Abstract
Rhizosphere bacterial communities of two transgenic potato lines which produce T4 lysozyme for protection against bacterial infections were analyzed in comparison to communities of wild-type plants and transgenic controls not harboring the lysozyme gene. Rhizosphere samples were taken from young, flowering, and senescent plants at two field sites in three consecutive years. The communities were characterized in a polyphasic approach. Cultivation-dependent methods included heterotrophic plate counts, determination of species composition and diversity based on fatty acid analysis of isolates, and community level catabolic profiling. Cultivation-independent analyses were based on denaturing gradient gel electrophoresis (DGGE) of 16S rRNA gene fragments amplified from rhizosphere DNA using primers specific for Bacteria, Actinomycetales, or α- or β-Proteobacteria. Several bands of the DGGE patterns were further characterized by sequence analysis. All methods revealed that environmental factors related to season, field site, or year but not to the T4 lysozyme expression of the transgenic plants influenced the rhizosphere communities. For one of the T4 lysozyme-producing cultivars, no deviation in the rhizosphere communities compared to the control lines was observed. For the other, differences were detected at some of the samplings between the rhizosphere community structure and those of one or all other cultivars which were not attributable to T4 lysozyme production but most likely to differences observed in the growth characteristics of this cultivar.
Rhizobacteria can significantly influence plant growth either positively or negatively (33). After the removal of crop plants, plant-induced changes in the soil microbial community can affect the follow-on crop (2). One possible reason for negative feedback is the enrichment of host-specific pathogens, which is familiar to agronomists (5). Positive feedback might be due to an increase in plant growth-promoting microbial populations. The compositions of compounds which are released from the roots and interact with bacteria in the rhizosphere vary depending on the plant species, cultivar, and physiological status of the plants (45). Genetically engineered plants might change the bacterial consortia in the rhizosphere due to the release of transgene products or an altered composition of root exudates (35). The transgenic potato plants investigated in this study were engineered to overcome limitations of conventional resistance breeding by heterologous expression of the T4 lysozyme gene, which is a strategy applicable to various crop plants and against a variety of economically important crop diseases (11, 30). T4 lysozyme affects gram-positive and gram-negative bacteria either by its muramidase activity against the bacterial cell wall component murein (46) or by a nonenzymatic mechanism which may involve disruption of membranes (14). In contrast to many other transgenic plants, this genetic modification targeted bacteria, and it was shown that plant-associated bacteria were indeed affected, in that the susceptibility of the transgenic potato plants to infection by Erwinia carotovora was significantly reduced (12, 13). Furthermore, it was demonstrated that a detectable amount of T4 lysozyme was released from the roots (6), causing bactericidal activity on the root surface (1). Many plant-associated bacterial species were susceptible in vitro (6, 10, 25). As the effects on different species varied significantly, the T4 lysozyme expression has the potential to change the structure of the bacterial community in the soil fraction which is influenced by the plant roots (i.e., the rhizosphere) and subsequently to affect the quality or function of the soil. Only a few studies of the effects of transgenic crop plants on the composition of bacterial communities in the rhizosphere under field conditions (26, 42) or in the greenhouse (7) have been published so far. In all three studies, only culturable bacteria were investigated.
The objective of this study was to characterize the structure and dynamics of bacterial consortia in the rhizospheres of potato plants under field conditions and to compare them to those of the transgenic plants. Two T4 lysozyme-producing lines, a transgenic control without the T4 lysozyme gene, and the parental line were investigated over 3 years at two distant field sites with different soil types. The bacterial communities were analyzed by three different approaches, which were intended to complement each other. In the first approach, the relative abundances of bacterial species in the rhizosphere were determined based on the cultivation and characterization of isolates by fatty acid analysis. Although this approach is restricted to axenically culturable bacteria, it allowed us to identify species and keep representative strains for further characterization. The second approach was the catabolic profiling of the communities as functional units using Biolog GN microplates (16; reviewed in reference 15). The third approach was based on the analysis of 16S rRNA gene fragments amplified from total rhizosphere DNA by denaturing gradient gel electrophoresis (DGGE) (32; reviewed in reference 31) or by cloning and sequencing. This made it possible to monitor changes in the bacterial rhizosphere consortia, including those bacteria not readily culturable or those in a nonculturable state.
MATERIALS AND METHODS
Plants and field design.
Tubers of genetically modified and unmodified potato plants (Solanum tuberosum L.) were provided by K. Düring (MPB Cologne, Cologne, Germany). The T4 lysozyme-producing plant lines DL4 and DL5 were derived from Solanum tuberosum cv. Désirée and harbor the T-DNA from the binary vector pSR8-30 (9) containing the nptII marker gene and the T4 lysozyme gene in a small polylinker site (11). The transgenic plants constitutively expressed the bacteriophage T4 lysozyme under control of the cauliflower mosaic virus 35S promoter (13). The lysozyme gene was fused to the barley α-amylase signal peptide gene. This leading sequence caused the secretion of the lysozyme into the intercellular spaces (13, 24). The transgenic control plant line DC1 was transformed with the same construct as DL4 and DL5 but without the T4 lysozyme gene. Plants derived from the parental cell line, DES, were taken as the nontransgenic control. Tubers of DES, DC1, DL4, and DL5 were planted at Groβ Lüsewitz near Rostock (field L) and Quedlinburg (field Q) in complete randomized block design in three consecutive years. Fields and tillage were provided by the Bundesanstalt für Züchtungsforschung (Quedlinburg, Germany). The fields and sampling times are described in Table 1. The soil was sandy loam with 1.1% organic matter in field L and silt loam with 2.2% organic matter in field Q; both soils had pH 5.9 (26).
TABLE 1.
Fields and samplings
| Field description | Sampling | Wk after planting |
|---|---|---|
| Field site, Quedlinburg; first field (8 blocks | Q1 | 5 |
| with 4 plots each, i.e., 8 replicate plots | Q2 | 13 |
| per plant line); first year of continuous | Q3 | 16 |
| potato cultivation; planting in May 1996 | ||
| Field site, Quedlinburg; second field (4 | Q4 | 7 |
| blocks with 4 plots each; the first field | Q5 | 10 |
| was devastated); second year of | Q6 | 13 |
| continuous potato cultivation; planting | Q7 | 16 |
| in April 1997 | Q8 | 19 |
| Field site, Quedlinburg; second field (4 | Q9 | 10 |
| blocks with 4 plots each); third year of | Q10 | 13 |
| continuous potato cultivation; planting | Q11 | 16 |
| in April 1998 | Q12 | 19 |
| Q13 | 22 | |
| Field site, Groβ Lüsewitz near Rostock; | L1 | 6 |
| first field (8 blocks with 4 plots each); | L2 | 11 |
| first year of continuous potato | L3 | 17 |
| cultivation; planting in May 1996 | ||
| Field site, Groβ Lüsewitz near Rostock; | L4 | 9 |
| first field (8 blocks with 4 plots each); | L5 | 17 |
| second year of continuous potato | L6 | 20 |
| Field site, Groβ Lüsewitz near Rostock; | L9 | 5 |
| second field (3 blocks with 4 plots each); | L10 | 10 |
| first year of continuous potato | L11 | 15 |
| cultivation; planting in April 1998 |
Sampling.
A rhizosphere sample consisted of 5 g (wet weight) of roots with adhering soil from three to five plants from one plot. All plots were sampled at least three times a year, from young plants (principal growth stage 1, according to Hack et al. [19]), flowering plants (principal growth stage 6), and senescent plants shortly before harvest of the tubers (principal growth stage 9). The samples were extracted three times for 1 min at 260 rpm in a stomacher blender with sterile solutions of (i) 0.3 g of Chelex-100 (Bio-Rad, Hercules, Calif.) in 20 ml of phosphate-buffered saline (40), (ii) 15 ml of SDP (0.02% sodium deoxycholate, 0.5% polyethylene glycol), and (iii) 15 ml of distilled water, according to a protocol slightly modified from that of Herron and Wellington (20). The three extracts were combined, 1 ml was used for serial dilution and plating, 2 ml were centrifuged (20 min; 14,000 × g; 4°C), and the pellets were stored at −20°C for later extraction of DNA. From the rest, most of the soil particles were removed by a low-speed centrifugation step (2 min; 500 × g; 20°C), and then microorganisms were harvested from the supernatant (20 min; 5,000 × g; 20°C) and resuspended in 5 ml of sterile saline (0.85% NaCl). An aliquot of 0.5 ml was diluted in 17 ml of sterile saline and mixed, and 0.15 ml was transferred to each well of a Biolog GN microplate for community level catabolic profiling.
Community level catabolic profiling in Biolog GN microplates.
Community level catabolic profiles were determined for eight replicate rhizosphere samples per plant line from 10 samplings of fields Q and L. The mixtures of microorganisms extracted from the rhizosphere samples were incubated in Biolog GN microplates in the dark at room temperature (20 to 25°C). The optical density (OD) was measured with a computer-linked microplate reader (Vmax; Molecular Devices Corp., Menlo Park, Calif.) at 595 nm at several time points after the first appearance of color. The data analysis was done as described by Glimm et al. (17). Shortly, the raw OD values in each microplate were corrected for the background color (OD0). The time point at which the highest number of wells over all microplates exceeded an OD0 of 0.1 (significant catabolic activity) was chosen for analysis. The OD0 was divided by the average well color of the plate to reduce the bias due to differing initial cell densities of the samples. The 95 variables were transformed to principal components (PCs) using the covariance of the variables. The first five PCs were used to test for significant differences between T4 lysozyme-producing plants and control plants (multivariate analysis of variance [MANOVA], P < 0.1).
Plating and characterization of bacterial isolates.
Aerobe heterotrophic plate counts were determined on R2A medium (Difco Laboratories, Detroit, Mich.), supplemented with 100 mg of cycloheximide (Sigma, Steinheim, Germany) liter−1, after 4 days' incubation at 20 to 23°C. The species compositions of the cultured rhizobacteria from the four plant lines were determined for all three samplings of field Q in the first year and for the last samplings of Q and L in the second year. For each sample, an equal number of colonies were randomly selected and purified on Trypticase soy broth (Becton Dickinson, Cockeysville, Md.) supplemented with 15 g of Bacto Agar (Difco Laboratories) liter−1 to obtain axenic cultures. A total of 2,010 strains were characterized by fatty acid analysis according to the standard procedure of the Microbial Identification System (MIS; MIDI Inc., Newark, N.J.).
Identification and classification of bacterial strains and analysis of species composition.
The MIS database TSBA (revision 3.9) was searched to identify the strains by their fatty acid profiles. To assign unnamed strains to known bacterial groups, their fatty acid profiles were compared to a database of 1,856 identified strains (MIS similarity index [SIM] > 0.2) which were isolated from the potato phyllosphere and rhizosphere. The strains were grouped into gram-positive bacteria, Cytophaga-Flavobacterium-Bacteroides (CFB) group bacteria, and gram-negative bacteria according to combinations of signature fatty acids (mainly with regard to hydroxylation or branching). The procedure FASTCLUS of the statistical package SAS (SAS Institute Inc., Cary, N.C.) was used to determine the most similar strain identified and to calculate the Euclidian distance, using the datasets of strains for which SIM was >0.2 as fixed cluster seeds. PC analysis (SAS procedure PRINCOMP, using the covariance matrix) was performed to compare the relative species abundances in the rhizospheres of the four plant lines. Shannon's diversity index H (41), which is a measure of both species richness and the degree of equitability of the relative species abundances, was calculated from the number of species, s, which were isolated from a plant line as determined by fatty acid analysis, and their proportion, p, of the total number of isolates: H = −Σ pi (log pi), where i = 1, . . . , s. To correct for differing numbers of isolates from the different plant lines at the five samplings, H was calculated repeatedly from random subsets of 47 strains until the average H did not change by further iterations (40 times).
Molecular analysis of the rhizosphere communities.
Total DNA was extracted by the method of Smalla et al., which included an enzymatic and a bead beating step for cell lysis, phenol-chloroform extractions, and a purification by glass milk (43). Amplification of the bacterial 16S ribosomal DNA (rDNA) fragment between positions 968 and 1401 (Escherichia coli numbering [3]) by PCR and separation of the products by DGGE were done as previously described (20a). A denaturing gradient of 3.2 to 4.6 M urea plus 16.0 to 23.2% (vol/vol) formamide was used for DGGE, which was performed in 1× Tris-acetate-EDTA buffer at 60°C and a constant voltage of 180 V for 4 h. A mixture of the 16S rDNA fragments from the following 11 bacterial species was applied to each DGGE gel as a marker: Clostridium pasteurianum DSM 525(a), E. carotovora DSM 30168(b), Agrobacterium tumefaciens DSM 30205(c), Pseudomonas fluorescens R2f(d), Pantoea agglomerans (e), Nocardia asteroides N3(f), Rhizobium leguminosarum DSM 30132(g), Actinomadura viridis DSM 43462(h), Kineosporia aurantiaca JCM 3230(i), Nocardiopsis atra ATCC 31511(j), and Actinoplanes philippinensis JCM 3001(k). Acid silver staining was used for detection of DNA in DGGE gels (39). Selective amplification of 16S rDNA from Actinomycetales in a first PCR followed by amplification of the nested fragment for DGGE analysis was done as previously described (23). The same approach was analogously applied to analyze the α- and β-proteobacterial populations in the rhizosphere, using F203α (CCGCATACGCCCTACGGGGGAAAGATTTAT) and F948β (CGCACAAGCGGTGGATGA) as taxon-specific primers (18). Two strategies were applied to characterize bands of the DGGE fingerprints by sequence analysis. The first approach was to excise bands from DGGE gels stained with SYBRgreen (BMA, Vallensbaek Strand, Denmark). The recovered DNA was reamplified and cloned with the pGEM-T kit (Promega, Mannheim, Germany). Cloned fragments were checked by DGGE to assess whether the desired band was cloned and were sequenced. The second approach was to amplify and clone large fragments of the 16S rDNA from total rhizosphere DNA for sequencing of ca. 1,400 bp between E. coli positions 28 and 1491. Sequences were analyzed using Chimera Check of the Ribosomal Database Project II (29) and the ARB software package (Department of Microbiology, Technical University of Munich, Munich, Germany [http://www.arb-home.de]).
Nucleotide sequence accession numbers.
Sequence accession numbers in GenBank are AY048885 to AY048918.
RESULTS
Phenotypes of the potato lines.
The plant lines DES, DC1, and DL5 showed normal growth in the field, whereas DL4 appeared to have a reduced stem length and smaller leaves when the plants were young (principal growth stages 1 to 5), and the progress of senescence during growth stage 9 was enhanced.
Community level catabolic profiling.
The mixtures of microorganisms extracted from the rhizospheres of the different plant lines were compared as functional units in their relative respiration rates of the 95 sole carbon sources which are supplied in Biolog GN microplates. To test sensitively for differences, the multivariate data sets were normalized and reduced in dimensionality to five PCs typically explaining more than 70% of the total variance and were compared by MANOVA. Significant differences (P < 0.1) in the community level catabolic potential were detected between rhizobacteria from DL4 and DC1 in six samplings, and in four of these samplings also between rhizobacteria from DL4 and DES (Table 2). The other T4 lysozyme-producing plant line, DL5, was never significantly different from the control lines. The deviation of DL4 did not correlate well with a particular plant growth stage, but it was most pronounced for senescent plants. A specific set of carbon sources correlated with the phenomenon could not be identified due to the variability of the Biolog pattern between samplings. The sets of utilized carbon sources were highly similar among all microplates of a sampling event irrespective of the plant line from which the inoculated sample originated. Three Biolog wells, where significant (univariate) differences in catabolic rates among samples from DL4 and both control lines were detected, were sampled for DGGE analysis after the experiment. For all plant lines, the DGGE fingerprints showed the same set of ribotypes being enriched by growth on the sole carbon source (data not shown). Only small differences in their relative abundances were observed. This indicated the sensitivity of the catabolic profiling and the detection of minor shifts in the community structure.
TABLE 2.
Pairwise statistical comparisons of community-level catabolic profiles (CLCP) of rhizosphere samples from T4 lysozyme-producing plants (DL4 and DL5) and control plants (DES and DC1) and aerobe bacterial plate counts
| Field | Plant growth stage | Significant difference in CLCP of rhizosphere samples in plant lines (8 replicates; P < 0.1)a:
|
Plate countb
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| DES-DL4 | DC1-DL4 | DES-DL5 | DC1-DL5 | DES | DC1 | DL4 | DL5 | ||
| L (1st yr) | Young | Yes | 7.2 | 7.1 | 7.0 | 7.2 | |||
| Flowering | 7.9 | 7.7 | 7.7 | 7.7 | |||||
| Senescent | Yes | Yes | 7.6 | 7.5 | 7.6 | 7.7 | |||
| Q (1st yr) | Young | 7.8 | 7.7 | 7.7 | 7.7 | ||||
| Flowering | Yes | 7.8 | 7.7 | 7.8 | 7.8 | ||||
| Senescent | Yes | Yes | 7.8A | 7.8A | 9.0B | 7.9A | |||
| L (2nd yr) | Young | Yes | Yes | 7.3 | 7.1 | 7.2 | 7.4 | ||
| Flowering | 8.2 | 8.2 | 8.3 | 8.2 | |||||
| Senescent | 8.6 | 8.7 | 8.6 | 8.5 | |||||
| Q (2nd yr) | Senescent | Yes | Yes | 8.2A | 8.0A,B | 7.9B | 8.1A | ||
MANOVA on the first five PCs (details in Materials and Methods).
Average log10 CFU g−1 (wet weight) of root. Significant differences within a sampling are indicated by different letters (ANOVA; P < 0.1).
Aerobe heterotrophic plate counts.
The numbers of aerobic bacteria that could be recovered from the rhizospheres of the four plant lines were determined in four samplings for field Q and six samplings for field L (Table 2). Only in the two samplings of senescent plants in field Q were significant differences (P < 0.1) between numbers of CFU of the potato lines apparent: samples from DL4 showed higher plate counts than all other plant lines in the first year but slightly less than DES and DL5 in the second year. This deviation of the DL4 samples coincided with reduced diversity indices (H) (Table 3). The average rhizosphere population sizes over the 10 samplings did not significantly differ between T4 lysozyme-producing and control plants (P > 0.1; paired Student's t tests).
TABLE 3.
Percentages of species and diversity of cultured rhizobacteria from plant lines DC1, DES, DL4, and DL5 in five samplings
| Identification and classification by fatty acid profiles | Percentage
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First year
|
Second year
|
||||||||||||||||||||
| Young plants (Q)
|
Flowering plants (Q)
|
Senescent plants (Q)
|
Senescent plants (Q)
|
Senescent plants (L)
|
|||||||||||||||||
| DC1 | DES | DL4 | DL5 | DC1 | DES | DL4 | DL5 | DC1 | DES | DL4 | DL5 | DC1 | DES | DL4 | DL5 | DC1 | DES | DL4 | DL5 | ||
| Chryseobacterium spp. | 6 | 2 | 3 | 3 | 1 | 3 | 1 | 1 | 5 | 3 | 2 | 21 | 24 | 14 | 15 | ||||||
| Flavobacterium spp. | 6 | 9 | 2 | 22 | 5 | 3 | 4 | 3 | 6 | 7 | 15 | 3 | 15 | 13 | 7 | 4 | 7 | 5 | 15 | 10 | |
| Sphingobacterium spp. | 2 | 2 | 2 | 7 | 2 | 1 | 4 | 3 | 3 | ||||||||||||
| Other CFB group bacteria | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 7 | 2 | 1 | ||||||||
| Agrobacterium spp. | 2 | 3 | 2 | 3 | 1 | 3 | 5 | 2 | 5 | 1 | 1 | 1 | 2 | 3 | 6 | 5 | |||||
| Bradyrhizobium japonicum | 1 | 2 | |||||||||||||||||||
| Brevundimonas vesicularis | 2 | 1 | 2 | 1 | |||||||||||||||||
| Methylobacterium spp. | 1 | 3 | 1 | ||||||||||||||||||
| Other α-proteobacteria | 2 | 3 | 1 | 1 | 1 | 6 | 2 | 2 | 2 | 1 | 1 | ||||||||||
| Acidovorax spp. | 2 | 2 | 1 | 1 | 1 | 2 | |||||||||||||||
| Comamonas acidovorans | 2 | 2 | 4 | 9 | 3 | 10 | 9 | 5 | 3 | 7 | 10 | 7 | 11 | 4 | 2 | 2 | 6 | 5 | 3 | ||
| V. paradoxus | 8 | 7 | 2 | 4 | 16 | 15 | 14 | 15 | 15 | 6 | 3 | 7 | 3 | 2 | 2 | 2 | 3 | 3 | 3 | ||
| Burkholderia spp. | 2 | 2 | 2 | 3 | 2 | 1 | 1 | 1 | |||||||||||||
| Ralstonia pickettii | 5 | 4 | 5 | 9 | 6 | ||||||||||||||||
| Other β-proteobacteria | 2 | 1 | 2 | 4 | 4 | 2 | 6 | 3 | 2 | 6 | |||||||||||
| Enterobacter spp. | 2 | 6 | 1 | 3 | 8 | 3 | 4 | 1 | 1 | 8 | 5 | ||||||||||
| Pectobacterium spp. | 1 | 2 | 1 | 1 | 2 | 1 | |||||||||||||||
| Pantoea spp. | 2 | 1 | 1 | 4 | 3 | 2 | 3 | 3 | 3 | 3 | |||||||||||
| Other Enterobacteriaceae | 4 | 2 | 2 | 2 | 2 | 3 | 5 | 2 | 2 | 5 | 3 | 1 | 8 | 1 | 3 | ||||||
| P. chlororaphis | 4 | 7 | 9 | 4 | 3 | 8 | 2 | 5 | 3 | 9 | 2 | 3 | 3 | 6 | 4 | 15 | 9 | 6 | 10 | ||
| P. fluorescens | 14 | 5 | 15 | 1 | 5 | 3 | 11 | 8 | 7 | 4 | 9 | 6 | 13 | 9 | 6 | 3 | 2 | 6 | |||
| P. putida | 2 | 2 | 2 | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 10 | 6 | 12 | 9 | 12 | 2 | 7 | 8 | |||
| P. syringae | 2 | 8 | 14 | 2 | 3 | 2 | 1 | 3 | 6 | 12 | 5 | 3 | 2 | 4 | 3 | 2 | 1 | ||||
| Other Pseudomonaceae | 8 | 2 | 6 | 10 | 7 | 14 | 14 | 14 | 13 | 24 | 1 | 2 | 1 | 6 | 4 | 2 | 1 | ||||
| Stenotrophomonas maltophilia | 8 | 3 | 2 | 8 | 8 | 5 | 3 | 2 | 5 | 6 | 2 | 1 | 3 | 4 | 13 | 9 | 5 | 7 | 7 | ||
| Xanthomonas campestris | 3 | 2 | 1 | 2 | 3 | 3 | 1 | 3 | 1 | ||||||||||||
| Acinetobacter; Moraxella | 3 | 2 | 3 | 2 | 1 | 1 | 3 | 1 | 2 | 2 | 2 | 3 | 2 | ||||||||
| Clavibacter michiganensis | 1 | 1 | 2 | 1 | 1 | 2 | |||||||||||||||
| Microbacterium spp. | 6 | 2 | 3 | 3 | 3 | 1 | 3 | 1 | 1 | 2 | 2 | 3 | 2 | 2 | 4 | 1 | 1 | ||||
| Rathayibacter tritici | 4 | 2 | 2 | 2 | 3 | 1 | 3 | 1 | 1 | 1 | |||||||||||
| Arthrobacter spp. | 7 | 2 | 6 | 4 | 3 | 3 | 7 | 2 | 1 | 3 | 1 | 2 | 1 | 1 | 2 | ||||||
| Kocuria spp. | 2 | 3 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 3 | 6 | 2 | 1 | 1 | |||||||
| Other Micrococcineae | 2 | 5 | 2 | 1 | 3 | 1 | 1 | 2 | 2 | 4 | 1 | 1 | |||||||||
| Corynebacterineae | 1 | 3 | 1 | 2 | 1 | 2 | 3 | 1 | 1 | 6 | 1 | ||||||||||
| Bacillus and related | 20 | 41 | 30 | 18 | 9 | 6 | 12 | 7 | 4 | 2 | 2 | 1 | 5 | 2 | 2 | 1 | 1 | ||||
| Other gram-positive bacteria | 6 | 5 | 6 | 8 | 12 | 17 | 6 | 16 | 6 | 3 | 7 | 1 | 2 | 6 | 2 | 1 | |||||
| Uncertain affiliation | 8 | 2 | 4 | 3 | 2 | 2 | 1 | 2 | 2 | 2 | 4 | 7 | 5 | 3 | 3 | 1 | 1 | ||||
| No. of isolates analyzed | 51 | 58 | 66 | 49 | 116 | 115 | 104 | 116 | 116 | 118 | 122 | 121 | 129 | 129 | 123 | 123 | 82 | 97 | 88 | 87 | |
| Diversity index (H) | 2.8 | 2.7 | 2.9 | 2.6 | 2.8 | 2.8 | 2.9 | 2.8 | 2.9 | 2.9 | 2.6 | 2.9 | 3.0 | 3.2 | 2.8 | 3.1 | 2.7 | 2.9 | 3.1 | 2.9 | |
Species compositions of axenically cultured rhizobacteria.
The species compositions of the cultured rhizobacteria from the four plant lines were determined for young, flowering, and senescent plants in field Q in the first year and for senescent plants in fields Q and L in the second year (Table 3). A diverse spectrum of species from the α, β and γ subdivision of the Proteobacteria, the CFB group, actinobacteria, and low-G+C gram-positive bacteria was isolated from the rhizospheres of all four plant lines, as determined by identification of strains based on their fatty acid profiles. A total of 156 species could be distinguished, of which 28 species represented 75% of the 2,010 strains analyzed. Most frequently isolated were Variovorax paradoxus (6.6%), P. fluorescens (6.4%), Flavobacterium johnsoniae (6.3%), and Pseudomonas chlororaphis (5.6%). Differences in species or genus composition of rhizosphere samples from T4 lysozyme-producing plants and control lines were apparent for single samplings but were not consistent (Table 3). For instance, Chryseobacterium was more frequently detected in samples from the senescent control plants from field L in the second year but was rarely found in samples from field Q in both years. Flavobacterium, Agrobacterium, Comamonas, and Enterobacter were more often cultured from T4 lysozyme-producing plants in some samplings but more often from control plants in other samplings. The higher abundance of Pseudomonas syringae and Acinetobacter baumannii in the rhizospheres of young T4 lysozyme plants and of Burkholderia cepacia in the rhizospheres of senescent control plants in the first year could not be confirmed in other samplings. All these deviations in species or genus abundance were the effect of natural variation rather than of T4 lysozyme production.
The taxonomic distribution of all strains retrieved from the five samplings is summarized in Table 4. This shows a slightly reduced occurrence of α-Proteobacteria, mainly represented by the genus Agrobacterium, for the samples from T4 lysozyme plants. For all other taxa, the differences within the groups of T4 lysozyme plants and control plants exceeded the difference between those groups. Compared to the other plant lines, the exceptionally high abundance of the genus Pseudomonas in the rhizosphere of DL4 was noticeable. Gram-positive bacteria, which on average might be more affected by the muramidase activity of T4 lysozyme than other bacteria, were more frequently recovered from the wild-type DES (27% of all strains from DES) than from the T4 lysozyme-producing lines DL4 and DL5 (23%). Nevertheless, the transgenic control line DC1 showed the same percentage of gram-positive strains as DL4 and DL5. The higher percentage of gram-positive bacteria in the rhizosphere of DES could be attributed exclusively to the sampling of young plants. The percentages of gram-positive bacteria decreased during the season (first year; field Q), from an average of 45% for young plants and 35% for flowering plants to 12% for senescent plants. The low proportion of gram-positive bacteria in the rhizospheres of senescent plants was also observed in the second year in fields Q and L (13 and 8%, respectively).
TABLE 4.
Taxonomic distribution of strains from the rhizospheres of T4 lysozyme-producing plants (DL4 and DL5) and control plants (DC1 and DES) in five samplings
| Taxonomic affiliation (most frequent genus or species of group) | Percentage of strains from plant line:
|
|||
|---|---|---|---|---|
| DES | DC1 | DL4 | DL5 | |
| α-Proteobacteria (Agrobacterium) | 4 | 6 | 3 | 2 |
| β-Proteobacteria (V. paradoxus) | 15 | 17 | 14 | 16 |
| Enterobacteriaceae (Enterobacter) | 7 | 5 | 4 | 8 |
| Pseudomonas (P. fluorescens) | 21 | 26 | 32 | 25 |
| CFB group (Flavobacterium) | 17 | 15 | 16 | 15 |
| Other gram-negative bacteria (Stenotrophomonas) | 7 | 8 | 8 | 9 |
| Actinomycetales (Arthrobacter) | 16 | 13 | 11 | 15 |
| Gram-positive low G+C (Bacillus) | 11 | 7 | 10 | 6 |
| Unidentified gram-positive bacteria | 1 | 3 | 2 | 2 |
| No. of strains analyzed | 517 | 494 | 503 | 496 |
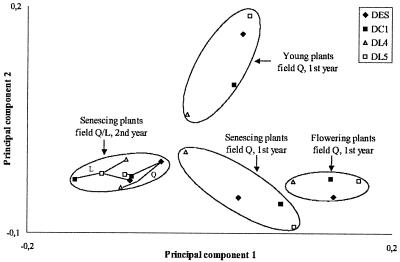
The distributions of the more abundant species (>5% of all isolates) were compared among the different plant lines and samplings by PC analysis in order to consider concerted effects on two or more species (Fig. 1). Each datum point represented the distribution of species which were isolated from the rhizosphere of a plant line at one sampling event. The species patterns clustered in the first and second PCs according to the plant growth stage and year of sampling, whereas the production of T4 lysozyme was not a major factor. The patterns from young plants were discriminated from the patterns of older plants mainly in the second PC, which was highly correlated with the prevalence of Bacillus megaterium (Pearson correlation, r2 = 0.75). The species patterns from flowering and senescent plants were mainly separated by the first PC due to the higher percentages of V. paradoxus (r2 = 0.76) and Streptoverticillium (r2 = 0.75) in the rhizospheres of flowering plants. The samples from DL4 had smaller values of the first PC than the other plant lines for all samplings of field site Q. The samples of the second year from both field sites were highly similar irrespective of the plant line they originated from. For all samplings, the average distance in the PC plots between control plants and T4 lysozyme plants was small compared to the average within-group distance. Thus, the differences in species composition could not be attributed to the T4 lysozyme factor but rather to other plant properties, where DL4 and DL5 especially differed.
FIG. 1.
PC analysis of the relative species compositions (based on identification by fatty acids) of cultured rhizobacteria from the plant lines DC1, DES, DL4, and DL5 in five samplings. The clustering of the species patterns was based on PCs 1 and 2, explaining 28 and 16% of the variance, respectively.
Genetic fingerprints of the bacterial rhizosphere communities by DGGE.
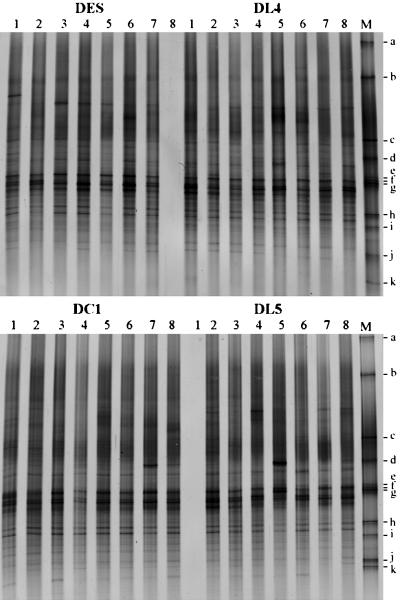
Molecular analysis of the bacterial rhizosphere communities was done by PCR amplification of 16S rRNA genes with bacterium-specific primers from total DNA of each sample and separation in DGGE based on sequence variability. The rhizosphere communities of the four plant lines were compared by DGGE profiling for all 22 samplings which were taken in three consecutive years at the two field sites and at various growth stages of the plants. For each sampling, community fingerprints were done in DGGE gels comparing the replicate samples of a T4 lysozyme-producing plant line to a control plant line. The DGGE fingerprints of rhizosphere communities from young plants, which were growing in separate plots of field L in the first year, are shown in Fig. 2. Despite some variable bands, all fingerprints from the different plots were highly similar. Bands specific for the T4 lysozyme plants or the control plants were not observed, which indicated that neither the plant line nor the production of T4 lysozyme significantly affected the abundant bacterial species in the rhizosphere. This result was observed for nearly all samplings. The only exception was sampling Q3 (field Q; first year; senescent plants), where a particular band was dominant in the DGGE fingerprints of seven of the eight replicate rhizosphere samples of plant line DL4. In the fingerprints from rhizosphere samples of the other plant lines, this band was much weaker, with the exception of a single sample from plant line DES. The sequence of the DNA from this band could be attributed to the species E. carotovora. The plants did not show symptoms of an infection with this species, but senescence was more pronounced than for the other plants. The other T4 lysozyme-producing plant line DL5 corresponded in the DGGE fingerprints with the control lines DES and DC1 in sampling Q3 as well as in all other samplings.
FIG. 2.
DGGE profiles of bacterial rhizosphere communities of young potato plants (first year; field L). Each lane represents a rhizosphere sample from one field plot generated by amplification of 16S rRNA gene fragments with bacterium-specific primers from rhizosphere DNA and electrophoretic separation of the PCR products in a denaturing gradient (insufficient PCR product was obtained from two samples). DL4 and DL5, T4 lysozyme-expressing cultivars; DES and DC1, control cultivars without the T4 lysozyme gene; M, standard with fragments a to k (see Materials and Methods).
Effects of environmental factors other than T4 lysozyme analyzed by DGGE.
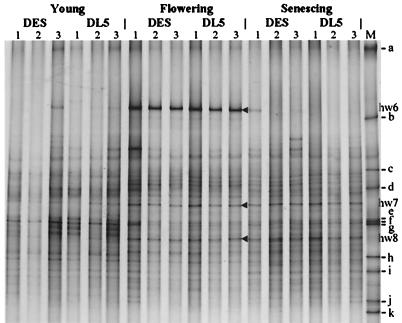
In order to draw conclusions about the biosafety of the transgenic plants, we had to analyze whether DGGE analysis is sensitive enough to detect relevant changes in the bacterial community structure. A relevant perturbation should cause deeper changes in the community structure than changes due to common environmental factors related to the season, field site, or year. To analyze this natural variability, samples representative of all samples of a particular sampling with regard to their DGGE fingerprints were selected for within-gel comparison between different samplings. In Fig. 3, a DGGE gel is shown which demonstrates seasonal changes of the bacterial community structure in the rhizospheres of DES and DL5 (field L; third year). Once again, it became obvious that the fingerprints from samples of the transgenic and the wild-type plants did not differ significantly. However, seasonal shifts in the community structure became apparent. Band hw6 was most intense for flowering plants. It represented a ribotype with a high sequence identity (99.5%) to that of Serratia ficaria JCM1241. Many band intensities changed when the plants reached the growth stage of flowering and remained stable until the harvest of the potatoes, e.g., bands hw7 and hw8. Band hw7 represented an α-proteobacterium with 95.7% sequence identity of the DGGE fragment to the type strain of Devosia riboflavina. Identical sequences were found in three samplings (L3, L10, and L11). Band hw8 originated from the 16S rRNA gene of a streptomycete (99.0% sequence identity to Streptomyces olivoreticuli). Also, for hw8 it was confirmed that the band represented identical sequences in fingerprints of samplings L10 and L11. In general, fingerprints of rhizosphere samples from senescent plants were more variable than those of samples from young or flowering plants with respect to strong bands in the upper part of the DGGE gels, some of which could be attributed to enterobacterial genera (Enterobacter, Serratia, and Kluyvera), as determined by sequencing of the DNA from the respective bands (B3, hw6, and hw3).
FIG. 3.
DGGE profiles showing the seasonal dynamics of bacterial communities in the rhizospheres of the potato plant lines DES and DL5 (field L; third year). Bands hw6, hw7, and hw8 were excised for DNA sequence determination. M, standard with fragments a to k (see Materials and Methods).
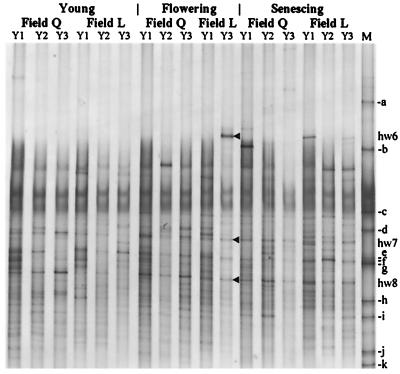
In Fig. 4, representative fingerprints of samples from the two field sites and from different years are shown for comparable growth stages of the plants. A significant natural variability could be detected. Many bands appeared only temporarily (e.g., hw6). The bacterial rhizosphere communities were more influenced by environmental factors related to the field site and the year of sampling than by those related to seasonal changes. Despite this variability, some bands were present in all fingerprints (e.g., hw8), and many others were present in samples from different years or different field sites (e.g., hw7). This showed that part of the community was maintained under very different environmental conditions with regard to the soil type, climate, and state of the plant roots.
FIG. 4.
Temporal and spatial variability of rhizosphere communities as determined by DGGE analysis of samples from plant line DES at three growth stages. Each lane represents a typical DGGE profile of the respective sampling of fields Q or L in three consecutive years (Y1, Y2, and Y3). M, standard with fragments a to k (see Materials and Methods); hw6 to hw8, sequenced DNA fragments.
DGGE analysis of α- and β-Proteobacteria and Actinomycetales.
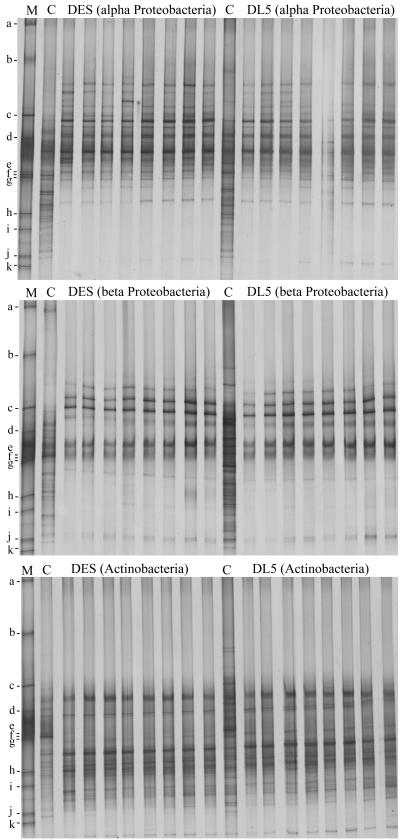
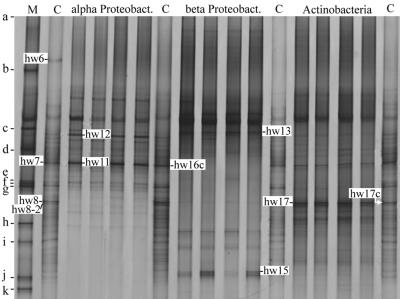
Specific primers were used to selectively amplify 16S rRNA genes of the taxa α-Proteobacteria, β-Proteobacteria, and Actinomycetales from total rhizosphere DNA in a first PCR. The diluted PCR products were used as targets for DGGE analyses in order to compare rhizosphere communities of T4 lysozyme-producing and control plants on the taxon level. It was apparent that an effect of T4 lysozyme on the bacterial rhizosphere populations belonging to the three taxa was not detectable (Fig. 5). The sensitivity was increased by the taxon-specific approach, because many bands of these profiles were not visible or hardly visible in the total bacterial profiles (Fig. 5 and 6, lanes C), especially bands of β-proteobacteria. In contrast, some of the α-proteobacteria (e.g., the population represented by band hw7) and many of the Actinomycetales (e.g., the population represented by band hw8) were among the most abundant community members, as the taxon-specific fingerprints partially agree with the total bacterial profiles (Fig. 6). The specificity of the approach was confirmed by recovery of DNA from bands of the taxon-specific fingerprints (Fig. 6) and sequence analyses. The sequences of the DNA recovered from the bands hw11 and hw12 of the α-proteobacterial profile were affiliated with the α-proteobacteria, hw13 and hw15 of the β-proteobacterial profile were affiliated with the β-proteobacteria, and hw17 of the actinomycete profile was affiliated with the genus Streptomyces (Fig. 7). The taxon-specific DGGE fingerprints of DES and DL5 were also virtually identical for other samplings, while seasonal shifts were easily detectable (data not shown).
FIG. 5.
Taxon-specific DGGE profiles of rhizosphere communities from plant lines DES and DL5 (senescent plants; field L; second year). C, bacterial community DGGE profiles; M, standard with fragments a to k (see Materials and Methods).
FIG. 6.
Sequenced DNA fragments (hw) excised from taxon-specific and bacterial DGGE profiles of rhizosphere communities from plant line DL5 (senescent plants; field L; third year). C, bacterial community DGGE profiles; M, standard with fragments a to k (see Materials and Methods).
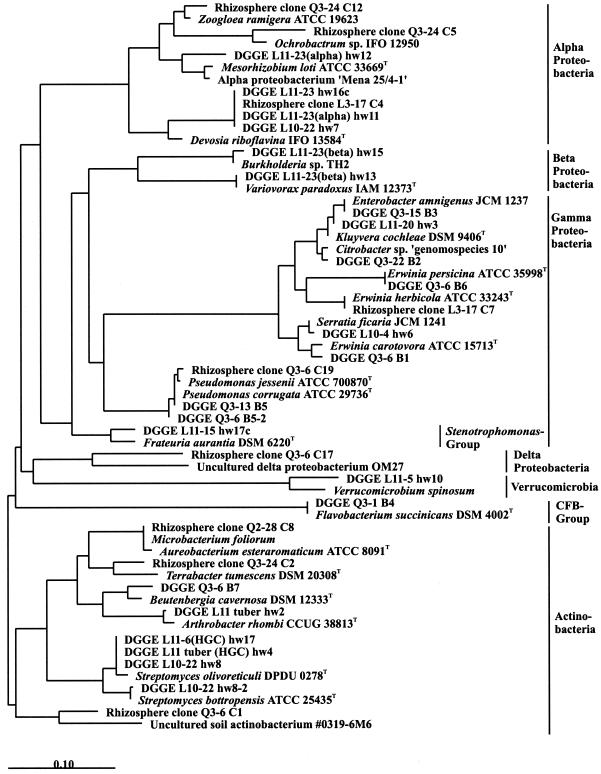
FIG. 7.
Phylogenetic sequence analysis of 16S rRNA gene fragments. C1 to C19, cloned 1.5-kb fragments amplified from rhizosphere DNA and selected to correspond to prominent bands of DGGE profiles from potato rhizospheres; B1 to B7 (AF060530 to AF060536 [20a]) and hw2 to hw17c, sequences of DNA extracted from prominent bands of potato rhizosphere DGGE profiles (see the text). The tree is based on a maximum-likelihood tree of full sequences (ARB package). New sequences were added by parsimony using filters (the original data and 50% conservation filters provided by ARB) without disturbing the general topology of the tree, and unnecessary sequences were removed.
Sequence analysis of amplified 16S rRNA genes.
Two strategies were applied to characterize bands of the DGGE fingerprints by sequence analysis. The first approach was to excise bands from DGGE gels and reamplify and clone the recovered DNA for sequencing. Altogether, 21 sequences were obtained which were not suspected to be chimeric (DGGE bands B1 to B7 (20a) and hw3 to hw17c [Fig. 7]). The second approach was to amplify and clone 1.5-kb fragments of the 16S rDNA from total rhizosphere DNA. A library of 80 clones from three samplings (Q2, Q3, and L3) was analyzed in DGGE for correspondence with bands of the rhizosphere community fingerprints. Nine clones were selected for sequence analysis (rhizosphere clones C1 to C19 [Fig. 7]). The rhizobacteria from which the sequences originated were phylogenetically affiliated with the α-Proteobacteria, the families Enterobacteriaceae and Pseudomonadaceae, the Stenotrophomonas group of the γ-Proteobacteria, the δ-Proteobacteria, verrucomicrobia, CFB group bacteria, and the Actinomycetales. Sequences of β-proteobacteria (Variovorax and Burkholderia) were found only if this group was specifically amplified from total rhizosphere DNA, providing more evidence for its lower abundance in the potato rhizosphere relative to other taxa. In contrast, the percentage of cultured β-proteobacterial strains from the potato rhizosphere was more than 10% (mainly Variovorax and Comamonas). From rhizosphere DNA of sampling L3, a 1.5-kb 16S rDNA fragment was cloned and sequenced in which the sequence of the DGGE fragment was identical with that of band hw7. This band was highly abundant in samples from most of the samplings. The nearly complete 16S rDNA had only 92.1% sequence identity with the most similar database sequence, which belongs to the α-proteobacterium D. riboflavina. Thus, this sequence probably originated from an abundant but still-uncultured rhizobacterium.
DISCUSSION
This extensive field study of the impact of T4 lysozyme-producing potato plants on bacterial communities in agricultural soil contributed to the biosafety issue of whether genetically engineered crop plants have the potential to unintentionally change the agroecosystem by selecting bacterial communities different from those selected by conventionally bred varieties. In contrast to many other transgenic plants, the genetic modification targeted bacteria (12, 13), the transgene product had the potential to affect many rhizobacterial species (6, 10), and the roots released it in an active form (1). Moreover, the rhizosphere is known to be a location of strong interaction between plants and microorganisms, and potential effects of the T4 lysozyme on the bacterial community structure were allowed to accumulate by using the same plots for up to 3 years. Despite all that, differences between the rhizosphere communities of the T4 lysozyme-expressing line DL5 and control plants were not detectable. One possible explanation for this result is that the rhizosphere communities were stable enough to resist T4 lysozyme effects. Evidence for that was provided by the highly similar community profiles, especially the DGGE fingerprints, of all samples taken from the different plants in a field at a certain time. Another explanation might be that an existing effect was not detected due to insufficient resolution and sensitivity of the methods or too few replicate samples, considering the natural variability. We tried to minimize the probability of the latter error by using a high number of replicate samples (n = 8 plots), by setting the significance level of statistical tests to 0.1 instead of 0.05, by taking composite samples from several plants from one plot to reduce variability, and by applying a range of complementary methods with differing levels of resolution and sensitivity. Biosafety as a relative term is reasonably associated with an acceptable risk which is not significantly different from accepted background levels. A relevant effect of the transgenic plants should cause deeper changes in the community structure than the commonly accepted changes due to environmental factors related to the season, field site, or year. In this study, the environmental factors, but not the T4 lysozyme expression of the transgenic plants, influenced the rhizosphere communities, as shown by the DGGE profiles, the compositions of bacterial isolates, and Biolog profiles of the different samplings. Thus, the risk associated with transgene effects on the bacterial community seems to be below accepted background levels. Furthermore, the resolutions and sensitivities of the applied methods were good enough to detect relevant community changes. In parallel studies by Lottmann et al., the distribution of potentially plant-beneficial bacteria, which were isolated from the transgenic and control roots and characterized phenotypically and genotypically, was not affected by T4 lysozyme expression (25, 26). Lukow et al. found seasonal and spatial changes in soil bacterial communities by terminal restriction fragment length polymorphism analysis in three soil plots planted with transgenic GUS (β-glucuronidase gene), transgenic GUS/Barnase/Barstar potato lines, or nontransgenic potato plants (28). The seasonal shifts were more prominent than the spatial differences. It remained unclear whether transgene effects contributed to the spatial effects because replicated plots were lacking. In a field experiment, Lottmann et al. monitored the survival of two bacterial strains, a T4 lysozyme-tolerant Pseudomonas putida strain and a T4 lysozyme sensitive Serratia grimesii strain, in the rhizospheres of DL5, DES, and DC1, where both strains were established after seed tuber inoculation (27). During flowering of the plants and coincidentally with the highest measured level of T4 lysozyme expression of DL5, significantly more colony counts of the T4 lysozyme-tolerant P. putida were recovered from transgenic T4 lysozyme plants than from the transgenic control and the parental line. In a previous study, phyllosphere bacterial communities of T4 lysozyme potato plants and control lines were compared in the greenhouse and a field sampling (22). Differences in the percentages of culturable species were detectable for the T4 lysozyme plants, namely, a higher percentage of isolates classified as Erwinia sp. and lower abundances of gram-positive isolates and of Agrobacterium, but the observed effects were minor relative to the natural phyllosphere community differences between field samplings.
The real community structure cannot be completely reflected by a single method but is best recovered by the applied polyphasic approach. By cultivation of bacteria on solid media and characterization of isolated pure cultures by the standard MIS procedure, species or populations that did not grow on the applied media were not detectable. Other unrecovered populations possibly entered a nonculturable state (47) or depended on biotic interactions. As DGGE profiles were derived independently of cultivation, they more accurately displayed abundant species. The high percentage of cultured β-proteobacterial strains from the potato rhizosphere was not confirmed by molecular detection (Fig. 5). Differential lysis efficiencies and preferential amplification of species may bias the relative intensities of bands within a genetic fingerprint (21), but this does not affect comparison between fingerprints.
In contrast to DL5, the T4 lysozyme-expressing line DL4 often differed from the control lines and also from DL5 in its rhizosphere community structure. Rhizosphere samples of senescent DL4 plants in field Q were characterized in the first year by higher plate counts, reduced diversity of cultured bacteria, higher percentages of isolated strains classified as Pseudomonas or Flavobacterium, and differences in community level catabolic profiles and specific bands in DGGE fingerprints. In several other samplings, differences of DL4 from control plants were detected mainly in community level catabolic profiles, which were not reflected in DGGE profiling. Obviously, the catabolic profiling sensitively detected differences in the relative abundance of a community fraction which grew well in the Biolog plates but was not among the most abundant community members. By analyses of Biolog wells inoculated with cell suspensions from the potato rhizosphere, Smalla et al. observed that, compared with the inoculum, there was a decrease in the number of 16S rRNA gene fragments obtained from various wells, as well as a concomitant loss of populations that had been numerically dominant in the inoculum, and they suggested that fast-growing bacteria adapted to high substrate concentrations may have been primarily responsible for the patterns of substrate use (44). The deviation of DL4 was most likely connected to a general weakness of these plants evidenced by a significantly reduced root biomass (26), shorter stems and smaller leaves in the young plants, and a faster progress of senescence in the old plants. This could be the consequence of multiple and irregular integration of the T-DNA into the plant genome, affecting the functions of flanking genetic regions (38).
In contrast to experiments in which DGGE fingerprints have been used to analyze bacterial rhizosphere communities of plants grown in microcosms (8, 34), seasonal shifts of bacterial rhizosphere communities were found for field-grown plants (references 18, 27, and 44a and this study). Compared to natural community shifts, the T4 lysozyme was only a minor factor—if any—in changing the bacterial community structure of the agroecosystem. However, the expression of T4 lysozyme significantly reduced the susceptibility of transgenic potato plants to infection by Erwinia carotovora subsp. atroseptica (12, 13), the causative agent of blackleg disease and soft rot of tubers (37). This effect could be achieved despite rather low expression levels (<18 ppm of total soluble protein [12]). The probable reason is that the T4 lysozyme is fused to a signal peptide, leading to secretion into the apoplast (13), where the initially small population of the plant pathogen has to multiply before it is able to attack the plant (4, 36). Thus, this genetic modification may be a suitable strategy for enhancing resistance of crops to bacterial disease, as counteractive effects on potentially beneficial populations are avoided.
Acknowledgments
We are grateful to Henrike Westphal for excellent technical assistance, to Johann de Vries and all others who helped to sample the roots, and to D. Felgentreu for his help with the gas chromatographic analysis of fatty acids.
This work was supported by BMBF grants 0310582A and 0311295.
REFERENCES
- 1.Ahrenholtz, I., K. Harms, J. de Vries, and W. Wackernagel. 2000. Increased killing of Bacillus subtilis on hair roots of transgenic T4 lysozyme-producing potatoes. Appl. Environ. Microbiol. 66:1862-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bever, J. D., K. M. Westover, and J. Antonovics. 1997. Incorporating the soil community into plant population dynamics: the utility of the feedback approach. J. Ecol. 85:561-573. [Google Scholar]
- 3.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organisation and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 4.Cui, Y., A. Chatterjee, Y. Liu, C. K. Dumenyo, and A. K. Chatterjee. 1995. Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-l-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. J. Bacteriol. 177:5108-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Boer, S. H. 1989. Bacteria and potatoes in crop rotation systems, p. 131-152. In J. Vos, C. D. van Loon, and G. J. Bollen (ed.), Effects of crop rotation on potato production in the temperate zones. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 6.de Vries, J., K. Harms, I. Broer, G. Kriete, A. Mahn, K. Düring, and W. Wackernagel. 1999. The bacteriolytic activity in transgenic potatoes expressing a chimeric T4 lysozyme gene and the effect of T4 lysozyme on soil- and phytopathogenic bacteria. Syst. Appl. Microbiol. 22:280-286. [Google Scholar]
- 7.Di Giovanni, G. D., L. S. Watrud, R. J. Seidler, and F. Widmer. 1999. Comparison of parental and transgenic alfalfa rhizosphere bacterial communities using Biolog GN metabolic fingerprinting and enterobacterial repetitive intergenic consensus sequence-PCR (ERIC-PCR). Microb. Ecol. 37:129-139. [DOI] [PubMed] [Google Scholar]
- 8.Duineveld, B. M., A. S. Rosado, J. D. van Elsas, and J. A. van Veen. 1998. Analysis of the dynamics of bacterial communities in the rhizosphere of the chrysanthemum via denaturing gradient gel electrophoresis and substrate utilization patterns. Appl. Environ. Microbiol. 64:4950-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Düring, K. 1994. Differential patterns of bacteriolytic activities in potato in comparison to bacteriophage T4 and hen egg white lysozymes. J. Phytopathol. 141:159-164. [Google Scholar]
- 10.Düring, K. 1994. A plant transformation vector with a minimal T-DNA. Transgen. Res. 3:138-140. [DOI] [PubMed] [Google Scholar]
- 11.Düring, K. 1996. Genetic engineering for resistance to bacteria in transgenic plants by introduction of foreign genes. Mol. Breed. 2:297-305. [Google Scholar]
- 12.Düring, K., and A. Mahn. 1999. Freisetzung und Resistenzprüfung transgener Lysozym-Kartoffeln, p. 39-44. In J. Schiemann (ed.), Freisetzungsbegleitende Sicherheitsforschung mit gentechnisch veränderten Pflanzen und Mikroorganismen. Proceedings zum BMBF-Workshop Biologische Sicherheit, 25.-26. Mai 1998, Braunschweig. BEO, Jülich, Germany.
- 13.Düring, K., P. Porsch, M. Fladung, and H. Lörz. 1993. Transgenic potato plants resistant to the phytopathogenic bacterium Erwinia carotovora. Plant J. 3:587-598. [Google Scholar]
- 14.Düring, K., P. Porsch, A. Mahn, O. Brinkmann, and W. Gieffers. 1999. The non-enzymatic microbicidal activity of lysozymes. FEBS Lett. 449:93-100. [DOI] [PubMed] [Google Scholar]
- 15.Garland, J. L. 1997. Analysis and interpretation of community-level physiological profiles in microbial ecology. FEMS Microbiol. Ecol. 24:289-300. [Google Scholar]
- 16.Garland, J. L., and A. L. Mills. 1991. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl. Environ. Microbiol. 57:2351-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glimm, E., H. Heuer, B. Engelen, K. Smalla, and H. Backhaus. 1997. Statistical comparisons of community catabolic profiles. J. Microbiol. Methods 30:71-80. [Google Scholar]
- 18.Gomes, N. C. M. C., H. Heuer, J. Schönfeld, R. Costa, R. Hagler-Mendonca, and K. Smalla. 2001. Bacterial diversity of the rhizosphere of maize (Zea mays) grown in tropical soil studied by temperature gradient gel electrophoresis. Plant Soil 232:167-180. [Google Scholar]
- 19.Hack, H., H. Gall, T. Klemke, R. Klose, U. Meier, R. Strauss, and A. Witzenberger. 1993. Phänologische Entwicklungsstadien der Kartoffel (Solanum tuberosum L.). Nachrichtenbl. Dtsch. Pflanzenschutzd. 45:11-19. [Google Scholar]
- 20.Herron, P. R., and E. M. H. Wellington. 1990. New methods for extraction of streptomycete spores from soil and application to the study of lysogeny in sterile amended and nonsterile soil. Appl. Environ. Microbiol. 56:1406-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Heuer, H., K. Hartung, G. Wieland, I. Kramer, and K. Smalla. 1999. Polynucleotide probes that target a hypervariable region of 16S rRNA genes to identify bacterial isolates corresponding to bands of community fingerprints. Appl. Environ. Microbiol. 65:1045-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heuer, H., and K. Smalla. 1997. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) for studying soil microbial communities, p. 353-373. In J. D. van Elsas, E. M. H. Wellington, and J. T. Trevors (ed.), Modern soil microbiology. Marcel Dekker, New York, N.Y.
- 22.Heuer, H., and K. Smalla. 1999. Bacterial phyllosphere communities of Solanum tuberosum L. and T4-lysozyme-producing transgenic variants. FEMS Microbiol. Ecol. 28:357-371. [Google Scholar]
- 23.Heuer, H., M. Krsek, P. Baker, K. Smalla, and E. M. H. Wellington. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hippe, S., K. Düring, and F. Kreuzaler. 1989. In situ localization of a foreign protein in transgenic plants by immunoelectron microscopy following high pressure freezing, freeze substitution and low temperature embedding. Eur. J. Cell Biol. 50:230-234. [Google Scholar]
- 25.Lottmann, J., and G. Berg. 2001. Phenotypic and genotypic characterization of antagonistic bacteria associated with roots of transgenic and non-transgenic potato plants. Microbiol. Res. 156:75-82. [DOI] [PubMed] [Google Scholar]
- 26.Lottmann, J., H. Heuer, K. Smalla, and G. Berg. 1999. Influence of transgenic T4-lysozyme-producing potato plants on potentially beneficial plant-associated bacteria. FEMS Microbiol. Ecol. 29:365-377. [Google Scholar]
- 27.Lottmann, J., H. Heuer, J. de Vries, A. Mahn, K. Düring, W. Wackernagel, K. Smalla, and G. Berg. 2000. Establishment of introduced antagonistic bacteria in the rhizosphere of transgenic potatoes and their effect on the bacterial community. FEMS Microbiol. Ecol. 33:41-49. [DOI] [PubMed] [Google Scholar]
- 28.Lukow, T., P. F. Dunfield, and W. Liesack. 2000. Use of the T-RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol. Ecol. 32:241-247. [DOI] [PubMed] [Google Scholar]
- 29.Maidak, B. L., G. J. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey, and C. R. Woese. 1997. The RDP (Ribosomal Database Project). Nucleic Acids Res. 25:109-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mourgues, F., M. N. Brisset, and E. Chevreau. 1998. Strategies to improve plant resistance to bacterial diseases through genetic engineering. Trends Biotechnol. 16:203-210. [DOI] [PubMed] [Google Scholar]
- 31.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 32.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nehl, D. B., S. J. Allen, and J. F. Brown. 1996. Deleterious rhizosphere bacteria: an integrating perspective. Appl. Soil Ecol. 5:1-20. [Google Scholar]
- 34.Normander, B., and J. I. Prosser. 2000. Bacterial origin and community composition in the barley phytosphere as a function of habitat and presowing conditions. Appl. Environ. Microbiol. 66:4372-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oger, P., A. Petit, and Y. Dessaux. 1997. Genetically engineered plants producing opines alter their biological environment. Nat. Biotechnol. 15:369-372. [DOI] [PubMed] [Google Scholar]
- 36.Perombelon, M. C. M. 1981. The ecology of Erwinias on aerial plant surfaces, p. 411-431. In J. P. Blakeman (ed.), Microbial ecology of the phylloplane. Academic Press, London, United Kingdom.
- 37.Perombelon, M. C. M., and A. Kelman. 1980. Ecology of the soft rot Erwinias. Annu. Rev. Phytopathol. 18:361-387. [Google Scholar]
- 38.Porsch, P., A. Jahnke, and K. Düring. 1998. A plant transformation vector with a minimal T-DNA. II. Irregular integration patterns of the T-DNA in the plant genome. Plant Mol. Biol. 37:581-585. [DOI] [PubMed] [Google Scholar]
- 39.Riesner, D., G. Steger, R. Zimmat, R. A. Owens, M. Wagenhöfer, W. Hillen, S. Vollbach, and K. Henco. 1989. Temperature-gradient gel electrophoresis of nucleic acids: analysis of conformational transitions, sequence variations, and protein-nucleic acid interactions. Electrophoresis 10:377-389. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Shannon, C. E. 1948. A mathematical theory of communication. Bell Syst. Technol. J. 27:379-423. [Google Scholar]
- 42.Siciliano, S. D., and J. J. Germida. 1999. Taxonomic diversity of bacteria associated with the roots of field-grown transgenic Brassica napus cv. Quest, compared to the non-transgenic B. napus cv. Excel and B. rapa cv. Parkland. FEMS Microbiol. Ecol. 29:263-272. [Google Scholar]
- 43.Smalla, K., N. Cresswell, L. C. Mendonca-Hagler, A. Wolters, and J. D. van Elsas. 1993. Rapid DNA extraction protocol from soil for polymerase chain reaction-mediated amplification. J. Appl. Bacteriol. 74:78-85. [Google Scholar]
- 44.Smalla, K., U. Wachtendorf, H. Heuer, W.-T. Liu, and L. Forney. 1998. Analysis of BIOLOG GN substrate utilization patterns by microbial communities. Appl. Environ. Microbiol. 64:1220-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Smalla, K., G. Wieland, A. Buchner, A. Zock, J. Parzy, S. Kaiser, N. Roskot, H. Heuer, and G. Berg. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67:4742-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sørensen, J. 1997. The rhizosphere as a habitat for soil microorganisms, p. 21-45. In J. D. van Elsas, E. M. H. Wellington, and J. T. Trevors (ed.), Modern soil microbiology. Marcel Dekker, New York, N.Y.
- 46.Tsugita, A., M. Inouye, E. Terzaghi, and G. Streisinger. 1968. Purification of bacteriophage T4 lysozyme. J. Biol. Chem. 243:391-397. [PubMed] [Google Scholar]
- 47.Wilson, M., and S. E. Lindow. 1992. Relationship of total viable and culturable cells in epiphytic populations of Pseudomonas syringae. Appl. Environ. Microbiol. 58:3908-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]