Abstract
We developed a new procedure for concentration of enteric viruses from water using a negatively charged membrane. Rinsing the membrane with 0.5 mM H2SO4 (pH 3.0) in order to elute cations prior to viral elution with 1 mM NaOH (pH 10.5) promoted poliovirus recovery yields from 33 to 95% when applied to pure water and 38 to 89% when applied to natural seawater from Tokyo Bay, Japan, respectively. This method showed average recovery yields of spiked poliovirus of 62% (n = 8) from 1 liter of artificial seawater. This method showed higher recovery yields (>61%) than that of the conventional method using positively charged membrane (6%) when applied to seawater. This method is also free from beef extract elution, which has an inhibitory effect in the subsequent viral genome detection by reverse transcription-PCR. Naturally occurring Norwalk viruses from 2 liters of Tokyo Bay water in winter and infectious enteroviruses from 2 liters of recreational coastal seawater in summer were detected by using this viral concentration method.
To determine the public health risk caused by human enteric viruses in water, a reliable, sensitive, and practical method for detecting small concentrations of viruses is needed. Concentrating viruses in water by adsorption to and subsequent elution from a positively charged membrane (38) is currently considered to be one of the most useful methods (3). This method has been applied to tap water (25, 39), groundwater (1), river water (22, 23), lake water (23), secondarily treated sewage (36) or marine water (29). The virus concentrations are determined by conventional plaque assays (22, 23, 25, 36, 38, 39). However, the recoveries from seawater are not always high enough because of low adsorption of viruses to the positively charged membrane due to the influence of salts (24). Most of the enteric viruses are known to adsorb to a negatively charged membrane in the presence of Mg2+ (40, 43) or other multivalent cations, or under acid conditions (37), while the recovery of viruses is not always easy.
According to the infectious disease weekly reports from the National Institute of Infectious Disease, Tokyo, Japan, infection with enteroviruses is common in the summer. The use of recreational seawater is suspected as one of the main pathways of infection. On the other hand, the outbreaks of Norwalk viruses have been occurring often in winter, and the consumption of molluscan shellfish fecally contaminated in the harvesting seawater has been suspected to be one of the main pathways of these viruses. Hence, the viral contamination of seawater is one of the important issues from the epidemiological point of view. The occurrence of these viruses in oysters or other seafood has been widely reported (4, 8, 10, 18), while the fate of viruses in seawater is unknown and the levels of the virus have not been quantitatively discussed (9).
In recent years, the PCR method has been used to detect enteric viruses in environmental samples (16, 19-21, 28, 31, 42). PCR is one of the most-sensitive methods available for viral monitoring (2, 5, 30). In conventional concentrating methods, beef extract was often used as an eluate from various adsorbents (22, 27, 38, 41, 43, 44). However, contents of beef extract are suspected to have some inhibitory effect on PCR detection for viruses, especially after reconcentration (1, 32). Many researchers have tried to reduce the inhibitory effects of the eluate (1, 15, 34) or of environmental inhibitors (13, 16, 17, 32, 33), although the proposed procedures were complicated and the recovery yields could not be clearly assessed. These studies suggest that beef extract might not be the best eluate prior to the PCR detection of viruses.
We have developed a new series of procedures to concentrate viruses by adsorption to and elution from a negatively charged membrane, with the insertion of an acid rinse step for removing cations and other inhibitors without eluting the viruses from the membrane between the adsorption and elution steps. An inorganic eluting medium was also tested as a better pretreatment for reverse transcription (RT)-PCR detection of viruses. The developed viral concentration method was applied to 2 liters of seawater to detect naturally occurring enteroviruses, hepatitis A virus (HAV), and Norwalk viruses.
MATERIALS AND METHODS
Comparative study of various concentration methods.
Poliovirus type I, LSc 2ab Sabin strain, was propagated on the BGM cell line and purified to obtain stock solution. The concentration of virus was determined by plaque assay using the BGM cell line.
A type HA negatively charged membrane (Nihon Millipore, Tokyo, Japan) with a 0.45-μm pore size and 47-mm diameter was used. The type 1MDS positively charged membrane (Cuno, Meriden, Conn.) with a 47-mm diameter was also used for comparison of recovery yields.
Poliovirus was spiked into pure water with 25 mM MgCl2 or into natural seawater in Tokyo Bay, Japan. Forty- or fifty-milliliter samples were passed through the membrane at a flow rate of 100 ml/min, and then the membrane was rinsed with 200 ml of 0.5 mM H2SO4 (pH 3.0) followed by elution with 5 ml of 1 mM NaOH (pH 10.5 to 10.8) or 5 ml of beef extract solution (pH 9.5). The concentrates were neutralized upon elution with 50 μl of 50 mM H2SO4 and 50 μl of 100× Tris-EDTA (TE) buffer (pH 8.0) in the sodium hydroxide elution cases. The recovery yields were compared to those of the concentrates without acid rinse. The type 1MDS positively charged membrane was also used for comparison of recovery yields, where no addition of MgCl2 to pure water was also compared. The amount of the virus recovered was determined by plaque assay, and the recovery yields were calculated.
In order to confirm the applicability of this method to practical volume, 1 liter each of artificial seawater made from sea salts (Sigma-Aldrich Japan, Tokyo, Japan) was spiked with poliovirus and was passed through the HA membrane at a flow rate of 100 ml/min. The membrane was then rinsed with 200 ml of 0.5 mM H2SO4 (pH 3.0) followed by elution with 5 ml of 1 mM NaOH (pH 10.5 to 10.8). The concentrates were neutralized upon elution by 50 μl of 50 mM H2SO4 and 50 μl of 100× TE buffer. The recovered poliovirus was determined by plaque assay using BGM cells.
Poliovirus stock solution was spiked into 1 liter each of natural seawater samples from three sampling points (Daiba Kaihin Park and Kasai Kaihin Park, Tokyo, Japan, and Enoshima Beach [west beach], Fujisawa City, Japan). These samples were concentrated as described above. The concentrates were then filtered at 1,500 × g for 10 min at 4°C with an ultrafiltration system (Centriprep Concentrator 50; Nihon Millipore) into 1 ml of the final volume. The recovered solution was added to 4 ml of Eagle's minimum medium, which was followed by determination of poliovirus by plaque assay.
Study area and sample collection.
Samples were collected from two points of Enoshima Beach (east beach and west beach) in summer (22, 23, and 24 August 2000) and in winter (23 November and 22 December 2000 and 6 January 2001) and two points in Tokyo Bay (Daiba Kaihin Park and Kasai Kaihin Park) in winter (19 November at Kasai, 23 November at Daiba, and 7 and 29 December 2000 and 8 January 2001 at both sites). Enoshima Beach receives water from a small river running through an urbanized area. This beach is one of the most popular bathing beaches in Japan and was crowded with people on the day of sampling in the summer. On the other hand, at the two points in Tokyo Bay swimming is not permitted, but these areas continuously receive discharges from the Tokyo metropolitan area, including treated wastewater after chlorination and the overflow of combined sewer system in periods of heavy rain. All samples were collected in the daytime.
All the samples were assayed for fecal coliforms by a double agar layer method using deoxycholic acid agar (14). Total coliforms were also tested by a double agar layer method using deoxycholic acid agar (14). F-specific phages (12) were also determined by the single agar layer method using 100 ml of the sample (7) on the host strain Salmonella enterica serovar Typhimurium WG49 (NCTC12484) (11) as described in ISO 10705 (3a).
Virus concentration method from field samples.
A type HA negatively charged membrane (Nihon Millipore) with a 0.45-μm pore size and a 90-mm diameter was used in a vacuum pump system. Two liters of collected seawater was filtered to adsorb the viruses to the membrane, 200 ml of 0.5 mM H2SO4 was passed through the membrane to rinse out the cation, and then 10 ml of 1 mM NaOH (pH 10.5 to 10.8) was poured on the membrane and the filtrate was recovered in a tube containing 0.1 ml of 50 mM H2SO4 and 0.1 ml of 100× TE buffer for neutralization. The samples were stored at −20°C until further processing.
All 10 ml of the eluate was added into Centriprep Concentrator 50 (Nihon Millipore) and filtered at 1,500 × g for 10 min at 4°C. After removing the filtrate, the concentrate was rinsed twice with 10 ml of MilliQ water by the same procedure, and a final volume of 2 ml was obtained.
Determination of occurrence of viruses by cell culture RT-PCR and by direct RT-PCR.
To detect infectious enteroviruses, two plates of BGM cell monolayer were inoculated with a mixture of the 300-μl portion of the concentrate and 700 μl of Eagle's minimum medium, followed by incubation for 48 h. The viral genomic RNA was extracted using SepaGene RV-R (Sanko Jun-yaku, Tokyo, Japan) from 300 μl of each supernatant and subjected to TaqMan RT-PCR detection of enteroviruses. Since the initial sample volume was 2 liters and the tested volume here was 300 μl out of 2 ml of a final concentrate, this single-plate test corresponded with the positive-negative test for infectious viruses from 300 ml of initial seawater.
SepaGene RV-R (Sanko Jun-yaku) was also used to directly extract RNA from 300 μl of concentrate to obtain a final volume of 12.5 μl. One microliter of DNase I (Wako, Jun-yaku, Osaka, Japan) and 1.5 μl of 5× DNase I buffer were added to the extracted RNA samples and incubated for 30 min at 37°C, followed by inactivation of DNase I at 75°C for 5 min, and then cooling down to 4°C.
An RNA-extracted sample was added to 1.5 μl of SuperScript II (Life Technologies Japan, Tokyo, Japan), 1.5 μl of 100 mM dithiothreitol, 6.0 μl of 5× first-strand buffer, 1.5 μl of deoxynucleoside triphosphates (10 mM each), 0.75 μl of RNase inhibitor (20 U/liter; Life Technologies Japan), 0.75 μl of 0.1 M random hexamer, and distilled water to obtain a final volume of 30 μl; incubated at 42°C for 60 min followed by 99°C for 5 min; and then cooled down to 4°C. Five microliters each of a cDNA sample was then applied for detection of enterovirus, Norwalk virus type G1, Norwalk virus type G2, and HAV, respectively. PCR was performed using a TaqMan PCR reagent kit (PE Biosystems Japan, Urayasu, Japan) with a 200 nM concentration of each primer and a 100 nM concentration of TaqMan probe. The primer pair used for detection of enteroviruses was the same one used by Shieh et al. (34), and the probe was elongated by five bases for a suitable length in the TaqMan PCR system. The number of mismatches to the sequence of the enterovirus genomes in the DNA Data Bank of Japan was surveyed as shown in Table 1. The number of mismatches of the developed TaqMan probe is less than that of the primer pair. The primer pairs and the probes used for Norwalk virus type G1 and type G2 were developed by Katayama Kazuhiko (private communication); the primer pair for HAV was the same one used by Tsai et al. (42). The oligonucleotide sequences are shown in Table 2. The tubes for detection of enterovirus or HAV were incubated as follows: 95°C for 10 min; 40 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min; and 72°C for 6.5 min, followed by cooling to 4°C. The tubes for detection of Norwalk viruses were incubated 50°C for 2 min, followed by 10 min at 95°C and 50 cycles at 95°C for 15 s and 56°C for 1 min.
TABLE 1.
Mismatches between genomes of enterovirus and the oligonucleotide sequences used
| Virus typea | No. of mismatches with:
|
||
|---|---|---|---|
| Pan- entero + | Pan- entero − | TaqMan probe | |
| Coxsackievirus A | 0 | 1 | 0 |
| Coxsackievirus A16 G-10 | 0 | 0 | 0 |
| Coxsackievirus A24 | 0 | 0 | 1 |
| Coxsackievirus A9 | 0 | 0 | 0 |
| Coxsackievirus B1 | 0 | 0 | 0 |
| Coxsackievirus B2 strain Ohio-1 | 0 | 0 | 0 |
| Coxsackievirus B3 | 0 | 0 | 0 |
| Coxsackievirus B4 | 0 | 1 | 0 |
| Coxsackievirus B5 | 0 | 1 | 0 |
| Coxsackievirus B6 | 0 | 1 | 0 |
| Echovirus 23 | 9 | 12 | 16 |
| Echovirus 5 | 0 | 0 | 0 |
| Echovirus 6 lytic strain | 0 | 0 | 0 |
| Echovirus 9 | 1 | 0 | 0 |
| Echovirus type 12, prototype Travis | 0 | 3 | 1 |
| Enterovirus 70 | 0 | 1 | 0 |
| Enterovirus 71 BrCr | 0 | 1 | 0 |
| Enterovirus 71 MS,7423,87 | 0 | 0 | 0 |
| Poliovirus type 2 genome strain Sabin 2 | 0 | 0 | 0 |
| Poliovirus type 3 strain 23127 | 0 | 1 | 0 |
| Poliovirus, strain Sabin 1 | 0 | 0 | 0 |
Viruses registered in the DNA Data Bank of Japan as a full sequence of enterovirus but excluding animal viruses.
TABLE 2.
Oligonucleotide sequences used for detection of enteric viruses
| Virus | Primer | Probe | Sequencea | Reference |
|---|---|---|---|---|
| Enterovirus | Pan-entero+(444-450) | CCTCCGGCCCCTGAATG | 34 | |
| Pan-entero−(621-638) | ACCGGATGGCCAATCCAA | 34 | ||
| TaqMan Probe | (FAM)CCGACTACTTTGGGTGTCCGTGTTTC (TAMRA) | This study | ||
| Norwalk virusa type G1 | COG1F | CTTAGACGCCATCATCATTYAC | K. Katayama et al., unpublished | |
| COG1R | CGYTGGATGCGNTTYCATGA | K. Katayama et al., unpublished | ||
| TaqMan Probe | (TET)AGATYGCGATCYCCTGTCCA(TAMRA) | K. Katayama et al., unpublished | ||
| Norwalk virus type G2 | COG2F | CARGARBCNATGTTYAGRTGGATGAG | K. Katayama et al., unpublished | |
| COG2R | TCGACGCCATCTTCATTCACA | K. Katayama et al., unpublished | ||
| TaqMan Probe | (FAM)TGGGAGGGCGATCGCAATCT(TAMRA) | K. Katayama et al., unpublished | ||
| HAV | HAV+ | CAGCACATCAGAAAGGTGAG | 42 | |
| HAV− | CTCCAGAATCATCTCCAAC | 42 |
Single-letter code: y stands for C or T; N, stands for A, C, G, or T; R stands for A or G; and B stands for B, T, or C. Abbreviations: FAM, 6-carboxy-fluorescein; TET, tetrachloro-6-carboxy-fluorescein; TAMRA, 6-carboxy-tetramethyl-rhodamine.
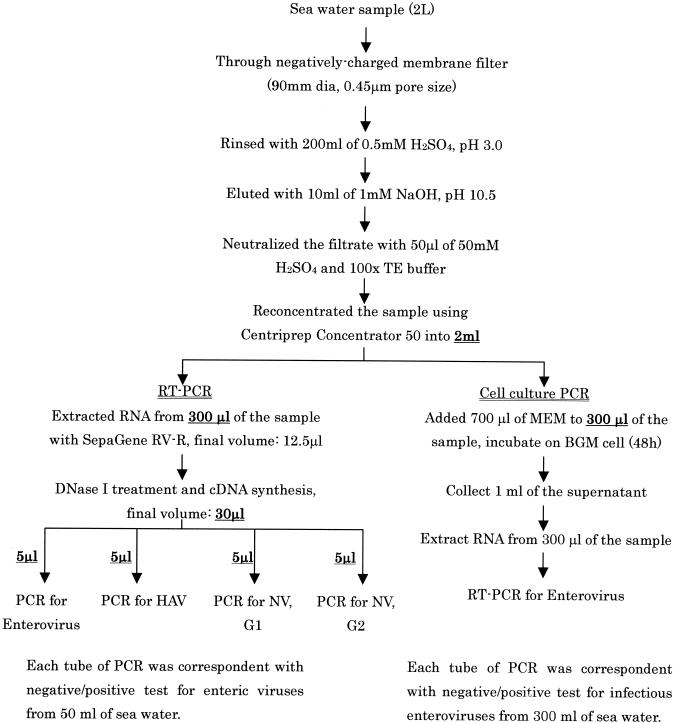
Since the 30 μl of a cDNA sample was obtained from 300 μl out of 2 ml of a final concentrate and a 5-μl portion of the cDNA was subjected to PCR, this single-tube test without cell culture corresponded with a positive-negative test for 50 ml of an initial seawater sample. The volumes obtained from 2 liters of seawater samples and the volumes tested for virus detection are schematically shown in Fig. 1.
FIG. 1.
Protocol for concentration and detection of enteric viruses from seawater samples. The volume of the sample during processing and the volume applied are specified. NV, Norwalk virus
Enterovirus and Norwalk virus types G1 and type G2 were detected by TaqMan SDS 7200(PE Biosystems Japan). Normalized reporter (Rn, defined as the ratio of reporter to passive reference) of each tube was determined using TaqMan SDS 7200 before and after PCR. The magnitude of the signal generated by PCR was calculated as the difference in Rn value (described as dRn). The positive results were obtained when the difference between the dRn values of no-template controls (n = 8) and that of the sample was significant (>t0.001).
The PCR products for HAV were analyzed by gel electrophoresis with ethidium bromide staining (21). A positive band of 192 bp on the gel was considered to be a positive result for HAV (42).
RESULTS
Comparative study of the virus concentration methods.
The recovery yields of viruses by various concentrating conditions are summarized in Table 3.
TABLE 3.
Recovery yields of poliovirus from freshwater and seawater using various concentration methods
| Membrane | Sample type | Acid rinse | Elution medium | Virus input
|
Virus recovered
|
% Recovery | ||
|---|---|---|---|---|---|---|---|---|
| Concn (PFU/ml) | Vol (ml) | Concn (PFU/ml) | Vol (ml) | |||||
| HA | Pure water with cationa | Applied | NaOHb | 710 | 40 | 5,400 | 5 | 95 |
| Not applied | NaOH | 710 | 40 | 1,900 | 5 | 33 | ||
| Applied | BEc | 710 | 40 | 4,700 | 5 | 83 | ||
| Not applied | BE | 710 | 40 | 4,500 | 5 | 79 | ||
| 1MDS | Pure water with cationa | Applied | NaOH | 300 | 1,000 | 480 | 5 | 0.8 |
| 1MDS | Pure water | Applied | NaOH | 350 | 1,000 | 394 | 5 | 0.6 |
| Not applied | NaOH | 350 | 1,000 | 43,200 | 5 | 62 | ||
| Not applied | BE | 710 | 40 | 2,850 | 5 | 50 | ||
| HA | Sea water | Applied | NaOH | 570 | 50 | 5,400 | 5 | 95 |
| Not applied | NaOH | 570 | 50 | 1,950 | 5 | 34 | ||
| Applied | BE | 570 | 50 | 5,350 | 5 | 94 | ||
| Not applied | BE | 570 | 50 | 2,400 | 5 | 42 | ||
| Applied | NaOH | 770 | 50 | 6,300 | 5 | 82 | ||
| Not applied | NaOH | 770 | 50 | 3,200 | 5 | 42 | ||
| 1MDS | Seawater | Not applied | BE | 770 | 50 | 460 | 5 | 6 |
MgCl2 was added to pure water to obtain a final concentration of 25 mM Mg2+.
10 mM NaOH, pH 10.5 to 10.8, was used for elution.
Beef extract (BE) solution, pH 9.5, was used for elution.
The acid rinse promoted the recovery yields from the HA membrane. In the cases with acid rinse, the recovery of poliovirus always exceeded 80%, while it was mostly less than 50% without the acid rinse. Especially in natural seawater cases, the acid rinse enhanced the elution by NaOH solution and by beef extract solution from 38% (average of 34 and 42%) to 89% (average of 82 and 95%) and from 42 to 94%, respectively. Sodium hydroxide solution provided almost the same recovery yields as beef extract solution from both pure water and seawater. The 1MDS positively charged membrane provided comparable recovery from pure water without the addition of MgCl2 and without acid rinse. On the other hand, the recovery yields after acid rinse were quite low. These results are probably due to the absence of attractive interaction between virions and membrane under the acid condition under which viruses were positively charged. The recovery from seawater was quite poor. This result agreed with the study indicating that multivalent salts help virus adsorption to the negatively charged membrane but inhibit virus adsorption to the positively charged membrane (24).
Recovery of spiked poliovirus from 1 liter of artificial seawater.
Table 4 shows the results of the poliovirus recovery from 1 liter of artificial seawater using HA membrane with acid rinse followed by alkaline elution. The recovery yields obtained were 50 to 73%, reproducible enough considering the variation of plaque assay of poliovirus. Since these results were not much different from those obtained from 50-ml samples of natural seawater (Table 3), increasing the volume of seawater did not affect the recovery in this method.
TABLE 4.
Recovery yields of viruses from 1 liter of artificial seawater
| Initial poliovirus concn in artificial seawatera (1,000 ml) (PFU/ml) | Poliovirus concn of the eluatea (5 ml) (PFU/ml) | % Recovery |
|---|---|---|
| 1.7 × 102 | 1.7 × 104 | 50 |
| 1.8 × 104 | 53 | |
| 1.7 × 104 | 50 | |
| 2.1 × 104 | 62 | |
| 1.3 × 102 | 1.6 × 104 | 62 |
| 1.9 × 104 | 73 | |
| 1.8 × 104 | 69 | |
| 1.9 × 104 | 73 | |
| Avg | 61.5 | |
| SD | 10 |
Average from two plates.
Recovery of spiked poliovirus in two-step concentration from 1 liter of natural seawater.
Three kinds of natural seawater (1 liter) were spiked with the poliovirus stock solution and were concentrated by the method developed here followed by reconcentration with the Centriprep Concentrator 50. The titers of recovered poliovirus together with titers of poliovirus from spiked samples are shown in Table 5. The recovery yields were almost the same regardless of the sampling points and almost as high as those obtained by one-step concentration from 1 liter of artificial seawater and from 50 ml of natural seawater. The recovery yields of the reconcentration with the Centriprep Concentrator 50 were high enough judging from these results. The applicability of the developed virus concentration method was confirmed.
TABLE 5.
Recovery yields of poliovirus spiked in 1 liter of natural seawater by negatively charged membrane followed by centrifugal ultrafiltration
| Seawatera sampling place | Total coliforms (CFU/ml) | Fecal coliforms (CFU/100 ml) | Spiked amountb (PFU) | Poliovirus concn recovered (PFU/ml) | Vol of concentratec (ml) | % Recovery |
|---|---|---|---|---|---|---|
| Enoshima | 0 | 0 | 1,650 | 240 | 5 | 73 |
| Daiba | 3 | 25 | 1,650 | 225 | 5 | 68 |
| Kasai | 77 | 1,815 | 1,650 | 200 | 5 | 61 |
Enteroviruses were not detected by cell culture RT-PCR from 2 liters of each seawater sample.
Spiked in 1 liter of natural seawater.
Volumes of the concentrates were adjusted with Eagle's minimum medium.
Occurrence of enteric viruses in seawater samples.
The results of the field survey are summarized in Table 6. Water temperatures were 28 to 30°C in summer at Enoshima Beach and 8 to 16°C in winter at all the sampling points. Since the levels of fecal coliforms at Enoshima Beach in summer were higher than those in winter and those in Tokyo Bay, it is suspected that on-site fecal pollution was occurring at the beach in the summer. The F-specific phages were detected only once in six samples tested from Enoshima Beach and also once in four samples tested from Tokyo Bay. From the results of these microbial parameters, the coastal seawater samples were not extremely contaminated by human or animal feces or wastewater from an urbanized area.
TABLE 6.
Results of detection of naturally occurring viruses from seawater and indicator profiles
| Sampling point | Season | Temp (°C) | Fecal coliforms (CFU/100 ml)
|
No. of positive samples/no. of samples tested byd:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F-specific phagea | Direct RT-PCRb
|
Cell culture PCRc (EV) | ||||||||||||
| Mean | Range | NV G1 | NV G2 | HAV | EV | |||||||||
| Enoshima | Summer | 28-30 | 215 | 19-4,000 | 0/2 | 0/6 | 0/6 | 0/6 | 0/6 | 4/6 | ||||
| Enoshima | Summer | |||||||||||||
| Tokyo Bay | Winter | 8-16 | 8 | 0-172 | 1/4 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | ||||
| Tokyo Bay | Winter | 8-16 | 62 | 4-800 | 1/4 | 3/8 | 1/8 | 0/8 | 0/8 | 0/8 | ||||
Plaque assay for 100 ml using S. enterica serovar Typhimurium WG49 as a host strain.
Equivalent with detection from 50 ml of seawater.
Equivalent with detection from 300 ml of seawater; at least one positive result from two samples tested.
Abbreviations: NV, Norwalk virus; EV, enterovirus.
From the 2-liter samples of the seawater collected, Norwalk virus types G1 and G2 were detected in one and three samples, respectively, out of eight samples tested from Tokyo Bay in winter. With regard to the volume tested (Fig. 1), occurrences of Norwalk virus in a 50-ml seawater sample were determined. On the other hand, enteroviruses were not detected in the same samples by direct RT-PCR or by cell culture PCR. When 2-liter samples of coastal seawater from Enoshima Beach taken in summer were tested, enteroviruses were detected in four out of six samples tested by cell culture PCR, though no enteroviruses were detected by the direct RT-PCR. These results suggested that one or more infectious enteroviruses were present in 300-ml aliquots of the four samples but enterovirus was absent in 50-ml aliquots of the same samples. HAV was not detected in any sample tested. F-specific phage was not detected in 100-ml aliquots of the samples containing the enteric viruses in 50- or 300-ml aliquots. The levels of fecal coliforms were not always high even when the enteric viruses were detected.
DISCUSSION
Since the levels of human enteric viruses in water environments are normally low, detection methods with a high sensitivity are needed. At the same time, however, the potential risk of contamination must be considered carefully. Since the processing of a PCR product might cause the carryover of contamination to the next samples, nested PCR was not applied in this study. It was also important that in this study TaqMan PCR was employed where the product of PCR were analyzed without opening the lid of the tube to prevent carryover of PCR products to the future samples.
The positive results of Norwalk virus detection by RT-PCR directly after the reconcentration step (Table 6) means that inhibitory effects on PCR carried from seawater were negligible. Since enteroviruses were not detected in the same samples although the recovery yields of the enteroviruses were supposed to be high enough according to the experimental results of recovery of poliovirus from spiked samples (Tables 3, 4, and 5), the levels of enteroviruses were lower than those of Norwalk viruses in Tokyo Bay in winter. These results agreed with the epidemiological reports from the National Institute of Infectious Disease that the reports of detection of enteroviruses from patients' feces were higher in the summer season and those of Norwalk-like viruses were higher in the winter season. HAV was not detected in any sample tested, but the level of HAV could not be discussed on the basis of these results because the recovery yields of HAV were unknown. Further study is necessary to investigate the recovery of HAV. Moreover, the recovery of viral genomic RNA in the RNA extraction step should be considered to assess the levels of enteric viruses from the PCR determination of viruses in water.
As for the indicators, the level of F-specific phages was not higher than that of enteric viruses and the levels of fecal coliforms were not correlated to the detection of enteric viruses. It was difficult, therefore, to assess the risk of infection with enteric viruses based on the level of fecal coliforms or F-specific phages in Tokyo Bay and at Enoshima Beach.
There have been a number of studies on improving the methods of concentrating virus from water in order to make the concentrates suitable for subsequent RT-PCR detection of viruses. It was of great concern to remove inhibitors from the beef extract concentrate used as an eluate in the conventional positively charged membrane virus concentrating methods, such as resin treatments (1), polyethylene glycol precipitation-resuspension techniques (15), immunomagnetic capture (17, 33), or glass purification (13). However, these procedures were expensive and too complicated to determine the overall recovery yields.
An acid rinse step was introduced between the adsorption and elution steps in the method developed in this study. The acid solution proved successful in eluting cations and keeping viruses on the negatively charged membrane. The acid rinse step may promote both elution and detection efficiency, because it may reduce inhibitors in the recovered eluates from original environmental water samples.
The hydrophobic interactions with organic substances in beef extract played an important role in the elution of viruses from various absorbents in previous studies (22, 27, 38, 41, 43, 44). However, the eluate obtained by using beef extract solution caused some inhibitory effect on PCR detection of viruses (1, 15, 32, 34). Since PCR is one of the best tools for detecting viruses, an inorganic solution may be a better eluate in the virus concentration process, where the electrostatic interactions must be considered as one of the main mechanisms. A negatively charged membrane may be appropriate because of the repulsive force between the membrane and the negatively charged viruses under alkaline conditions.
The mechanism of enhancement of virus recovery with the acid rinse step can be explained as follows. In the adsorption process, cations link viruses to the negatively charged membrane (6), and they may inhibit the elution of viruses in the same manner. This explanation is supported by the fact that addition of cation (43) to a freshwater sample was necessary in the adsorption to a negatively charged membrane. In the acid rinse procedure at pH 3.0, in which viruses become positively charged since they normally have a higher isoelectric point, viruses are expected to attach themselves directly to the negatively charged membrane, while the cations are eluted from the membrane. This explanation was supported by the fact that acidification (35, 37) of freshwater samples increased virus adsorption to negatively charged membrane. In the subsequent alkali elution, the charge of virus surfaces was converted from positive to negative, which allowed viruses to be eluted from the negatively charged membrane.
Detection of viruses in seawater has been difficult because the virus adsorption to the positively charged membrane has been poor (24) and huge volumes of seawater were collected (9). Recently, an attractive technique was developed to concentrate viruses from seawater on a negatively charged membrane and culture viruses by placing the membrane on the host cell (26), although the virus types applicable to this method are limited to the culturable ones.
In this study we have developed a method applicable to nonculturable viruses as well. Norwalk viruses are of great concern and have been widely surveyed and frequently detected in oysters (4, 8, 10, 18), while reports of detection of Norwalk viruses from seawater have been limited. The virus concentration method developed in this study has enough potential to be applied to the detection of Norwalk viruses from seawater, including waters from oyster-harvesting areas.
REFERENCES
- 1.Abbaszadegan, M., M. S. Huber, C. P. Gerba, and I. L. Pepper. 1993. Detection of enteroviruses in groundwater with the PCR. Appl. Environ. Microbiol. 59:1318-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abbaszadegan, M., P. Stewart, and M. LeChevallier. 1999. A strategy for detection of viruses in groundwater by PCR. Appl. Environ. Microbiol. 65:444-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Public Health Association. 1995. Standard methods for the examination of water and wastewater, 19th ed. American Public Health Association, Washington, D.C.
- 3a.Anonymous. 1995. ISO 10705-1 (first edition): Water quality. Detection and enumeration of bacteriophages—part 1. Enumeration of F-specific RNA bacteriophages. International Organization for Standardization, Geneva, Switzerland.
- 4.Atmar, R. L., F. H. Neill, J. L. Romalde, F. L Guyader, C. M. Woodley, T. G. Metcalf, and M. K. Estes. 1995. Detection of Norwalk virus and hepatitis A virus in shellfish tissues with the PCR. Appl. Environ. Microbiol. 61:3014-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung, H., L. A. Jaykus, and M. D. Sobsey. 1996. Detection of human enteric viruses in oysters by in vivo and in vitro amplification of nucleic acid. Appl. Environ. Microbiol. 62:3772-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerba, C. P. 1984. Applied and theoretical aspects of virus adsorption to surfaces. Adv. Appl. Microbiol. 30:133-168. [DOI] [PubMed] [Google Scholar]
- 7.Grabow, W. O. K., and P. Coubrough. 1986. Practical direct plaque assay for coliphage in 100-ml samples of drinking water. Appl. Environ. Microbiol. 52:430-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green, J., K. Henshilwood, C. I. Gallimore, D. W. G. Brown, and D. N. Lees. 1998. A nested reverse transcriptase PCR assay for detection of small round-structured viruses in environmentally contaminated molluscan shellfish. Appl. Environ. Microbiol. 64:858-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffin, D. W., C. J. Gibson III, E. K. Lipp, K. Riley, J. H. Paul III, and J. B. Rose. 1999. Detection of viral pathogens by reverse transcriptase PCR and of microbial indicators by standard methods in the canals of the Florida Keys. Appl. Environ. Microbiol. 65:4118-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guyader, F. L., L. Haugarreau, L Miossec, E. Dubois, and M. Pommepuy. 2000. Three-year study to assess human enteric viruses in shellfish. Appl. Environ. Microbiol. 66:3241-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Havelaar, A. H., W. M. Hogeboom, and R. Pot. 1984. F-specific RNA bacteriophages in sewage: methodology and occurrence. Water Sci. Technol. 17:645-655. [Google Scholar]
- 12.IAWPRC Study Group on Health Related Microbiology. 1991. Bacteriophages as model viruses in water quality control. Water Sci. Technol. 25:529-545. [Google Scholar]
- 13.Ijzerman, M. M., D. R. Dahling, and G. S. Fout. 1997. A method to remove environmental inhibitors prior to the detection of waterborne enteric viruses by reverse transcriptase-PCR. J. Virol. Methods 63:145-153. [DOI] [PubMed] [Google Scholar]
- 14.Japanese Water Works Association. 1993. Japanese standard method for examination of water. Japanese Water Works Association, Tokyo, Japan. (In Japanese.)
- 15.Jaykus, L. A., R. De Leon, and M. D. Sobsey. 1996. A virion concentration method for detection of human enteric viruses in oysters by PCR and oligoprobe hybridization. Appl. Environ. Microbiol. 62:2074-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jothikumar, N., K. Aparna, S. Kamatchiammal, R. Paulmurugan, S. Saravanadevi, and P. Khanna. 1993. Detection of hepatitis E virus in raw and treated wastewater with the PCR. Appl. Environ. Microbiol. 59:2558-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jothikumar, N., D. O. Cliver, and T. W. Mariam. 1998. Immunomagnetic capture PCR for rapid concentration and detection of hepatitis A virus from environmental samples. Appl. Environ. Microbiol. 64:504-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawamoto, H., S. Hasegawa, S. Sawatari, C. Miwa, O. Morita, T. Hosokawa, and H. Tanaka. 1993. Small, round-structured viruses (SRSVs) associated with acute gastroenteritis outbreak in Gifu, Japan. Microbiol. Immunol. 37:991-997. [DOI] [PubMed] [Google Scholar]
- 19.Kopecka, H., S. Dubrou, J. Prevot, J. Marechal, and Lopez-Pila. 1993. Detection of naturally occurring enteroviruses in water by reverse transcription, PCR, and hybridization. Appl. Environ. Microbiol. 59:1213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Limsawat, S., and S. Ohgaki. 1997. Fate of liberated viral RNA in wastewater determined by PCR. Appl. Environ. Microbiol. 63:2932-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Limsawat, S., N. Kamiko, and S. Ohgaki. 1995. Application of PCR to detect coliphage Qβ in environmental water sample. Water Sci. Technol. 31:383-390. [Google Scholar]
- 22.Logan, K. B., G. E. Rees, N. D. Seely, and S. B. Primrose. 1980. Rapid concentration of bacteriophages from large volumes of freshwater: evaluation of positively charged, microporous filters. J. Virol. Methods 1:87-97. [DOI] [PubMed] [Google Scholar]
- 23.Logan, K. B., G. E. Scott, N. D. Seely, and S. B. Primrose. 1981. A portable device for the rapid concentration of viruses from large volumes of natural freshwater. J. Virol. Methods 3:241-249. [DOI] [PubMed] [Google Scholar]
- 24.Lukasik, J., T. M. Scott, D. Andryshak, and S. R. Farrah. 2000. Influence of salts on virus adsorption to microporous filters. Appl. Environ. Microbiol. 66:2914-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nupen, E. M., and B. W. Bateman. 1985. The recovery of viruses from drinking water by means of an in-line electropositive cartridge filter. Water Sci. Technol. 17:63-69. [Google Scholar]
- 26.Papageorgiou, G. T., L. M. Llivina, C. G. Christodoulou, F. Lucena, D. Akkeidou, E. Ioannou, and J. Jofre. 2000. Simple methodological approach for counting and identifying culturable viruses adsorbed to cellulose nitrate membrane filter. Appl. Environ. Microbiol. 66:194-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Payment, P., and M. Trudel. 1985. Detection and health risk associated with low virus concentration in drinking water. Water Sci. Technol. 17:97-103. [Google Scholar]
- 28.Puig, M., J. Jofre, F. Lucena, A. Allard, G. Wadell, and R. Girones. 1994. Detection of adenoviruses and enteroviruses in polluted water by nested PCR amplification. Appl. Environ. Microbiol. 60:2963-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds, K. A., J. B. Rose, and A. T. Giordano. 1993. Comparison of methods for the recovery and quantification of coliphage and indigenous bacteriophage from marine water and sediments. Water Sci. Technol. 27:115-117. [Google Scholar]
- 30.Rose, J. B., X. Zhou, D. W. Griffin, and J. H. Paul. 1997. Comparison of PCR and plaque assay for detection and enumeration of coliphage in polluted marine waters. Appl. Environ. Microbiol. 63:4564-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwab, K. J., R. De Leon, and M. D. Sobsey. 1993. Development of PCR methods for enteric virus detection in water. Water Sci. Technol. 27:211-218. [Google Scholar]
- 32.Schwab, K. J., R. De Leon, and M. D. Sobsey. 1995. Concentration and purification of beef extract mock eluates from water samples for detection of enteroviruses, hepatitis A viruses, and Norwalk viruses by reverse transcription-PCR. Appl. Environ. Microbiol. 61:531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwab, K. J., R. De Leon, and M. D. Sobsey. 1996. Immunoaffinity concentration and purification of waterborne enteric viruses for detection by reverse transcriptase PCR. Appl. Environ. Microbiol. 62:2086-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shieh, Y.-S. C., D. Wait, L. Tai, and M. D. Sobsey. 1995. Method to remove inhibitors in sewage and other fecal wastes for enterovirus detection by the PCR. J. Virol. Methods 54:51-66. [DOI] [PubMed] [Google Scholar]
- 35.Shields, P. A., S. A. Berenfeld, and S. R. Farrah. 1985. Modified membrane-filter procedure for concentration of enteroviruses from tap water. Appl. Environ. Microbiol. 49:453-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shri, N., and C. P. Gerba. 1983. Concentration of coliphage from water and sewage with charge-modified filter aid. Appl. Environ. Microbiol. 45:232-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sobsey, M. D., C. Wallis, M. Hendersen, and J. L. Melnick. 1973. Concentration of enteroviruses from large volumes of water. Appl. Microbiol. 26:529-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobsey, M. D., and B. L. Jones. 1979. Concentration of poliovirus from tap water using positively charged microporous filters. Appl. Environ. Microbiol. 37:588-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobsey, M. D., S. E. Oglesbee, D. A. Wait, and A. I. Cuenca. 1985. Detection of hepatitis A virus in drinking water. Water Sci. Technol. 17:23-38. [Google Scholar]
- 40.Sobsey, M. D. 1995. Male-specific coliphages as indicators of viral contamination of drinking water. American Water Works Association, Denver, Colo.
- 41.Stetler, R. E., S. C. Waltrip, and C. J. Hurst. 1992. Virus removal and recovery in the drinking water treatment train. Water Res. 26:727-731. [Google Scholar]
- 42.Tsai, Y., M. D. Sobsey, L. R. Sangermano, and C. J. Palmer. 1993. Simple method of concentrating enteroviruses and hepatitis A virus from sewage and ocean water for rapid detection by reverse transcriptase PCR. Appl. Environ. Microbiol. 59:3488-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallis, C., and J. L. Melnick. 1967. Concentration of enteroviruses on membrane filters. J. Virol. 1:472-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yano, K., Y. Yoshida, T. Shinkai, and M. Kaneko. 1993. A practical method for the concentration of viruses from water using fibriform cellulose and organic coagulant. Water Sci. Technol. 27:295-298. [Google Scholar]