Abstract
Background
Interleukin (IL)-13 is a central mediator of airway inflammation in asthma, and gene polymorphisms may play a role in asthma pathogensis. However, the relationship between IL-13 gene polymorphisms and asthma susceptibility remain unclear. Therefore, we conducted this meta-analysis to evaluate this relationship and better understand the risk of asthma in Chinese children.
Methods and Results
PubMed, cnki.net, China Wanfang, and Embase were searched to identify relevant case–control studies in Chinese children published until July 2021. In total, 217 Chinese and English articles were searched; 23 articles were included (19 Chinese and 4 English). Compared with genotype CC and allele C, genotypes TT, TC, and TT + TC at the rs1800925 (C-1112 T) loci and allele T, respectively, were not associated with the risk of bronchial asthma. At the rs1295686 (C + 1923 T) loci, compared with genotype CC, there were significant associations between genotypes TT, TC, and TT + TC and the risk of bronchial asthma. Compared with allele C, allele T increased the risk of bronchial asthma. At the rs20541 (G + 2044A) loci, compared with genotype GG, the genotypes AA and AA + GA were associated with the risk of bronchial asthma, but the genotype GA was not. Compared with allele G, allele A increased the risk of bronchial asthma.
Conclusions
rs1295686 (C + 1923 T) and rs20541 (G + 2044A) gene polymorphisms are associated with asthma susceptibility, whereas rs1800925 (C-1112 T) gene polymorphisms are not.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12887-025-05963-4.
Keywords: Bronchial asthma, Children, Gene polymorphism, Interleukin-13, Meta-analysis
Introduction
Bronchial asthma, referred to as asthma, is a polygenic genetic disease affected by both genetic and environmental factors. With increasing research on the molecular genetic epidemiology of asthma, asthma susceptibility genes have become the focus of research worldwide. The relationship between genetic polymorphisms of some inflammatory cytokines (such as interleukin (IL)−4 and IL-13) and the risk of asthma has recently attracted significant attention. IL-13 is an important inflammatory cytokine produced following T-cell activation. Some studies have suggested that IL-13 is a central inflammatory mediator of airway inflammation in asthma and that gene polymorphism plays an important role in asthma pathogenesis. There are multiple single nucleotide polymorphisms in the IL-13 gene region, which in the Chinese population, are mainly concentrated in rs1800925 (C-1112 T), rs1295686 (C + 1923 T), and rs20541 (G + 2044 A). However, the relationship between asthma and rs1800925 (C-1112 T), rs1295686 (C + 1923 T), and rs20541 (G + 2044 A) gene polymorphisms remains controversial. We conducted a meta-analysis of case–control studies on the relationship between genetic polymorphisms in rs1800925 (C-1112 T), rs1295686 (C + 1923 T), and rs20541 (G + 2044 A) and susceptibility to bronchial asthma in Chinese children and subsequently evaluated the relationship between the above-mentioned three types of SNPs and the risk of asthma.
Materials and methods
Literature retrieval
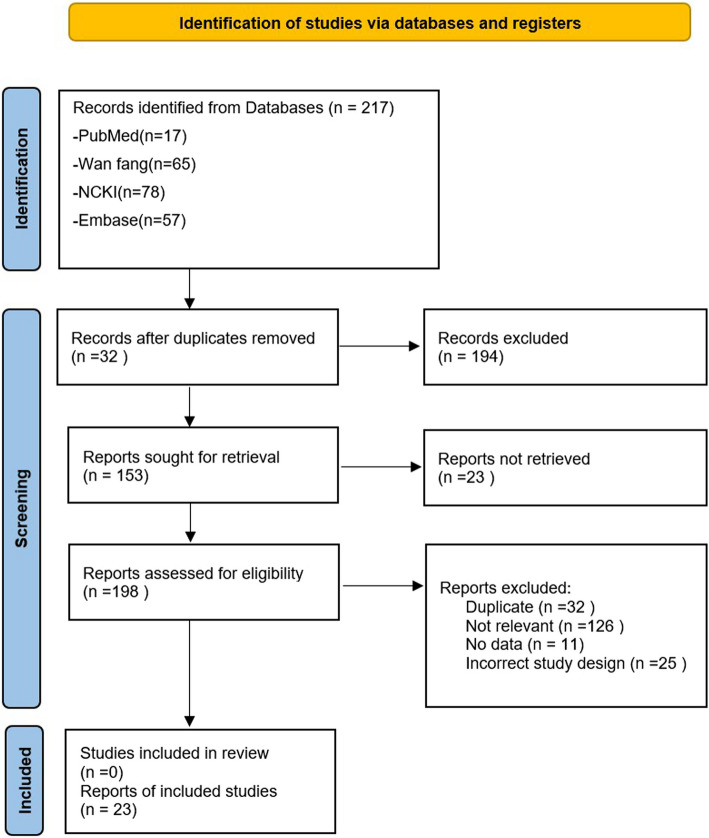
Literature was retrieved from the Wanfang sci-tech periodical full-text database, cnki.net database, PubMed, and Embase database network resources (Fig. 1). The retrieval time was from the establishment of the database until July 2021. Databases were searched using the following keywords: IL-13 gene, interleukin-13 gene polymorphism, asthma and child.
Fig. 1.
PRISMA diagram. Abbreviations: PRISMA, Preferred Reporting Items for Systematic Reviews and Meta analysis; SLR, systematic literature review
Data inclusion and exclusion criteria
The data inclusion criteria were as follows: (1) studies on the relationship between genetic polymorphisms of rs1800925 (C-1112 T), rs1295686 (C + 1923 T), and rs20541 (G + 2044 A) loci and susceptibility to asthma, (2) case–control studies in which the subjects were Chinese children with asthma and were compared to a control group of unrelated healthy children, (3) the study participants based on the GINA (Global Initiative for Asthma) guidelines for asthma diagnosis and age cutoff < 18 years, (4) articles published in Chinese or English, (5) the genotype frequency of the control group was in accordance with the Hardy–Weinberg (H–W) genetic equilibrium, and (6) data were complete with a corresponding genotype frequency distribution or the required data to calculate the distribution. The exclusion criteria were as follows: (1) studies published in languages other than Chinese or English, (2) the genotypes in the control group did not conform to the laws of H–W genetic equilibrium, (3) case reports, reviews, or summaries of a meeting, conference abstracts, meeting minutes, preprints, (4) the quality evaluation was less than three points, (5) the description of the detection methods and means was not clear, and (6) information was lacking or the data were incomplete.
Data extraction
Literature screening and data extraction were performed independently by the two authors according to the set retrieval strategy and inclusion criteria. Any conflicts were judged by a third author to reach a consensus. The contents of data extraction included the author, publication year, description of asthma group and control group, population area, frequency of each genotype, and the sample numbers the of asthma and control groups.
Literature quality evaluation
The methodological quality evaluation included in the study was based on the STREGA criteria, including: (1) whether the sample size was sufficient, (2) whether the diagnostic criteria were clearly explained, (3) matching of the groups, (4) whether there was comparability between the control and case groups, and whether the genotypic distribution in the control group conformed to the law of H–W genetic equilibrium, (5) whether the gene detection method was reasonable, and (6) whether the data were sufficient. Using the scoring criteria listed above, each study was scored and given one point for each criterion that was met. A total score greater than or equal to three points indicated reliable quality.
Statistical processing
Statistical analysis was carried out using Revman5.3 and Stata14 software of the Cochrane collaboration network, and the effect indices of genotype and gene frequency at each gene locus were calculated, which were expressed by the OR value and 95% confidence interval (CI). The genetic models calculated for rs1800925 (C-1112 T) and rs1295686 (C + 1923 T) included the TT vs CC, CT vs CC, and TT + CT vs CC, and gene frequency included T ratio C. The genetic models calculated for rs20541 (G + 2044 A) included the AA vs GG, GA vs GG, and AA + GA vs GG, and gene frequency included A vs G. Between-study heterogeneity was evaluated using the Q-test (significance threshold p < 0.1) and I2 statistic (I2 > 50% indicates moderate-to-high heterogeneity). If the results are inconsistent (e.g., Q-test p = 0.09 with I2 = 45%), prioritize the I2 value to determine the degree of heterogeneity: When I2 ≤ 50%: Accept the heterogeneity level as tolerable. Even if the Q-test p-value is close to 0.1 (e.g., 0.05 < p < 0.1), a fixed-effect model is applied. When I2 > 50%: Regardless of Q-test significance (e.g., p > 0.1 but I2 = 55%), a random-effects model is used to provide a conservative estimate of effect sizes.. According to the data of each study, the studies with the largest weight reduction were used for the sensitivity analysis. The symmetry of the funnel chart was used to observe the publication bias, and a p value of 0.05 was considered significant. The Egger regression method was used to quantitatively detect publication bias. Stata14.0 was used to analyze publication bias. When there was publication bias, the trim and fill method in Stata14.0 was used to evaluate the bias and study stability.
Results
Characteristics of the included studies
A total of 217 Chinese and English articles were retrieved based on the proposed keywords. By reviewing the topics, abstracts and keywords, reviews, review abstracts, non-case–control studies, and studies with incomplete data, we excluded data that could not be extracted or did not conform to the H–W equilibrium law. A total of 23 articles that met the inclusion criteria were retrieved [1–23], of which 19 were published in Chinese and four in English. Five articles investigated rs1295686 (C + 1923 T), 12 articles discussed the correlation between rs20541 (G + 2044 A) polymorphism and asthma, two articles performed rs1800925 (C-1112 T) genotype analysis, two articles described the relationship between rs1800925 (C-1112 T) and rs20541 (G + 2044 A) genotypes and asthma, and two articles described the correlation between rs1295686 (C + 1923 T) and rs20541 (G + 2044 A) genotypes and asthma.
Characteristics of the included literature
Data characteristics of the literature included in the analysis are shown in Table 1. In all studies, the subjects of the included literature were Chinese children, asthma diagnosis was performed in the GINA (Global Initiative for Asthma) guidelines, the methodology was clear and reliable, and the results were clearly defined.
Table 1.
Characteristics of references cited in this meta-analysis
| First Author | Year | Genoty ping method | asthmas/controls | genetic locus | 1112(asthmas/control) | 1923(asthmas/controls) | 2044(asthmas/cont trols) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | TC | CC | TT | TC | CC | AA | GA | GG | |||||
| Ding | 2016 | PCR–RFLP | 90/82 | + 2044 | - | - | - | - | - | - | 9/4 | 47/22 | 34/56 |
| Liao | 2014 | PCR–RFLP | 300/200 | −1112 | 39/24 | 146/70 | 115/106 | - | - | - | - | - | - |
| Cao | 2012 | PCR–RFLP | 122/144 | + 2044 | - | - | - | - | - | - | 30/60 | 69/67 | 23/17 |
| Zhou | 2016 | DNA Direct PCR-sequen cing method | 38/30 | + 2044 | - | - | - | - | - | - | 20/14 | 13/12 | 5/4 |
| Ma | 2021 | PCR | 35/35 | + 2044 | - | - | - | - | - | - | 21/4 | 11/14 | 8/10 |
| Feng | 2009 | PCR、CELIF | 45/43 | + 2044 | - | - | - | - | - | - | 10/3 | 18/10 | 17/30 |
| Jia | 2013 | PCR–RFLP | 77/50 | + 1923 | - | - | - | 13/3 | 42/22 | 22/25 | - | - | - |
| Zhang | 2016 | Sequen om MassARRAY®SNP | 153/103 | + 2044 | - | - | - | - | - | - | 20/6 | 75/41 | 58/55 |
| Sun | 2003 | PCR–RFLP | 96/53 | + 1923 | - | - | - | 12/0 | 43/14 | 41/39 | - | - | - |
| Xi | 2003 | PCR–RFLP | 43/31 | + 2044 | - | - | - | - | - | - | 10/16 | 25/13 | 8/2 |
| Pu | 2013 | PCR–RFLP | 96/96 | + 1923 | - | - | - | 12/5 | 45/24 | 39/67 | - | - | - |
| Wang | 2016 | PCR–RFLP | 173/56 | + 1923 | - | - | - | 77/9 | 58/23 | 38/24 | - | - | - |
| Wang | 2014 | SNaPshot | 435/601 | + 1923 | - | - | - | 62/64 | 191/246 | 182/291 | - | - | - |
| Yan | 2015 | PCR–RFLP | 34/30 | + 2044 | - | - | - | - | - | - | 10/1 | 1/7 | 23/22 |
| Yang | 2010 | PCR–RFLP | 178/158 | + 2044 | - | - | - | - | - | - | 47/19 | 60/66 | 71/73 |
| Zhao | 2005 | PCR–RFLP | 130/100 | + 2044 | - | - | - | - | - | - | 52/50 | 60/42 | 18/8 |
| Guo | 2017 | PCR–RFLP | 80/112 | −1112 + 2044 | 3/5 | 31/50 | 46/57 | - | - | - | 15/15 | 47/46 | 18/51 |
| Deng | 2015 | PCR–RFLP | 250/200 | −1112 | 40/24 | 124/70 | 86/106 | - | - | - | - | - | - |
| Wang | 2004 | PCR–RFLP | 45/52 | + 2044 | - | - | - | - | - | - | 17/20 | 17/18 | 11/14 |
| Chan | 2006 | PCR–RFLP | 238/126 | + 2044 | - | - | - | - | - | - | 42/15 | 117/57 | 79/54 |
| Liu | 2013 | PCR–RFLP | 384/384 | + 1923 + 2044 | - | - | - | 46/36 | 164/169 | 174/179 | 50/21 | 154/164 | 180/199 |
| Wang | 2009 | PCR–RFLP | 446/505 | −1112 + 2044 | 12/18 | 113/136 | 321/357 | - | - | - | 49/59 | 194/234 | 203/212 |
| Wu | 2010 | PCR–RFLP | 252/227 | + 1923 + 2044 | - | - | - | 32/16 | 114/85 | 106/126 | 36/18 | 111/84 | 105/125 |
Genotype frequency analysis of the rs1800925 (C-1112 T) locus
Meta-analysis was performed using a fixed-effects model for all genotypes. For genotypes TT/CC, TC/CC, TC + TT/CC, and T/C, the results of the heterogeneity test were I2 = 42%, p = 0.16 (Fig. 2a), I2 = 84% (p < 0.05) (Fig. 2b), I2 = 85% (p < 0.05) (Fig. 2c), and I2 = 79%, (p < 0.05) (Fig. 2d), respectively. The results of meta-analysis showed that in rs1800925 (C-1112 T) loci, genotypes TT and CC: OR = 1.38, 95% CI: 0.98–1.94, P = 0.06, TC and CC: OR = 1.33, 95% CI: 0.81–2.19, P = 0.26; TT + TC and CC: OR = 1.30, 0.80, 2.12, P = 0.29; allele T/C: OR = 1.17, 95% CI: 0.84–1.63, p = 0.35, respectively. This analysis showed no significance in any of the genetic models, indicating no significant correlation between genotypes TT, TC, TC + TT, and the risk of bronchial asthma. However, compared with allele C, allele T was not significantly correlated with the risk of asthma.
Fig. 2.
a Forest map of the association between rs1800925 (C-1112 T) TT/CC gene polymorphisms and asthma. b Forest map of the association between rs1800925 (C-1112 T) TC/CC gene polymorphisms and asthma. c Forest map of the association between rs1800925 (C-1112 T) TT + TC/CC gene polymorphisms and asthma. d Forest map of the association between rs1800925 (C-1112 T) T/C gene polymorphisms and asthma
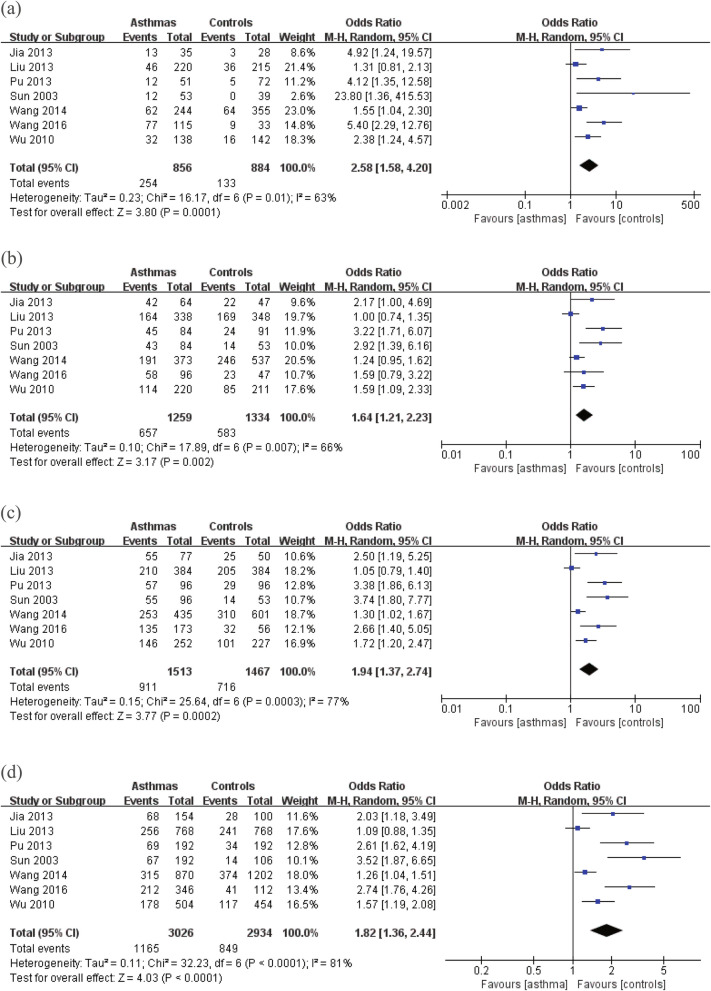
Genotype frequency analysis of the rs1295686 (C + 1923 T) locus
For genotypes TT/CC, TC/CC, TC + TT/CC, and T/C, the heterogeneity test results using the random-effect models were I2 = 63%, p = 0.01 (Fig. 3a), I2 = 66%, p = 0.007 (Fig. 3b), I2 = 77%, p < 0.05 (Fig. 3c), I2 = 81%, p < 0.05, (Fig. 3d), respectively. The results of the meta-analysis were: genotypes TT and CC: OR = 2.58, 95% CI: 1.58–4.20, p < 0.001; genotypes TC and CC: OR = 1.64, 95% CI: 1.21–2.23, p < 0.05; genotypes TT + TC and CC: OR = 1.94%, 95% CI: 1.37–2.74, p < 0.001; allele T/C: OR = 1.82%, 95% CI: 1.36–2.44, p < 0.001. The above results showed that all genetic models were significant. Thus, compared with genotype CC, genotypes TT, TC, and TT + TC were associated with the risk of bronchial asthma, while compared with allele C, individuals with allele T had an increased risk of bronchial asthma.
Fig. 3.
a Forest map of the association between rs1295686 (C + 1923 T) TT/CC gene polymorphisms and asthma. b Forest map of the association between rs1295686 (C + 1923 T) TC/CC gene polymorphisms and asthma. c Forest map of the association between rs1295686 (C + 1923 T) TT + TC/CC gene polymorphisms and asthma. d Forest map of the association between rs1295686 (C + 1923 T) T/C gene polymorphisms and asthma
Genotype frequency analysis of the rs20541 (G + 2044 A) locus
Using the random-effect model, the heterogeneity test results of genotypes AA/GG, GA/GG, AA + GA/GG, and A/G were: I2 = 74%, p < 0.001 (Fig. 4a), I2 = 63%, p < 0.001 (Fig. 4b), I2 = 70%, p < 0.001 (Fig. 4c), and A I2 = 81%, p < 0.001 (Fig. 4d), respectively. The results of meta-analysis were: genotype AA and GG: OR = 1.70, 95% CI: 1.10–2.62, p = 0.02; genotype GA and GG: OR = 1.27, 95% CI: 0.98–1.64, p = 0.07; genotype AA + GA and GG: OR = 1.40, 95% CI: 1.08–1.83, p = 0.01; allele A and G: OR = 1.34, 95% CI: 1.07–1.67, p = 0.01. The results showed that the genotypes AA, AA + GA, and allele A were significantly associated with increased asthma risk, while genotype GA had no association with the risk of asthma.
Fig. 4.

a Forest map of the association between rs20541 (G + 2044 A) AA/GG gene polymorphisms and asthma. b Forest map of the association between rs20541 (G + 2044 A) GA/GG gene polymorphisms and asthma. c Forest map of the association between rs20541 (G + 2044 A) GA + AA/GG gene polymorphisms and asthma. d Forest map of the association between rs20541 (G + 2044 A) A/G gene polymorphisms and asthma
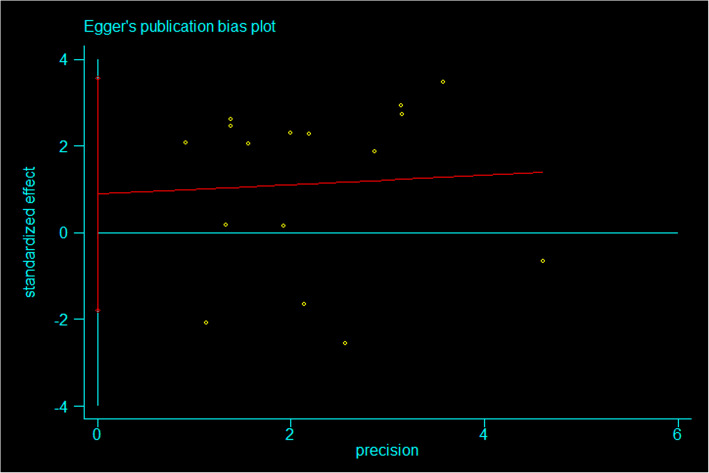
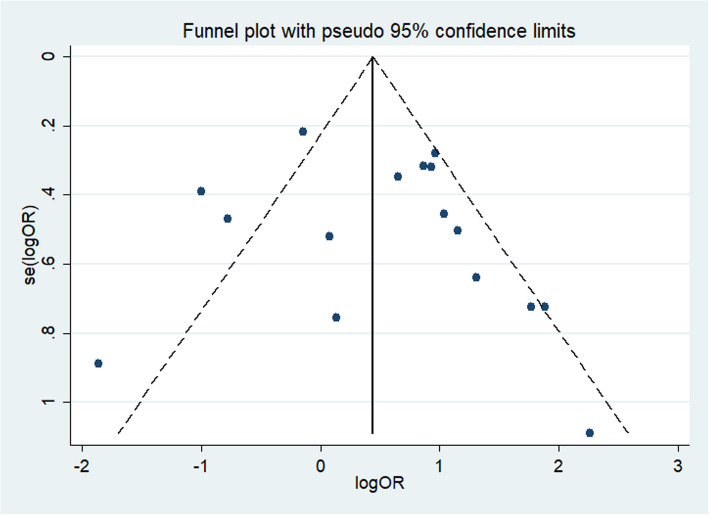
Analysis of publication bias
The funnel shape of the genetic model of each gene locus constructed using stata14.0 software did not show any evident asymmetry (Fig. 5). Using the Egger linear regression method, the included literature was evaluated for publication bias (Fig. 6).The results of Egger's test showed that there was no publication bias in the genetic models of each gene locus (p > 0.05), except for rs1295686 (C + 1923 T) (Table 2).
Fig. 5.
Genotype AA/GG funnel diagram of the rs20541 (G + 2044 A) locus
Fig. 6.
Publication bias detection of genotype AA/GG in the rs20541 (G + 2044 A) locus
Table 2.
Detection of publication bias between gene polymorphisms at various loci and asthma genotypes
| genetic locus | Begg’s Test | Egger’s Test | ||
|---|---|---|---|---|
| t | P | t | P | |
| rs1800925 (C-1112T) | ||||
| TT/CC | 0.34 | 0.734 | −1.15 | 0.369 |
| TC/CC | 0.34 | 0.734 | 0.20 | 0.857 |
| TT + TC/CC | 0.34 | 0.734 | 0.14 | 0.903 |
| T/C | 0.34 | 0.734 | −0.31 | 0.786 |
| rs1295686(C + 1923 T) | ||||
| TT/CC | 1.50 | 0.133 | 4.19 | 0.009 |
| TC/CC | 1.20 | 0.23 | 3.22 | 0.023 |
| TT + TC/CC | 1.20 | 0.23 | 4.81 | 0.005 |
| T/C | 1.50 | 0.133 | 5.29 | 0.003 |
| rs20541(G + 2044 A) | ||||
| AA/GG | 0.32 | 0.753 | 0.71 | 0.489 |
| GA/GG | 0.77 | 0.444 | 0.28 | 0.786 |
| AA + GA/GG | −0.05 | 1.000 | 0.66 | 0.517 |
| A/G | 0.77 | 0.444 | 1.08 | 0.297 |
Sensitivity analysis
Meta-analysis of each genetic model was performed again after the largest weighted studies were removed. Compared with the original results, there was no significant change in the effect indices of the genetic models, including the OR, 95% CI, and p values, indicating the stability of the study (Table 2). As publication bias was detected in the genetic models of the rs1295686 (C + 1923 T) gene locus (p < 0.05) and visual verification: asymmetric point distribution in the original funnel plot, the bias was evaluated using the pruning method. Before and after pruning, the OR value and 95% CI did not change significantly, and the p values of each genetic model of the rs1295686 (C + 1923 T)gene locus were all lower than 0.05. For example, before pruning, the genotypes TT + TC and CC: OR = 1.94%, 95% CI: 1.37–2.74, p < 0.001 and OR = 2.00%, 95% CI: 1.45–2.74, p < 0.001 after pruning. Therefore, publication bias was considered to have no substantial influence on the conclusion.
Discussion
Bronchial asthma is a chronic inflammatory disease involving a variety of cells and inflammatory cytokines and is characterized by chronic airway inflammation and airway hyperresponsiveness. Genetic inheritance plays an important role in the onset, severity, and response to treatment of asthma [24]. As gene testing technology has developed, research on asthma susceptibility genes has made great breakthroughs, and an increasing number of studies have confirmed that genes and genetic polymorphisms are associated with asthma susceptibility [25–27]. Among them, research on IL-13 and IL-4 gene polymorphisms is the most extensive. IL-13 is an important inflammatory cytokine primarily produced following T cell activation, and secreted by CD8 + T cells, activated monocytes, and B cells. IL-13 participates in the development of asthma in a variety of ways. It plays an important role in the late asthmatic reaction and in the occurrence and development of allergic airway inflammation [28–33] Studies have shown that mutations of the IL-13 gene locus primarily create obstacles that prevent IL-13 binding to its receptor, thus affecting its function and expression, and increasing the carrier’s susceptibility to asthma [34].
In this meta-analysis, we identified gene polymorphisms in the IL-13 gene locus that comprised multiple single nucleotide polymorphisms (SNPs). SNP refers to the mutation of a single base at the genomic level, resulting in a change in the DNA. SNPs can be found in various areas of the genome, including the promoter region, exon, and intron region. SNPs in the promoter region can affect the expression of the whole gene, those in exons can modify the encoded protein, and those in introns can affect the splicing mode of the gene sequence [35]. Some articles studied the SNP of the IL-13 gene and found that it is a single copy gene with four exons, three introns, and 2937 nucleotides, located on the long arm 5q31 of human chromosome 5 [36, 37]. Among the multiple SNPs of the IL-13 gene, the correlation between rs1800925 (C-1112 T), rs1295686 (C + 1923 T), and rs20541 (G + 2044 A) gene polymorphisms and asthma has attracted the most attention.
In this study, a meta-analysis was performed to study the correlation between IL-13 gene polymorphisms and asthma. The results showed that genotype AA, GA + AA, and allele A in the rs20541 (G + 2044 A) locus were significantly more common in the asthma group compared with the control group (p < 0.05). Therefore, the rs20541 (G + 2044 A) gene locus may be a susceptibility gene for asthma, and allele A might increase the risk of asthma. Studies have shown that individuals with A variants of this gene locus have higher IgE levels than G variants [38, 39]. Allele A can enhance the activity of IL-13 and the contractility of airway smooth muscles in asthmatic patients [40, 41]. Bottema et al. [42] conducted a case–control study on Dutch Caucasians and found that the 2044 gene polymorphisms were closely related to asthma airway hyperresponsiveness, skin allergic reactions, and total serum IgE levels. Genotypes TT, TC, and TT + TC of the rs1295686 (C + 1923 T) locus in the asthma group were statistically significant compared with the control group, suggesting that the rs1295686 (C + 1923 T) locus may be a susceptibility gene for asthma, and allele A can increase the risk of asthma. Pu et al. used a case–control study to prove that this gene locus polymorphism may increase the levels of serum IgE and IL-13 by enhancing the expression of the IL-13 gene, ultimately increasing the risk of asthma [7, 11]. However, at the rs1800925 (C-1112 T) locus, genotypes TT, TC, TT + TC, and alleles in the asthma group were statistically insignificant compared to the control group, suggesting that polymorphisms in this gene locus are unrelated to the risk of asthma, and this gene locus may not be an asthma susceptibility gene.
In terms of publication bias, we observed no evident asymmetry in the funnel diagram, but considering that the funnel diagram is a qualitative judgment, the quantitative measurement of publication bias was carried out through Egger linear regression. The results showed that there was publication bias only between genetic models of rs1295686 (C + 1923 T) (p < 0.05), and there was no significance or publication bias in the genetic models of other gene loci, indicating that the results are reliable. The publication bias of each genetic model at the rs1295686 (C + 1923 T) gene locus was evaluated using the pruning method. No significant change was found in the OR, 95% CI, and p values before and after pruning. Therefore, the research results are relatively stable, indicating that publication bias had no substantive impact on the conclusion. We removed one of the most weighted studies for each genetic model. There was no significant change in the research effect indices of each study, which also verified the research stability. We analyzed the reasons for publication bias of rs1295686 (C + 1923 T).
This study did have some limitations. First, both authors and publishers are more willing to publish articles with positive results, introducing inherent bias. The articles were limited to Chinese and English, and some unpublished studies were not included if the data were incomplete or did not provide genotype frequency, potentially introducing selection and publication bias. Additionally, the quality of some studies was low, and the number of included studies was relatively small. When Egger linear regression was used to detect publication bias, its sensitivity could be reduced. Therefore, we needed to constantly update the data, include more high-quality and unbiased literature, and continue to verify the research results. we explored subgroup analyses or meta-regression analyses based on age groups to investigate potential effect modification. However, due to the limited number of included studies or small sample sizes in some subgroups, such analyses may lead to imprecise estimations or unreliable results. Therefore, we did not formally report the results of these subgroup analyses, but we suggest that larger-scale meta-analyses or studies targeting specific subgroups should include such analyses. In addition, focusing on differences in the age range and geographical distribution of participants, and differences in environmental exposure levels (such as variations in air pollution in different regions) and lifestyle on the potential impact of asthma risk, which might be the cause of the variability in research results.
In conclusion, this meta-analysis of the literature on the relationship between rs1800925 (C-1112 T), rs1295686 (C + 1923 T), and rs20541 (G + 2044 A), and asthma susceptibility revealed that rs20541 (G + 2044 A) and rs1295686 (C + 1923 T) gene polymorphisms are related to asthma pathogenesis. These results can help clinicians gain a deeper understanding of the pathogenesis of bronchial asthma and provide information to aid in the diagnosis, treatment, and prognosis of bronchial asthma. In the future, genetic studies of bronchial asthma could guide clinical work. However, it is important to note that asthma is affected by the interactions between genes and environmental factors. Asthma is a complex polygenic genetic disease, but environmental factors may affect gene susceptibility in many ways. The correlation between genes and the environment and between genes has been investigated. The above conclusions still need to be confirmed by more large-scale and high-quality studies to obtain more specific results.
Supplementary Information
Authors’ contributions
S.S. wrote the main manuscript text; T.Z. H.Z. and Y.R. prepared figures; Y.W. and W.H. collated data; M.W. and F.Z.used software; H.Z. checked manuscript. All authors reviewed the manuscript.
Funding
None.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent to publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Song Su, Tong Zhang and Yaping Wang contributed equally to this work and share first authorship.
References
- 1.Ding BD, Wang MW, Zhao YH, et al. Association of IL-13 gene + 2044-G/A polymorphism to the risk of pediatric asthma and its relevancy to serum levels of IL-13. Clin Pediatr. 2016;3:838–41. 10.3969/j.issn.1000-3606.2016.11.009. [Google Scholar]
- 2.Liao XJ, Zhu XP, Li JF, et al. Analysis on polymorphisms of IL-4-590C/T, IL-13-1112C/T and IL-4RαQ576R of asthmatic children in Guiyang. Chin J Immunol. 2014;30:523–7. [Google Scholar]
- 3.Cao YL, Cui QH, Tang CH, et al. Association between polymorphisms interleukin-4,interleukin-4 receptor and interleukin-13 gene and childhood asthma. China J Mod Med. 2012;22:31–5. [Google Scholar]
- 4.Zhou GH, Zhang JH, Xu PR. The relationship between Arg130Gln polymorphism of IL-13 gene and serum IL-13 level in Uygur asthmatic children in Xinjiang. J Xinjiang Med Univ. 2016;39:622–6. [Google Scholar]
- 5.Ma ZN, Gu LH. Study on the correlation between polymorphism of IL-4 and IL-13 gene and the occurrence of asthma in children. Contemp Med Forum. 2021;19:21–2. 10.3969/j.issn.2095-7629.2021.11.012. [Google Scholar]
- 6.Feng D. Research on the polymorphism of gene IL-13 in asthma and their first degree relatives. Heilongjiang Med J. 2009;33(481–482):485. 10.3969/j.issn.1004-5775.2009.07.001. [Google Scholar]
- 7.Jia CM, Liu XM, Wang DM, et al. Relationship between polymorphisms of IL-13 gene Intron 3 + 1923 and bronchial asthma in children. Chin J Appl Clin Pediatr. 2013;28:682–5. 10.3760/cma.j.issn.2095-428X.2013.09.013. [Google Scholar]
- 8.Zhang JF, Yang J, Li B, et al. The polymorphism of G+2044A and A-1512C on IL-13 peptide chain among 153 children with asthma in Guiyang. Chin J Microbiol Immunol. 2016;36:144–8. 10.3760/cma.j.issn.0254-5101.2016.02.012. [Google Scholar]
- 9.Sun HP, Chen JQ, Guo XR, Chen RH. The relationship between IL-13 gene polymorphism and the levels of serum IL-13 and serum eosinophil cation protein in asthmatic children. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2003;20:547–8. 10.3760/j.issn:1003-9406.2003.06.021. [PubMed] [Google Scholar]
- 10.Xi D, Pan SX, Cui TP, et al. Association between IL-13 gene polymorphism and asthma in Han nationality in Hubei Chinese population. Chin J Immunol. 2003;19:757–60. [DOI] [PubMed] [Google Scholar]
- 11.Pu HP, Liu H, Lv ZH, et al. Study on the correlation between asthma and IL-13 gene polymorphism. Pract J Med Pharm. 2013;30:289–92. 10.3969/j.issn.1671-4008.2013.04.001. [Google Scholar]
- 12.Wang HL. The influence of IL-4 and IL-13 gene polymorphism on IgE level in children with asthma. J Pediatr Pharm. 2016;22:7–9. 10.13407/j.cnki.jpp.1672-108X.2016.04.003.
- 13.Wang Y, Li TX, Deng Y, et al. Association study of rs1295686 polymorphisms in IL13 gene with pediatric asthma in a northeastern Han Chinese population. J Harbin Med Univ. 2014;48:1–4. [Google Scholar]
- 14.Yan M, Yang KX. Association of IL-4 and IL-13 gene polymorphisms with asthma in children in Huizhou, Guangdong Province. Guangdong Med J. 2015;8: 1203–1205. [Google Scholar]
- 15.Yang LF, Zhang Y, Genetic Argl LQL. 44Gln polymorphism of interleukin-13 and asthma in children. Mod Med J China. 2010;12:46–7. 10.3969/j.issn.1672-9463.2010.03.016. [Google Scholar]
- 16.Zhao KS, Lu JR, Li SY, et al. Correlationship between interleukin-13 genotype and phenotype in children with bronchial asthma. J Clin Pediatr. 2005;23:312–4. 10.3969/j.issn.1000-3606.2005.05.020. [Google Scholar]
- 17.Guo YX, Lin N, Liu YG, et al. Relationship of gene polymorphism of interleukin-13 with susceptibility to asthma, serum IgE and interleukin-13 levels in children with Zhuang nationality. Guangxi Med J. 2017;39:1288–91. 10.11675/j.issn.0253-4304.2017.09.02
- 18.Deng SL, Li B, Yang J, et al. The analysis of the polymorphism of IL-4 gene −590C/T, IL-13 gene −1112C/T in asthmatic children. Chin J Microbiol Immunol. 2015;4:276–80. 10.3760/cma.j.issn.0254-5101.2015.04.008. [Google Scholar]
- 19.Wang ZH, Li LH, Zhao FX. Study on the correlation between plasma levels of IL-13 and asthma. Matern Child Health Care China. 2004;19:76–7. 10.3969/j.issn.1001-4411.2004.21.045. [Google Scholar]
- 20.Chan IH, Leung TF, Tang NL, et al. Gene-gene interactions for asthma and plasma total IgE concentration in Chinese children. J Allergy Clin Immunol. 2006;117:127–33. 10.1016/j.jaci.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 21.Liu Q, Hua L, Fang D, et al. Interleukin-13 and RANTES polymorphisms in relation to asthma in children of Chinese Han nationality. Asian Pac J Allergy Immunol. 2013;31:247–52. 10.12932/AP0298.31.3.2013 [DOI] [PubMed]
- 22.Wang JY, Liou YH, Wu YJ, et al. An association study of 13 SNPs from seven candidate genes with pediatric asthma and a preliminary study for genetic testing by multiple variants in Taiwanese population. J Clin Immunol. 2009;29:205–9. 10.1007/s10875-008-9256-6. [DOI] [PubMed] [Google Scholar]
- 23.Wu X, Li Y, Chen Q, et al. Association and gene-gene interactions of eight common single-nucleotide polymorphisms with pediatric asthma in middle China. J Asthma. 2010;47:238–44. 10.3109/02770900903509099. [DOI] [PubMed] [Google Scholar]
- 24.Leung TF, Ko FW, Sy HY, et al. Differences in asthma genetics between Chinese and other populations. J Allergy Clin Immunol. 2014;133:42–8. 10.1016/j.jaci.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 25.Slager RE, Hawkins GA, Li X, et al. Genetics of asthma susceptibility and severity. Clin Chest Med. 2012;33:431–43. 10.1016/j.ccm.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.March ME, Sleiman PM, Hakonarson H. Genetic polymorphisms and associated susceptibility to asthma. Int J Gen Med. 2013;6:253–65. 10.2147/IJGM.S28156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cookson W. The alliance of genes and environment in asthma and allergy. Nature. 1999;402(Suppl):B5-11. 10.1038/35037002. [DOI] [PubMed] [Google Scholar]
- 28.Schnyder B, Lugli S, Feng N, et al. Interleukin-4 (IL-4) and IL-13 bind to a shared heterodimeric complex on endothelial cells mediating vascular cell adhesion molecule-1 induction in the absence of the common gamma chain. Blood. 1996;87:4286–95. 10.1182/blood.V87.10.4286.bloodjournal87104286. [PubMed] [Google Scholar]
- 29.Shao Q, Ning H, Lv J, et al. Regulation of Th1/Th2 polarization by tissue inhibitor of metalloproteinase-3 via modulating dendritic cells. Blood. 2012;119:4636–44. 10.1182/blood-2011-08-376418. [DOI] [PubMed] [Google Scholar]
- 30.Wills-Karp M. The gene encoding interleukin-13: a susceptibility locus for asthma and related traits. Respir Res. 2000;1:19–23. 10.1186/rr7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen KP. Recent development of research in asthma gene in children and adults. Med Recapitulate. 2012;18:38–40. 10.3969/j.issn.1006-2084.2012.01.014. [Google Scholar]
- 32.Liao LF, Chen W, Dai JH, et al. Interleukin-13 and interleukin-18 on the expression of nerve growth factor mRNA in bronchial asthma model of rats. J Chongqing Med Univ. 35;(8):1208–11.
- 33.Li HJ, Wang SJ, Li YL, et al. Effects of different doses of lipopolysaccharide pretreatment on lung inflammation in asthmatic mice. Chin J Cell Mol Immunol. 2008;24:1008–10. 10.3321/j.issn:1007-8738.2008.10.021. [Google Scholar]
- 34.Wan S. Progress in interleukin and pathogenesis of asthma. Med Recapitulate. 2011;17:820–2. 10.3969/j.issn.1006-2084.2011.06.008. [Google Scholar]
- 35.Wang R, Qin HM, Huang HT, et al. Research progress of interleukin-13 gene polymorphism. Youjiang Med J. 2017;45:231–4. 10.3969/j.issn.1003-1383.2017.02.028. [Google Scholar]
- 36.Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mckenzie AN, Li X, Largaespada DA, et al. 150 (12) Structural comparison and chromosomal localization of the human and mouse IL-13 genes. J Immunol. Baltimore, MD:5436–5444. [PubMed]
- 38.Graves PE, Kabesch M, Halonen M, et al. A cluster of seven tightly linked polymorphisms in the IL-13 gene is associated with total serum IgE levels in three populations of white children. J Allergy Clin Immunol. 2000;105:506–13. 10.1067/mai.2000.104940. [DOI] [PubMed] [Google Scholar]
- 39.Chen H, Liu HP, Bao YX, et al. Correlation of asthma susceptibility gene polymorphisms with API positive infantile wheezing. Clin Pediatr. 2013;6:547–50. 10.3969/j.issn.1000-3606.2013.06.014. [Google Scholar]
- 40.Chen W, Ericksen MB, Levin LS, Khurana Hershey GK. Functional effect of the R110Q IL13 genetic variant alone and in combination with IL4RA genetic variants. J Allergy Clin Immunol. 2004;114:553–60. 10.1016/j.jaci.2004.04.044. [DOI] [PubMed] [Google Scholar]
- 41.Syed F, Panettieri RA Jr, Tliba O, et al. The effect of IL-13 and IL-13R130Q, a naturally occurring IL-13 polymorphism, on the gene expression of human airway smooth muscle cells. Respir Res. 2005;6:9. 10.1186/1465-9921-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bottema RW, Nolte IM, Howard TD, et al. Interleukin 13 and interleukin 4 receptor-α polymorphisms in rhinitis and asthma. Int Arch Allergy Immunol. 2010;15:259–67. 10.1159/000314366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.