Abstract
The National Institutes of Health’s All of Us Research Program is an accessible platform that hosts genomic and phenotypic data collected from one million participants in the United States. Its mission is to accelerate medical research and clinical breakthroughs with a special emphasis on diversity.
Single Sentence Summary:
The NIH’s All of Us Research Program hosts genomic and phenotypic data collected from one million diverse participants in the United States.
Diversity at the core of All of Us
In 2015, the Precision Medicine Initiative Working Group of the National Institutes of Health’s Advisory Committee to the Director delivered an initial blueprint and scientific strategy for the All of Us Research Program (1). Launched in May 2018, the All of Us Research Program is expected to enroll at least one million diverse participants from across the United States. Simply stated, the goal of the All of Us Research Program is to enable better care for all of us. From its inception, All of Us has been committed to collecting five foundational data streams from participants in the program: biometrics data, survey data, electronic health records, genomics, and digital health data from wearables. To achieve its vision, the program depends on three strategic pillars: Nurturing the partnership with one million participants, assembling one of the largest and most diverse biomedical research data sets that is accessible to researchers worldwide, and catalyzing an ecosystem for researchers, communities and funders to enable the advancement of precision health and medicine.
A distinctive feature of All of Us is its commitment to diversity and health equity (2). It is an explicit goal for the program to increase representation of diverse participants and to encourage other longitudinal population studies to do so. Enrollment in All of Us is open to any eligible adult in the United States who wishes to participate, but the focus of the program’s engagement efforts is on underrepresented groups and communities. Among those who have completed the initial steps of the protocol, including biospecimen collection, about 51% self-identified as White; 18% as Black, African American, or African; 16% as Hispanic or Latino; and 3% as Asian. In addition, among those who identify as White, more than 60% are from groups that are underrepresented owing to their sexual orientation or gender identity, disability status, or other socio-demographic features (e.g., rural residence, age, educational attainment, or economic status). The challenge of diverse participant enrollment is being addressed by funding enrollment centers and engagement partners (3) covering a wide range of geography and demography, complemented by strategies to reach people where they live using mobile engagement vehicles, blood banks, mailed saliva kits, and home visits for recruitment. Strategic enrollment and engagement partnerships (4) have been established with national, state, and local organizations (5) to build a network of trusted leaders that raise awareness about the program among African Americans, Asian Americans, Native Hawaiians and Pacific Islanders, Hispanics and Latinos, LGBTQ communities, disability communities, and rural and older adults.
Numerous studies have highlighted the lack of diversity in publicly available genomic data sets (6). Overrepresentation of individuals of European descent in genome-wide association studies (GWAS) has been used as a benchmark for the gaps in our scientific knowledge about underrepresented communities. These biases in genomic data have also led to poor replication of findings and false or erroneous associations in non-European populations, some of which may be the basis of therapeutic decision making. All of Us released its first set of genomic data in March 2022, and had a second release in April 2023 that now includes more than 245,000 whole genome sequences and more than 312,000 genotyping arrays. Nearly 50% of the genomic data comes from participants who self-identify with a racial or ethnic minority group making this among the most diverse genomic data sets at this scale available to researchers (7). All of Us will complement, collaborate with, and augment ongoing work by other cohorts and consortia ---such as The Human Heredity and Health in Africa (H3Africa), The Trans-Omics for Precision Medicine (TOPMed), Polygenic Risk Methods in Diverse Populations (PRIMED), African Population Cohort Consortium (APCC), International Hundred K+ Cohorts Consortium (IHCC), and International Common Disease Alliance (ICDA) ---to close the diversity gaps in genomic and non-genomic data as well as support research directed to previously underrepresented populations.
The structure of All of Us
Since its inception in 2016, All of Us has sought to provide access to high-quality, standardized data to broad research communities including international researchers. This has been achieved through several key strategic approaches that can be viewed as guidance for large-scale population studies that have similar goals of accelerating scientific breakthroughs and making an impact on human health (8–11) (Table 1). A Core Protocol (12) was developed to set the foundational standards for data and the requirements for recruiting, consenting, and retaining participants, as well as the types of data that they would be asked to contribute.
Table 1. Selected challenges and solutions since the inception of All of Us.
| Challenges | Solutions |
|---|---|
| Recruitment of 1 million participants by the end of 2026 | Increased recruitment sites nationwide (5); orderly shutdown and enrollment restart during the COVID-19 pandemic; virtual/app and web-based enrollment, with saliva samples sent by mail instead of blood collected in person; diverse enrollment partnerships; enrollment “surge strategy” in 2022 (8) |
| Diversity of participants | >50 engagement partners; a diversity of enrollment sites; mobile enrollment vehicles; enrollment from Federally Qualified Health Centers; participant ambassadors (3,5) |
| Broad data collection protocol | Commitment and consent (9) to capture five data streams |
| Data access and privacy | Workbench workshops and tools; tiered access requiring adherence to privacy rules |
| Data harmonization | Use of OMOP as a standard for most phenomic data, and GA4GH standards for genomic data (10) |
| Regulatory | Central Institutional Review Board; early partnership with FDA on return of genetic results and investigational device exemption approval (11) |
OMOP, Observational Medical Outcomes Partnership; GA4GH, Global Alliance for Genomics and Health
The Researcher Workbench (13) was created as a data platform that provides readily accessible tiered data (Figure 1). The Public Tier contains only anonymized, aggregate data that is available to anyone without logging in. The Public Tier tools include a Data Browser, Research Projects Directory, Publications, Data Snapshots, and the Survey Explorer. The Registered Tier contains curated, anonymized, individual-level data. Registered Tier tools include Cohort Builder, Dataset Builder, Workspaces, Notebooks, and the Support Hub. Registered Tier users require data use agreements between the program and their home institutions that specify the access and training requirements. The Controlled Tier is available to approved institutions whose researchers have taken additional steps and training to access these more granular data. Controlled Tier data include genomic data, additional clinical fields from electronic health records, and additional demographic data from surveys that are suppressed or generalized in the Registered Tier. Educational modules and tools are included in the Researcher Workbench and have been developed to encourage the greatest access and use of the program data (Figure 1).
Figure 1. Data and tools hosted on the All of Us Researcher Workbench.
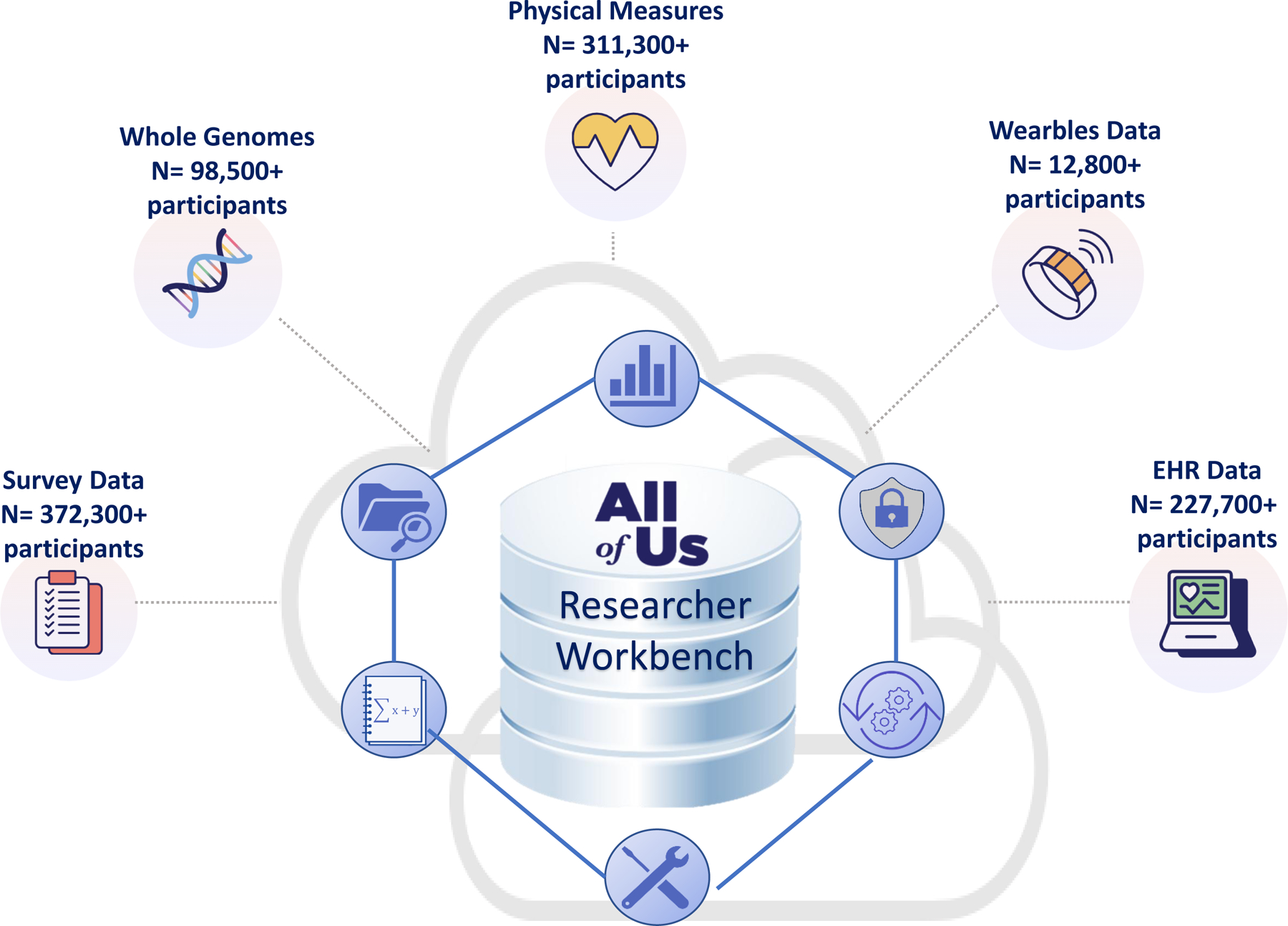
The All of Us Researcher Workbench integrates five major sources of data collected from participants. The five data streams include survey data provided by participants, whole genome sequence data from participant DNA samples, physical measures collected directly from participants or electronic health records (EHR), and wearable data primarily from Fitbits ( https://www.researchallofus.org/data-tools/data-sources/). Shown are the number of participants from which curated data are available as of November 2023. The Researcher Workbench is a cloud-based platform that allows tiered access to these data types by researchers and the public through the All of Us Research Hub (https://www.researchallofus.org/). Each data type has tools to facilitate use in research. The public has access to summary data through the public browser, and researchers have access to more granular data in the registered tier and controlled tier.
CREDIT: A. FISHER/SCIENCE TRANSLATIONAL MEDICINE
Data quality standards have been established as have cross-cutting solutions to overcome the potential consequences of biases resulting from non-random missing values in participants’ data. Data quality efforts include utilizing the Observational Medical Outcomes Partnership (OMOP) common data model for data harmonization (14), ongoing quality and missing value monitoring, filling data gaps proactively, engagement of participants in finding solutions and offering incentives, and communicating expectations to researchers through multiple channels. Currently, nearly 60% of recruitment sites are within clinical organizations that utilize the Epic electronic health record system; other electronic health record systems include Cerner, Allscripts, NextGen, eClinicalWorks, and VHA GeneISIS. The program requires each site to harmonize the data to OMOP prior to sending it to the Data and Research Center. The variability in electronic health record vendors and availability of records for some participants has led to heterogeneity in data capture and a data gap for some participants. The program is closing this gap through Fast Healthcare Interoperability Resources and accessing data in health information exchanges.
Early challenges and solutions
The COVID-19 pandemic had a major impact on All of Us recruitment and enrollment (8). Prior to the pandemic, the program enrolled ~ 12,500 participants per month at more than 400 clinical sites. In March 2020, all in-person activity at program sites was paused for safety, and new processes and procedures for virtual engagement, and remote data and biospecimen collection were implemented. Reactivation of in-person enrollment began in the Fall of 2021; by February 2022, 224 clinical sites had been reactivated, and all enrollment partners had adopted new remote data collection methods. For example, recruitment switched from in person to phone calls; consent was remote using videoconferencing or screen sharing; surveys were completed remotely; biosampling was done with saliva kits by mail; and physical measures were captured from electronic health records. Today, the program is approaching the pre-pandemic number of recruitment sites, and enrollment is close to pre-pandemic rates, with new safety protocols in place to ensure staff and participant safety.
All of Us has a strong commitment to health equity (2) including participant recruitment and engagement, data access, and hiring of program personnel. All of Us has created a health equity research group with the explicit goal to address health equity questions in all of the program’s research activities. The initial goal for diversity among those from whom we obtain biospecimens was >75% from individuals not previously represented in biomedical research (13). The program has thus far been able to sustain that percentage and continues to set a standard for longitudinal population health studies. For example, in January 2023, the US government proposed the use of the Middle Eastern North African (MENA) descriptor and the use of a single combined race-ethnicity for the Office of Management and Budget (15). Both recommendations have been implemented in All of Us since the program’s launch in 2018. In March 2023, the National Academies of Sciences, Engineering and Medicine issued recommendations for the use of race, ethnicity, and ancestry as population descriptors in genomics research. All of Us co-sponsored this effort and is assisting in evaluating its practices around diversity, equity, and inclusion. All of Us is working with the NIH (16), the National Academies of Medicine (17), and international consortia to establish culturally responsive methods for the use of race, ethnicity, and genetic ancestry as population descriptors across genomic studies and biomedical research studies.
All of Us prioritizes the protection of participant privacy through rigorous standards and data safeguards (18), while at the same time balancing the need for privacy with the granularity of data required to investigate social and environmental determinants of health using geospatial data. Return of research findings of value to participants is a core characteristic of the program (19). This principle recognizes the partnership between participants and the program, in which participants share data with the program with the expectation that the program will provide the opportunity for participants to receive information about their health in return. All of Us has worked closely with the U.S. Food and Drug Administration (20) to develop an investigational device exemption application process for whole genome sequencing that will allow for supplementary submissions to support new genetic variant classes and reportable results. The purpose of the investigational device exemption is to demonstrate that the sequencing test has analytical validity and to protect the interests of study participants who might receive test results that could affect their clinical care.
The All of Us Data Roadmap
The program recently reached a milestone of more than 700,000 consented participants. The number of core data sets collected so far include participant answered surveys, whole genome sequences, physical measures, wearable data, and electronic health records (Figure 1). To date, 427,000 participants have donated a biospecimen (14).
The five core data types collected by All of Us (13) are diverse and offer a rich longitudinal reservoir of clinical phenotypes that can be computed and linked to enrich the data. Now with an established process for collecting, curating, and delivering the foundational data streams to researchers, the program has developed a data release schedule for researchers (Table S1). With this schedule, intended as a living document (these dates are estimates and are subject to change), researchers can anticipate and plan investigations that will use All of Us data.
Anchored in the recommendations from the Report to the NIH Director’s Advisory Committee (1) and aiming to address the CDC’s top ten causes of morbidity and mortality for the United States (21), the initial scientific agenda for the program is focused on two questions. Will the outcome of the research have potential (or is likely) to impact the human condition in health or disease? Can the research be done with the current data that is available to researchers (in terms of data types and sample sizes with complete data)? Although the collection of data started in 2017, the electronic health records, wearable data, and some survey questions have retrospective time frames for allowing researchers to access the data. Some electronic health records data go back more than 40 years and therefore include pediatric data; some wearable data records began almost 10 years ago (22).
In 2023, the first version of an evergreen scientific framework for the program was completed and is now being used to guide decisions for research priorities. With input from broad stakeholder groups including the program’s funded investigators, NIH institutes and centers, government agencies, and participants, this plan focuses on research objectives that will drive new discoveries in health and disease biology. As a platform that supports large-scale 21st century epidemiological research, the expectation is that All of Us will provide new insights across the lifespan including health risks, disease diagnosis, therapeutic strategies, and iterative monitoring methods to optimize health and minimize morbidity and mortality. The data streams from electronic health records and their longitudinal capabilities, participant surveys, wearable device data, and future assays from banked and temporally acquired biospecimens will enable these studies. The scientific priorities of the program intentionally recognize a scientific agenda that can be carried out today by the research community. The Data Roadmap informs and enables the All of Us program’s overarching focus to advance precision health and medicine through new risk assessments, diagnostics, and treatments for all people. The roadmap has nine strategic focus areas including development of new methods to improve utilization of All of Us data by researchers. Four focus areas concentrate on drivers of health and disease including lifestyle and behavioral health; the environment; diversity, equity, inclusion and accessibility; genetics and biology. The final four focus areas are: Common and rare health conditions; maternal and child health; healthy aging and resilience; impact of return of results. Together, these nine areas cut across the NIH research portfolio helping its institutes achieve their scientific objectives, while also catalyzing a vibrant research portfolio beyond the NIH.
Short-term and long-term scientific opportunities
Having >1 million whole genome sequences integrated with longitudinal data from questionnaires and electronic health records will allow a comprehensive molecular epidemiological approach across the lifespan. Genetic, environmental, and lifestyle data will be integrated and accessible promoting an understanding of how their interactions drive transitions from health to disease and enabling a robust assessment of vulnerabilities and resilience for an individual or population. The genetic data will specifically support return of findings that may impact health care decisions using the recommendations of the American College of Medical Genetics and Genomics (ACMG) (23) and the Clinical Pharmacogenomics Implementation Consortium (CPIC) for return of results. Genetic data will also drive the discovery of new genetic risk factors (e.g., polygenic risk scores) that can be defined by ancestry, addressing a valid critique that these are biased and, to date, can only be applied to individuals of European descent whose data currently dominate large genomic data sets. Genomic data may also enhance our ability to understand the concept of gene penetrance, genetics of emerging infections, drug and vaccine responses, and more.
Return of Results
The All of Us program began reporting genetic ancestry (7 populations and 20 subpopulations) and genetic traits (such as lactose intolerance, cilantro preference, earwax type) in 2020. Since then, more than 182,000 participants have received these results. Beginning in December 2022, health-related DNA results are also being returned to participants including a Hereditary Disease Risk report for 59 genes (24) informed by ACMG recommendations, and a Medicine and Your DNA report for variants in 7 genes involved in drug metabolism (25) informed by CPIC guidelines (26). To date, more than 68,000 “Hereditary Disease Risk” reports and 65,000 “Medicine and Your DNA” reports have been returned to participants. The results are returned through the All of Us secure participant portal and participants who elect to see their results can access a free genetic counseling visit through the program’s Genetic Counseling Resource with materials and counseling available in both English and Spanish. The number of participants who are invited to receive health-related DNA results continues to increase and includes new participants who join the program. The program is currently designing a broad outcomes research strategy, as a focus area in the scientific priorities roadmap, to assess the impact of results downstream of their return on the participants, their families, their providers, and their healthcare. Importantly, this research agenda will seek to better understand the methods for return of results and their impact on historically marginalized populations to optimize their value to all participants.
All of Us, by design, is disease-agnostic enabling cross-cutting thematic research. Data collected on environmental and geospatial data linkages, mental health, sleep, substance use and abuse, lifestyle, diet, physical activity, social determinants of health, and other factors will allow researchers to address how these ubiquitous health influences impact disease development and trajectories. At the same time, there will be an opportunity for disease-focused research in areas such as the impact of social determinants of health and immune status on the development of COVID-19 or post-acute COVID-19 sequelae. Research efforts will also focus on mental health, Mendelian diseases, pain, cancer, cardiovascular diseases, oral and skin diseases, as well as rare diseases.
What does success look like 10 years from now?
In the next decade, we hope that All of Us will be an indispensable part of health research, particularly across the NIH strategic portfolio, with one of the largest, most diverse data sets accessible to researchers globally. Just as the program’s cohort is diverse, so we hope that the network of researchers using All of Us data will be diverse, encompassing individuals from different organizational settings, demographics, and scientific backgrounds, including groups that are underrepresented in the biomedical workforce. Each researcher will bring unique questions and perspectives to the program’s platform, driving new insights into foundational health challenges. Ultimately, these advances will help to deliver on the promise to participant partners, their families, and communities to “accelerate health research and medical breakthroughs, enabling individualized prevention, treatment, and care for all of us.”
Supplementary Material
Acknowledgements:
The authors acknowledge the entire senior leadership team of the All of Us Research Program for their contributions. We also thank our participant community for their commitment to the program and for allowing sharing of their data. We acknowledge the contributions of Andrea Ramirez and Katrina Theisz regarding the Data Roadmap.
References and Notes
- 1.Precision Medicine Initiative Working Group, The Precision Medicine Initiative Cohort Program – Building a Research Foundation for 21st Century Medicine: Precision Medicine Initiative Working Group Report to the Advisory Committee to the Director, NIH https://www.nih.gov/sites/default/files/research-training/initiatives/pmi/pmi-working-group-report-20150917-2.pdf (2015).
- 2.Mapes BM, Foster CS, Kusnoor SV et al. , Diversity and inclusion for the All of Us Research Program: A scoping review. PLoS ONE (2020) 15(7): e0234962. 10.1371/journal.pone.0234962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.https://allofus.nih.gov/funding-and-program-partners/health-care-provider-organizations (accessed Nov 17, 2023)
- 4.https://www.rand.org/pubs/research_reports/RR2578.html (accessed Nov 17, 2023)
- 5.Fair A, Watson KS, Cohn EG et al. , Innovation in Large-Scale Research Programs: Elevating Research Participants to Governance Roles Through the All of Us Research Program Engagement Core. Acad Med (2022) 97:1794–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fatumo S, Chikowore T, Choudhury A et al. , A roadmap to increase diversity in genomic studies. Nat Med 28, 243–250 (2022). 10.1038/s41591-021-01672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.https://allofus.nih.gov/news-events/announcements/all-us-research-program-makes-nearly-250000-whole-genome-sequences-available-advance-precision-medicine (accessed Nov 17, 2023).
- 8.Hedden Sarra L, McClain James, Mandich Allison, Baskir Rubin, Caulder Mark S, Denny Joshua C, Hamlet Michelle R J, Das Irene Prabhu, Ford Nicole McNeil, Lopez-Class Maria. The Impact of COVID-19 on the All of Us Research Program. American Journal of Epidemiology, Volume 192, Issue 1, January 2023, Pages 11–24, 10.1093/aje/kwac169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doerr M, Moore S, Barone V et al. , AJOB Empir Bioeth (2021) 12: 72–83. [DOI] [PubMed] [Google Scholar]
- 10.https://www.researchallofus.org/data-tools/methods/ (Accessed Nov 17, 2023)
- 11.https://allofus.nih.gov/news-events/research-highlights/allofus-meets-fda-standards-dna-results-delivery#:~:text=All%20of%20Us%20received%20an,data%2C%20and%20creating%20participant%20reports (Accessed Nov 17, 2023)
- 12.The All of Us Research Program Investigators, The “All of Us” Research Program. New Engl. J. Med 381, 668–676 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Website: All of Us Research Program, Research Hub https://www.researchallofus.org/ (accessed Nov 17, 2023)
- 14.https://www.researchallofus.org/data-tools/data-snapshots/ (Accessed Nov 17, 2023)
- 15.https://www.federalregister.gov/documents/2023/01/27/2023-01635/initial-proposals-for-updating-ombs-race-and-ethnicity-statistical-standards (Accessed Nov 17, 2023)
- 16.https://allofus.nih.gov/news-events/announcements/nih-launches-unite-effort-end-structural-racism-research (accessed Nov 17, 2023).
- 17.https://www.nationalacademies.org/our-work/use-of-race-ethnicity-and-ancestry-as-population-descriptors-in-genomics-research (accessed Nov 17, 2023)
- 18.https://allofus.nih.gov/protecting-data-and-privacy (accessed Nov 17 27, 2023)
- 19.https://allofus.nih.gov/about/core-values (Accessed Nov 17, 2023)
- 20.Venner E, Muzny D, Smith JD, Walker K, Neben CL, Lockwood CM, Empey PE, Metcalf GA, Kachulis C; All of Us Research Program Regulatory Working Group, Mian S, Musick A, Rehm HL, Harrison S, Gabriel S, Gibbs RA, Nickerson D, Zhou AY, Doheny K, Ozenberger B, Topper SE, Lennon NJ. Whole-genome sequencing as an investigational device for return of hereditary disease risk and pharmacogenomic results as part of the All of Us Research Program. Genome Med 2022. Mar 28;14(1):34. doi: 10.1186/s13073-022-01031-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.https://www.cdc.gov/nchs/fastats/leading-causes-of-death.htm (Accessed Nov 17, 2023)
- 22.Ramirez AH, Sulieman L, Schlueter DJ, Halvorson A, Qian J, Ratsimbazafy F, Loperena R, Mayo K, Basford M, Deflaux N, Muthuraman KN, Natarajan K, Kho A, Xu H, Wilkins C, Anton-Culver H, Boerwinkle E, Cicek M, Clark CR, Cohn E, Ohno-Machado L, Schully SD, Ahmedani BK, Argos M, Cronin RM, O’Donnell C, Fouad M, Goldstein DB, Greenland P, Hebbring SJ. The All of Us Research Program: Data quality, utility, and diversity. Patterns 2022. 3:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, Herman GE, Hufnagel SB, Klein TE, Korf BR, McKelvey KD, Ormond KE, Richards CS, Vlangos CN, Watson M, Martin CL, Miller DT. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med 2017. Feb;19(2):249–255. doi: 10.1038/gim.2016.190. Epub 2016 Nov 17. Erratum in: Genet Med. 2017 Apr;19(4):484. [DOI] [PubMed] [Google Scholar]
- 24.https://www.joinallofus.org/what-participants-receive/hereditary-disease-risk (accessed Nov 17, 2023)
- 25.https://www.joinallofus.org/what-participants-receive/medicine-and-your-dna (accessed Nov 17, 2023)
- 26.https://cpicpgx.org/ (accessed Nov 17, 2023)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


