Abstract
XYL3, which encodes a d-xylulokinase (EC 2.7.1.17), was isolated from Pichia stipitis CBS 6054 genomic DNA by using primers designed against conserved motifs. Disruption of XYL3 eliminated d-xylulokinase activity, but d-ribulokinase activity was still present. Southern analysis of P. stipitis genomic DNA with XYL3 as a probe confirmed the disruption and did not reveal additional related genes. Disruption of XYL3 stopped ethanol production from xylose, but the resulting mutant still assimilated xylose slowly and formed xylitol and arabinitol. These results indicate that XYL3 is critical for ethanol production from xylose but that P. stipitis has another pathway for xylose assimilation. Expression of XYL3 using its P. stipitis promoter increased Saccharomyces cerevisiae d-xylulose consumption threefold and enabled the transformants to produce ethanol from a mixture of xylose and xylulose, whereas the parental strain only accumulated xylitol. In vitro, d-xylulokinase activity in recombinant S. cerevisiae was sixfold higher with a multicopy than with a single-copy XYL3 plasmid, but ethanol production decreased with increased copy number. These results confirmed the function of XYL3 in S. cerevisiae.
d-Xylulose is phosphorylated to d-xylulose-5-phosphate by d-xylulokinase (EC 2.7.1.17) before it enters glycolysis through the nonoxidative pentose phosphate pathway. d-Xylulokinase is the first step in the metabolism of xylose that is common to both bacteria and eukarya (14, 20, 21).
The bacterial gene for d-xylulokinase (xylB) was first cloned from Escherichia coli in 1984 (26, 34). In 1989 Ho and Chang (19) reported cloning the Saccharomyces cerevisiae gene for d-xylulokinase by complementing an Escherichia coli xylB deficiency. It was also sequenced as a part of the S. cerevisiae genome project and named XKS1 (32). Of the few putative eukaryotic genes for d-xylulokinases, only the S. cerevisiae (31) and the presently reported Pichia stipitis genes have been characterized (45). The present report is the first physiological characterization of a d-xylulokinase-encoding gene from a eukaryote that uses xylose for growth and ethanol production.
Among the native xylose-fermenting yeasts, Pachysolen tannophilus, Candida shehatae, and Pichia stipitis can ferment xylose to ethanol with high yields (8, 9, 43). However, relative to glucose fermentation by Saccharomyces, these xylose-fermenting yeasts display lower ethanol production rates. Moreover, yeast xylose fermentations require low levels of aeration for optimal ethanol production (10, 15). In an attempt to overcome these problems, researchers have cloned and expressed the genes for xylose reductase (XR) and xylitol dehydrogenase (XDH) (XYL1 and XYL2, respectively) from P. stipitis in S. cerevisiae (2, 25, 47, 49). The resulting transformants can grow on xylose aerobically and produce ethanol under oxygen-limited conditions in low yield (23, 46).
The S. cerevisiae xylulokinase gene, which corresponds to XKS1 or YGR194C (32), increased ethanol and decreased xylitol production in a Saccharomyces sp. that had been engineered with P. stipitis XYL1 and XYL2 (17, 18). Richard et al. (31), along with Stevis and Ho (44), showed that XKS1 is the only route for xylulose metabolism in this yeast. Since the first report by Chang and Ho (4), others have overexpressed Saccharomyces XKS1 along with the P. stipitis genes for XR and XDH in S. cerevisiae (12, 17, 48). These efforts have enabled higher ethanol yields from d-xylose in recombinant Saccharomyces strains, but these heterologous transformants generally do not perform as well as native P. stipitis or C. shehatae (10).
It is not clear that overexpression of XKS1 is entirely beneficial. In fact, overexpression of XKS1 in a different Saccharomyces genetic background was reported to inhibit growth on d-xylulose (32). Other studies by Toivari et al. (48) and Richard et al. (31) did not confirm this finding, but Johansson et al. (24) found that overexpression of S. cerevisiae XKS1 reduced xylose consumption by 50 to 80% in S. cerevisiae transformants even while it increased the yield of ethanol from xylose, and they cautioned against unmodulated overexpression of this gene. Thus, some controversy surrounds this approach. The S. cerevisiae d-xylulokinase is also active on d-ribulose (31), whereas the P. stipitis d-xylulokinase is not (13).
The objective of our work was to clone and characterize the P. stipitis xylulokinase gene (XYL3) so that it might be used for engineering xylulose metabolism without the side activity seen with S. cerevisiae XKS1 (51).
MATERIALS AND METHODS
Strains.
Microbial strains used in this study are listed in Table 1. P. stipitis CBS 6054 was the source of all sequenced DNA. Michael Culbertson (Department of Genetics, University of Wisconsin) kindly provided S. cerevisiae 679 (MATα trp1-2). Escherichia coli XL-1 Blue MRF and SOLR (Stratagene, La Jolla, Calif.) were used in conjunction with the λ-ZAPII genomic DNA library of P. stipitis (28).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| P. stipitis CBS 6054 | Wild type | CBSa |
| P. stipitis UC7 | ura3-2 | 27 |
| P. stipitis YS30 | Isogenic to P. stipitis UC7 except xyl3::URA3 | This study |
| S. cerevisiae 679 | MATα trp1-289 | M. Culbertson |
| S. cerevisiae 679C | S. cerevisiae 679(pRS424) | This study |
| S. cerevisiae 679[pYS31N] | S. cerevisiae 679(pYS31N) | This study |
| S. cerevisiae 679[pYS32N] | S. cerevisiae 679(pYS32N) | This study |
| Plasmids | ||
| pRS314 | TRP1, CEN/ARS | 42 |
| pRS424 | TRP1, 2μm origin | 6 |
| pX3 | XYL3 in pBluescript KS(+) | This study |
| pYS03 | XYL3 disruption cassette containing xyl3::URA3 | This study |
| pYS31N | XYL3 in pRS314 | This study |
| pYS32N | XYL3 in pRS424 | This study |
CBS, Centraalbureau voor Schimmelcultures (Delft, The Netherlands).
Media and culture conditions.
E. coli was grown in Luria-Bertani (LB) medium. Ampicillin (50 μg/ml) was added to the medium when required. Yeast strains were grown in yeast peptone (YP) medium (10 g of yeast extract and 20 g of Bacto peptone per liter) or yeast synthetic complete (YSC) medium containing 6.7 g of yeast nitrogen base (YNB) without amino acids (Difco, Detroit, Mich.) per liter, which was complemented with appropriate amino acids and nucleotides according to Rose et al. (33). Glucose (20 g/liter) or xylose (40 g/liter) was used as a carbon source. For xylulose fermentation, a 20:80 xylulose-xylose mixture was prepared using xylose isomerase (Novo Nordisk, Copenhagen, Denmark). Yeast cells were cultivated at 30°C in 50 ml of medium in a 125-ml Erlenmeyer flask, and oxygen transfer rates were determined by sulfite oxidation (7).
Enzymes, primers, and chemicals.
Restriction enzymes, DNA-modifying enzymes, and other molecular reagents were obtained from New England Biolabs (Beverly, Mass.), Promega (Madison, Wis.), Stratagene (La Jolla, Calif.), and Roche Biochemical (Indianapolis, Ind.). Reaction conditions were as recommended by the suppliers. All general chemicals were purchased from Sigma (St. Louis, Mo.). Primers for PCR and sequencing were synthesized by Sigma-Genosys (The Woodlands, Tex.).
Yeast transformation.
A yeast EZ-Transformation kit (Bio 101, Carlsbad, Calif.) was used for all yeast transformations according to the manufacturer's instructions. Transformants were selected on YSC medium containing 20 g of glucose per liter. Necessary amino acids were added if required.
Cloning of P. stipitis xylulokinase gene.
Homologous segments of protein sequences from bacterial d-xylulokinases, along with other putative eukaryotic proteins having similarity to that coded by the S. cerevisiae YGR194C open reading frame, were aligned against the S. cerevisiae protein sequence to identify the highly conserved regions at the N terminus, LGFDLSTQQ and QQHGSVYWS. Primers were designed against these motifs based on a P. stipitis codon usage table (39). A 300-bp fragment was amplified by Pfu polymerase (Stratagene, La Jolla, Calif.) from the genomic DNA of P. stipitis CBS 6054 using primers 1 (5′-CTATCTTGGGTTTGATCTTTCGACC-3′) and 2 (5′-GACCAGTAGACAGACCGTGCTGCT-3′). The amplicon was used as a probe to screen a λ-ZAPII P. stipitis genomic DNA library (28). A total of 3 × 106 plaques were initially screened. After the tertiary screening, one plaque remained positive. The single-stranded DNA was converted to double-stranded DNA in a phagemid, pBluescript vector as described by Lu et al. (28).
DNA sequencing.
XYL3 was sequenced by using an Applied Biosystems model 371 DNA sequencer (Perkin-Elmer). Sequence analysis was performed using Genetics Computer Group sequence analysis (GCG, Madison, Wis.) and DNAman software (Lynnon BioSoft, Vaudreuil, Quebec, Canada).
Multiple alignment and phylogenetic analysis of protein sequence.
Homologues of P. stipitis xylulokinase were identified using Blast search program at the NCBI web site (http://www.ncbi.nlm.nih.gov/BLAST/). They were then aligned using ClustalW with default settings at the EBI website (http://www.ebi.ac.uk/clustalw). A phylogenetic tree was generated from the alignments using the neighbor-joining method (36). By throwing out the sequences in the gap, ambiguous parts of the alignment were excluded from the phylogenetic analysis. For bootstrap analysis, a multiple alignment was sampled 10,000 times. Bootstrap values are expressed in percentages and placed at nodes.
Plasmid construction.
Plasmids used in this study are summarized in Table 1. Plasmid DNA was isolated by QIAprep spin plasmid kit (Qiagen Inc., Chatsworth, Calif.). We isolated yeast genomic DNA using a genome DNA isolation kit (Bio 101, Vista, Calif.). PstI digestion of pX3 produced a 2.9-kbp fragment containing the XYL3 gene with its native promoter. A single-copy vector (pYS31N) was constructed by inserting the 2.9-kbp PstI-PstI fragment of XYL into the PstI site of pRS314 (42). Plasmid pRS32N (multicopy vector) was made by inserting the same 2.9-kbp fragment into the PstI site of pRS424 (6).
The disruption cassette, pYS03, was constructed by inserting a BamHI-BamHI fragment carrying P. stipitis URA3 into the BglII site located in the middle of the XYL3 open reading frame in pYS31N. The 1.2-kbp BamHI-BamHI fragment carrying URA3 was amplified from P. stipitis genomic DNA by PCR using primers 3 (5′-CGGGATCCGCTTATGTTGCATCTCAAA-3′) and 4 (5′-CGGGATCCCGGAATAGGCCTCTGCTTGT-3′). The 2.9-kbp fragment containing the disruption cassette was amplified from pYS31N by PCR with primers 5 (5′-GTTGACACTCACAATGACC-3′) and 6 (5′-CCGATCCATTCCTTCTTGG-3′).
PCR screening of XYL3 disruptants.
Putative XYL3 disruption mutants were screened by colony PCR. Yeast colonies were lysed at 37°C for 1 h using the whole cell yeast PCR kit (Bio 101, Vista, Calif.). Primers 5 and 6 were used in the PCR screening. The resulting lysate was mixed with 200 μM deoxynucleoside triphosphates, 10 μM primers, and 1.5 mM MgCl2 to a final volume of 100 μl; then, the mixture was incubated at 95°C for 15 min. Five units of Taq DNA polymerase (Roche Biochemicals, Indianapolis, Ind.) was added to the mixture, and the reaction was performed at 95°C for 1 min, 50°C for 1 min, and 72°C for 1 min for 40 cycles, followed by 72°C for 4 min. The product was stored at 4°C.
Southern blot hybridization.
Genomic DNA preparations digested with restriction enzymes were electrophoresed in 8 g of agarose per liter. A Southern transfer was then performed by capillary action according to the method of Sambrook et al. (37). Randomly primed DNA labeling with the purified XYL3 coding region as a template generated a probe labeled with digoxigenin-11-dUTP (Roche Biochemicals, Indianapolis, Ind.). DIG Easy Hyb solution (Roche, Indianapolis, Ind.) was used for hybridization at 37°C.
Preparation of crude extracts.
S. cerevisiae and P. stipitis were grown to exponential phase in YSC medium supplemented with appropriate amino acids and nucleotides and harvested by centrifugation. The pellet was washed and suspended in the buffer (100 mM phosphate buffer, 1 mM EDTA, 5 mM β-mercaptoethanol, pH 7.0). The suspended cells were mixed with glass beads (Sigma, St. Louis, Mo.), vortexed at the maximum rate in bursts of 30 to 120 s, and then cooled on ice for a similar period. This procedure was repeated for up to 8 min of vortexing or until microscopic examination showed >80% cell breakage. The crude extract, collected after centrifugation for 10 min at 15,000 × g, was used in the enzyme assay.
Xylulokinase and ribulokinase assays.
Xylulokinase and ribulokinase activities were measured according to the method of Shamanna and Sanderson (38). For the P. stipitis xylulokinase assay, another blank without pyruvate kinase and lactate dehydrogenase was used to minimize the interference of xylitol dehydrogenase activity in P. stipitis. We used a photodiode array spectrophotometer (Hewlett Packard, Wilmington, Del.) to monitor the reaction by absorbance at 340 nm. All assays were performed within 2 to 4 h of cell breakage. Control homogenates with protease inhibitor cocktail (Sigma, St. Louis, Mo.) did not show significant differences in activity. Our assay could detect 0.001 U/mg of protein. One unit of activity is defined as the amount of enzyme that phosphorylates 1 μmol of xylulose per min at 30°C. Protein concentration was determined by the BCA method (Pierce, Rockford, Ill.).
Analytical methods.
Glucose, xylose, xylitol, xylulose, and ethanol concentrations were determined by high-pressure liquid chromatography (HPLC) (HP, Wilmington, Del.) with an Ion 300 column (Interaction Chromatography, San Jose, Calif.). The variance was approximately 5% in replicate samples. For the gas chromatography-mass spectrometry (GC-MS) analysis, fermentation samples were concentrated by rotary evaporation, dissolved with pyridine, and converted into a trimethylsilyl derivative. Gas chromatography (Finnigan, Austin, Tex.) with a ZB-5-MS column (Phenomenex) was used with the following conditions: 100°C for 1 min, then increasing to 300°C at 20°C/min. Cell growth was monitored by optical density at 600 nm (OD600). One OD600 unit was equivalent to 0.17 g of cells/liter for S. cerevisiae and 0.21 g of cells/liter for P. stipitis.
Nucleotide sequence accession numbers.
The sequences of the coding region and 5′- and 3′-flanking regions were deposited in GenBank (AF127802 and AAF72328).
RESULTS
Cloning of XYL3 from P. stipitis.
To clone XYL3, we identified two highly conserved regions at the N terminus, LGFDLSTQQLK and SGSCQQHGSVYWS, by aligning ungapped Blast segments (1) of protein sequences from bacterial xylulokinases reported in the literature and putative d-xylulokinases from unidentified open reading frames of eukaryotic proteins (Caenorhabditis elegans [accession no. YNE7__CAEEL] and Arabidopsis thaliana [BAA97229]) against the S. cerevisiae sequence from the S. cerevisiae sequencing project, YGR194C (Fig. 1). LGFDLSTQQLK is involved in phosphate binding, which is a conserved region common to sugar kinases (3). The function of the second region is not known.
FIG. 1.
Alignment of xylulokinase sequences from several bacteria and eukaryotes against the P. stipitis XYL3 sequence. Highly conserved sequences are in bold. Sequences used for primer design are underlined.
Using primers designed to these two regions, we successfully amplified a 300-bp fragment from P. stipitis genomic DNA. One open reading frame of the nucleotide sequence of this fragment showed homology to previously cloned xylulokinase genes. This fragment was then used as a probe to screen phage plaques from the λ-ZAPII genome library of P. stipitis CBS 6054. After three rounds of screenings, a positive clone was identified and confirmed by Southern blots. This 5.3-kbp clone was named pX3.
XYL3 sequence analysis.
The XYL3 sequence (GenBank accession no. AF127802 and AAF72328) from pX3 contained an open reading frame of 1,872 nucleotides, coding for a polypeptide of 623 amino acids with a total calculated molecular mass of 69,198 Da, which agreed well with the previous result (71 kDa) from a P. stipitis xylulokinase purification study (13). Sequence analysis of the 900 bp 5′ to the open reading frame identified three putative TATA elements located at −180 to −188, −286 to −294, and −335 to −342.
A phylogenetic analysis showed that the P. stipitis d-xylulokinase protein had diverged significantly from the 16 other known yeast, bacterial, plant, and animal xylulokinases (Fig. 2). The XYL3 open reading frame was only 28% identical at the DNA level to its nearest neighbor, S. cerevisiae d-xylulokinase XKS1 (YGR194C) (19, 32), while at the amino acid level, the P. stipitis xylulokinase displayed 44% identity to the S. cerevisiae homologue. The P. stipitis xylulokinase protein belongs to the FGGY, FGGY family of carbohydrate kinases (Prosite PDOC00408), which includes d-xylulokinase (EC 2.7.1.17), l-xylulokinase (EC2.7.1.53), glycerol kinase (EC 2.7.1.30), glucokinase (EC 2.7.1.12), and l-fuculokinase (EC 2.7.1.51) (30).
FIG. 2.
Phylogenetic tree of P. stipitis xylulokinase protein homologues found in GenBank. Homologues of xylulokinase were identified by Blast search, and found protein sequences were then aligned. A phylogenetic tree was generated from the aligned regions after excluding sequences creating gaps.
Disruption of XYL3.
To investigate the function of the XYL3 gene product, we disrupted XYL3 in a P. stipitis strain, FPL-UC7 (ura3) (27), by gene replacement (35) with P. stipitis URA3 in the middle of the XYL3 coding region (Fig. 3A). Ura+ strains were selected on YSC-glucose without uracil and then replica plated on YSC with xylose medium to screen for colonies deficient in xylose assimilation. All Ura+ transformants could grow on YSC medium with xylose as a sole carbon source, but 2 of the 50 selected colonies grew very slowly.
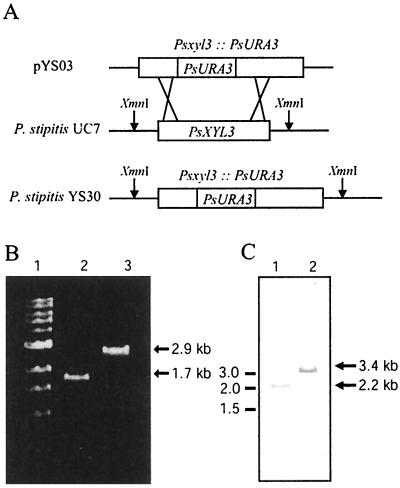
FIG. 3.
Construction of XYL3 gene disruption cassette and confirmation of XYL3 disruption by PCR and Southern blotting. (A) Systematic representation of the disruption cassette (pYS03). The linear disruption cassette was amplified by PCR from pYS03. It was then introduced into the chromosome by gene replacement. The pYS03 contains 1.2 kbp of the P. stipitis URA3 gene in the middle of the XYL3 coding region. (B) PCR with specific primers for amplification of the 1.7-kbp XYL3. The templates were used as follows: lane 1, markers; lane 2, P. stipitis FPL-UC7 genomic DNA; lane 3, P. stipitis FPL-YS30 genomic DNA. (C) Southern blot analysis of XYL3 disruption with the probe made from the XYL3 coding region. The genomic DNAs from P. stipitis FPL-UC7 (lane 1) and P. stipitis YS30 (lane 2) were digested with XmnI. The parental strain, P. stipitis FPL-UC7, showed a 2.2-kbp band and the XYL3 disruptant, P. stipitis FPL-YS30 (xyl3::URA3), showed a 3.4-kbp band. Positions of size markers are shown to the left.
Colony PCR and Southern blot analyses were used to determine whether these were XYL3 disruptants. In colony PCR, the slow-growing transformants displayed a 2.9-kbp fragment corresponding to the URA3 gene inserted into the coding region of XYL3. In contrast, the Ura+ colonies resulting from random integration of the disruption cassette or from reversion showed a 1.7-kbp fragment that corresponded to the coding region of XYL3 (Fig. 3B). One of the XYL3 disruptants was named FPL-YS30 (xyl3::URA3). Genomic DNA from the parental strain, FPL-UC7 (ura3), or the XYL3 disruptant, FPL-YS30, was digested with XmnI. After probing the Southern blot with an XYL3 fragment, FPL-YS30 displayed a 3.4-kbp band corresponding to insertion of the 1.2-kbp fragment of URA3 into XYL3. However, the parental strain showed only a single 2.2-kbp band (Fig 3C). This confirmed the disruption and indicated that only a single isozyme of XYL3 was present in the genome.
The frequency of site-specific integration of the disruption cassette into the P. stipitis chromosome was low. Only 4% of the Ura+ transformants were true disruptants and showed slower growth phenotypes on xylose. This frequency was in line with our previous disruption studies of the P. stipitis ADH (5) and CYC1 (40) genes.
Growth experiments on various carbon sources.
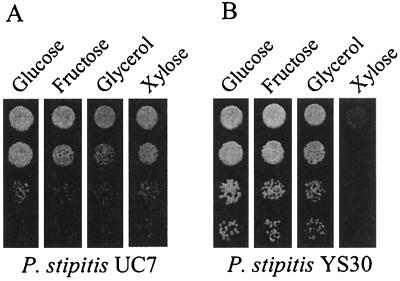
Because xylulokinase belongs to a family that includes glycerol kinase and because there are structural similarities between xylulose and fructose, we tested FPL-YS30 for growth on fructose or glycerol as a sole carbon source (Fig. 4). FPL-YS30 showed extremely slow growth on d-xylose, but growth on the other carbon sources was comparable to that of the parental strain.
FIG. 4.
Growth of the parental and xyl3::URA3 disruptant strains on various carbon sources. (A) P. stipitis FPL-UC7 (ura3); (B) P. stipitis FPL-YS30 (xyl3::URA3). YP medium (10 g of yeast extract, 20 g of Bacto peptone, and 20 g of agar per liter) with 20 g of each carbon source per liter was used, and cells were serially diluted. Approximately 104 cells were spotted in the undiluted (top) droplet.
XYL3 disruption mutant FPL-YS30 (xyl3::URA3) accumulates polyols.
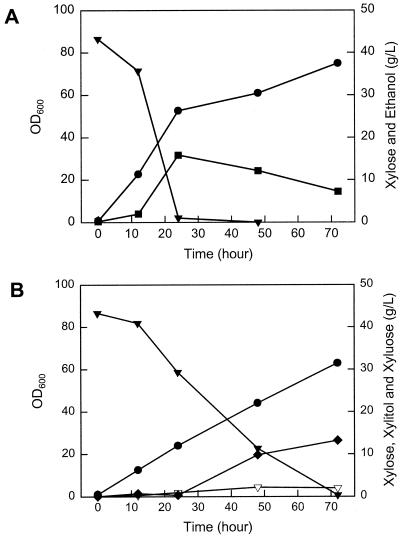
We could not detect any significant differences in growth, sugar consumption, or ethanol production between the parental strain and FPL-YS30 when glucose was used as the carbon source (data not shown). The main products from both strains were biomass and ethanol. In contrast, FPL-YS30 displayed significant differences from the parental strain, FPL-UC7, in xylose fermentation (Fig. 5A and B). FPL-UC7 consumed 42 g of xylose/liter within 24 h and produced 16 g of ethanol/liter, with a negligible amount of xylitol production. However, xylose assimilation in the disruptant FPL-YS30 was severely hindered (0.98 g of xylose per g of cells per h compared with 4.2 g of xylose per g of cells per h for FPL-UC7). Cells of P. stipitis FPL-YS30 required about 70 h to consume 43 g of xylose/liter. Specific growth rates of the disruptant and parental strains on xylose were 0.06 and 0.28 h−1, respectively. In contrast to the parental strain, FPL-YS30 did not produce detectable amounts of ethanol from xylose, and it accumulated up to 13.2 g of xylitol per liter in the medium.
FIG. 5.
Fermentation of xylose by P. stipitis FPL-UC7 (ura3) (A) and P. stipitis FPL-YS30 (xyl3::URA3) (B). Cells were cultured in YPX with 40 g of xylose per liter, with an oxygen transfer rate of 5.3 mM h−1. Symbols: ▾, xylose; ▿, xylulose; ⧫, xylitol; •, cell density; ▪, ethanol. Results in A and B are each from a single representative trial.
The XYL3 disruptant also produced an unknown product that eluted adjacent to xylitol during HPLC analysis. GC-MS analysis and sample loading with standard compounds showed it to be arabinitol, although our analytical methods could not determine its stereochemistry. Thus, the XYL3 disruptant produced both arabinitol and xylitol, whereas the parental strain did not accumulate significant amounts of these two polyols. These results showed that the XYL3 disruptant could grow on xylose, but the fate of consumed xylose is different in FPL-YS30 and its parent.
In vitro xylulokinase and ribulokinase assays of P. stipitis FPL-UC7 and P. stipitis FPL-YS30.
P. stipitis FPL-YS30 could grow slowly on xylose. However, our enzyme assays from the culture of the XYL3 disruptant could not detect d-xylulokinase activity. By contrast, a control culture of the parental strain, FPL-UC7, showed xylulokinase activity of 0.24 U/mg under the same conditions. Assays performed with and without protease inhibitor did not show significant differences in activity. The specific activity of d-ribulose was unaltered in the mutant and was approximately 0.07 U/mg of protein for FPL-UC7 and 0.11 U/mg of protein for FPL-YS30.
XYL3 is actively expressed in S. cerevisiae under the native XYL3 promoter.
We confirmed the function of the XYL3 gene product by heterologous expression in S. cerevisiae. A 2.9-kbp PstI-PstI fragment containing 900 bp of 5′ untranslated region and 200 bp of 3′ untranslated region in addition to the d-xylulokinase coding region was subcloned into a CEN/ARS-based (pYS31N) or a 2μm-based vector (pYS32N). Both plasmids were introduced into S. cerevisiae 679, and transformants were selected on YSC-glucose medium without tryptophan. After growing the cells in YSC-glucose medium, we measured xylulokinase activities in the crude extracts from the 679[pYS31N] centromeric and 679[pYS32N] 2μm-based strains as well as the parental strain. The 679[pYS31N] and 679[pYS32N] strains showed much higher xylulokinase activities (0.019 ± 0.002 U/mg and 0.121 ± 0.008 U/mg, respectively) than the parental strain, which did not have detectable activity. We also observed that xylulokinase activity in the extract from the cells bearing the 2μm-based vector, 679[pYS32N], were sixfold higher than that from cells bearing the centromeric vector, 679[pYS31N].
Xylulose fermentation by recombinant S. cerevisiae.
S. cerevisiae naturally ferments d-xylulose, a keto-isomer of xylose, but it uses xylulose at one-tenth the rate it uses glucose (52). Xylulose phosphorylation has been discussed as a possible rate-limiting step in xylose fermentation by recombinant S. cerevisiae (17). We tested whether XYL3 expression can enhance xylulose fermentation by S. cerevisiae under respirofermentative conditions (oxygen transfer rate, 5.3 mM O2/h). For the xylulose fermentation experiment, a mixture of xylose and xylulose was used.
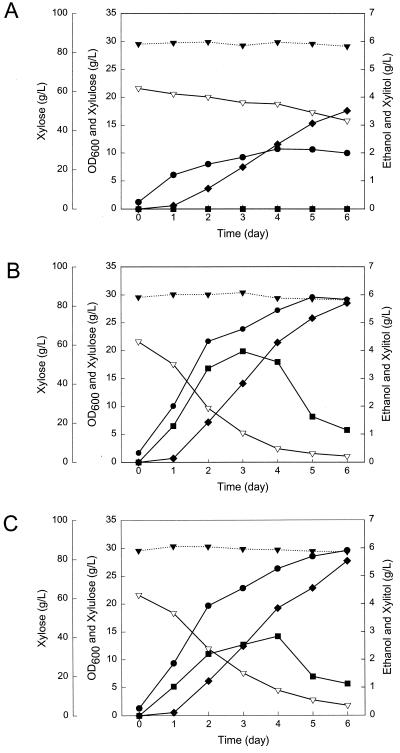
S. cerevisiae 679[pYS31N] and 679[pYS32N] used the d-xylulose about three times faster than the parental strain transformed with the control vector, pRS424, which carries TRP1 without the XYL3 insert (Table 2). The parental strain consumed only a fraction of the available d-xylulose and produced no ethanol during entire fermentation trials (Fig. 6A), whereas both recombinant strains carrying XYL3 completely utilized the added 20 g of d-xylulose per liter within 6 days and produced ethanol from d-xylulose (Fig. 6B and C). The maximum ethanol concentration observed was 4.0 g/liter in the transformant bearing the single-copy vector (679[pYS31N]) and 2.8 g/liter in the case of the transformant bearing the multicopy vector (679[pYS32N]). Even though we saw a sixfold increase in d-xylulokinase activity in the strain bearing the multicopy plasmid, this strain showed a slight decrease (ca. 15%) in the volumetric uptake rate of d-xylulose and a moderate decrease (ca. 30%) in volumetric ethanol production compared to the strain bearing a single-copy vector. The specific xylulose consumption rates of the two strains were comparable (0.61 and 0.62 g of xylulose per g of cells per h, respectively).
TABLE 2.
Growth and fermentation characteristics between host and recombinant strains during xylulose fermentation
| Strain | Specific growth rate (h−1) | Specific xylulose consumption rate (g of xylose [g of cells h−1]) | Max ethanol concn (g of ethanol/liter) |
|---|---|---|---|
| S. cerevisiae 679C | 0.026 | 0.198 | 0 |
| S. cerevisiae 679[pYS31N] | 0.037 | 0.611 | 3.97 |
| S. cerevisiae 679[pYS32N] | 0.039 | 0.622 | 2.83 |
FIG. 6.
Fermentation of xylose-xylulose mixture by (A) parental strain (S. cerevisiae 679) transformed with the multicopy control vector pRS424 and recombinant S. cerevisiae carrying the XYL3 gene in the single-copy vector pYS31N (B) or the multicopy vector pYS32N (C). Cells were cultured in YSC medium with xylose-xylulose, with an oxygen transfer rate of 5.3 mM O2 h−1. Symbols: ▾, xylose; ▿, xylulose; ⧫, xylitol; •, cell density; ▪, ethanol. Results in A, B, and C are each from a single representative trial.
DISCUSSION
The P. stipitis gene for xylulokinase (XYL3) differs significantly from its nearest homologue, and its heterologous expression enhances xylulose fermentation by S. cerevisiae. This is the only xylulokinase characterized from a eukaryote that is capable of converting xylose to ethanol. The divergent structure of XYL3 is somewhat surprising because the genes for CYC1, URA3, LEU2, ADH1, ADH2, PDC1, and PDC2 cloned previously from P. stipitis show between 81 and 65% identity to their S. cerevisiae homologues at the protein level (5, 27-29, 40). Genes for carbohydrate metabolism, such as the P. stipitis sugar transporters XYL1 and XYL2 (2, 25, 50), show between 60 and 53% identity to their nearest known S. cerevisiae homologues. Of the 16 P. stipitis enzymes with known homologues in S. cerevisiae, the XYL3 product shows the least similarity to its corresponding Saccharomyces protein. Some P. stipitis genes, such as those for d-arabinitol dehydrogenase (16) and STO1 (39), have no known S. cerevisiae homologues.
The amino acid sequence deduced from the open reading frame of XYL3 had all five sequence motifs, phosphate 1 (residues 17 to 22), connect 1 (residues 304 to 310), phosphate 2 (residues 323 to 327), adenosine (residues 527 to 529), and connect 2 (residues 559 to 563), that are found in the ATP-binding pattern as defined from the multiple sequence alignment of the known members of the sugar kinases, actin, and hsp70 class (3). Especially, the invariant GT in phosphate 2, which is present only in sugar kinases, was also found in the deduced protein sequence of XYL3.
The FPL-YS30 xyl3::URA3 disruptant could grow on xylose, but only at a much lower rate than its parent, FPL-UC7 (ura3). On the other hand, the mutant produced no ethanol from xylose. We did not expect it to use xylose at all because disruption of XYL3 should have eliminated xylulose phosphorylation. Its unexpected capacity for growth on xylose led us to consider two possibilities: the presence of a second isozyme for xylulokinase, and an alternative pathway for xylulose metabolism.
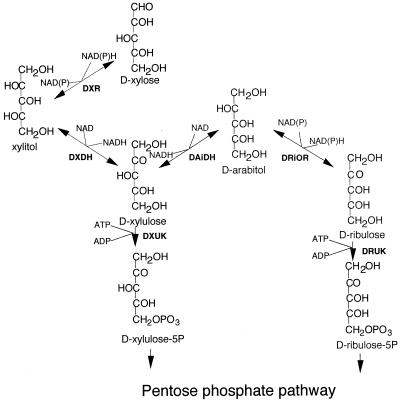
The presence of multiple isozymes is very common in yeasts and other higher eukaryotes. However, when we used the cloned XYL3 fragment to probe the genomic DNA by Southern blot, only one band was observed (Fig. 3C) and the mutant did not exhibit d-xylulokinase activity. Given the divergence of XYL3 from other known d-xylulokinases, it is possible that a second enzyme would not have been picked up in a Southern hybridization. However, because polyols, particularly arabitol, accumulated in the culture of the XYL3 disruptant, we suspect that xylulose phosphorylation can be bypassed via formation of arabinitol and ribulose (Fig. 7). This latter route was described by Shi et al. (41) through the characterization of a P. stipitis xyl2 mutant that could not use d-xylose or l-arabinose but could grow on d-arabinitol, and is further evidenced by the presence of d-arabinitol dehydrogenase in P. stipitis (16) and by the reported production of ribitol from xylose by P. stipitis strains blocked with respiration inhibitors (22). Eliasson et al. (11) reported that S. cerevisiae engineered to overexpress XKS1 also secretes arabinitol, but it is unclear how that observation is related to the current findings.
FIG. 7.
Proposed bypass of d-xylulose metabolism through d-arabinitol and d-ribulose.
Even though our analytical method could not determine the stereochemistry of the arabinitol produced, the observations cited above suggest that it was d-arabinitol. Using the proposed bypass, d-xylulose could be reduced to d-arabinitol by d-arabinitol dehydrogenase, and subsequently d-arabinitol could be oxidized to d-ribulose by d-ribulose reductase. Finally d-ribulose would be phosphorylated to ribulose-5-phosphate by ribulokinase, which then would enter the pentose phosphate pathway.
This route is not functional in S. cerevisiae, because an XKS1 disruption mutant could not grow on xylulose and growth on xylulose was restored after complementation of XKS1 (32). Moreover, Blast and tblastn (http://www3.ncbi.nlm.nih.gov/BLAST/) searches of the S. cerevisiae genome did not reveal any proteins with structures similar to the d-arabinitol dehydrogenase of P. stipitis, so it is unlikely that the bypass pathway could exist in S. cerevisiae. The d-xylulokinase from S. cerevisiae is also known to possess d-ribulokinase activity (31). The P. stipitis d-xylulokinase differs in that it does not possess d-ribulokinase activity and hence is more specific for d-xylulose (13).
The XYL3 gene under its native P. stipitis promoter was actively expressed in S. cerevisiae, as shown by the observed significant increase in in vitro d-xylulokinase activity. This is similar to the observations for the native promoters for P. stipitis XYL1 and XYL2 expressed in S. cerevisiae (2, 25). We observed a copy number effect. d-Xylulokinase activity increased sixfold in the multicopy versus single-copy vector transformant, but the capacities of multi- and single-copy strains were very similar in their fermentation of d-xylulose. Ethanol production decreased with increasing copy number and increasing enzyme activity, but our results are not comparable to the deleterious effect previously observed with the overexpression of XKS1 in S. cerevisiae (32). Recombinant S. cerevisiae strains bearing XYL3 produced ethanol from a mixture of xylulose and xylose, whereas the parental strain could not. These results, along with the structural homologies to known bacterial d-xylulokinases and the absence of d-xylulokinase activity in the xyl3 disruption mutant, strongly suggest that the gene product of XYL3 is a d-xylulokinase.
Acknowledgments
We thank M. Culbertson for kind donation of some plasmids and S. cerevisiae strains, A. Gargas for advice on phylogenetic analysis, R. Pettersen for GC-MS analysis, and K. Sydney for assistance in manuscript preparation.
This research was supported by USDA NRICGP grants 96-3550-3172 and 98-35504-6966.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amore, R., P. Kötter, C. Kuster, M. Ciriacy, and C. P. Hollenberg. 1991. Cloning and expression in Saccharomyces cerevisiae of the NAD(P)H-dependent xylose reductase-encoding gene (XYL1) from the xylose-assimilating yeast Pichia stipitis. Gene 109:89-97. [DOI] [PubMed] [Google Scholar]
- 3.Bork, P., C. Sander, and A. Valencia. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA 89:7290-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, S. F., and N. W. Y. Ho. 1988. Cloning the yeast xylulokinase gene for the improvement of xylose fermentation. Appl. Biochem. Biotechnol. 17:313-318. [DOI] [PubMed] [Google Scholar]
- 5.Cho, J. Y., and T. W. Jeffries. 1998. Pichia stipitis genes for alcohol dehydrogenase with fermentative and respiratory functions. Appl. Environ. Microbiol. 64:1350-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 7.Cooper, C. M., G. A. Fernstrom, and S. A. Miller. 1944. Performance of agitated gas-liquid contactors. Ind. Eng. Chem. 36:504-509. [Google Scholar]
- 8.Dellweg, H., M. Rizzi, H. Methner, and D. Debus. 1984. Xylose fermentation by yeasts. 3. Comparison of Pachysolen tannophilus and Pichia stipitis. Biotechnol. Lett. 6:395-400. [Google Scholar]
- 9.Du Preez, J. C., and J. P. van der Walt. 1983. Fermentation of d-xylose to ethanol by a strain of Candida shehatae. Biotechnol. Lett. 5:357-362. [Google Scholar]
- 10.Du Preez, J. C., B. Vandriessel, and B. A. Prior. 1989. Ethanol tolerance of Pichia stipitis and Candida shehatae strains in fed-batch cultures at controlled low dissolved oxygen levels. Appl. Microbiol. Biotechnol. 30:53-58. [Google Scholar]
- 11.Eliasson, A., E. Boles, B. Johansson, M. Osterberg, J. M. Thevelein, I. Spencer Martins, H. Juhnke, and B. Hahn-Hägerdal. 2000. Xylulose fermentation by mutant and wild-type strains of Zygosaccharomyces and Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 53:376-382. [DOI] [PubMed] [Google Scholar]
- 12.Eliasson, A., C. Christensson, C. F. Wahlbom, and B. Hahn-Hägerdal. 2000. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl. Environ. Microbiol. 66:3381-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flanagan, T., and M. J. Waites. 1992. Purification and characterization of d-xylulokinase from the pentose-fermenting yeast Pichia stipitis NCYC-1541. Enzyme Microb. Technol. 14:975-979. [Google Scholar]
- 14.Gong, C.-S., L. F. Chen, M. C. Flickinger, and G. T. Tsao. 1981. Conversion of hemicellulose carbohydrates. Adv. Biochem. Eng. 20:93-118. [Google Scholar]
- 15.Grootjen, D. R. J., R. Vanderlans, and K. Luyben. 1990. Effects of the aeration rate on the fermentation of glucose and xylose by Pichia stipitis CBS 5773. Enzyme Microb. Technol. 12:20-23. [Google Scholar]
- 16.Hallborn, J., M. Walfridsson, M. Penttila, S. Keranen, and B. Hahn-Hagerdal. 1995. A short-chain dehydrogenase gene from Pichia stipitis having d-arabinitol dehydrogenase activity. Yeast 11:839-847. [DOI] [PubMed] [Google Scholar]
- 17.Ho, N. W., Z. Chen, and A. P. Brainard. 1998. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl. Environ. Microbiol. 64:1852-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho, N. W., Z. Chen, A. P. Brainard, and M. Sedlak. 1999. Successful design and development of genetically engineered Saccharomyces yeasts for effective cofermentation of glucose and xylose from cellulosic biomass to fuel ethanol. Adv. Biochem. Eng. Biotechnol. 65:163-192. [DOI] [PubMed] [Google Scholar]
- 19.Ho, N. W. Y., and S. F. Chang. 1989. Cloning of yeast xylulokinase gene by complementation of Escherichia coli and yeast mutations. Enzyme Microb. Technol. 11:417-421. [Google Scholar]
- 20.Jeffries, T. W. 1983. Utilization of xylose by bacteria, yeasts, and fungi. Adv. Biochem. Eng. Biotechnol. 27:1-32. [DOI] [PubMed] [Google Scholar]
- 21.Jeffries, T. W., and N. Q. Shi. 1999. Genetic engineering for improved xylose fermentation by yeasts. Adv. Biochem. Eng. Biotechnol. 65:117-161. [DOI] [PubMed] [Google Scholar]
- 22.Jeppsson, H., N. J. Alexander, and B. Hahn-Hägerdal. 1995. Existence of cyanide-insensitive respiration in the yeast Pichia stipitis and its possible influence on product formation during xylose utilization. Appl. Environ. Microbiol. 61:2596-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin, Y. S., T. H. Lee, Y. D. Choi, Y. W. Ryu, and J. H. Seo. 2000. Conversion of xylose to ethanol by recombinant Saccharomyces cerevisiae containing genes for xylose reductase and xylitol dehydrogenase from Pichia stipitis. J. Microbiol. Biotechnol. 10:564-567. [Google Scholar]
- 24.Johansson, B., C. Christensson, T. Hobley, and B. Hahn-Hägerdal. 2001. Xylulokinase overexpression in two strains of Saccharomyces cerevisiae also expressing xylose reductase and xylitol dehydrogenase and its effect on fermentation of xylose and lignocellulosic hydrolysate. Appl. Environ. Microbiol. 67:4249-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kötter, P., R. Amore, C. P. Hollenberg, and M. Ciriacy. 1990. Isolation and characterization of the Pichia stipitis xylitol dehydrogenase gene, XYL2, and construction of a xylose-utilizing Saccharomyces cerevisiae transformant. Curr. Genet. 18:493-500. [DOI] [PubMed] [Google Scholar]
- 26.Lawlis, V. B., M. S. Dennis, E. Y. Chen, D. H. Smith, and D. J. Henner. 1984. Cloning and sequencing of the xylose isomerase and xylulose kinase genes of Escherichia coli. Appl. Environ. Microbiol. 47:15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu, P., B. P. Davis, J. Hendrick, and T. W. Jeffries. 1998. Cloning and disruption of the beta-isopropylmalate dehydrogenase gene (LEU2) of Pichia stipitis with URA3 and recovery of the double auxotroph. Appl. Microbiol. Biotechnol. 49:141-146. [DOI] [PubMed] [Google Scholar]
- 28.Lu, P., B. P. Davis, and T. W. Jeffries. 1998. Cloning and characterization of two pyruvate decarboxylase genes from Pichia stipitis CBS 6054. Appl. Environ. Microbiol. 64:94-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Passoth, V., B. Schafer, B. Liebel, T. Weierstall, and U. Klinner. 1998. Molecular cloning of alcohol dehydrogenase genes of the yeast Pichia stipitis and identification of the fermentative ADH. Yeast 14:1311-1325. [DOI] [PubMed] [Google Scholar]
- 30.Reizer, A., J. Deutscher, M. H. Saier, and J. Reizer. 1991. Analysis of the gluconate (gnt) operon of Bacillus subtilis. Mol. Microbiol. 5:1081-1089. [DOI] [PubMed] [Google Scholar]
- 31.Richard, P., M. H. Toivari, and M. Penttila. 2000. The role of xylulokinase in Saccharomyces cerevisiae xylulose catabolism. FEMS Microbiol. Lett. 190:39-43. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez-Pena, J. M., V. J. Cid, J. Arroyo, and C. Nombela. 1998. The YGR194c (XKS1) gene encodes the xylulokinase from the budding yeast Saccharomyces cerevisiae. FEMS Microbiol. Lett. 162:155-160. [DOI] [PubMed] [Google Scholar]
- 33.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Rosenfeld, S. A., P. E. Stevis, and N. W. Y. Ho. 1984. Cloning and characterization of the xyl genes from Escherichia coli. Mol. Gen. Genet. 194:410-415. [DOI] [PubMed] [Google Scholar]
- 35.Rothstein, R. J. 1983. One-step gene disruption in yeast. Methods Enzymol. 101:201-211. [DOI] [PubMed] [Google Scholar]
- 36.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Shamanna, D. K., and K. E. Sanderson. 1979. Uptake and catabolism of d-xylose in Salmonella typhimurium LT2. J. Bacteriol. 139:64-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi, N. Q. 2000. Dissecting the respiratory machinery in the xylose-metabolizing yeast Pichia stipitis. Ph.D. thesis. University of Wisconsin, Madison, Wis.
- 40.Shi, N. Q., B. Davis, F. Sherman, J. Cruz, and T. W. Jeffries. 1999. Disruption of the cytochrome c gene in xylose-utilizing yeast Pichia stipitis leads to higher ethanol production. Yeast 15:1021-1030. [DOI] [PubMed] [Google Scholar]
- 41.Shi, N. Q., K. Prahl, J. Hendrick, J. Cruz, P. Lu, J. Y. Cho, S. Jones, and T. Jeffries. 2000. Characterization and complementation of a Pichia stipitis mutant unable to grow on d-xylose or l-arabinose. Appl. Biochem. Biotechnol. 84-86:201-216. [DOI] [PubMed] [Google Scholar]
- 42.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slininger, P. J., R. J. Bothast, J. E. van Cawenberg, and C. P. Kurtzman. 1982. Conversion of d-xylose to ethanol by the yeast Pachysolen tannophilus. Biotechnol. Bioeng. 26:371-384. [DOI] [PubMed] [Google Scholar]
- 44.Stevis, P. E., and N. W. Y. Ho. 1989. Construction of yeast xylulokinase mutant by recombinant DNA techniques. Appl. Biochem. Biotechnol. 20-21:327-334. [Google Scholar]
- 45.Stevis, P. E., J. J. Huang, and N. W. Y. Ho. 1987. Cloning of the Pachysolen tannophilus xylulokinase gene by complementation in Escherichia coli. Appl. Environ. Microbiol. 53:2975-2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tantirungkij, M., T. Izuishi, T. Seki, and T. Yoshida. 1994. Fed-batch fermentation of xylose by a fast-growing mutant of xylose-assimilating recombinant Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 41:8-12. [Google Scholar]
- 47.Tantirungkij, M., N. Nakashima, T. Seki, and T. Yoshida. 1993. Construction of xylose-assimilating Saccharomyces cerevisiae. J. Ferm. Bioeng. 75:83-88. [Google Scholar]
- 48.Toivari, M. H., A. Aristidou, M. Ruohonen, and M. Penttila. 2001. Conversion of xylose into ethanol by recombinant Saccharomyces cerevisiae: importance of xylulokinase (XKS1) and oxygen availability. Metabol. Eng. 3:236-349. [DOI] [PubMed] [Google Scholar]
- 49.Walfridsson, M., M. Anderlund, X. Bao, and B. Hahn-Hägerdal. 1997. Expression of different levels of enzymes from the Pichia stipitis XYL1 and XYL2 genes in Saccharomyces cerevisiae and its effects on product formation during xylose utilisation. Appl. Microbiol. Biotechnol. 48:218-224. [DOI] [PubMed] [Google Scholar]
- 50.Weierstall, T., C. P. Hollenberg, and E. Boles. 1999. Cloning and characterization of three genes (SUT1-3) encoding glucose transporters of the yeast Pichia stipitis. Mol. Microbiol. 31:871-883. [DOI] [PubMed] [Google Scholar]
- 51.Yang, V. W., and T. W. Jeffries. 1997. Regulation of phosphotransferases in glucose- and xylose-fermenting yeasts. Appl. Biochem. Biotechnol. 63-65:97-108. [DOI] [PubMed] [Google Scholar]
- 52.Yu, S., H. Jeppsson, and B. Hahn-Hägerdal. 1995. Xylulose fermentation by Saccharomyces cerevisiae and xylose-fermenting yeast strains. Appl. Microbiol. Biotechnol. 44:314-320. [DOI] [PubMed] [Google Scholar]