12). 10.3390/nu15122760. [DOI] [PMC free article] [PubMed]
- 95.Lombardi APG, Cavalheiro RP, Porto CS, Vicente CM. Estrogen receptor signaling pathways involved in invasion and colony formation of Androgen-Independent prostate cancer cells PC-3. Int J Mol Sci. 2021;22(3):1153. 10.3390/ijms22031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rapuano R, Riccio A, Mercuri A, Madera JR, Dallavalle S, Moricca S, et al. Proliferation and migration of PC-3 prostate cancer cells is counteracted by PPARγ-cladosporol binding-mediated apoptosis and a decreased lipid biosynthesis and accumulation. Biochem Pharmacol. 2024;222:116097. 10.1016/j.bcp.2024.116097. [DOI] [PubMed] [Google Scholar]
- 97.Sabbatino F, Liguori L, Polcaro G, Salvato I, Caramori G, Salzano FA, et al. Role of human leukocyte antigen system as A predictive biomarker for Checkpoint-Based immunotherapy in cancer patients. Int J Mol Sci. 2020;21(19):7295. 10.3390/ijms21197295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Walker L, Millena AC, Strong N, Khan SA. Expression of TGFβ3 and its effects on migratory and invasive behavior of prostate cancer cells: involvement of PI3-kinase/AKT signaling pathway. Clin Exp Metastasis. 2013;30(1):13–23. 10.1007/s10585-012-9494-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boreddy SR, Nair R, Pandey PK, Kuriakose A, Marigowda SB, Dey C, et al. BCA101 is a tumor-Targeted bifunctional fusion antibody that simultaneously inhibits EGFR and TGFβ signaling to durably suppress tumor growth. Cancer Res. 2023;83(11):1883–904. 10.1158/0008-5472.Can-21-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Poria DK, Sheshadri N, Balamurugan K, Sharan S, Sterneck E. The STAT3 inhibitor stattic acts independently of STAT3 to decrease histone acetylation and modulate gene expression. J Biol Chem. 2021;296:100220. 10.1074/jbc.RA120.016645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zheng L, Zhang Y, Mei S, Xie T, Zou Y, Wang Y, et al. Discovery of a potent dual son of sevenless 1 (SOS1) and epidermal growth factor receptor (EGFR) inhibitor for the treatment of prostate cancer. J Med Chem. 2024;67(9):7130–45. 10.1021/acs.jmedchem.3c02433. [DOI] [PubMed] [Google Scholar]
- 102.Choi SY, Jeon JM, Na AY, Kwon OK, Bang IH, Ha YS, et al. SIRT5 directly inhibits the PI3K/AKT pathway in prostate cancer cell lines. Cancer Genom Proteom. 2022;19(1):50–9. 10.21873/cgp.20303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chang KS, Chen ST, Sung HC, Hsu SY, Lin WY, Hou CP, et al. Androgen receptor upregulates Mucosa-Associated lymphoid tissue 1 to induce NF-κB activity via Androgen-Dependent and -Independent pathways in prostate carcinoma cells. Int J Mol Sci. 2023;24(7). 10.3390/ijms24076245. [DOI] [PMC free article] [PubMed]
- 104.Su P, Zhang M, Kang X. Targeting c-Met in the treatment of urologic neoplasms: current status and challenges. Front Oncol. 2023;13:1071030. 10.3389/fonc.2023.1071030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yin Y, Wang Z, Hu Y, Wang J, Wang YI, Lu Q. Caffeic acid hinders the proliferation and migration through Inhibition of IL-6 mediated JAK-STAT-3 signaling axis in human prostate cancer. Oncol Res. 2024;32(12):1881–90. 10.32604/or.2024.048007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.T RR, Saharay M, Smith JC, Krishnan M. Correlated response of protein Side-Chain fluctuations and conformational entropy to ligand binding. J Phys Chem B. 2021;125(34):9641–51. 10.1021/acs.jpcb.1c01227. [DOI] [PubMed] [Google Scholar]
- 107.National Cancer Institute (NCI)| Cancer Biology Research. https://www.cancer.gov/research/areas/biology Accessed 26 December 2024.
- 108.Chen J, Potlapalli R, Quan H, Chen L, Xie Y, Pouriyeh S, et al. Exploring DNA damage and repair mechanisms: A review with computational insights. BioTech. 2024;13(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fahrer J, Christmann M. DNA alkylation damage by nitrosamines and relevant DNA repair pathways. Int J Mol Sci. 2023;24(5). 10.3390/ijms24054684. [DOI] [PMC free article] [PubMed]
- 110.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Molecular Operating Environment (MOE). 2022.02 Chemical Computing Group ULC, 910–1010 Sherbrooke St. W., Montreal, QC H3A 2R7, 2022.
- 112.Zhao Y, Zhang X, Chen Y, Lu S, Peng Y, Wang X, et al. Crystal structures of PI3Kalpha complexed with PI103 and its derivatives: new directions for inhibitors design. ACS Med Chem Lett. 2013;5(2):138–42. 10.1021/ml400378e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cheng H, Orr STM, Bailey S, Brooun A, Chen P, Deal JG, et al. Structure-Based drug design and synthesis of PI3Kα-Selective inhibitor (PF-06843195). J Med Chem. 2021;64(1):644–61. 10.1021/acs.jmedchem.0c01652. [DOI] [PubMed] [Google Scholar]
- 114.Sabbah DA, Vennerstrom JL, Zhong HA. Binding selectivity studies of phosphoinositide 3-kinases using free energy calculations. J Chem Inf Model. 2012;52:3213–24. 10.1021/ci3003057. [DOI] [PubMed] [Google Scholar]
- 115.Sharma J, Bhardwaj V, Purohit R. Structural perturbations due to mutation (H1047R) in Phosphoinositide-3-kinase (PI3Kα) and its involvement in oncogenesis: an in Silico insight. ACS Omega. 2019;4(14):15815–23. 10.1021/acsomega.9b01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Janku F, Wheler JJ, Naing A, Falchook GS, Hong DS, Stepanek VM, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res. 2013;73(1):276–84. 10.1158/0008-5472.CAN-12-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files). The NCI calculated IC50 for compound **11** is embedded in the Supplementary Table S1 (Excel Spreadsheet) and the 2D calculated descriptors for the synthesized analogues are shown in Supplementary Table S2 (Excel Spreadsheet).