Abstract
There is a clear need for new approaches in the field of microbial community analyses, since the methods used can be severely biased. We have developed a DNA array-based method that targets16S ribosomal DNA (rDNA), enabling the direct detection and quantification of microorganisms from complex communities without cultivation. The approach is based on the construction of specific probes from the 16S rDNA sequence data retrieved directly from the communities. The specificity of the assay is obtained through a combination of DNA array hybridization and enzymatic labeling of the constructed probes. Cultivation-dependent assays (enrichment and plating) and cultivation-independent assays (direct fluorescence microscopy and scanning electron microscopy) were used as reference methods in the development and evaluation of the method. The description of microbial communities in ready-to-eat vegetable salads in a modified atmosphere was used as the experimental model. Comparisons were made with respect to the effect of storage at different temperatures for up to 12 days and with respect to the geographic origin of the crisphead lettuce (Spanish or Norwegian), the main salad component. The conclusion drawn from the method comparison was that the DNA array-based method gave an accurate description of the microbial communities. Pseudomonas spp. dominated both of the salad batches, containing either Norwegian or Spanish lettuce, before storage and after storage at 4°C. The Pseudomonas population also dominated the batch containing Norwegian lettuce after storage at 10°C. On the contrary, Enterobacteriaceae and lactic acid bacteria dominated the microbial community of the batch containing Spanish lettuce after storage at 10°C. In that batch, the Enterobacteriaceae also were abundant after storage at 4°C as well as before storage. The practical implications of these results are that microbial communities in ready-to-eat vegetable salads can be diverse and that microbial composition is dependent both on the origin of the raw material and on the storage conditions.
Currently, only a minor part of the diversity of microorganisms in nature is known (2, 24). In addition, most of the microorganisms in natural habitats are probably not cultivable under the conditions typically used (1). Thus, cultivation-independent methods are important in the description of microbial communities (1, 28). All methods used for the description of microbial communities are associated with certain artifacts. That is why modern taxonomy is based on the comparison of several different approaches—a polyphasic taxonomy (22). Unfortunately, most of the taxonomic markers used today are designed for the analysis of microorganisms in pure cultures and cannot be adapted to the analysis of these organisms in natural complex communities (28).
Open-field vegetable crops, such as crisphead lettuce, are directly exposed to the environment. The microbial communities on the plant surfaces are affected by both the climate and the growth conditions of the vegetables (20). The plant material may contain bacteria commonly present in the environment, in fertilizers, and in irrigation water. Several cases where vegetables have been responsible for the transmission of disease (13, 21) have been documented.
Usually, plant tissues are trimmed, cut, and washed during processing for ready-to-eat salads. The washing process reduces the bacterial load approximately 10-fold (14), while treatment with chlorine (giving a free, available chlorine level of between 2 and 10 mg/liter) may reduce the load 100-fold (19). Recently, methods have been developed to increase the shelf-life of lettuce and other vegetables through packaging in a modified atmosphere (MA) having reduced levels of O2 and increased levels of CO2 compared to air (8, 13). Although the MA also has a bactericidal or bacteriostatic effect for some bacterial genera (6, 14), most of the extension of shelf-life is due to the retardation of the physiological aging of the salads themselves. Microorganisms in retail fresh-cut lettuce and salads are kept at low numbers by the combined effect of washing, MA packaging, and low temperature (13). A concern with such production, however, is the possible growth of psychrophilic facultative anaerobic human pathogens, or opportunistic pathogens (13). Fresh-cut salads must be considered a potential safety hazard, since the occurrence of pathogens cannot be excluded and the product is consumed without heating.
In order to describe the microbial composition and communities on plant tissues (13), numerous studies have been conducted. These studies have mainly been based on the detection and quantification of cultivable microorganisms. The methods used in these studies alone may give a biased picture of microbial communities (2).
The aim of the work presented here was to develop and evaluate a DNA array-based method for the direct description of microbial communities in MA-packaged vegetable salads. We have developed a method that enables the direct description of microorganisms in complex communities without cultivation. The approach is based on retrieving DNA sequence information directly from the communities and using this information in a DNA array-based detection and quantification system. For reference, traditional plate counts and enrichments were compared to direct detection by fluorescence microscopy and 16S ribosomal DNA (rDNA) analyses. Furthermore, the colonization and distribution of the bacteria on the vegetable surfaces were investigated by scanning electron microscopy (SEM). We determined the dominating microbial flora with respect to the effect of storage conditions and the geographic origin of the lettuce used. Salad made with Spanish lettuce (grown in a Mediterranean temperate climate) was compared to salad made with Norwegian lettuce (grown in a boreal temperate climate). The microbial flora in salad stored at 4°C was compared to that in salad stored at 10°C for up to 12 days after production.
MATERIALS AND METHODS
Salad composition and processing.
The ready-to-eat salads used in this study were taken from the regular production run in a commercial factory in Norway. The salad contained approximately 5% red cabbage, 20% carrot, and 75% crisphead lettuce by weight. The carrots and the red cabbage were produced in Norway and stored prior to use. The crisphead lettuce, being more perishable, was used within 1 week after harvest. Two experimental series were conducted with salads containing lettuce grown in Norway or Spain.
For the production of ready-to-eat salads, crisphead lettuce from two farmers in southeastern Norway was used. The lettuce was harvested early in the morning 1 to 4 days before production and then stored at 2 to 3°C. After trimming, 44 to 77% of the lettuce could be used for production. The two batches were mixed. The Spanish lettuce was harvested and transported to Norway in a truck at 2 to 3°C. The lettuce was used for the production of ready-to-eat salads 1 week after harvest. After trimming, 66% could be used for production.
The lettuce, carrots, and cabbage were trimmed manually, cut in an industrial cutter, and then washed in cold water. The different components of the salad were mixed during the washing step. The salad was centrifuged to remove excess water, and portions of 325 g were packed in polymer film bags flushed with a gas of unknown composition. The production process is schematically shown in a report by Yildiz (36). Fifteen bags were randomly selected for analysis from each production run; 5 bags were analyzed before storage, 5 were analyzed after storage at 4°C, and 5 were analyzed after storage at 10°C. Ten to 12 days of storage was used. A thorough description of the research material is given by Flateland (12a).
Measurements of CO2, O2, and pH.
The CO2 and O2 concentrations in the packages were measured with 10-ml gas samples and gas analyzing equipment (CO2 gas analyzer PG-100 and oxygen gas analyzer LC 700F; Toray Engineering Co., Ltd., Osaka, Japan). The samples were drawn through septa with a syringe to prevent gas leakage from the packages.
The pH in the salads was measured both after the salads were rinsed and after the salads were treated for 1 min in double-distilled water (1:10 [wt/wt]) in a stomacher. The pH was measured with a φ 10 pH meter (Beckman Instruments, Inc., Fullerton, Calif.) according to the manufacturer's recommendations.
SEM.
The vegetable pieces analyzed were fixed in 2.5% glutaraldehyde in cacodylate buffer (0.137 M; pH 7) for 24 h and then washed three times in cacodylate buffer. The samples were dehydrated by ethanol treatment (once in 70% ethanol, once in 90% ethanol, once in 96% ethanol, and four times in 100% ethanol) and shaken for 15 min. The samples were then dehydrated by critical point dehydration with an apparatus from Balzers Union (Balzers, Liechtenstein; CPD 020) and mounted on aluminum pins with colloidal silver (Electron Microscopy Sciences, Fort Washington, Pa.). The samples were dried for 1 h before being coated with palladium-platina (Fine Coat Ion Sputter JFC-1100; JEOL, Ltd., Tokyo, Japan). Finally, the samples were analyzed with a scanning electron microscope (JSM 840; JEOL).
Fluorescence microscopy with acridine orange staining.
Samples of 25 g of salad were homogenized for 3 min with 225 g of 0.9% NaCl in water by using a Waring Blender (Waring Product Division, Dynamics Corporation of America). The homogenates were filtered through a sterile compress (Norsk Medisinaldepot AS, Oslo, Norway). The samples were stained for 15 min with 0.02% acridine orange, diluted 1:10 with water, and filtered by using black 0.2-μm-pore-size glass fiber filters (Whatman International Ltd., Maidstone, England). The filters were examined in a fluorescence microscope (DM RE; Leica,Wetzlar, Germany) connected to a digital camera (MPS 48; Leica).
Separation of microorganisms from the salad matrix.
Twenty-five grams of salad was mixed with 225 g of peptone water (pH 7; 0.8% NaCl and 1% Bacto Peptone from Difco, Detroit, Mich.) in a stomacher bag with a filter (Bagfilter; Interscience, Saint-Nom, France). The samples were gently washed, and the filtered liquid phase was used for the later applications. The bacteria in this fraction were only loosely attached to the salad matrix. The samples were then dried with a sterile compress, and 225 g of peptone water was added to the stomacher bag once more. The bag was then treated with a stomacher for 60 s (Colworth Stomacher 400; A. J. Seward, UAC House, London, England), and the filtrate was used for the later applications. The bacteria in this fraction represent the population more tightly associated with the salad matrix.
Culture-dependent characterization of microorganisms.
Dilution series with peptone water were spread on nutrient agar (NA) for total counts; cephaloridine-fuidine-cetrime (CFC) agar, based on type CM 559 and selective supplement SR 103, for Pseudomonas; and Man-Rogosa-Sharpe (MRS) agar for lactic acid bacteria (LAB). The agars used were from Oxoid Ltd., Hampshire, England. Petrifilm (3 M, St. Paul, Minn.) was used for the selection of molds and yeasts and for the selection of coliform bacteria. Parallel platings were done for washed and stomacher-treated samples. In addition, an enrichment-based method for the detection of Listeria monocytogenes (23) was used. The colonies from the different plates were visually inspected, and the morphology and motility were determined by microscopy. Traditional microbiological examinations, such as Gram staining, an oxidase test (BR 64A; Oxoid), and a catalase test with H2O2 (E. Merck AG, Darmstadt, Germany), were also carried out.
DNA purification.
To isolate DNA from the bacteria, 50 ml of the rinse water obtained after either wash or stomacher treatment was centrifuged at 4,300 rpm for 10 min at 4°C (Sorvall RC Plus rotor 29; Du Pont Co., Wilmington, Del.). The pellet was resuspended in 1 ml of H2O in a microcentrifuge tube and pelleted at 10,000 rpm for 5 min at 4°C (Biofuge Fresco; Kendro Laboratory Products, Osterodes, Germany). Finally, the samples were resuspended in 50 μl of TE buffer (pH 8; 10 mM Tris-HCl, 1 mM EDTA) and stored at −80°C.
Depending on the sample size obtained following centrifugation, 5 to 50 μl of the suspension was used for DNA purification. Lysozyme (Sigma Chemical Co., St. Louis, Mo.; 120 μg) was added to a total volume of 50 μl, and the samples were incubated at room temperature for 10 min in TE buffer. Subsequently, 180 μl of 4 M guanidine thiocyanate, 1% Sarkosyl, and 20 μg of Dynabeads DNA DIRECT beads (Dynal AS, Oslo, Norway) were added. The samples were incubated at 65°C for 10 min, after which 500 μl of 96% ethanol was added and incubation was continued for 5 min. DNA bound to the magnetic beads was attracted to the side of the microcentrifuge tube with a magnet (MPC-E; Dynal), and the supernatant was removed. The samples were washed twice with 1 ml of 70% ethanol, with the magnet being applied between the washes. Finally, the beads with the purified DNA were resuspended in 50 μl of water and heated to 65°C with an open lid to remove excess alcohol. The colonies were analyzed by resuspension directly in 4 M guanidine thiocyanate buffer following the purification steps described above.
PCR amplification, cloning, restriction enzyme cutting, and sequencing.
The genes encoding the small-subunit rRNA (16S rDNA) were amplified through the application of primers targeted to universally conserved regions at positions 10 to 34 and 1485 to 1507 relative to the published 16S rRNA sequence for Escherichia coli (7). The primers have the following sequences: forward, 5′-TGG CTC AGA TTG AAC GCT GGC GGC-3′ (KR1), and reverse 5′-TAC CTT GTT ACG ACT TCA CCC CA-3′ (KR2). Amplifications were done with a GeneAmp 9600 PCR system (Applied Biosystems, Norwalk, Conn.) and 50-μl volumes containing 10 pmol of primers, 200 μM each deoxynucleotide triphosphate, 10 mM Tris-HCl (pH 8.8), 1.5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, 2 U of DynaZyme thermostable DNA polymerase (Finnzymes OY, Espoo, Finland), and 1 to 5 μl of bead-DNA complex. The PCR was initiated with a denaturation step at 94°C for 4 min. This step was followed by 30 cycles with the following denaturation, annealing, and synthesis parameters: 95°C for 30 s, 55°C for 30 s, and 72°C for 90 s. An extension step at 72°C for 7 min was included at the end of the PCR.
TA cloning (Promega Technical Manual no. 042) separated the different amplified 16S rDNA species. To remove the polymerase from the aqueous phase, 10 μl of chloroform-isoamyl alcohol (24:1) was added to the reaction mixture, which was briefly vortexed. Approximately 0.06 pmol of the purified amplification product was ligated into pGEM-T Easy Vector with 1× Rapid Ligation Buffer (Promega) overnight at 4°C. Competent E. coli JM109 cells (Promega, Madison, Wis.) were used in the transformation. The transformation was done as described in Promega Technical Manual No. 042. Plasmids from the positive colonies were isolated by resuspending a colony in 30 μl of water, heating the mixture to 99°C for 5 min, removing the cell debris by centrifugation at 13,000 rpm (Biofuge Fresco) for 1 min, and transferring 25 μl to a new tube. The insert was amplified with primers HU (5′-CGC CAG GGT TTT CCC AGT CAC GAC G-3′) and HR (5′-GCT TCC GGC TCG TAT GTT GTG TGG-3′), which are specific for the Bluescript vector. The following amplification reaction was used: 94°C for 4 min and then 30 cycles at 95°C for 15 s, 65°C for 30 s, and 72°C for 1 min. The reaction was completed with an extension step at 72°C for 7 min. A nested PCR with primers KR1 and KR2 was used to remove the vector sequences. The template was diluted 1:10,000 prior to the nested amplification, and the cycling was done for 25 cycles. Subsequently, the amplification products were cut with RsaI (GT ↓ AC) in a 21-μl reaction volume containing 1× Buffer C (Promega), 1 μl of RsaI (Promega), and 5 μl of template from the nested amplification. The DNA was cut for 1 h and then analyzed by agarose gel electrophoresis. Selected amplification products representing different restriction patterns were sequenced by using ABI PRISM BigDye terminator cycle sequencing chemistry in accordance with the manufacturer's (Applied Biosystems) instructions. The sequencing fragments were separated on an ABI PRISM 310 genetic analyzer (Applied Biosystems) and analyzed with the accompanying software.
Sequence comparisons and probe identification.
The sequences were analyzed for homology to other known sequences through a homology search of the EMBL database (release 61, December 1999) with the program FASTA in the Genetics Computer Group (Madison, Wis.) package. The sequences determined in this work were aligned with a selection of the sequences in the database. The alignment was constructed with the PILEUP program in the Genetics Computer Group package and manually edited with the GeneDoc program (GeneDoc Multiple Sequence and Alignment Editor and Shading Utility, version 2.5.002, www.cris.com/∼ketchup/genedoc.shtml). The alignment was then used to reconstruct a phylogenetic distance tree with LogDet distances and neighbor-joining analysis (31). The statistical support for the branches in the tree was tested by bootstrap analysis (11) with 1,000 replicates.
The alignment was manually inspected in order to identify regions specific for the dominating groups of bacteria in the ready-to-eat vegetable salads. The dominating microbial floras were quantified through the construction of probes for LAB, the Oxalobacter group, Pseudomonas, and Enterobacteriaceae. Probes for chloroplasts were also constructed in order to determine the background contamination of plant material in the samples (Table 1).
TABLE 1.
Probes used for sequence-specific labeling
| Notation | Sequence | Melting temp (°C) |
|---|---|---|
| ENT AG | 5′ CAG CGG GGA GGA AGG CGA 3′ | 65.6 |
| ENTEROB | 5′ GGC YCA CCT AGG CGA CGA TCC 3′ | 68.6 |
| PROTEOB | 5′ CCA RAC TCC TAC GGG AGG CAG 3′ | 64.1 |
| PSEUD | 5′ CCT TGC GCT ATC AGA TGA GCC TAG GT 3′ | 65.5 |
| OXALOB | 5′ CAT CGA TCA GTA GCT GGT CTG AGA GGA 3′ | 64.8 |
| LAB | 5′ ATG GGA AGA ACA GCT AGA GTA GGG AAT GA 3′ | 64.2 |
| CHLOROPLAST | 5′ CGC TGT GCG TAT CGA CCC GTG 3′ | 67.6 |
Sequence-specific labeling of oligonucleotide probes.
An approach that enabled us to determine the relative composition of the entire microbial community in a single reaction was also used. The sequence-specific labeling was performed as previously described (29, 30). Twenty-microliter volumes of PCR amplification products were used in cyclic labeling reactions. The deoxynucleotide triphosphates were dephosphorylated, and excess primers were degraded through the addition of 4 U of shrimp alkaline phosphatase (U.S. Biochemical Corp., Cleveland, Ohio) and 20 U of exonuclease I (U.S. Biochemical Corp.). The samples then were incubated at 37°C for 30 min. Finally, the enzymes were inactivated by heating the solution to 95°C for 10 min.
Cyclic labeling reactions were carried out with 60-μl volumes containing 10 pmol of each of the probes listed in Table 1 and 100 pmol of ddATP, 100 pmol of ddGTP, and 100 pmol of ddTTP (all from Roche Molecular Biochemicals, Manheim, Germany); 100 pmol of fluorescein-12-ddCTP (NEN, Boston, Mass.); 1× Thermo Sequenase reaction buffer (Amersham Pharmacia, Little Chalfont, Buckinghamshire, England); and 24 μl of PCR product treated with phosphatase and exonuclease I. The labeling was done with 25 cycles at 95°C for 15 s and 60°C for 1 min, 25 cycles at 95°C for 15 s and 55°C for 1 min, and finally 25 cycles at 95°C for 15 s and 50°C for 1 min.
Fifty picomoles of probes, complementary to those used in the labeling reactions, were spotted on GeneScreen Plus nylon membranes (NEN) and cross-linked for 25 min with a UV transilluminator (model TL33; UVP Inc., San Gabriel, Calif.). The membranes were prehybridized in 0.5 M Na2HPO4 (pH 7.2)-1% sodium dodecyl sulfate (SDS) for 2 h. The labeled probes were added to 300 μl of 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate)-6% polyethylene glycol 6000 heated to 80°C for 5 min. Hybridization was done overnight at room temperature with agitation in a Cross Blot Dot Blot hybridization chamber (Sebia, Moulinaux, France). The membranes then were rinsed in 1× SSC-1% SDS for 5 for min, in 0.1× SSC-0.1% SDS for 5 min, and finally in 0.1 M Tris-HCl (pH 7.5)-0.15 M NaCl (antibody buffer) for 5 min. The membranes then were blocked for 2 h in antibody buffer containing 1% skim milk (Difco) (blocking buffer). Then, blocking buffer containing1/500 anti-fluorescein-horseradish peroxidase conjugate was added, and hybridization was continued at room temperature for 2 h. Finally, the membranes were rinsed for 30 min in antibody buffer, and signals were detected with 4 CN Plus chromogenic substrate in accordance with the manufacturer's (NEN) recommendations.
RESULTS
Microbial colonization and physiological properties of the salad matrix.
The CO2 concentration in the salad packages increased from approximately 0.2% at the production date to 5.5% after approximately 10 days of storage at 4°C. The O2 concentrations were low during the whole experiment (<0.5%). In the control packages, without salads, CO2 was undetectable, while the O2 concentrations increased to 2 to 8% during the storage period. The gas composition was not measured for the packages stored at 10°C.
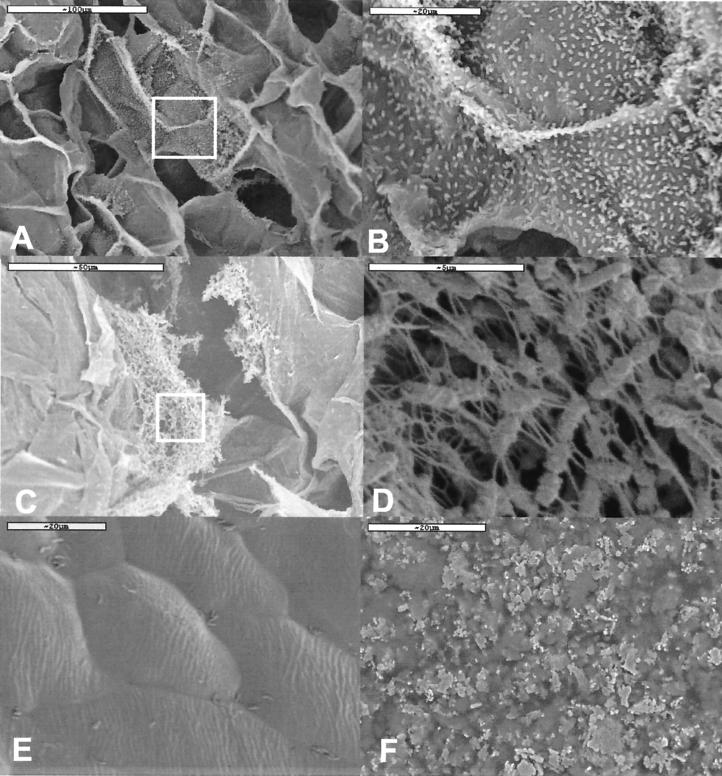
Visual inspection and SEM examination showed that the salad samples had a fresh appearance after storage at 4°C (Fig. 1). This was also the case for the salad samples containing the Norwegian lettuce stored at 10°C, although these were a little more brownish at the cut surfaces. The pH in these samples was in the range of 6.0 to 6.5. SEM showed that microbial colonization was predominant in the salads containing Spanish lettuce stored at 10°C (Fig. 1A to D). Figure 1E shows the intact lettuce surface after storage at 4°C, while the surface was heavily decayed after storage at 10°C (Fig. 1F). The packages stored at 10°C were also expanded due to gas production (probably CO2). The pH in these samples was between 4 and 4.5.
FIG. 1.
Scanning electron micrographs of bacterial colonization of plant matrixes. (A and B) Colonization of sliced carrot in salad manufactured with Spanish lettuce and stored at 10°C. Inset in panel A is magnified in panel B. (C and D) Colonization of cut surfaces of lettuce. Inset in panel C is magnified in panel D. (E) Intact surface of Spanish lettuce stored at 4°C. (F) Degraded surface of lettuce in salad stored at 10°C.
Microbial counts determined by plating and fluorescence microscopy.
The total mesophile aerobic bacterial counts of the salad mixture on NA were on the order of 104 to 105 CFU/g before storage (Table 2). Storage at 4°C for 10 days resulted in approximately 107 CFU/g, while approximately108 CFU/g was found after storage at 10°C. There was no major difference (<1 log unit) between the total CFU of the salads containing Norwegian lettuce versus Spanish lettuce.
TABLE 2.
Log CFU per gram of salad after washing (W) and after treatment in a stomacher (S)
| Origin | Storage conditionsa | Mean ± SD countsb of the following organisms with the indicated treatment:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total
|
Pseudomonas (CFC medium)
|
Lactic acid bacteria (MRS medium)
|
Yeasts and molds (Petrifilm)
|
Coliforms (Petrifilm)
|
|||||||||
| W | S | W | S | W | S | W | S | W | Sc | ||||
| Norwegian | BS | 4.6 ± 0.4 | 4.7 ± 0.3 | 4.2 ± 0.6 | 4.3 ± 0.4 | <1 | <1 | 1.6 ± 0.2 | 1.6 ± 0.3 | NT | 2.5 | ||
| Norwegian | 4°C | 7.2 ± 0.2 | 7.1 ± 0.2 | 7.0 ± 0.1 | 7.0 ± 0.1 | 4.9 ± 0.5 | 5.0 ± 0.3 | 3.6 ± 0.1 | 3.1 ± 0.2 | NT | 2.3 | ||
| Norwegian | 10°C | 8.2 ± 0.1 | 8.5 ± 0.1 | 7.8 ± 0.1 | 7.6 ± 0.2 | 6.3 ± 0.4 | 6.2 ± 0.1 | 4.1 ± 0.2 | 3.6 ± 0.2 | NT | <1 | ||
| Spanish | BS | 4.2 ± 0.2 | 4.5 ± 0.6 | 4.0 ± 0.3 | 4.0 ± 0.5 | <1 | <1 | 1.6 ± 0.4 | 1.1 ± 0.2 | NT | 2 | ||
| Spanish | 4°C | 7.4 ± 0.2 | 7.4 ± 0.2 | 7.3 ± 0.2 | 7.4 ± 0.2 | 5.5 ± 0.8 | 5.6 ± 0.9 | 3.7 ± 0.3 | 3.6 ± 0.4 | NT | 3 | ||
| Spanish | 10°C | 8.1 ± 0.1 | 7.8 ± 0.1 | 8.1 ± 0.2 | 6.8 ± 0.2 | 9.0 ± 0.1 | 7.6 ± 0.1 | 4.1 ± 0.2 | 3.6 ± 0.4 | NT | >5 | ||
BS, before storage.
Each log value is based on duplicate analyses of five independent bags, except as otherwise noted. The standard deviations are based on the log values, not the actual counts. NT, not tested.
Single samples.
Direct counts determined by fluorescence microscopy yielded approximately the same cell numbers as the total CFU for samples stored at 4°C. The direct counts, however, were approximately 10-fold higher than the CFU for salads stored at 10°C (Table 3). The freshly produced salads had bacterial numbers too low to be determined on the basis of direct counts.
TABLE 3.
Log number of bacteria (CFU) counted by fluorescence microscopy per gram of salad
| Origin | Storage conditions (°C) | Mean ± SD total countsa |
|---|---|---|
| Norwegian | 4 | 8.1 ± 0.1 |
| Norwegian | 10 | 8.9 ± 0.1 |
| Spanish | 4 | 8.3 ± 0.2 |
| Spanish | 10 | 8.7 ± 0.1 |
Each log count is based on analyses of five independent bags. The standard deviations are based on the log values, not the actual counts.
L. monocytogenes was not detected in any of the samples. The levels of yeasts and molds were low in the fresh salads but increased throughout the storage period at both temperatures (Table 2). The Pseudomonas counts were in the range of 104 to 105 CFU/g for the freshly produced salads. After storage, the counts increased to approximately 107 CFU/g at 4°C and between 107 and 108 CFU/g at 10°C. The number of LAB was very low just after production (<10 CFU/g). LAB cell counts of 105 and 106 CFU/g for the Norwegian salads and 106 and 109 CFU/g for the salads containing Spanish lettuce were found after storage at 4 and 10°C, respectively (Table 2). The salad mixtures contained approximately 102 CFU of coliform bacteria/g before storage. In salads made with Norwegian lettuce, the numbers of coliforms were the same before and after storage at 4°C. The numbers of coliforms were below the detection limit at 10°C. In salads made with Spanish lettuce, the levels of coliform bacteria increased from approximately 102 CFU/g just after production to 103 CFU/g when the salads were stored at 4°C, but at 10°C the coliform counts reached more than 105 CFU/g (Table 2).
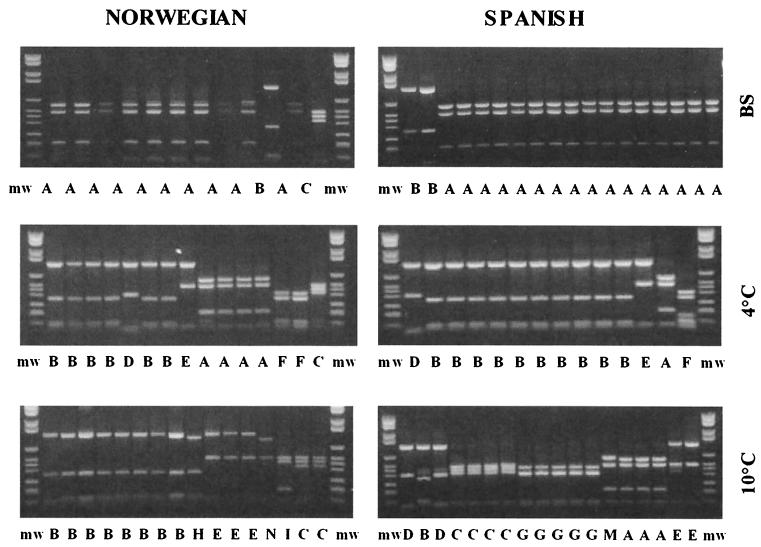
Biodiversity determined by 16S rDNA restriction fragment length polymorphism analyses.
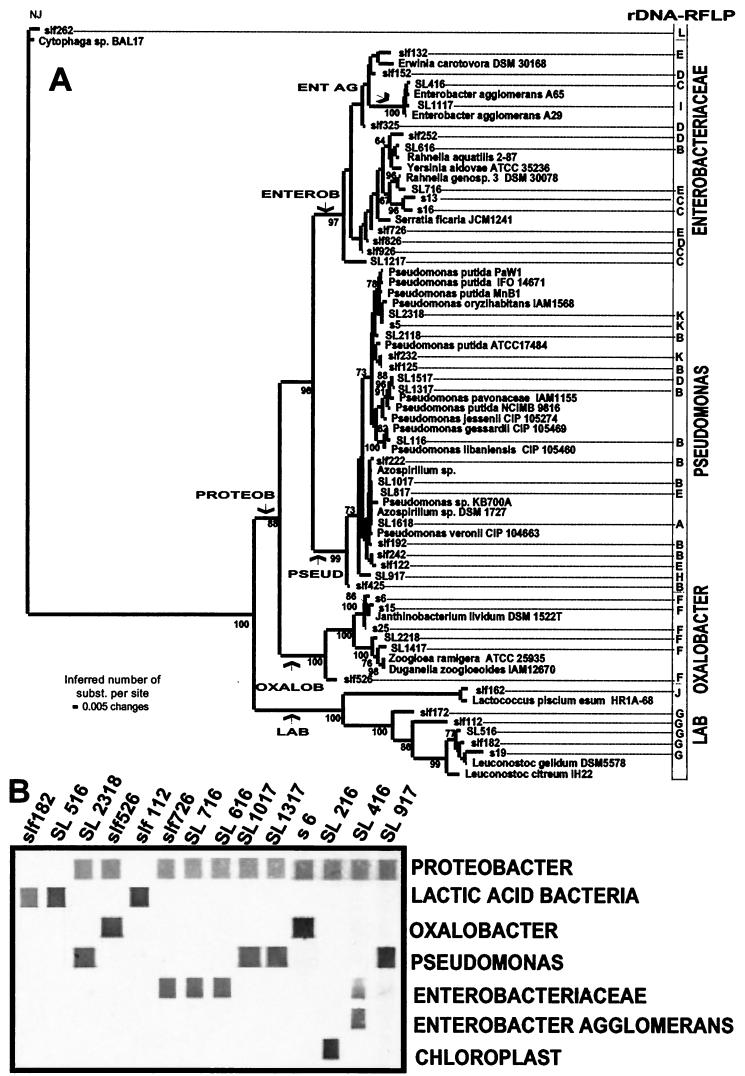
Eleven different restriction patterns, denoted A to K, were identified after the KR1- and KR2-amplified 16S rDNA was cut with restriction enzyme RsaI (Table 4 and Fig. 2). The phylogenetic distance tree for the DNA sequences of the clones representing the different cutting patterns revealed that the main bacterial groups encountered were Enterobacteriaceae, Pseudomonas, Oxalobacter, and LAB (Fig. 3A). A relatively high frequency of chloroplast sequences was also retrieved for fresh lettuce from both Norway and Spain (restriction pattern A). Clones with restriction patterns K, B, and H, which are specific for the genus Pseudomonas, were isolated from all samples investigated, indicating that Pseudomonas species were abundant in the MA-packed salads. Pseudomonas was the dominant genus both in freshly produced salads and after storage at 4°C. There was a noticeable difference in the microbial compositions of the salads produced with Spanish lettuce and Norwegian lettuce after storage at 10°C. The main difference was that clones with restriction pattern G (LAB) were retrieved only from the salads containing Spanish lettuce.
TABLE 4.
Restriction fragment length polymorphism (RFLP) patterns, origins, and EMBL accession numbers of the samples analyzed in this work
| Code | Clone or colony | Origin | Storage conditionsa | RFLP pattern | EMBL accession no. | Phylogenetic group |
|---|---|---|---|---|---|---|
| slf 262 | NA | Spanish | 4°C | L | AJ314667 | Cytophaga |
| slf 132 | Clone | Spanish | 10°C | E | AJ314668 | Enterobacteriaceae |
| slf 152 | Clone | Spanish | 10°C | D | AJ314672 | |
| SL 416 | Clone | Norwegian | BS | C | AJ314677 | |
| SL 1117 | Clone | Norwegian | 10°C | I | AJ314678 | |
| slf 325 | Clone | Spanish | 4°C | D | AJ314673 | |
| slf 252 | NA | Spanish | 10°C | D | AJ314669 | |
| SL 616 | Clone | Norwegian | 10°C | Bb | AJ314670 | |
| SL 716 | Clone | Norwegian | 10°C | E | AJ314671 | |
| s 13 | CFC medium | Spanish | 10°C | C | AJ314679 | |
| s 16 | NA | Spanish | 10°C | C | AJ314680 | |
| slf 726 | Clone | Spanish | 4°C | E | AJ314674 | |
| slf 826 | Clone | Spanish | 10°C | D | AJ314675 | |
| slf 926 | Clone | Spanish | 10°C | C | AJ314676 | |
| SL 1217 | Clone | Norwegian | 10°C | C | AJ314681 | |
| SL 2318 | CFC medium | Norwegian | NT | K | AJ314683 | Pseudomonas |
| s 5 | NA | Spanish | BS | K | AJ314682 | |
| SL 2118 | NA | Norwegian | NT | B | AJ314684 | |
| slf 232 | NA | Spanish | BS | K | AJ314691 | |
| slf 125 | Clone | Spanish | BS | B | AJ314697 | |
| SL 1517 | Clone | Norwegian | 4°C | Db | AJ314685 | |
| SL 1317 | Clone | Norwegian | 10°C | B | AJ314686 | |
| SL 116 | Clone | Norwegian | BS | B | AJ314687 | |
| slf 222 | CFC | Spanish | 4°C | B | AJ314688 | |
| SL 1017 | Clone | Norwegian | 10°C | B | AJ314692 | |
| SL 817 | Clone | Norwegian | 10°C | Eb | AJ314693 | |
| SL 1618 | Clone | Norwegian | 4°C | Ab | AJ314694 | |
| slf 192 | CFC | Spanish | 10°C | B | AJ314689 | |
| slf 242 | NA | Spanish | 4°C | B | AJ314690 | |
| slf 122 | Clone | Spanish | 10°C | Eb | AJ314695 | |
| SL 917 | Clone | Spanish | 10°C | H | AJ314696 | |
| slf 425 | Clone | Spanish | 4°C | B | AJ314698 | |
| s 6 | NA | Spanish | BS | F | AJ314699 | Oxalobacter |
| s 15 | NA | Spanish | 10°C | F | AJ314700 | |
| s 25 | NA | Spanish | 4°C | F | AJ314701 | |
| SL 2218 | NA | Norwegian | NT | F | AJ314702 | |
| SL 1417 | Clone | Norwegian | 4°C | F | AJ314703 | |
| slf 526 | Clone | Spanish | 4°C | F | AJ314704 | |
| slf 162 | MRS | Spanish | BS | J | AJ314705 | Lactic acid bacteria |
| slf 172 | MRS | Spanish | 4°C | G | AJ314706 | |
| slf 112 | Clone | Spanish | 4°C | G | AJ314707 | |
| SL 516 | MRS | Norwegian | NT | G | AJ314708 | |
| slf 182 | MRS | Spanish | 4°C | G | AJ314709 | |
| s 19 | MRS | Spanish | 10°C | G | AJ314710 |
BS, before storage; NT, not tested.
Recombinant clone.
FIG. 2.
Restriction fragment length polymorphism pattern for RsaI-digested 16S rDNA. The 16S rDNA was amplified, cut as described in Materials and Methods, and separated on gels. The samples were analyzed at the beginning of the storage period (BS), after storage at 4°C, and after storage at 10°C. The different restriction patterns are designated with letters. mw, molecular weight standards.
FIG.3.
Phylogenetic distance tree for 16S rDNA sequences (A) and specific detection with taxonomic probes (B). The distance tree (A) was built by using LogDet distances and neighbor-joining analysis for tree construction (31). The statistical support of the branches in the tree was tested by bootstrap analysis (11). The distance between two organisms, expressed in substitutions (subst.) per nucleotide, is obtained by adding the horizontal branches connecting them. The numbers at the nodes indicate the percentages of 500 bootstrap trees in which the cluster descending from the node was found. The arrows indicate the phylogenetic positions of the constructed probes. A selection of clones representing the major lineages in the phylogenetic tree shown in panel A was used for the probe labeling assay (B). RFLP, restriction fragment length polymorphism.
A problem with a direct approach using gene cloning is the generation of chimeric sequences (17). We found one chimeric sequence within the sequenced genome region (approximately 400 bp in the 5′ region of the 16S rDNA gene). We also identified potential recombinant clones from a comparison of the sequence phylogeny and the restriction enzyme cutting patterns. We identified one potential type of recombinant clone (9% of the clones) with restriction pattern B in the Enterobacteriaceae—the rest of the B clones were found in Pseudomonas. Four out of 10 clones were found to be recombinant for the genus Pseudomonas, whereas for the LAB and the Oxalobacter group, no chimeras were identified.
Relative community compositions determined by sequence-specific labeling of oligonucleotide probes.
The probes used (Table 1) were constructed from variable sites identified in the multiple sequence alignment from which the phylogenetic tree in Fig. 3A was constructed. Substitutions defining the branches leading to Enterobacter agglomerans (probe ENT AG), the clones belonging to the Enterobacteriaceae (probe ENTEROB), Pseudomonas (probe PSEUD), Oxalobacter (probe OXALOB), Proteobacter (probe PROTEOB), and LAB (probe LAB) were identified. The specificities of the probes were determined through analysis of both clones and pure cultures (Fig. 3B). These analyses showed that the constructed probes were specific for their selected targets. The labeling efficiencies were approximately similar for the different probes, and no background signal could be detected (Fig. 3B). For instance, there was only one base difference between E. agglomerans and other species of Enterobacteriaceae.
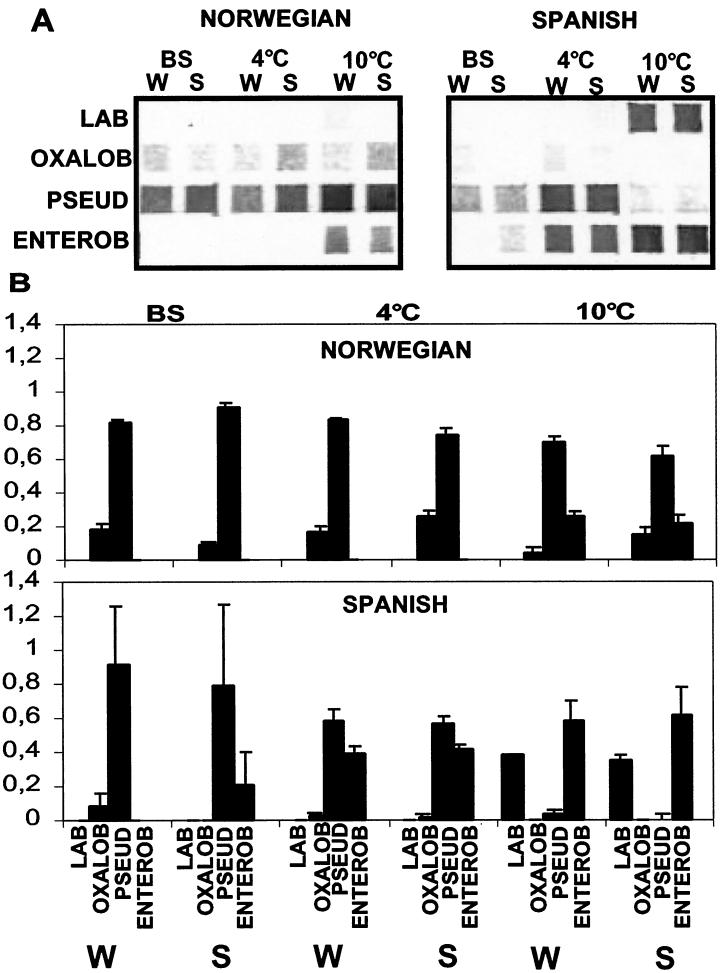
The analysis of the intact communities (Fig. 4) gave very reproducible results, with low standard deviations for most of the independent experiments (Fig. 4B). Pseudomonas spp. dominated in the freshly produced salads made with both Spanish lettuce and Norwegian lettuce. Organisms belonging to the Oxalobacter group could also be retrieved from both samples. There was a marked difference in the compositions of the bacterial communities for the two salads. Pseudomonas spp. dominated the salads made with Norwegian lettuce after storage at 4°C, while there were approximately equal numbers of Pseudomonas spp. and bacteria belonging to the Enterobacteriaceae in the salads made with Spanish lettuce. There was a marked increase in the population of Enterobacteriaceae after storage at 10°C compared to storage at 4°C, but Pseudomonas still dominated the microbial population in the salads made with Norwegian lettuce. The Pseudomonas population was nearly absent in the salads containing Spanish lettuce stored at 10°C, while a population of both LAB and Enterobacteriaceae dominated.
FIG. 4.
Relative compositions of microbial communities as determined by sequence-specific labeling of oligonucleotide probes. The relative compositions of the microbial communities of the salads containing Norwegian lettuce and Spanish lettuce were determined by sequence-specific labeling of oligonucleotide probes. (A) Signals obtained with the different probes used before storage (BS) and after storage at 4 and 10°C. The bacteria were separated from the salad matrix either by washing the sample (W) or by treating the sample in a stomacher (S). Triplicate samples from a single bag were analyzed for each time. (B) Signal intensities for the different probes relative to the total signal intensity (addition of all of the signals) for each sample. Signal intensities were measured as pixel density with an 8-bit gray scale. The error bars are standard deviations based on three replicates of the experiment shown in panel A.
DISCUSSION
The aim of this work was to develop and evaluate a new DNA array-based approach for describing complex microbial communities. Bacterial populations on MA-packed ready-to-eat vegetable salads were used as a model.
Evaluation of the methods applied with respect to an accurate description of microbial communities.
There have been discussions of whether the bacteria that can be detected by culture-based methods actually reflect real biodiversity (1). By comparing CFU with direct counts from microscopy, we found a good correspondence for the vegetable salads stored at 4°C. The correspondence may be due to equal errors introduced in both approaches or may simply indicate that most bacteria on the salads also formed colonies on NA plates. The latter hypothesis was supported by the finding that mainly the same bacterial genera could be detected by analyzing the colonies and by sequencing the clones directly retrieved from the salads. However, we obtained 10-times-higher cell counts from MRS agar plates (selective for LAB) than from NA plates (supporting the growth of aerobic bacteria) after storage at 10°C for the Spanish lettuce. This result indicates that numbers of CFU do not always reflect the total biodiversity in MAs.
Apparent limitations with the plating approach are that one has to decide which bacteria to search for and that one has to know under which conditions these bacteria can grow. The example from our work is the population of bacteria in the Oxalobacter group, for which we do not have a selective medium. Furthermore, selective media may also be a problem for damaged bacteria, which may not survive the selective conditions. However, media may also protect bacteria by allowing nucleic acid synthesis and growth and thus enabling the detection of cells missed by the epifluorescence approach.
A potential source of error with the direct cloning approach is differential lysis of the bacteria in the sample. This is the reason why we used rigid lysis conditions in our work. Also, gene copy numbers and not cell numbers are determined. One genome may contain from 1 to about 10 copies of ribosomal operons. In addition, the numbers of genomes may also differ according to the growth conditions. However, these uncertainties are probably less common than the uncertainties introduced in the direct plating approach, e.g., bacterial aggregates (Fig. 2).
A challenge with genetic analyses is, however, to develop methods which provide gene information for complex mixtures. The combination of DNA array hybridization with enzymatic labeling of DNA probes (29, 30) has solved the problem of low signal-to-noise ratios in standard hybridization assays and thus led to the possibility of obtaining quantitative results. Furthermore, the linking of gene cloning with the construction of specific probes allows quantitative analyses of uncultivated microorganisms. This factor is important in community analyses, since several of the microorganisms in natural communities cannot be easily cultivated or enriched.
The construction of probes at different taxonomic levels leads to a new possibility for the description of microbial communities. The direct hierarchical classification system allows a more accurate description of microbial communities at different taxonomic levels. For instance, as demonstrated in our work, we can monitor the populations of both Enterobacteriacea and E. agglomerans simultaneously to provide important information about the population structure.
Effect of storage temperature and the origin of crisphead lettuce on microbial populations.
There were already differences in the microbial communities before storage of the batches of salads containing Norwegian lettuce versus Spanish lettuce (Fig. 4). The most marked difference was the presence of the population of Enterobacteriaceae in the salads containing Spanish lettuce. This population seemed to be tightly associated with the salad matrixes (20) (the relative populations were higher in the stomacher-treated samples than in the wash water), indicating that these bacteria originated from the raw materials and not from the water used in processing the salads. We also found relatively high frequencies of chloroplast sequences in the samples before storage. The reason for this finding is probably that the microbial load was so low that the chloroplasts in the traces of plant material were amplified.
The differences between the microbial populations in the salad batches made with Norwegian lettuce versus Spanish lettuce after storage were probably due to different bacterial loads before storage, since the other conditions were similar. The conclusion from our study is that microbial compositions in different batches can be relatively diverse, with respect both to storage temperature and to the origin of the lettuce used.
Origin, human health significance, and spoilage potential of the dominating microbial flora.
Generally, microorganisms belonging to the genus Pseudomonas and to the family Enterobacteriaceae dominated the samples. Thus, the emphasis here is on these two groups of microorganisms.
The three main genera of the Enterobacteriaceae identified in the samples in this work, Erwinia, Enterobacter, and Rahnella, all belong to groups of plant pathogens (3, 15, 33) and may thus have originated from the raw materials. Some of the bacterial species of these genera may also be opportunistic pathogens, for instance, for immunocompromised persons (12, 15, 22, 27). However, since all of these bacteria are common both in the environment and in fresh vegetables, they probably do not represent a health risk in ready-to-eat vegetable salads. Still, they may be a problem for the quality of the vegetable salads, both through tissue disruption and through the production of polysaccharides (3, 22, 27, 33).
A broad diversity of bacteria closely related to Pseudomonas spp. was isolated from the salads. Bacteria classified as Azospirillum (32, 34) formed a separate cluster together with Pseudomonas veronii (9). The Azospirillum spp. identified in the salads were atypical for the genus (10, 35). Azospirillum, however, can grow under microaerophilic conditions and utilize cellulose as a carbon source. Packaging of salads in an MA may therefore favor bacteria with these characteristics. The other Pseudomonas cluster contained the species P. putida, P. oryzhabitans, P. pavonaceae, P. jessenii, P. gessardii, and P. libaniensis. Pseudomonas spp. are widely distributed in the environment, and several of the species are potential plant pathogens (16, 25).
The Oxalobacter group of microorganisms is not well characterized, but it has recently been shown that this group can be associated with plant pathogens (18) and may contain opportunistic human pathogens (26).
Finally, a relatively large population of LAB was encountered in the salads containing Spanish lettuce after storage at 10°C. LAB are not pathogenic but may be involved in the spoilage processes for food (4, 5).
Genera containing plant-pathogenic species dominated the flora in the MA-packed vegetable salads. These bacteria could have been selected during the extensive washing procedures used in the manufacturing of the salads, since plant-pathogenic bacteria are tightly associated with plant matrixes, while the other bacteria commonly found in the environment are removed during washing (20).
Polyphasic analytic system for bacterial communities.
The combination of methods for the direct description of microbial communities with traditional methods for enrichment and isolation of important strains will be a powerful tool in future research. Obtaining and investigating pure cultures are essential for studies of bacterial properties such as pectinolytic activity, spoilage potential, and pathogenicity. On the other hand, one cannot understand these bacteria without tools to study them in their natural complex environments.
Acknowledgments
We are very grateful to Dordi Strand, who provided the salad samples. We also thank Ellen S. Tronrud and Helga Næs for carefully reading and commenting on the manuscript.
This work was financed by the research levy on certain agricultural products.
REFERENCES
- 1.Amann, R. I. 1995. Fluorescently labelled rRNA-targeted oligonucleotide probes in the study of microbial ecology. Mol. Ecol. 4:543-554. [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, R. A., A. R. Eriksson, R. Heikinheimo, A. Mae, M. Pirhonen, V. Koiv, H. Hyytiainen, A. Tuikkala, and E. T. Palva. 2000. Quorum sensing in the plant pathogen Erwinia carotovora subsp. carotovora: the role of expR (Ecc). Mol. Plant Microbe Interact. 13:384-393. [DOI] [PubMed] [Google Scholar]
- 4.Bjorkroth, K. J., R. Geisen, U. Schillinger, N. Weiss, P. De Vos, W. H. Holzapfel, H. J. Korkeala, and P. Vandamme. 2000. Characterization of Leuconostoc gasicomitatum sp. nov., associated with spoiled raw tomato-marinated broiler meat strips packaged under modified-atmosphere conditions. Appl. Environ. Microbiol. 66:3764-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjorkroth, K. J., P. Vandamme, and H. J. Korkeala. 1998. Identification and characterization of Leuconostoc carnosum, associated with production and spoilage of vacuum-packaged, sliced, cooked ham. Appl. Environ. Microbiol. 64:3313-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brocklehurst, T. F., C. M. Zaman-Wong, and B. M. Lund. 1987. A note on the microbiology of retail packs of prepared salad vegetables. J. Appl. Bacteriol. 63:409-415. [DOI] [PubMed] [Google Scholar]
- 7.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Day, B., and R. Wiktorowicz. 1999. MAP goes online. Food Manuf. 6:40-42. [Google Scholar]
- 9.Elomari, M., L. Coroler, B. Hoste, M. Gillis, D. Izard, and H. Leclerc. 1996. DNA relatedness among Pseudomonas strains isolated from natural mineral waters and proposal of Pseudomonas veronii sp. nov. Int. J. Syst. Bacteriol. 46:1138-1144. [DOI] [PubMed] [Google Scholar]
- 10.Fani, R., C. Bandi, M. Bazzicalupo, M. T. Ceccherini, S. Fancelli, E. Gallori, L. Gerace, A. Grifoni, N. Miclaus, and G. Damiani. 1995. Phylogeny of the genus Azospirillum based on 16S rDNA sequence. FEMS Microbiol. Lett. 129:195-200. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 12.Ferguson, R., C. Feeney, and V. A. Chirurgi. 1993. Enterobacter agglomerans-associated cotton fever. Arch. Intern. Med. 153:2381-2382. [PubMed] [Google Scholar]
- 12a.Flateland, S. L. 2000. Microbial flora in fresh-cut salads—importance of storage conditions and geographical origin. C.S. thesis. Agricultural University of Norway, Ås. (In Norwegian.)
- 13.Francis, G., C. Thomas, and T. O'Beirne. 1999. The microbiological safety of minimally processed vegetables. Int. J. Food Sci. Technol. 34:1-22. [Google Scholar]
- 14.Garcia-Gimeno, R. M., and G. Zurera-Cosano. 1997. Determination of ready-to-eat vegetable salad shelf-life. Int. J. Food Microbiol. 36:31-38. [DOI] [PubMed] [Google Scholar]
- 15.Geere, I. W. 1977. Enterobacter agglomerans: the clinically important plant pathogen. Can. Med. Assoc. J. 116:517-519. [PMC free article] [PubMed] [Google Scholar]
- 16.Hazlewood, G. P., and H. J. Gilbert. 1998. Structure and function analysis of Pseudomonas plant cell wall hydrolases. Prog. Nucleic Acid Res. Mol. Biol. 61:211-241. [DOI] [PubMed] [Google Scholar]
- 17.Liesack, W., H. Weyland, and E. Stackebrandt. 1991. Potential risk of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb. Ecol. 21:191-198. [DOI] [PubMed] [Google Scholar]
- 18.Lincoln, S. P., T. R. Fermor, and B. J. Tindall. 1999. Janthinobacterium agaricidamnosum sp. nov., a soft rot pathogen of Agaricus bisporus. Int. J. Syst. Bacteriol. 49:1577-1589. [DOI] [PubMed] [Google Scholar]
- 19.Lund, B. 1999. Bacteriology of fresh, stored and minimally processed vegetables and fruit. School of Biological Sciences, University of Surrey, Surrey, United Kingdom.
- 20.Lund, B. 1992. Ecosystems in vegetable foods. J. Appl. Bacteriol. Symp. Suppl. 72:115S-126S. [DOI] [PubMed] [Google Scholar]
- 21.Lund, B. 1993. The microbiological safety of prepared salad vegetables. Food Technol. Int. Eur. 1993:196-200.
- 22.Matsukura, H., K. Katayama, N. Kitano, K. Kobayashi, C. Kanegane, A. Higuchi, and S. Kyotani. 1996. Infective endocarditis caused by an unusual gram-negative rod, Rahnella aquatilis. Pediatr. Cardiol. 17:108-111. [DOI] [PubMed] [Google Scholar]
- 22a.Murray, R., D. Brenner, R. Colwell, P. De Vos, M. Goodfellow, P. Grimont, N. Pfenning, E. Stackebrandt, and G. Zavarin. 1990. Report of the ad hoc commitee on approaches to taxonomy within the proteobacteria. Int. J. Syst. Bacteriol. 40:213-215. [Google Scholar]
- 23.Nordisk Metodikkommitte för Livsmedel. 1990. Detection of Listeria monocytogenes in foods. No 136. Statens Tekniska Forskningscentral, Livsmedelslaboratoriet, Espoo, Finland.
- 24.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 25.Rahme, L. G., F. M. Ausubel, H. Cao, E. Drenkard, B. C. Goumnerov, G. W. Lau, S. Mahajan-Miklos, J. Plotnikova, M. W. Tan, J. Tsongalis, C. L. Walendziewicz, and R. G. Tompkins. 2000. Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. USA 97:8815-8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Redkar, R., J. Kalns, W. Butler, L. Krock, F. McCleskey, A. Salmen, E. Piepmeier, and V. DelVecchio. 2000. Identification of bacteria from a non-healing diabetic foot wound by 16 S rDNA sequencing. Mol. Cell. Probes. 14:163-169. [DOI] [PubMed] [Google Scholar]
- 27.Reina, J., and A. Lopez. 1996. Clinical and microbiological characteristics of Rahnella aquatilis strains isolated from children. J. Infect. 33:135-137. [DOI] [PubMed] [Google Scholar]
- 28.Rudi, K. Application of nucleic acid for microbial identification and quantiation. In W. Olson (ed.), Automated microbial identification and quantitation, in press. Davis Horwood International Publishing, Centennial, Conn.
- 29.Rudi, K., O. M. Skulberg, F. Larsen, and K. S. Jakobsen. 1998. Quantification of toxic cyanobacteria in water by use of competitive PCR followed by sequence-specific labeling of oligonucleotide probes. Appl. Environ. Microbiol. 64:2639-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rudi, K., O. M. Skulberg, R. Skulberg, and K. S. Jakobsen. 2000. Application of sequence-specific labeled 16S rRNA gene oligonucleotide probes for genetic profiling of cyanobacterial abundance and diversity by array hybridization. Appl. Environ. Microbiol. 66:4004-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 32.Skvortsov, I. M., and V. V. Ignatov. 1998. Extracellular polysaccharides and polysaccharide-containing biopolymers from Azospirillum species: properties and the possible role in interaction with plant roots. FEMS Microbiol. Lett. 165:223-229. [DOI] [PubMed] [Google Scholar]
- 33.Tallgren, A. H., U. Airaksinen, R. von Weissenberg, H. Ojamo, J. Kuusisto, and M. Leisola. 1999. Exopolysaccharide-producing bacteria from sugar beets. Appl. Environ. Microbiol. 65:862-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong, P. P., N. E. Stenberg, and L. Edgar. 1980. Characterization of a bacterium of the genus Azospirillum from cellulolytic nitrogen-fixing mixed cultures. Can. J. Microbiol. 26:291-296. [DOI] [PubMed] [Google Scholar]
- 35.Xia, Y., T. Embley, and A. Odonell. 1994. Phylogenetic analysis of Azospirillum by direct sequencing of PCR amplified 16S rDNA. Syst. Appl. Microbiol. 17:197-201. [Google Scholar]
- 36.Yildiz, F. 1994. Initial preparation, handling and distribution of minimally processed refrigerated fruits and vegetables, p. 15-65. In R. Wiley (ed.), Minimally processed refrigerated fruits and vegetables. Chapman & Hall, New York, N.Y.