Abstract
A chitinase gene was cloned on a 2.8-kb DNA fragment from Stenotrophomonas maltophilia strain 34S1 by heterologous expression in Burkholderia cepacia. Sequence analysis of this fragment identified an open reading frame encoding a deduced protein of 700 amino acids. Removal of the signal peptide sequence resulted in a predicted protein that was 68 kDa in size. Analysis of the sequence indicated that the chitinase contained a catalytic domain belonging to family 18 of glycosyl hydrolases. Three putative binding domains, a chitin binding domain, a novel polycystic kidney disease (PKD) domain, and a fibronectin type III domain, were also identified within the sequence. Pairwise comparisons of each domain to the most closely related sequences found in database searches clearly demonstrated variation in gene sources and the species from which related sequences originated. A 51-kDa protein with chitinolytic activity was purified from culture filtrates of S. maltophilia strain 34S1 by hydrophobic interaction chromatography. Although the protein was significantly smaller than the size predicted from the sequence, the N-terminal sequence verified that the first 15 amino acids were identical to the deduced sequence of the mature protein encoded by chiA. Marker exchange mutagenesis of chiA resulted in mutant strain C5, which was devoid of chitinolytic activity and lacked the 51-kDa protein in culture filtrates. Strain C5 was also reduced in the ability to suppress summer patch disease on Kentucky bluegrass, supporting a role for the enzyme in the biocontrol activity of S. maltophilia.
Biological sources for plant disease control remain an important potential alternative to the use of pesticides. Biological controls that are based on introduced microbes, however, have been slow to develop due to inconsistencies in their performance (70). Such inconsistencies often result from a lack of understanding the mechanisms by which individual microorganisms function to control disease. It is often difficult to gain a complete understanding of how biocontrol agents control diseases, since many function through a variety of mechanisms. In such cases, identifying contributing mechanisms often requires a systematic approach that directly evaluates individual traits and their contributing roles to the overall operating mechanisms.
The lytic activity of bacteria is one of a number of mechanisms that has been implicated in biocontrol for several years (5, 13, 23, 26, 35, 38, 41, 45). Investigative studies on lytic activity among biocontrol agents have focused largely on the characterization of enzyme systems capable of degrading fungal cell wall components, of which chitinases are among the most intensively studied (4, 23, 26, 45, 58, 72, 73).
Molecular and biochemical characterizations have revealed that chitinases, similar to other glycosyl hydrolases, are modular in nature and can differ according to their structural organization. Enzymes can vary both within and between microbes, depending on the numbers, types, and positions of discrete binding and catalytic domains (reviewed in references 16 and 66). Domains that have been described within bacterial chitinases include chitin-binding domains (22, 42, 60, 61, 65), cadherin-like domains (42), and fibronectin type III domains (14, 65, 69).
Stenotrophomonas maltophilia represents a rhizosphere bacterial species of potential agronomic importance (7, 10, 11, 12, 15, 30, 31, 33, 34, 39). Traits of S. maltophilia associated with biocontrol mechanisms include antibiotic production (25, 44), extracellular enzyme activities such as protease and chitinase (10, 30, 44, 72), and rhizosphere colonization potential (10, 12, 30, 33, 34). To date, however, only a few biocontrol traits expressed by S. maltophilia strains have been molecularly characterized (10), and therefore information concerning gene structure and evaluation of biocontrol traits through direct mutagenesis procedures for this group is lacking.
S. maltophilia strain 34S1 was identified as a biocontrol agent for summer patch disease of Kentucky bluegrass (Poa pratensis) caused by the root-infecting fungus Magnaporthe poae (30). Several traits expressed by S. maltophilia strain 34S1 are thought to contribute to its biocontrol activity; however, the contributing roles for the various traits in disease control remain unclear. In this study, we describe the molecular characterization and structural organization of a single chitinase gene responsible for chitinolytic activity in S. maltophilia 34S1 and evaluate its contribution to biocontrol of summer patch disease.
MATERIALS AND METHODS
Bacterial and fungal strains, vectors, and media.
All bacteria were grown and maintained on Luria-Bertani (LB) agar (Difco). S. maltophilia 34S1 and Burkholderia cepacia M53 were grown at 30°C as described (30). The genomic cosmid library, constructed in pLAFR3 (59), was maintained in Escherichia coli HB101 (Gibco-BRL). Subcloning experiments were performed using the vectors pUC118 (New England Biolabs) and pRK415 (27) and E. coli DH5α (Gibco-BRL) as the host strain. Strains containing plasmids were maintained on LB agar supplemented with appropriate antibiotics at the following levels unless otherwise noted: tetracycline (Tc), 12.5 μg/ml; kanamycin, 50 μg/ml; ampicillin, 50 μg/ml; and rifampin 100 μg/ml.
M. poae strain 73-15 (ATCC 64411) was maintained on potato dextrose agar (PDA) (Difco).
Construction of S. maltophilia genomic library and screening for chitinase activity.
Total genomic DNA of S. maltophilia 34S1 was isolated using the sodium dodecyl sulfate (SDS) lysis procedure described previously (59). DNA was purified twice on a cesium chloride gradient, and 60 μg of DNA was partially digested with Sau3A before size-fractionating on a 10 to 40% glycerol gradient centrifuged at 194,000 × g for 33 h in a Beckman SW40 rotor. DNA fragments greater than 15 kb were collected, ligated to the BamHI site in pLAFR3, and packaged into lambda phage using Gigapak Gold extracts (Stratagene).
No positive clones for chitinase activity were detected when the entire genomic library was initially screened in E. coli or any subsequent subcloning experiments regardless of fragment orientation and IPTG (isopropyl thiogalactopyranoside) induction. Therefore, chitinase activity was detected by mobilizing all clones into B. cepacia M53 by triparental matings (9), plating onto M9 minimal salts (53) in 0.7% agarose supplemented with 0.25% yeast extract (Difco), 2% colloidal chitin (36), and Tc (200 μg/ml), and screening for clearing zones around colonies.
Molecular characterizations and sequence analysis.
Restriction digests, electrophoresis, ligations, and Southern hybridizations were performed as described (53). The nucleotide sequence of both strands was determined by fluorescence labeling with the Amplitaq FS fluorescence labeling kit (Applied Biosystems Inc.) run on an ABI model 373 automated sequencer. All nucleotide sequences and trace plots were analyzed using the DNA analysis programs of DNAStar (Lasergene).
Chitinase activity from the cosmid clone pXM7B5 was subcloned to a 2.8-kb XhoI-SacI fragment by first ligating into the SalI and SacI sites located in the polylinker of pUC118 to form plasmid pXMC131. This subclone was excised as an HindIII-EcoRI fragment using restriction sites located in the vector and ligated into the low-copy-number, broad-host-range vector pRK415.
Saturation transposon mutagenesis of cosmid pXM7B5 contained in E. coli was performed using λ::Tn5 as described (8). A cosmid clone containing Tn5 in the coding region of the chitinase gene, designated pXM7B5::Tn5-1, was identified by screening for loss of chitinase activity in B. cepacia M53 on colloidal chitin agar. Restriction map and Southern blot analyses of this plasmid using sequences internal to Tn5 as a probe positioned the transposon insertion in a 250-bp KpnI fragment internal to the chiA open reading frame (see Fig. 1B). The mutation in pXM7B5::Tn5-1 was introduced into the genome of S. maltophilia 34S1 by marker exchange mutagenesis by the method of Ruvkun and Ausubel (51). A resulting mutant, designated strain C5, was verified by Southern blot analysis to contain Tn5 in the 250-bp KpnI fragment.
FIG. 1.
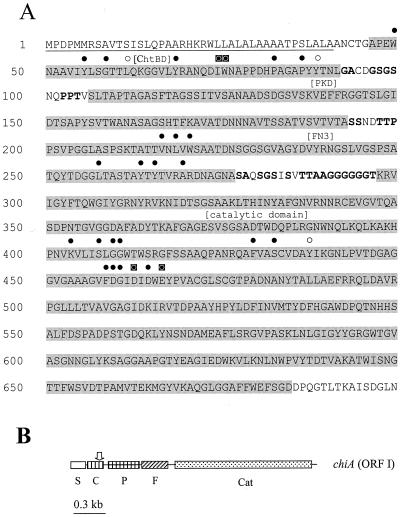
(A) Deduced amino acid sequence of the chiA gene from S. maltophilia. Underlined residues represent signal peptide sequence; shaded residues represent specific domain regions identified in brackets above the sequence as the chitin binding domain (ChtBD), PKD domain (PKD), fibronectin type III domain (Fn3), and catalytic domain. Residues in bold represent amino acids common to domain linker regions. Solid circles above amino acid positions represent conserved identical residues, open circles represent conserved similar residues, and boxed circles represent residues essential for function. (B) Map of the chiA open reading frame (ORF I) with relative positions of the signal peptide sequence (S), chitin binding domain (C), PKD domain (P), fibronectin type III domain (F), and catalytic domain (Cat). The vertical arrow above the chitin binding domain represents the position of Tn5 insertion.
Isolation and characterization of extracellular chitinase.
To characterize extracellular enzymes and chitinase activity in culture filtrates, all bacteria were grown in M9 salts broth supplemented with 0.25% yeast extract and 2% colloidal chitin. S. maltophilia transconjugants and B. cepacia transconjugants were grown in medium supplemented with Tc at 100 and 200 μg/ml, respectively. After 3 days of growth at 30°C, cells were pelleted by centrifugation (10 min, 7,500 × g), and culture filtrates were precipitated with 80% ammonium sulfate at room temperature for 60 min before centrifuging for 15 min at 16,900 × g. Pellets were resuspended in 1/1,000 the original volume and dialyzed in 10 mM Tris-HCl (pH 8.0).
Chitinase activity in culture filtrates was assayed by using p-nitrophenyl-N,N′-diacetylchitobiose (pNP) as a substrate similar to the method described (50). Values were standardized based on the cell densities of cultures.
SDS-polyacrylamide gel electrophoresis (PAGE) was performed as previously described (32). Nondenaturing (native) gels consisted of 10% acrylamide in 0.375 M Tris-HCl (pH 6.8). Chitinase activity was detected by UV light exposure of native gels containing 0.1 mM 4-methylumbelliferyl-β-d-N,N′-diacetylchitobioside [4-MU-(GlcNAc)2] or 4-methylumbelliferyl-β-d-N,N′-triacetylchitobioside [4-MU-(GlcNAc)3] (Sigma).
Chitinase was purified from S. maltophilia cultures by first centrifuging cells (10 min at 5,000 × g) and passing the culture fluid through a 0.45-μm membrane to remove particulates. The filtrate was buffered with 20 mM Tris-HCl (pH 8.0) with 1 mM phenylmethylsulfonyl fluoride included to inhibit proteases. Ammonium sulfate was added to a final concentration of 1 M, and the filtrate was applied to a 1-ml phenyl-Sepharose column (Amersham Pharmacia Biotech) equilibrated with 20 mM Tris-HCl (pH 8.0) buffer containing 1 M ammonium sulfate. Following washing with 20 ml of equilibration buffer, fractions were eluted by three × 1-ml washes of 20 mM Tris-HCl (pH 8.0), with stepwise 0.25 M decreases in ammonium sulfate, followed by three × 1-ml washes in 25% acetonitrile and a final elution in three washes with 1 ml of 50% acetonitrile. The protein concentration of each fraction was determined, and aliquots were run on SDS-PAGE. The purified protein was subjected to N-terminal Edman degradation peptide sequencing and confirmed as the chiA gene product by using Fastp web software (University of Virginia).
In vitro antagonism and biocontrol assays.
For all biological assays, mutant strain C5 was compared directly with the wild-type strain 34S1; strain C5 complemented with the chiA subclone in pXM431 was not included in experiments because of plasmid instability over long periods of time. In vitro antagonism assays were conducted as previously described (29). Biocontrol assays for summer patch disease were conducted on Kentucky bluegrass var. Baron grown in a 4:1 mix of pasteurized sand to peat in 9-cm conical containers (30). Containers were inoculated with the fungal pathogen at a depth of 1.5 cm below the soil surface at the time that seeds were sown (approximately 60 seeds per container).
Bacteria were prepared as treatments by pelleting cells from overnight cultures grown in LB and suspending them in sterile H2O to a concentration of 5 × 108 CFU/ml; 25 ml of the bacterial solution was applied to each container as a soil drench at 2 and 3 weeks after seeding. Plants were moved to the growth chamber set at 30°C and 70% humidity with 500 μE of light for 14 h, 4 weeks after seeding to induce disease.
Plants were rated for disease severity based on a visual assessment of the percentage of necrotic plants within a container, as described (30). Ratings were conducted every 3 to 4 days following movement of plants into the growth chamber (day 0). Each treatment was replicated 10 times (with each container representing a replicate) within an experiment, and the experiment was repeated five times.
The disease progress data followed a logistic pattern of increase over a 14- to 17-day period. For data analysis, disease ratings were transformed using the logit transformation, and a simple linear regression equation was calculated for each replicate. A slope and y intercept were estimated for each replicate. In addition, disease severity at day 10 was estimated for each replicate from the regression equations. The disease incidence at day 10, slope, and y- intercept were analyzed in a two-way analysis of variance using the three treatments as the first factor and the five experiments as the second factor.
S. maltophilia populations in the Kentucky bluegrass var. Baron rhizosphere were determined as previously described (30). Populations were sampled 1, 4, 7, 10, and 14 days after application of bacteria. All samples were dilution plated onto LB agar supplemented with rifampin. Samples were replicated three or five times, depending on the experiment, and the experiment was repeated three times.
Nucleotide sequence accession number.
The nucleotide sequence of chiA was deposited in the Genbank database under accession no. AF014950.
RESULTS AND DISCUSSION
Cloning and nucleotide sequence analysis of the chiA gene from S. maltophilia.
Screening the S. maltophilia 34S1 genomic library for chitinase activity in B. cepacia M53 resulted in the identification of two cosmid clones that caused bacterial colonies to clear colloidal chitin in agar within 5 days. Comparisons of restriction digest banding patterns indicated that the two cosmids had a region of overlap that was later confirmed to be homologous by Southern hybridizations (data not shown). One cosmid clone, designated pXM7B5, was selected for further characterization.
Chitinase activity from pXM7B5 was subcloned to a 2.8-kb XhoI-SacI fragment in pXMC431 as described in Materials and Methods. The nucleotide sequence of the 2.8-kb DNA fragment revealed a large open reading frame, designed chiA, which encoded a predicted protein of 700 amino acids with a molecular mass of 72.4 kDa (Fig. 1A). Seven bases upstream of the predicted start ATG was the purine-rich sequence GGAG, resembling a putative ribosome-binding site. An exact 12-base inverted repeat, separated by a single nucleotide, that resembles a transcription termination sequence (AACGCCCCGGCACTGCCGGGGCGTT) was identified 51 bases after the stop codon.
Structural organization of the chiA gene from S. maltophilia strain 34S1.
Analysis of the first 41 residues of the deduced sequence of chiA indicated the presence of a signal peptide sequence (47) with a predicted cleavage site between two alanine residues (Fig. 1). Cleavage at this site results in a predicted protein with a molecular mass of 68 kDa. An RPS-BLAST search of the conserved-domain database identified a catalytic domain belonging to family 18 glycosyl hydrolases (20) at the C-terminal end of the protein between residues 298 and 685.
The web-based analysis tool SMART (http://smart.embl-heidelberg.de), which identifies domains based on sequence homology and tertiary structure (55, 56), was used to identify three additional domains within the deduced ChiA sequence: a type 3 chitin-binding domain (ChtBD3) located between residues 47 and 92, a polycystic kidney disease (PKD) domain located between residues 107 and 194, and a fibronectin type III (Fn3) domain between residues 201 and 278 (Fig. 1). Spanning each of the four domains were short stretches of sequences found to be rich in Gly, Ala, Pro, Ser, and Thr (Fig. 1A) that typically resemble domain linker regions (63).
Because of the modular nature of chitinases, similarity of the chiA sequence to other chitinase genes at the whole-gene level was low. Therefore, chiA was also compared to other database sequences by domain regions. For each domain, the six sequences showing the highest amino acid identity with the deduced chiA protein are listed in Table 1. Comparisons of related sequences between domains clearly showed variation in species origin, gene source, and mean percent identity to the deduced chiA sequence. These observations are consistent with the proposal that the modular nature of glycosyl hydrolases contributes to domain shuffling (14, 16, 60) and suggests that the various domains of chiA were recruited from a range of different species and gene sources.
TABLE 1.
Pairwise comparison of deduced ChiA domains to homologous protein sequences
| Domain and organism | Protein | Accession no. | Reference | Residue positions | % Identity |
|---|---|---|---|---|---|
| ChtB3 domain | |||||
| Xanthomonas sp. | Chitinase A | BAA36460 | 52 | 34-74 | 49 |
| Deinococcus radiodurans | Hypothetical protein | D75274 | 71 | 176-210 | 34 |
| Vibrio cholerae | Chitodextrinase | AAF96599 | 19 | 38-75 | 34 |
| Arthrobacter sp. | Chitinase B | CAB62499 | 37 | 529-570 | 31 |
| Vibrio furmissii | Chitodextrinase | P96156 | 28 | 39-75 | 30 |
| Pseudoalteromonas sp. | Chitinase C | AAC79667 | 62 | 785-815 | 30 |
| PKD domain | |||||
| Xanthomonas sp. | Chitinase A | BAA36460 | 52 | 104-159 | 66 |
| Vibrio cholerae | Chitinase A | AAC72236 | 6 | 339-408 | 54 |
| Vibrio cholerae | Chitinase A | AAC72236 | 6 | 182-250 | 48 |
| Thermobifida fusca | Exocellulase E6 | CAA20643 | 24 | 154-223 | 40 |
| Streptomyces coelicolor | Cellulase | AAD39947 | 48 | 151-220 | 40 |
| Vibrio cholerae | Chitinase | AAC72236 | 6 | 88-156 | 39 |
| Fn3 domain | |||||
| Xanthomonas sp. | Chitinase A | BAA36460 | 52 | 185-244 | 66 |
| Cellulomonas fimi | Exoglucanase B | AAB00822 | 57 | 798-874 | 60 |
| Cellulomonas fimi | Exoglucanase B | AAB00822 | 57 | 895-971 | 60 |
| Cellulomonas fimi | Exoglucanase A | P50401 | 57 | 577-654 | 59 |
| Cellulomonas fimi | Endoglucanase D | AAA23089 | 40 | 454-530 | 56 |
| Cellulomonas fimi | Endoglucanase D | AAA23089 | 40 | 550-626 | 56 |
| Catalytic domain | |||||
| Janthinobacterium lividum | Chitinase 69 | AAA83223 | 17 | 246-652 | 54 |
| Doohwaniella chitinasigens | Chitinase 67 | AAF21468 | 221-619 | 52 | |
| Streptomyces peucetius | Chitinase C | AAF43629 | 264-614 | 47 | |
| Streptomyces coelicolor | Chitinase C | BAA75644 | 48 | 243-594 | 47 |
| Streptomyces lividans | Chitinase C | BAA02168 | 14 | 243-594 | 47 |
| Streptomyces thermoviolaceus | Chitinase 40 | JC2135 | 64 | 51-398 | 46 |
Comparison of the deduced ChiA sequence with other catalytic domains revealed several conserved residues previously identified within family 18 glycosyl hydrolases, including those essential for function (60, 68) (Fig. 1A). The identity of ChiA with the six most related catalytic domains ranged between 46 and 54% in pairwise comparisons. In contrast, ChtBD3 had the lowest range of amino acid identity to related sequences in pairwise comparisons among the four domains found in ChiA, ranging between 30 and 49% (Table 1). Despite the lower identity, ChtBD3 also contained identical or related residues (Fig. 1A) considered well conserved for this domain type (2, 22). Higher similarities were observed to both the PKD and Fn3 domains than to ChtBD3. Overall, the Fn3 domain was observed to have the highest range of similarity to related sequences, ranging between 56 and 66% identity, and contained several previously identified conserved amino acids (1) (Fig. 1A).
PKD domains were first identified in PKD1, the primary gene responsible for polycystic kidney disease in humans. PKD domains are structurally characterized as β-sheets containing an immunoglobulin (Ig)-like fold, similar to Ig-like folds found in proteins and domains grouped within the Ig superfamily, which includes Fn3 (3). Thus, PKD domains are structurally similar to Fn3 domains and have been previously identified in bacterial sequences (46). Within chitinases, however, Fn3 domains are quite prevalent, while PKD-like domains have not been reported in the literature. Our results with Blast searches suggest that many glycosyl hydrolases, including chitinases, contain sequences similar to the PKD region from ChiA (Table 1).
Fn3 and PKD domains have been proposed to originate from animals (1, 3), suggesting the possibility that both domains may have been recruited through similar events by S. maltophilia. The likelihood of this event is further supported by the prevalence of clinical strains of S. maltophilia. Since the functions of the putative binding domains found in ChiA were not evaluated in this study, their roles in chitinolytic activity remain unclear. However, previous studies have demonstrated that such binding domains in chitinases, including ChtBD3 and Fn3, enhance chitinolytic activity (18, 42, 60, 61, 67). The structural similarities between Fn3 and PKD suggest that the latter would also contribute to such a role, but this remains to be experimentally proven.
Characterization of chitinase activity produced by S. maltophilia strain 34S1 and construction of chitinase mutant strain C5.
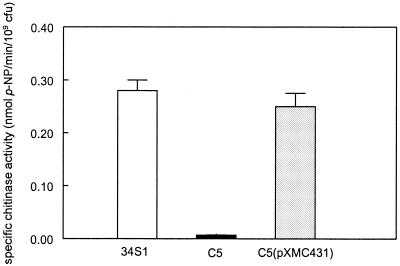
Strain C5, mutated within the chiA locus by marker exchange mutagenesis, was devoid of visible chitinolytic activity when screened on chitin agar medium. Quantitative analysis of chitinase activity in culture filtrates using pNP as a substrate verified that strain C5 was significantly reduced in activity. Activity was restored to levels comparable to that of the wild-type strain 34S1 when the mutant strain C5 was complemented with pXMC431 (Fig. 2).
FIG. 2.
Specific chitinase activity of S. maltophilia culture filtrates grown in M9 minimal salts medium supplemented with yeast extract and 2% colloidal chitin for 72 h. Activity was assayed using pNP as the substrate and standardized to 109 CFU for each culture. Data represents the mean of three replicates with standard deviation.
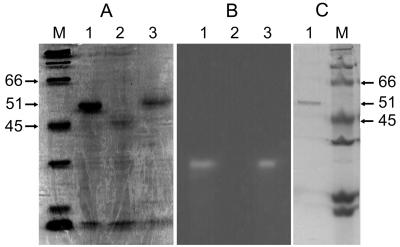
Chitinase activity was analyzed in culture filtrates of S. maltophilia strains grown for 72 h in chitinase-inducing medium. Denaturing gels resolved a major protein band of approximately 51 kDa observed in culture filtrates of strains 34S1(pRK415) and C5(pXMC431) that was absent in the culture filtrate of strain C5(pRK415) (Fig. 3A). Native protein gels containing either 4-MU-(GlcNAc)2 or 4-MU-(GlcNAc)3 resolved an identical major band of chitinase activity in culture filtrates of strains 34S1(pRK415) and C5(pXMC431), but not in culture filtrates of strain C5(pRK415) (Fig. 3B). Excision of the active band from native gels followed by electrophoresis on denaturing gels verified that chitinase activity corresponded in size with the 51-kDa protein observed in culture filtrates (data not shown).
FIG. 3.
Protein gel electrophoresis of chiA gene product expressed in S. maltophilia strains. (A) Coomassie-stained SDS-PAGE of extracellular proteins isolated from 3-day-old culture filtrates. Lane M, broad-range molecular size markers (in kilodaltons). Lane 1, wild-type strain 34S1; lane 2, chitinase mutant strain C5; lane 3, strain C5(pXMC431). (B) Chitinase active protein bands from culture filtrates detected in native gels using 4-MU-(GlcNAc)2. (Gel is not in alignment with gels in panels A and B.) Native gels stained with 4-MU-(GlcNAc)3 (not shown) produced identical activity banding patterns. Lane 1, wild-type strain 34S1; lane 2, chitinase mutant strain C5; lane 3, strain C5(pXMC431). (C) Coomassie-stained SDS-PAGE of chitinase protein purified from 3-day-old culture filtrate of S. maltophilia strain 34S1 by hydrophobic interaction chromatography. Lane 1, purified chitinase; lane M, broad-range molecular size markers.
To verify that the 51-kDa protein is associated with chitinolytic activity and is indeed encoded by chiA, the protein was purified by hydrophobic interaction chromatography. Fractions containing chitinolytic activity resolved a single band identical in size to the major 51-kDa protein band observed in culture filtrates (Fig. 3C). N-terminal sequencing resulted in an identical sequence of the first 15 amino acids of the mature protein encoded by chiA, beginning at the proposed signal peptide sequence cleavage site at position 42. The size, however, is not consistent with the predicted size of the mature protein of 68 kDa based on the sequence, suggesting that degradation of the mature protein occurs in culture.
The observations resulting from mutagenesis and complementation studies are consistent with the indication that chitinolytic activity produced by S. maltophilia strain 34S1 is likely encoded solely by chiA. In another study investigating chitinases produced by S. maltophilia, chitinolytic activity appears to be due to a multienzyme system (73). However, this particular strain, C3, is physiologically distinct from S. maltophilia type strains, including strain 34S1; strain C3 produces multiple isoforms of chitinase and also expresses β-1,3-glucanase activity (72, 73). In our own investigations, strain C3 is taxonomically distinct from type strains of S. maltophilia and likely is positioned in a taxon separate from Stenotrophomonas (21; D. Y. Kobayashi, unpublished data).
Biological control of summer patch and rhizosphere populations by S. maltophilia strains 34S1 and C5.
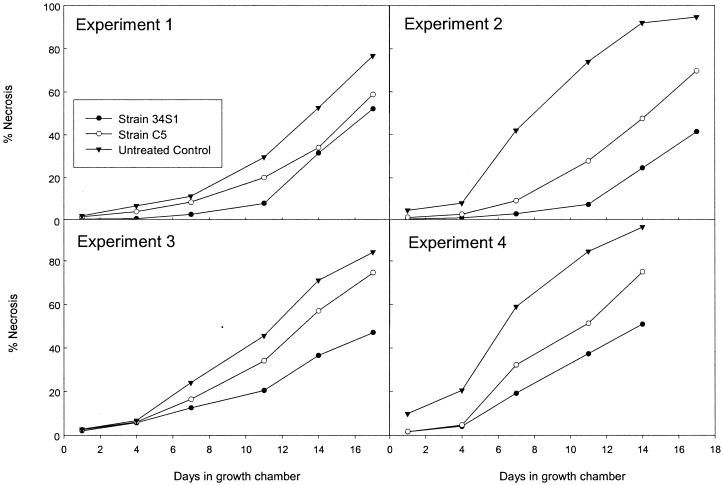
Comparison of the chiA mutant strain C5 with the wild-type strain 34S1 in in vitro antagonism assays using the fungus M. poae resulted in no obvious differences in growth inhibition of the fungus. In contrast, differences were observed between disease progress curves of summer patch on Kentucky bluegrass plants treated with mutant strain C5 compared with wild-type strain 34S1 over 14- to 17-day periods in five different experiments. Actual disease severity ratings are presented in Fig. 4 (for convenience, only four of the five experiments are presented). In all experiments, disease severity over time in plants treated with mutant strain C5 was less than in untreated control plants, but greater than in plants treated with strain 34S1. This intermediate response suggested that biocontrol activity was reduced but not completely lost in mutant strain C5.
FIG. 4.
Progression of summer patch disease severity on Kentucky bluegrass plants treated with S. maltophilia strain 34S1 and mutant strain C5. Progression of disease severity was plotted as average values estimated for percentage of necrosis within a container of plants over time. Four of five experiments are presented; statistical analysis conducted on the linearized data from all five experiments is presented in Table 2.
The increase in disease severity, depicted as percent necrosis over time, followed a nonlinear relationship (Fig. 4). A logit transformation on disease severity values for all five experiments combined linearized the data, and simple linear regression equations were calculated. These transformed data fit a linear relationship very well (r2 = 0.75 to 0.97 for all replications), and no interactions were found between experiment and treatment for any of the factors examined (disease severity, slope, or y intercept). Regressions resulting from the combined results of the five experiments confirmed that disease severity of plants treated with strain C3 was at an intermediate level between untreated control plants and plants treated with strain 34S1. Disease increased (as indicated by the slope of the regression) at a greater rate in untreated control plants compared to plants treated with the wild-type strain 34S1 or mutant strain C5; rates of disease increase for the latter two treatments were not significantly different (Table 2).
TABLE 2.
Results of linear regressions of summer patch disease on Kentucky bluegrass in greenhouse studiesa
| Treatment | Slope | y intercept | Disease severity |
|---|---|---|---|
| Strain 34S1 | 0.36∗ | −5.14∗ | −1.78∗ |
| Strain C5 | 0.41∗ | −4.63† | −0.86† |
| None (control) | 0.61† | −4.26† | 1.26‡ |
Slope was averaged over the five experiments and describes the slope of the logit-transformed line. The y intercept was averaged over the five experiments and describes the logit-transformed line. Disease severity was calculated from the regression equation and represents logit-transformed disease severity after 10 days. Similar symbols within columns represent no significant difference.
In contrast to the slope of the regression, the y intercept of the disease progress curve for plants treated with strain 34S1 was significantly less than that for plants treated with mutant strain C5 or the untreated control plants (Table 2). This observation indicated that onset of disease symptom production was significantly delayed in plants treated with strain 34S1 compared with strain C5 and untreated control plants. Since the rate of disease progression (slope) was not significantly different in plants treated with either strain, the difference in initial disease onset was the major factor contributing to differences in disease control. Similar results in disease progress curves were observed in previous studies when plants were treated with different concentrations of strain 34S1; rates of disease progress were not significantly different, but plants treated with higher concentrations of strain 34S1 were delayed in disease onset compared with plants treated with lower concentrations (30).
Disease severity at day 10 estimated from the regression equations showed significant differences among all three treatments, with the greatest disease severity occurring in untreated control plants, an intermediate level in plants treated with strain C5, and the lowest level of disease severity in plants treated with strain 34S1 (Table 2).
In contrast to disease suppression, mutant strain C5 did not appear to be drastically affected in ability to colonize the rhizosphere of Kentucky bluegrass compared with strain 34S1 during sampling times known to influence biocontrol activity (30). After both strains 34S1 and C5 initially established at 108 CFU/g (dry weight) of rhizosphere sample, populations steadily decreased to similar populations below 106 CFU/g of sample on five observation dates over a 2-week period beginning at the time of application. No significant differences were detected between strains C5 and 34S1 on any of the five observation dates during this period in three separate experiments (data not shown).
Chitinolytic activity has been implicated to play a role in the biocontrol activity of several biocontrol bacteria, including S. maltophilia strains. However, direct molecular evidence for its role is lacking for many of these biocontrol systems. Mutation of chiA in S. maltophilia strain 34S1 abolishes chitinolytic activity and affects the ability of the strain to suppress summer patch disease, providing direct evidence for the role of this enzyme in biocontrol activity. Populations of mutant strain C5 were not significantly impaired in the turfgrass rhizosphere compared with the wild-type strain 34S1, providing further support that the enzyme functions directly in fungal antagonism. Because of plasmid instability problems, we were unable to evaluate biocontrol activity to mutants restored with chitinase. As a result, the possibility that the mutation had an indirect or polar effect(s) on other traits contributing to biocontrol cannot be excluded. The latter is less likely, since molecular analysis of chiA suggests that it is transcribed monocistronically.
Combining evidence that chitinolytic activity functions in direct fungal antagonism with the observation that rate of disease progression was not affected in C5-treated plants compared with 34S1-treated plants, but also that disease onset was significantly delayed for the latter, it seems likely that 34S1 is better capable of reducing initial pathogen colonization of turfgrass plants. However, once initial infection occurs, disease progresses at similar rates.
Mutant strain C5 was not completely reduced in biocontrol activity, and thus it is evident that other biocontrol mechanisms are operating in this system. Other traits produced by S. maltophilia strain 34S1 could be contributing to antifungal antagonism in this system, including additional enzyme activities and antibiotic-like compounds. Stenotrophomonas spp. are known to produce several different types of extracellular enzymes and antibiotics that likely contribute to biocontrol activity among strains within this bacterial group. For example, Dunne et al. (10) found that the proteolytic activity of S. maltophilia contributes to biocontrol activity of sugar beet damping-off caused by Pythium ultimum. Nakayama et al. (43) characterized a new group of antibiotic-like compounds, xanthobactins, produced by Stenotrophomonas sp. strain SB-K88. In this study, purified xanthobactin A directly suppressed Pythium damping-off of sugar beet. As demonstrated with Trichoderma harzianum (54), it is possible that a synergistic, antifungal response results from the combined effect of chitinase with antibiotics produced by S. maltophilia.
Acknowledgments
This project was funded in part by the Rutgers University Center for Turfgrass Research and the New Jersey Agricultural Research Station.
We thank M. Holtman, N. El-Barrad, and G. MacDonald for technical assistance and Partec Peat Corp. for the generous donation of golf course top-dressing to conduct growth chamber assays. Peptide sequencing was conducted by Steve Smith at the UTMB protein chemistry laboratory, Galveston, Tex.
REFERENCES
- 1.Bork, P., and R. F. Doolittle. 1992. Proposed acquisition of an animal protein domain by bacteria. Proc. Natl. Acad. Sci. USA 89:8990-8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brun, E., F. Moriaud, P. Gans, M. J. Blackledge, F. Barras, and D. Marion. 1997. Solution structure of the cellulose-binding domain of the endoglucanase Z secreted by Erwinia chrysanthemi. Biochemistry 36:16074-16086. [DOI] [PubMed] [Google Scholar]
- 3.Bycroft, M., A. Bateman, J. Clarke, S. J. Hamill, R. Sandford, R. L. Thomas and C. Chothia. 1999. The structure of a PKD domain from polycystin-1: implications for polycystic kidney disease. EMBO J. 18:297-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chernin, L. S., L. De La Fuente, V. Sobolev, S. Haran, C. E. Vorgias, A. B. Oppenheim, and I. Chet. 1997. Molecular cloning, structural analysis, and expression in Escherichia coli of a chitinase gene from Enterobacter agglomerans. Appl. Environ. Microbiol. 63:834-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chernin, L., Z. Ismailov, S. Haran, and I. Chet. 1995. Chitinolytic Enterobacter agglomerans antagonistic to fungal plant pathogens. Appl. Environ. Microbiol. 61:1720-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connell, T. D., D. J. Metzger, J. Lynch, and J. P. Folster. 1998. Endochitinase is transported to the extracellular milieu by the eps-encoded general secretory pathway of Vibrio cholerae. J. Bacteriol. 180:5591-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Debette, J., and R. Blondeau. 1980. Présence de Pseudomonas maltophilia dans la rhizosphère de quelques plantes cultivées. Can. J. Microbiol. 26:460-463. [PubMed] [Google Scholar]
- 8.De Bruijn, F. J., and J. R. Lupski. 1984. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids—a review. Gene 27:131-149. [DOI] [PubMed] [Google Scholar]
- 9.Ditta, G., T., Schmidhauser, E. Yakobson, P. Lu, X.-W. Liang, D. R. Finlay, D. Guiney, and D. R. Helinski. 1985. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid 13:149-153. [DOI] [PubMed] [Google Scholar]
- 10.Dunne, C., J. J. Crowley, Y. Monne-Loccoz, D. N. Dowling, F. J. de Bruijn, and F. O'Gara. 1997. Biological control of Pythium ultimum by Stenotrophomonas maltophilia W81 is mediated by an extracellular proteolytic activity. Microbiology 143:3921-3931. [DOI] [PubMed] [Google Scholar]
- 11.Elad, Y., I. Chet, and R. Baker. 1987. Increased growth response of plants induced by rhizobacteria antagonistic to soilborne pathogenic fungi. Plant Soil 98:325-330. [Google Scholar]
- 12.Elliott Juhnke, M., D. E. Mathre, and D. C. Sands. 1987. Identification and characterization of rhizosphere-competent bacteria of wheat. Appl. Environ. Microbiol. 53:2793-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fridlender, M., J. Inbar, and I. Chet. 1993. Biological control of soilborne plant pathogens by a β-1,3 glucanase-producing Pseudomonas cepacia. Soil Biol. Biochem. 25:1211-1221. [Google Scholar]
- 14.Fujii, T., and K. Miyashita. 1993. Multiple domain structure in a chitinase gene (chiC) of Streptomyces lividans. J. Gen. Microbiol. 139:677-686. [DOI] [PubMed] [Google Scholar]
- 15.Giesler, L. J., and G. Y. Yuen. 1998. Evaluation of Stenotrophomonas maltophilia strain C3 for biocontrol of brown patch disease. Crop Protection 17:509-513. [Google Scholar]
- 16.Gilkes, N. R., B. Henrissat, D. G. Kilburn, R. C. Miller, Jr., and R. A. J. Warren. 1991. Domains in microbial glycanases: sequence conservation, function, and enzyme families. Microbiol. Rev. 55:303-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gleave, A. P., R. K. Taylor, R. A. M. Morris, and D. R. Greenwood. 1995. Cloning and sequencing of a gene encoding the 69-kDa extracellular chitinase of Janthinobacterium lividum. FEMS Microbiol. Let. 131:279-288. [DOI] [PubMed] [Google Scholar]
- 18.Hashimoto, M., T. Ikegami, S. Seino, N. Ohuchi, H. Fukada, J. Sugiyama, M. Shirakawa, and T. Watanabe. 2000. Expression and characterization of the chitin-binding domain of chitinase A1 from Bacillus circulans WL-12. J. Bacteriol. 182:3045-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, D. M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henirissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holtman, M. A. 1998. Ph.D. thesis. Rutgers, The State University of New Jersey, New Brunswick, N.J.
- 22.Ikegami, T., T. Okada, M. Hashimoto, S. Seino, T. Watanabe, and M. Shirakawa. 2000. Solution structure of the chitin-binding domain of Bacillus circulans WL-12 chitinase A1. J. Biol. Chem. 275:13654-13661. [DOI] [PubMed] [Google Scholar]
- 23.Inbar, J., and I. Chet. 1991. Evidence that chitinase produced by Aeromonas caviae is involved in the biological control of soil-borne plant pathogens by this bacterium. Soil Biol. Biochem. 23:973-978. [Google Scholar]
- 24.Irwin, D. C., S. Zhang, and D. B. Wilson. 2000. Cloning, expression and characterization of a family 48 exocellulase, Ce148A, from Thermobifida fusca. Eur. J. Biochem. 267:4988-4997. [DOI] [PubMed] [Google Scholar]
- 25.Jakobi, M., and G. Winkelmann. 1996. Maltophilin: a new antifungal compound produced by Stenotrophomonas maltophilia R3089. J. Antibiot. 49:1101-1104. [DOI] [PubMed] [Google Scholar]
- 26.Jones, J. D. G., K. L. Grady, T. V. Suslow, and J. R. Bedbrook. 1986. Isolation and characterization of genes encoding two chitinase enzymes from Serratia marcescens. EMBO J. 5:467-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keen, N., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 28.Keyhani, N. O., and S. Roseman. 1996. The chitin catabolic cascade in the marine bacterium Vibrio furnissii. Molecular cloning, isolation, and characterization of a periplasmic chitodextrinase. J. Biol. Chem. 271:33414-33424. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi, D. Y., and N. El-Barrad. 1996. Selection of bacterial antagonists using enrichment cultures for the control of summer patch disease of Kentucky bluegrass. Curr. Microbiol. 32:106-110 [Google Scholar]
- 30.Kobayashi, D. Y., M. Guglielmoni, and B. B. Clarke. 1995. Isolation of the chitinolytic bacteria Xanthomonas maltophilia and Serratia marcescens as biological control agents for summer patch disease of turfgrass. Soil Biol. Biochem. 27:1479-1487. [Google Scholar]
- 31.Kwok, O. C. H., P. C. Fahy, H. A. J. Hoitink, and G. A. Kuter. 1987. Interactions between bacteria and Trichoderma hamatum in suppression of Rhizoctonia damping-off in bark compost media. Phytopathology 77:1206-1212. [Google Scholar]
- 32.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 33.Lambert, B., F. Leyns, L. Van Rooyen, F. Gosselé, Y. Papon, and J. Swings. 1987. Rhizobacteria of maize and their antifungal activities. Appl. Environ. Microbiol. 53:1866-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambert, B., P. Meire, H. Joos, P. Lens, and J. Swings. 1990. Fast-growing, heterotrophic bacteria from the rhizosphere of young sugar beet plants. Appl. Environ. Microbiol. 56:3375-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim, H.-S., Y.-S. Kim, and S.-D Kim. 1991. Pseudomonas stutzeri YPL-1 genetic transformation and antifungal mechanism against Fusarium solani, an agent of plant root rot. Appl. Environ. Microbiol. 57:510-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lingappa, Y., and J. L. Lockwood. 1961. Chitin media for selective isolation and culture of actinomycetes. Phytopathology 52:317-323. [Google Scholar]
- 37.Lonhienne, T., K. Mavromatis, C. E. Vorgias, L. Buchon, C. Gerday and V. Bouriotis. 2001. Cloning, sequences, and characterization of two chitinase genes from the antarctic Arthrobacter sp. strain TAD20: isolation and partial characterization of the enzymes. J. Bacteriol. 183:1773-1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mavingui, P., and T. Heulin. 1994. In vitro chitinase and antifungal activity of a soil, rhizosphere and rhizoplane population of Bacillus polymyxa. Soil Biol. Biochem. 26:801-803. [Google Scholar]
- 39.Mazzola, M., P. W. Stahlman, and J. E. Leach. 1995. Application method affects the distribution and efficacy of rhizobacteria suppressive of downy brome (Bromus tectorum). Soil Biol. Biochem. 27:1271-2178. [Google Scholar]
- 40.Meinke, A., N. R. Gilkes, D. G. Kilburn, R. C. Miller Jr., and R. A. Warren. 1993. Cellulose-binding polypeptides from Cellulomonas fimi: endoglucanase D (CenD), a family Aβ-1,4-glucanase. J. Bacteriol. 175:1910-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitchell, R., and E. Hurwitz. 1964. Suppression of Pythium debaryanum by lytic rhizosphere bacteria. Phytopathology 55:156-158. [Google Scholar]
- 42.Morimoto, K., S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 1997. Cloning, sequencing and expression of the gene encoding Clostridium paraputrificum chitinase ChiB and analysis of the functions of novel cadherin-like domains and a chitin binding domain. J. Bacteriol. 179:7306-7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakayama, T., Y. Homma, Y. Hashidoko, J. Mizutani, and S. Tahara. 1999. Possible role of xanthobaccins produced by Stenotrophomonas sp. strain SB-K88 in suppression of sugar beet damping-off disease. Appl. Environ. Microbiol. 65:4334-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Brien, M., and G. H. G. Davis. 1982. Enzymatic profile of Pseudomonas maltophilia. J. Clin. Microbiol. 16:417-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ordentlich, A., Y. Elad, and I. Chet. 1988. The role of chitinase of Serratia marcescens in biocontrol of Sclerotium rolfsii. Phytopathology 78:84-88. [Google Scholar]
- 46.Ponting, C. P., L. Aravind, J. Schultz, P. Bork, and E. V. Koonin. 1999. Eukaryotic signaling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J. Mol. Biol. 289:729-745. [DOI] [PubMed] [Google Scholar]
- 47.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Redenbach, M., H. M. Kieser, D. Denapaite, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77-96. [DOI] [PubMed] [Google Scholar]
- 49.Robbins, P. W., K. Overbey, C. Albright, B. Benfield, and J. Pero. 1992. Cloning and high-level expression of chitinase-encoding gene of Streptomyces plicatus. Gene 111:69-76. [DOI] [PubMed] [Google Scholar]
- 50.Roberts, W. K., and C. P. Selitrennikoff. 1988. Plant and bacterial chitinases differ in antifungal activity. J. Gen. Microbiol. 134:169-176. [Google Scholar]
- 51.Ruvkun, G. B., and F. M. Ausubel. 1981. A general method for site-directed mutagenesis in prokaryotes. Nature 289:85-88. [DOI] [PubMed] [Google Scholar]
- 52.Sakka, K., R. Kusaka, A. Kawano, S. Karita, J. Sukhumavasi, T. Kimura, and K. Ohmiya. 1998. Cloning and sequencing of the gene encoding chitinase ChiA from Xanthomonas sp. strain AK and some properties of ChiA. J. Ferment. Bioeng. 86:527-533. [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.Schirmböck, M., M. Lorito, Y.-L. Wang, C. K. Hayes, I. Arisan-Atac, F. Scala, G. E. Harman, and C. P. Kubicek. 1994. Parallel formation and synergism of hydrolytic enzymes and peptaibol antibiotics, molecular mechanisms involved in the antagonistic action of Trichoderma harzianum against phytopathogenic fungi. Appl. Environ. Microbiol. 60:4364-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schultz, J., R. R. Copley, T. Doerks, C. P. Ponting, and P. Bork. 2000. SMART: a Web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28:231-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen, H., N. R. Gilkes, D. G. Kilburn, R. C. Miller, Jr., and R. A. Warren. 1995. Cellobiohydrolase B, a second exo-cellobiohydrolase from the cellulolytic bacterium Cellulomonas fimi. Biochem. J. 311:67-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sitrit, Y., C. E. Vorgias, I. Chet, and A. B. Oppenheim. 1995. Cloning and primary structure of the chiA gene from Aeromonas caviae. J. Bacteriol. 177:4187-4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1986. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Svitil, A. L., and D. L. Kirchman. 1998. A chitin-binding domain in a marine bacterial chitinase and other microbial chitinases: implications for the ecology and evolution of 1,4-β-glycanases. Microbiology 144:1299-1308. [DOI] [PubMed] [Google Scholar]
- 61.Tanaka, T. S. Fujiwara, S. Nishikori, T. Fukui, M. Takagi, and T. Imanaka. 1999. A unique chitinase with dual active sites and triple substrate binding sites from the hyperthermophilic archaeon Pyrococcus kodadaraensis KOD1. Appl. Environ. Microbiol. 65:5338-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Techkarnjanaruk, S., S. Pongpattanakitshote, and A. E. Goodman. 1997. Use of a promoterless lacZ gene insertion to investigate chitinase gene expression in the marine bacterium Pseudoalteromonas sp. strain S9. Appl. Environ. Microbiol. 63:2989-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tomme, P., R. A. J. Warren, and N. R. Gilkes. 1995. Cellulose hydrolysis by bacteria and fungi. Adv. Microb. Physiol. 37:1-81. [DOI] [PubMed] [Google Scholar]
- 64.Tsujibo, T., H. Endo, K. Minoura, K. Miyamoto, and Y. Inamori. 1993. Cloning and sequence analysis of the gene encoding a thermostable chitinase from Streptomyces thermoviolaceus OPC-520. Gene 134:113-117. [DOI] [PubMed] [Google Scholar]
- 65.Tsujibo, H., H. Orikoshi, D. Shiotani, M. Hayashi, J. Umeda, K. Miyamoto, C. Imada, Y. Okami, and Y. Inamori. 1998. Characterization of Chitinase C from a marine bacterium,Altermonas sp. strain O-7, and its corresponding gene and domain structure. Appl. Environ. Microbiol. 64:472-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Warren, R. A. J. 1996. Microbial hydrolysis of polysaccharides. Annu. Rev. Microbiol. 50:183-212. [DOI] [PubMed] [Google Scholar]
- 67.Watanabe, T., Y. Ito, T. Yamada, M. Hashimoto, S. Sekine, and H. Tanaka. 1994. The roles of C-terminal domain and type III domains of chitinase A1 from Bacillus circulans WL-12 in chitin degradation. J. Bacteriol. 176:4465-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watanabe T., K. Kobori, K. Miyashita, T. Fujii, H. Sakai, M. Uchida, and K. Tanaka. 1993. Identification of glutamic acid 204 and aspartic acid 200 in chitinase A1 of Bacillus circulans WL-12 as essential residues for chitinase activity. J. Biol. Chem. 268:18567-18572. [PubMed] [Google Scholar]
- 69.Watanabe, T., K. Suzuki, W. Oyanagi, K. Ohnishi, and H. Tanaka. 1990. Gene cloning of chitinase A1 from Bacillus circulans WL-12 revealed its evolutionary relationship to Serratia chitinase and to the type III homology units of fibronectin. J. Biol. Chem. 265:15659-15665. [PubMed] [Google Scholar]
- 70.Weller, D. 1988. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu. Rev. Phytopathol. 26:379-407. [Google Scholar]
- 71.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, K. W. Minton, R. D. Fleischmann, K. A. Ketchum, K. E. Nelson, S. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, Z., and G. Y. Yuen. 2000. The role of chitinase production by Stenotrophomonas maltophilia strain C3 in biological control of Bipolaris sorokiniana. Phytopathology 90:384-389. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, Z., and G. Y. Yuen. 2001. Chitinases from the plant disease biocontrol agent, Stenotrophomonas maltophilia C3. Phytopathology 91:204-211. [DOI] [PubMed] [Google Scholar]