Abstract
The aim of this study was to develop a protocol for the simultaneous extraction from bacterioplankton of RNA and DNA suitable for quantitative molecular analysis. By using a combined mechanical and chemical extraction method, the highest RNA and DNA yield was obtained with sodium lauryl sarcosinate-phenol or DivoLab-phenol as the extraction mix. The efficiency of extraction of nucleic acids was comparatively high and varied only moderately in gram-negative bacterial isolates and bacterioplankton (RNA, 52 to 66%; DNA, 43 to 61%); significant amounts of nucleic acids were also obtained for a gram-positive bacterial isolate (RNA, 20 to 30%; DNA, 20 to 25%). Reverse transcription-PCR and PCR amplification products of fragments of 16S rRNA and its genes were obtained from all isolates and communities, indicating that the extracted nucleic acids were intact and pure enough for community structure analyses. By using single-strand conformation polymorphism of fragments of 16S rRNA and its gene, community fingerprints were obtained from pond bacterioplankton. mRNA transcripts encoding fragments of the enzyme nitrite reductase gene (nir gene) could be detected in a pond water sample, indicating that the extraction method is also suitable for studying gene expression. The extraction method presented yields nucleic acids that can be used to perform structural and functional studies of bacterioplankton communities from a single sample.
The vast majority (>99%) of bacterial cells from aquatic systems do not grow on culture plates (1). This hampered the investigation of the biodiversity and taxonomic structure of bacteria until culture-independent methods were developed. Several nucleic-acid-based methods are now available to investigate the community structure of bacteria (for a compilation of methods, see, e.g., reference 11). Frequently, RNA in subunits of the ribosome and their genes are the target molecules of these techniques.
Approaches have also been developed for the functional analysis of bacteria, such as studying gene expression and mRNA (17, 21). Since the number of mRNA transcripts is related to activity whereas sequence heterogeneity may be related to phylogenetic distance, studies of functional genes may provide information on the activity of particular functional genes and on the phylogenetic affiliation of the bacterial populations expressing the genes (23). Linking community structure to activity and functionality is a central but poorly studied issue in microbial ecology.
For many of the methods for studying the structure and function of natural bacterial communities, nucleic acids have to be extracted from the cells before analyses can be performed. A variety of nucleic acid extraction methods have been described for bacterioplankton (16); however, the extraction efficiencies of these methods were usually not tested rigorously. We present a protocol for the parallel extraction of RNA and DNA from a single sample in a two-step procedure. We found a comparatively high extraction efficiency for total RNA and DNA and showed that the extracted nucleic acid is sufficiently intact for PCR amplification of the 16S rRNA gene and community fingerprinting, as well as for gene expression at the mRNA level.
MATERIALS AND METHODS
Sampling and sample preparation.
Five-liter water samples were collected with a bucket from the pond of the German Research Center for Biotechnology (GBF pond) and prefiltered through a 10-μm-mesh-size nylon net and a 90-mm-diameter 3.0-μm-pore-size polycarbonate filter (Nuclepore). Bacterioplankton from prefiltered 1.2-liter aliquots was harvested onto a filter sandwich consisting of a precombusted (4 h at 450°C) 90-mm-diameter glass fiber filter (GF/F; Whatman) on top of a MilliQ water-rinsed 90-mm-diameter 0.2-μm-pore-size polycarbonate filter (Nuclepore) as described by Dominik and Höfle (3). Bacterial strains were suspended in physiological saline, and aliquots (ca. 100 ml) were collected on the filter sandwich. The filter sandwiches were folded, wrapped in aluminum foil, and stored frozen at −70°C. For quantification of bacterial DNA and RNA, aliquots of bacterial cultures and pond water were filtered onto 25-mm-diameter 0.2-μm-pore-size polycarbonate filters. The filters were placed in a 2-ml Eppendorf tube along with 1 ml of Tris-Ca2+ buffer (0.1 M NaCl, 0.1 M Tris [pH 7.5], 0.1324 g of CaCl2O · 2H2O liter−1) and kept frozen at −70°C (12).
Strains.
Type strains from distantly related taxonomic groups were used in this study: the gram-negative bacterium Ralstonia eutropha DSM 531 (β subclass of Proteobacteria), Escherichia coli DSM 613 (γ subclass of Proteobacteria), and Flavobacterium johnsoniae DSM 2064 (Cytophaga-Flavobacteria group) and the high G+C-content gram-positive bacterium Arthrobacter globiformis DSM 20124. All strains were grown on nutrient broth agar (8 g liter−1; Difco Corp.), transferred to liquid nutrient broth medium and regrown at 30°C, and collected during exponential growth.
Extraction of nucleic acids.
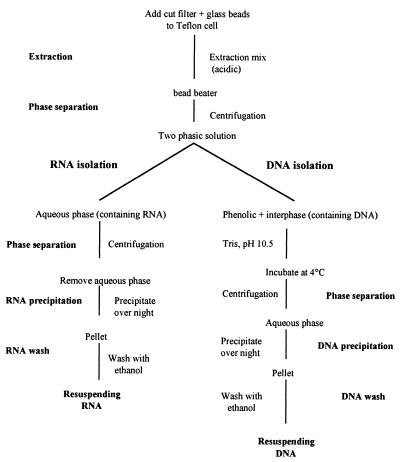
A combined mechanical and chemical extraction method (10) was used following the protocol of Dominik and Höfle (3). We expanded this protocol to the simultaneous extraction of DNA. A flowchart of this modified extraction method is shown in Fig. 1. The filter sandwich with the bacteria was cut into small pieces with a sterile scalpel and transferred to 20-ml Teflon extraction cells (no. 854495/6; Braun Corp., Melsungen, Germany) containing 2 g of 2- and 3-mm-diameter precombusted and siliconized glass beads. RNA was extracted with equal volumes (5 ml) of sodium lauryl sarcosinate (SLS; 0.5% in 50 mM sodium acetate and 10 mM EDTA [pH 4.2]) and phenol and 2 min of vibration in a high-speed cell disrupter (Microdismembrator II [no. 893162/4]; Braun Corp.) set at an amplitude of 15 mm. The homogenate was transferred to a 50-ml Falcon tube, mixed by vortexing, and centrifuged for 20 min at 7,200 × g at 4°C, and the aqueous phase containing RNA was removed. Following a second extraction, both aqueous phases were combined, purified by two chloroform-isoamyl alcohol (24:1) washing steps (10 min at 11,000 × g and 4°C), and precipitated with isopropanol (1 volume) and 3 M sodium acetate (0.1 volume; pH 4) at −20°C overnight. The RNA pellets were washed twice with ice-cold ethanol (70%), dried in a SpeedVac for 10 min, and resuspended in 300 μl of autoclaved MilliQ water. The filter remnants were precipitated by centrifugation (10 min at 20,000 × g and 4°C), and the aqueous RNA solution was mixed with precipitation mix (0.2 M sodium acetate and 10 mM MgCl2 in 100% ethanol). For more details of the protocol, consult the work of Dominik and Höfle (3).
FIG. 1.
Flowchart of method used for extracting DNA and RNA from bacterioplankton. Note that extraction steps are repeated to increase the yields of DNA and RNA.
DNA was eluted from the organic phase and interphase by establishing an alkaline pH of 8 by adding 5 ml of 1 M Tris base (pH 10.5), following the protocol of Majumdar et al. (13) as cited by Farrell (6). DNA was extracted for 40 min at 4°C. Samples were vortexed, and the phases were separated by centrifugation (2,000 × g for 15 min at 4°C). This extraction was repeated, and both aqueous phases were combined. The DNA was purified, precipitated, and washed as described for RNA.
Nucleic acids were also extracted by using equal volumes of 9.6% DivoLab No. 1 (chemical no. 004564F; DiverseyLever Ltd., Northampton, United Kingdom) and phenol in 120 mM sodium acetate (pH 4.0) (14) instead of SLS-phenol. Moreover, we tested two commercially available extraction kits, InstaPure (TRI InstaPure KP-0130; Eurogentec, Seraing, Belgium) and RNA-DNA isolators (RNA-ISO and DNA-ISO; Genosys, Cambridge, United Kingdom) in combination with the mechanical extraction. For details of these methods, see the protocols provided by the manufacturers. For all extraction kits and mixes, a filter sandwich consisting of a MilliQ water-rinsed polycarbonate filter and a precombusted GF/F filter was extracted to check for possible DNA or RNA contamination during the extraction procedure (negative control).
Quantification of nucleic acids.
RNA and DNA in bacteria were quantified by using the method described by Jeffrey et al. (12) with SYBRGreen II (see below) as the dye (24). Quantification was done as described by Weinbauer and Höfle (24) with the following specifications. Bacteria were homogenized on ice with a cell disrupter (4-mm needle diameter; Labsonic U 2000) set at 70 W and 0.5-s pulses. The optimum sonication time was determined by increasing the sonication time in 10-s intervals and determining the SYBRGreen II fluorescence. A maximum fluorescence was obtained after 30 to 60 s of sonication for gram-negative and gram-positive bacteria, as well as the pond water sample. The concentration of total nucleic acids and DNA (after RNase digestion) was determined using SYBRGreen II (10,000× in dimethyl sulfoxide [chemical no. S-7568; Molecular Probes]). RNA concentrations were calculated as the fluorescence of total nucleic acids minus the DNA fluorescence determined after RNase digestion. DNase digestion resulted in only a slight reduction of detectable DNA concentrations in cells, and thus we could not check the efficiency of nucleic acid digestion in cells by combined RNase and DNase treatments. The reasons for the failure of DNA digestion in cells remains unknown but was observed before (2).
The extracted RNA and DNA were quantified using RiboGreen (RNA quantitation kit [chemical no. R-11490]; Molecular Probes) and PicoGreen (double-stranded-DNA quantitation kit [chemical no. P-7581]; Molecular Probes) and a microtiter plate reader as described previously (24).
Primers.
The primer set F-27 and R-1492 was used to amplify ca. 1,450 bp of the 16S rRNA gene (Table 1). The primer set GC-F-984 and R-1385 amplifies a 16S rRNA gene fragment, and the primer GC-F-984 attaches a GC (denaturing gradient gel electrophoresis [DGGE] primer). The fragment amplified by the primers F-536 and R-907 was used for single-strand conformation polymorphism (SSCP) analyses (SSCP primer).
TABLE 1.
Primers used for amplification of 16S rDNA of the domain Bacteria and a central region of the nir gene
| Primera | Positionsb | Sequencec |
|---|---|---|
| F-27 | 8-27 | 5′-AGAGTTTGATCMTGGCTCAG-3′ |
| R-1492 | 1492-1513 | 5′-TACGGYTACCTTGTTACGACTT-3′ |
| GC-F-984 | 968-984 | 5′-GC-clamp-AACGCGGAAGAACCTTAC-3′ |
| R-1385 | 1385-1401 | 5′-CGGTGTGTACAAGAAGACCC-3′ |
| F-536 | 519-536 | 5′-CAGCAGCCGCGGTAATAC-3′ |
| R-907 | 907-926 | 5′-(Ph)-CCGTCAATTCCTTTRAGTTT-3′ |
| F-nir | NA | 5′-CGCCAGAGTTCTCCCTGCAG-3′ |
| R-nir | NA | 5′-TTGCCGGTCTTGGTGTCGAC-3′ |
F, forward primer; R, reverse primer; primers GC-F-984 and R-1385 were also termed U-968-GC-1 and L-1401, respectively, whereas primers F-536 and R-907 were also termed GM5f and 907R (or COM1), respectively.
E. coli numbering; NA, not applicable.
GC-clamp, 5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG G-3′. For SSCP, the primer R-907 was phosphorylated at the 5′ end (Com2-Ph).
For mRNA analyses, primers targeted against a central region of the nir gene, the structural gene of nitrite reductase, were used (21) (Table 1).
Preparation of crDNA.
A 2.5-μl portion of the undiluted sample RNA was mixed with 10× DNase buffer (400 mM Tris-HCl [pH 7.5], 60 mM MgCl2, 20 mM CaCl2) and DNase (10 U μl−1) (RNase-free DNase I [chemical no. 776 785]; Boehringer, Mannheim, Germany) and incubated for 3.5 h at 37°C. Contamination of RNA with DNA was checked by using PCR amplification. RNA was transcribed into complementary ribosomal DNA (crDNA) by using random hexamers as described by Teske et al. (20).
PCR and reverse transcription (RT)-PCR amplification.
DNAs from the SLS-phenol and DivoLab-phenol extractions of the bacteria were used for PCR, which was performed as described in the protocol provided with the AmpliTaq DNA polymerase Stoffel fragment (chemical no. N808-0038; Perkin-Elmer) and by Engelen et al. (5) for the primer set GC-F-984 and R-1385. A “touchdown PCR” approach including a “hot-start” technique was performed as described by Muyzer et al. (15). The total number of PCR cycles was 30 for the primer set GC-F-984 and R-1385.
PCR and RT-PCR amplifications of the SSCP fragment were performed using the Qiagen OneStep RT-PCR kit (catalog no. 210210) and the protocol provided by the manufacturer. For RT-PCR, DNA was digested in the RNA extracts as described above. The efficiency of DNA removal in the RNA extracts was checked by performing PCR after DNase treatment. To get a PCR amplification product from DNA extracts, reverse transcriptase was omitted. The number of PCR cycles was 35.
PCR and RT-PCR of mRNA.
To detect the presence and expression of the nir gene, PCR and RT-PCR amplification were performed using the Qiagen OneStep RT-PCR kit and the protocol provided by the manufacturer. DNA in RNA extracts was digested as described above. The efficiency of DNase digestion was tested by performing PCR on DNase-treated samples.
Gel electrophoresis.
Aliquots of PCR and RT-PCR amplification products were run on 3% (wt/vol) agarose gels, and the DNA was stained with ethidium bromide. The protocol of Schwieger and Tebbe (18) was used for SSCP community fingerprinting.
Fingerprint analysis.
16S rDNA fingerprints were analyzed using the software package GelCompare II (Applied Maths, Kortrijk, Belgium). The background was subtracted by using a rolling circle (circle diameter, 30 relative units), and the lanes were normalized. Only bands with a relative intensity of 2% or more of the total lane intensity were considered for this analysis.
RESULTS AND DISCUSSION
Extraction efficiency.
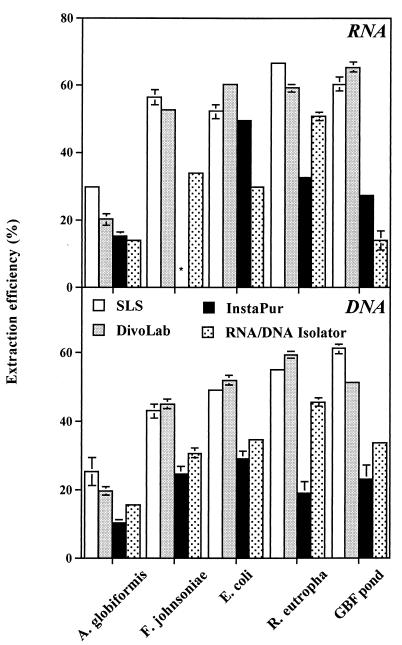
The efficiency of extraction of RNA and DNA was considerably higher for the SLS-phenol and DivoLab-phenol methods than for InstaPure or the RNA-DNA isolator (Fig. 2). The RNA extraction efficiency of the SLS-phenol versus the DivoLab-phenol method was 30 versus 20% for A. globiformis, ranged from 52 to 66% (average, 58%) versus 52 to 59% (average, 57%) for the gram-negative bacteria, and was 60 versus 65% for the pond community (Fig. 2). The DNA extraction efficiency of the SLS-phenol versus the DivoLab-phenol method was 25 versus 20% for A. globiformis, ranged from 43 to 55% (average, 49%) versus 45 to 60% (average, 52%) for the gram-negative bacteria, and was 61 versus 51% for the pond community (Fig. 2).
FIG. 2.
Efficiencies of extraction of RNA and DNA for gram-positive and gram-negative bacterial isolates and a bacterioplankton pond water community using various extraction protocols. The error bars are standard deviations from triplicate measurements. When error bars are not shown, they are smaller than the symbol. The asterisk indicates the RNA extract that was lost during handling.
Extraction efficiencies have not been estimated frequently. Comparatively high extraction efficiencies (64 to 87%) were reported for bacterial isolates using RNA-specific extraction methods (17, 22). Fuhrman et al. (7) estimated the extraction efficiency of DNA from marine bacterioplankton to be in the range of 23 to 54%. Thus, our extraction efficiencies were similar to values reported before. More important is the finding that the extraction efficiency varied only moderately within the gram-negative bacteria and the pond water sample. When the present study was performed, the view was that the abundance of gram-positive bacteria is low in pelagic environments. However, a recent study showed that gram-positive bacteria might be more abundant in lake water than previously assumed (8). Our data also showed that significant amounts of cellular DNA and RNA could be extracted from the gram-positive A. globiformis. Comparable extraction efficiencies have been demonstrated for RNA from Enterococcus faecalis and Bacillus megaterium using a similar extraction method (3). This indicates that this method could also be useful in ecosystems with an important fraction of gram-positive bacteria.
PCR and RT-PCR products of 16S rDNA.
Using extracted DNA obtained by the SLS-phenol and the DivoLab-phenol methods, a PCR product of the entire 16S rRNA gene was obtained for all reference strains, but not for the GBF pond. A PCR product of the entire 16S rRNA gene from the GBF pond samples was obtained for DivoLab-phenol extraction only after dilution of the samples, and the highest concentration of the PCR product was found at a dilution of 1:100. Using the DGGE and SSCP primer sets, a PCR product was amplified from DNA extracted from the GBF pond sample with both extraction mixes, and the PCR product yield was higher for DivoLab-phenol-extracted DNA than for the SLS-phenol method. No PCR product was obtained for the negative control (filter without cells).
An RT-PCR product of the entire 16S rRNA was obtained for only two isolates (E. coli and F. johnsoniae). Modified nucleotides, such as nucleotides 966 and 967 of the 16S rRNA, can lead to premature termination of reverse transcriptase activity (20, 25). This could be the reason why we did not get an RT-PCR product of the entire 16S rRNA for all strains. Using the DGGE and SSCP primer sets, we were able to amplify 16S rRNA in all isolates and in the pond water sample. Pond water occasionally had to be diluted before an amplification product could be obtained. The yield of RT-PCR product was stronger for the DivoLab-phenol method than for the SLS-phenol method. No PCR product was obtained in the negative control (filter without cells). To check whether DNA instead of crDNA was amplified, i.e., whether the DNase digestion was complete, PCR was performed after removal of the DNase. No PCR amplification products were detected, indicating that DNA digestion was complete.
Nucleic acid concentrations were similar in SLS-phenol and DivoLab-phenol extracts; however, the PCR product yield was typically higher for DivoLab-phenol- than for SLS-phenol-extracted nucleic acids, indicating that DivoLab might be the preferable extraction mix. One of the reasons for this might be that DivoLab yields purer nucleic acids. DivoLab-phenol in combination with mechanical extraction was the only tested method yielding sufficient RNA in microorganisms refractory to disruption, such as Mycobacterium bovis (14). The finding that a dilution of extracted nucleic acids from the pond water occasionally increased the yield of PCR and RT-PCR products suggests that inhibitory substances, such as humic and fulvic acids, which can inhibit Taq polymerases, were present (26, 27).
Functional analysis based on mRNA.
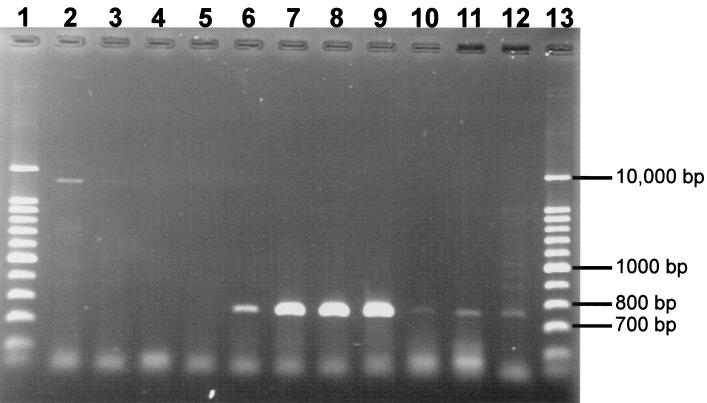
Using nucleic acids extracted from bacterial communities from pond water, we did not obtain a PCR product with primers used for the detection of the central region of the nir gene. However, we were able to get an RT-PCR amplification product from the mRNA. A possible reason for this is that the number of DNA templates was considerably lower than that of mRNA templates, since mRNA can be present in large copy numbers. Consequently, the number of DNA templates could have been too small to allow for detectable amplification. However, the fact that we were able to detect the nir gene expression and to affiliate the sequence also indicates the presence of this gene. Product yields were highest for the 1:10-diluted samples by the SLS-phenol method and for undiluted samples by the DivoLab-phenol method (Fig. 3). This further supports the notion that DivoLab yields purer nucleic acids or removes inhibitory substances more efficiently. The amplification product had a size of ca. 750 bp; the sequence similarity of various excised bands with sequences of the nir gene from databases as determined by a FASTA search was >98% (closest match, nir gene of Pseudomonas stutzeri [accession no. X56813]). The negative control showed a slight amplification product, but it was much larger than 750 bp. The data show that the extracted RNA is suitable for functional studies of bacterioplankton.
FIG. 3.
Ethidium-stained agarose gel (1.5%) of RT-PCR products (the entire mRNA of the nir gene) from dilutions of RNA extracted from the pond water sample using the SLS-phenol and the DivoLab-phenol methods [(a) and (b) in the lane descriptions refer to duplicate extracted filters]. Lanes 1 and 13, DNA ladders; lane 2, control without RT step; lane 3, control without template; lane 4, SLS-phenol undiluted (a); lane 5, SLS-phenol undiluted (b); lane 6, DivoLab-phenol undiluted (a); lane 7, DivoLab-phenol undiluted (b); lane 8, SLS-phenol diluted 1:10 (a); lane 9, SLS-phenol diluted 1:10 (b); lane 10, DivoLab-phenol diluted 1:10 (a); lane 11, DivoLab-phenol diluted 1:10 (b); lane 12, DivoLab-phenol diluted 1:100.
Community structure analysis.
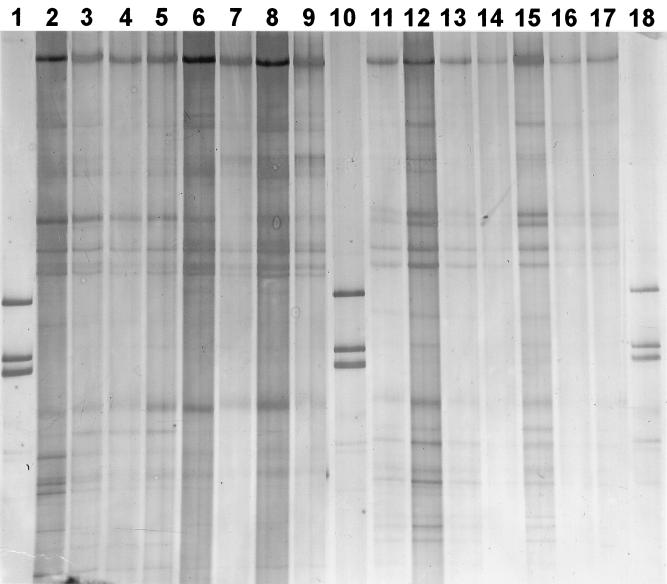
SSCP band patterns for PCR and RT-PCR products from pond water communities are shown in Fig. 4. The community fingerprints obtained by both extraction methods were essentially the same. Moreover, SSCP of RT-PCR and PCR products showed similar band patterns as well. For example, intense bands at the same position in the lanes were generally found for DNA- and RNA-based methods. However, there were also some differences. Most notably, the number of bands was higher with the RNA-based method (>10 bands) than in the DNA-based fingerprints (<10 bands). RNA-based fingerprints likely represent the community structure of active members, whereas DNA-based fingerprints aim at numerically abundant members. Differences of community structure between DNA- and RNA-based fingerprints as shown in this study were reported before (20). Variation of the concentrations of template nucleic acids over 1 order of magnitude in PCR and RT-PCR had only a comparatively small effect on the community profiles. Typically, due to dilution, some bands could not be detected any more; however, the more intense bands could be found at all target concentrations. The extraction method presented here was also successfully applied to 16S rDNA-based SSCP and temperature gradient gel electrophoresis community fingerprinting of other freshwater and marine bacterioplankton samples (B. Engelen and M. G. Höfle, unpublished data; D. F. Wenderoth and M. G. Höfle, unpublished data), confirming the utility of this extraction method for studying bacterial community structure based on molecular techniques.
FIG. 4.
SSCP patterns obtained with single-stranded PCR products of 16S rRNA genes (lanes 2 to 9) and single-stranded RT-PCR products (lanes 11 to 17) amplified from pond bacterioplankton extracted by the SLS-phenol and DivoLab-phenol methods [(a) and (b) in the lane descriptions refer to duplicate extracted filters]. Lanes 1, 10, and 18, DNA ladders; lane 2, SLS-phenol undiluted (a); lane 3, SLS-phenol diluted 1:10 (a); lane 4, SLS-phenol undiluted (b); lane 5, SLS-phenol diluted 1:10 (b); lane 6, DivoLab-phenol undiluted (a); lane 7, DivoLab-phenol diluted 1:10 (a); lane 8, DivoLab-phenol undiluted (b); lane 9, DivoLab-phenol diluted 1:10 (b); lane 11, SLS-phenol undiluted (a); lane 12, SLS-phenol diluted 1:10 (a); lane 13, SLS-phenol undiluted (b); lane 14, SLS-phenol diluted 1:10 (b); lane 15, DivoLab-phenol undiluted (a); lane 16, DivoLab-phenol diluted 1:10 (a); lane 17, DivoLab-phenol undiluted (b). Between lanes 17 and 18, a lane with a different marker was excised by using Adobe Photoshop.
Conclusions.
The extraction method presented has the following benefits. First, DNA and RNA are obtained from a single filter, allowing a direct comparison of community fingerprints based on numerically abundant and on active members. Such methods have been developed for sediments and soils as well (e.g., references 4 and 9); however, here we show that functional studies based on mRNA can be done from the same filter. Second, we show that the extraction efficiencies are comparatively high and comparatively constant for gram-negative bacteria (note, however, the lower extraction efficiencies for gram-positive bacteria). Quantitative PCR methods are about to be developed which circumvent the problem of differential amplification of target sequences and allow assessment of the numerical significance of sequences in situ (19). These methods would provide information on the number of different sequences and the abundance of single sequences, as well as on the number of transcripts from target mRNAs, which could be used for a quantification of gene expression. For such studies, a comparatively constant extraction efficiency is a prerequisite. Thus, the method presented here could form the basis for integrated molecular studies of the structure and function of aquatic bacteria and their role in the global biogeochemical cycles.
Acknowledgments
DivoLab No. 1 was kindly provided by DiverseyLever Ltd. The excellent technical assistance of Silke Pretzer and Julia Bötel is appreciated. We also thank Ines Pöhler for help with some nucleic acid extractions from pond water and B. Engelen for comments on an earlier version of the manuscript. The comments of two anonymous reviewers improved the manuscript significantly.
This work was supported by a grant (01SF9815/1) from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie to M.G.H. and partially by a TMR project grant (MAS3-CT97-5042) provided by the European Commission to M.G.W.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binder, B. J., and Y. C. Liu. 1998. Growth rate regulation of rRNA content of a marine Synechococcus (cyanobacterium) strain. Appl. Environ. Microbiol. 64:3346-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dominik, K., and M. G. Höfle. 1999. Extraction of total RNA from bacterioplankton, p. 1-9. In A. Akkermans, J. van Elsas, and F. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 4.Duarte, G. F., A. S. Rosado, L. Seldin, A. C. Keijzer-Wolters, and J. D. van Elsas. 1998. Extraction of ribosomal RNA and genomic DNA from soil for studying the diversity of the indigenous bacterial community. J. Microbiol. Methods 32:21-29. [Google Scholar]
- 5.Engelen, B., K. Meinken, F. von Wintzingerode, H. Heuer, H.-P. Malkomes, and H. Backhaus. 1998. Monitoring impact of a pesticide treatment on bacterial soil communities by metabolic and genetic fingerprinting in addition to conventional testing procedures. Appl. Environ. Microbiol. 64:2814-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrell, R. E. 1993. RNA methodologies. A laboratory guide for isolation and characterization. Academic Press, San Diego, Calif.
- 7.Fuhrman, J. A., D. E. Comeau, Å. Hagström, and A. M. Chan. 1988. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl. Environ. Microbiol. 54:1426-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glöckner, F. O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Höfle, M. G. 1992. Bacterioplankton community structure and dynamics after large-scale release of nonindigenous bacteria as revealed by low-molecular-weight-RNA analysis. Appl. Environ. Microbiol. 58:3387-3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurst, C. J., G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter. 1997. Manual of environmental microbiology. ASM Press, Washington, D.C.
- 12.Jeffrey, W. H., R. von Haven, M. P. Hoch, and R. B. Coffin. 1996. Bacterioplankton RNA, DNA, protein content and relationships to rates of thymidine and leucine incorporation. Aquat. Microb. Ecol. 10:87-95. [Google Scholar]
- 13.Majumdar, D., Y. J. Avissar, and J. H. Wyche. 1991. Simultaneous and rapid isolation of bacterial and eukaryotic DNA and RNA: a new approach for isolating DNA. BioTechniques 11:94-101. [PubMed] [Google Scholar]
- 14.Mangan, J. A., K. M. Sole, D. A. Mitchison, and P. D. Butcher. 1997. An effective method of RNA extraction from bacteria refractory to disruption, including mycobacteria. Nucleic Acids Res. 25:675-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muyzer, G., S. Hottenträger, A. Teske, and C. Wawer. 1996. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA--a new molecular approach to analyse the genetic diversity of mixed microbial communities, p. 1-23. In A. Akkermans, J. van Elsas, and F. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 16.Paul, J. H., and S. L. Pichard. 1995. Extractions of DNA and RNA from aquatic environments, p. 153-177. In J. T. Trevors and J. D. van Elsas (ed.), Nucleic acids in the environment. Springer, Berlin, Germany.
- 17.Pichard, S. L., and J. H. Paul. 1991. Detection of gene expression in genetically engineered microorganisms and natural phytoplankton populations in the marine environment by mRNA analysis. Appl. Environ. Microbiol. 57:1721-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwieger, F., and C. C. Tebbe. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teske, A., C. Wawer, G. Muyzer, and N. B. Ramsing. 1996. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl. Environ. Microbiol. 62:1405-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward, B. B., A. R. Cockcroft, and K. A. Kilpatrick. 1993. Antibody and DNA probes for detection of nitrate reductase in seawater. J. Gen. Microbiol. 139:2285-2293. [DOI] [PubMed] [Google Scholar]
- 22.Ward, D. M., A. L. Ruff-Roberts, and R. Weller. 1995. Methods for extracting RNA or ribosomes from microbial mats and cultivated microorganisms, p. 1-14. In A. Akkermans, J. van Elsas, and F. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 23.Wawer, C., and G. Muyzer. 1995. Genetic diversity of Desulfovibrio spp. in environmental samples analyzed by denaturing gradient gel electrophoresis of [NiFe] hydrogenase gene fragments. Appl. Environ. Microbiol. 61:2203-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinbauer, M. G., and M. G. Höfle. Quantification of nucleic acids from aquatic environments by using green-fluorescent dyes and microtiter plates. In A. Akkermans, J. van Elsas, and F. de Bruijn (ed.), Molecular microbial ecology manual, 5th supplement, in press. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 25.Weller, R., J. W. Weller, and D. M. Ward. 1991. 16S rRNA sequences of uncultivated hot spring cyanobacterial mat inhabitants retrieved as randomly primed cDNA. Appl. Environ. Microbiol. 57:1146-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]