Abstract
The microbial populations in no-till agricultural soil and casts of the earthworm Lumbricus rubellus were examined by culturing and molecular methods. Clone libraries of the 16S rRNA genes were prepared from DNA isolated directly from the soil and earthworm casts. Although no single phylum dominated the soil library of 95 clones, the largest numbers of clones were from Acidobacteria (14%), Cytophagales (13%), Chloroflexi (8%), and γ-Proteobacteria (8%). While the cast clone library of 102 clones was similar to the soil library, the abundances of several taxa were different. Representatives of the Pseudomonas genus as well as the Actinobacteria and Firmicutes increased in number, and one group of unclassified organisms found in the soil library was absent in the cast library. Likewise, soil and cast archaeal 16S rRNA gene libraries were similar, although the abundances of some groups were different. Two hundred and thirty aerobic bacteria were also isolated on general heterotrophic media from casts, burrows, and soil. The cast isolates were both phenotypically and genotypically different from the soil isolates. The cast isolates were more likely to reduce nitrate, grow on acetate and Casamino Acids, and utilize fewer sugars than the soil isolates. On the basis of their ribotypes, the cast isolates were dominated by Aeromonas spp. (28%), which were not found in the soil isolates, and other γ-Proteobacteria (49%). In contrast, the soil isolates were mostly Actinobacteria (53%), Firmicutes (16%), and γ-Proteobacteria (19%). Isolates obtained from the sides of earthworm burrows were not different from the soil isolates. Diversity indices for the collections of isolates as well as rRNA gene libraries indicated that the species richness and evenness were decreased in the casts from their levels in the soil. These results were consistent with a model where a large portion of the microbial population in soil passes through the gastrointestinal tract of the earthworm unchanged while representatives of some phyla increase in abundance.
Earthworm activity profoundly affects the physical and chemical composition of soil. Earthworm burrowing creates pore spaces, thus increasing water conductivity and benefitting root growth (19, 44). Earthworms break down large soil particles and leaf litter, which increases the availability of organic matter for microbial degradation (18). They promote macroaggregate formation by depositing casts rich in glycoproteins, polysaccharides, bacteria, and clay (12, 18, 25, 75). Earthworms also affect soil profile formation by mixing litter layers with surface soil layers, which acts to deepen humus layers within soil (18, 44).
The microbial community of soil may be profoundly affected by the presence of earthworms. Studies of various earthworm species have generally shown an increase in microbial numbers or activity either during or after passage through the gut and in the drilosphere (2, 12, 13, 20, 42, 54, 57, 66, 75, 78, 83), although corresponding increases in microbial biomass were not always observed (12, 65). Karsten and Drake (38) found more anaerobes and cellobiose utilizers in the earthworm alimentary canal than in soil. Specific phylogenetic groups of organisms have also been found in higher numbers in earthworm guts, casts, or burrows, such as Aeromonas hydrophila in Eisenia foetida (76), fluorescent pseudomonads in Lumbricus terrestris (13), and Actinobacteria in Lumbricus rubellus (41). The differences in microbial activity, bacterial numbers, and certain bacterial species between the earthworm alimentary canal or burrow and bulk soil indirectly support the hypothesis that the bacterial community structures of these habitats are different from those of the soil.
In this study we present the combined results of culture and culture-independent studies on earthworm casts, burrows, and the surrounding agricultural soil over a 3-year period. The phenotypic properties of 204 isolates were examined as well as the partial 16S ribosomal DNA (rDNA) sequences of the isolates and of over 200 clones constructed from earthworm casts and bulk soil. Of considerable interest was determining if the cast and soil populations were significantly different and whether the clone libraries would contain sequences similar to the isolates'.
MATERIALS AND METHODS
Sample site.
All samples were obtained from no-tilled agricultural plots at the Horseshoe Bend Long Term Environmental Research site, which is located in Athens, Georgia. This site is located on a river floodplain on Hiwassee series soil (fine loamy, siliceous, thermic, and Rhodic Kanhapludult). Clover and rye have been the winter cover crops in the plots since 1978 (3, 30, 31, 72). Maize (Zea mays L.) was the summer crop in 1996 and 1997. Kenaf (Hibiscus cannabinus) and cotton (Gossypium hirsutum) were planted during the summers of 1998 and 1999, respectively. Additional soil properties can be found in Beare et al. (3).
Sample collection for isolates.
In April and May 1996 a shovel was used to expose 15 soil shelves, approximately 1 m long and 10 cm deep. Fifty-five soil samples were taken at various depths within the top 5 cm of these shelves. In May 1999 three soil shelves were made and 15 soil samples were taken in the same manner. Sampling was performed by horizontally inserting a sterile Pasteur pipette 5 mm into the soil. Care was taken to avoid earthworm burrows and roots. These samples were defined as “bulk” soil. Upon removal from the soil, the tip of the pipette containing the soil sample was broken off into a microcentrifuge tube containing 0.5 ml of 0.85% (wt/vol) NaCl and crushed with a sterile inoculating stick. In a separate experiment the wet weight of 50 samples was measured, and the average wet weight was 5.2 mg (standard deviation = 1.2 mg). Using the same methods, 16 samples were also taken in March and April of 1996 from the sides of burrows found in the soil shelves or on the soil surface. All soil and burrow samples were taken 3 or more days following the last rainfall. Samples were also taken from the fresh casts of 29 and 12 L. rubellus worms in April 1997 and May 1999, respectively. These worms were either collected from the soil surface after the application of an electrical current to the soil (64) or from burrows exposed after excavation of soil shelves. The worms were rinsed with sterile water and placed in sterile petri plates containing a small amount of water. The plates were checked every 2 h. When a cast was observed, it was processed as described above for the soil samples.
Sample collection for clone libraries.
Soil and earthworm samples were collected in April 1999 from a no-till plot (main plot, subplot 7) which was overgrown with clover. Soil samples were collected from five randomly chosen sites from a depth of 5 to 10 cm below the litter layer and were put immediately on ice. The samples were stored at −20°C until processed. Two L. rubellus earthworms were collected from each of the five soil sample sites, placed into separate sterile petri dishes, and taken to the laboratory. Earthworms were washed with sterile water and placed into new sterile petri dishes until casts were collected. The first five casts were collected within 4 h of sampling, combined into a single sample, and frozen at −20°C.
Bacterial isolation.
Each soil and cast sample was vortexed for 30 s in 0.85% (wt/vol) NaCl to disperse bacteria and was serially diluted to a factor of 10−9 in two types of media. The dilution and growth media were MMSA (1 mM K2HPO4, 2 mM NH4NO3, 1 mM MgSO4, 1% [vol/vol] trace minerals [82], 1% [vol/vol] iron solution [82], 10% [vol/vol] soil extract, and 10 mM sodium acetate) at a pH of 7.0 and 50% (vol/vol) Difco nutrient broth (DNB) (Difco manual, Difco Laboratories, Detroit, Mich.). The soil extract was prepared by the method of Stackebrandt and Prauser (71), except that it was sterilized by filtration (0.2 μm pore size) rather than by autoclaving. Each soil, burrow, and cast sample taken in 1996 and 1997 was diluted and grown in MMSA and DNB. The samples taken in May 1999 were diluted and grown only in MMSA. All samples were incubated in the dark at 25°C. After 2 weeks, the highest dilutions showing growth in each medium were streaked onto solid medium of the same composition (MMSA or DNB with 1.5% [wt/vol] agar) for isolation of pure cultures. A representative of each colony type on each plate was then subcultured in 0.5 ml of DNB. In 1999, however, only one colony type per sample was selected. After turbidity became visible, 0.5 ml of sterile 15% (vol/vol) glycerol was added and the culture was stored at −70°C.
Phenotypic characterization of isolates.
Each isolate was cultured from frozen stocks on Difco nutrient agar (Difco manual) plates, and the colonial morphology was recorded. The presence of catalase was determined by a standard procedure (70). Each isolate was then subcultured in undiluted DNB. The cellular morphology was observed when turbidity became visible. Motility was tested using Difco Motility Test medium and was scored according to the Difco procedure (Difco manual). Substrate utilization was tested in 5.0 ml of minimal MMSA, prepared without acetate and soil extract. Potential substrates (cellulose, acetate, lactose, Casamino Acids, mannose, and cellobiose) were tested at a concentration of 0.2% (wt/vol). Test media were inoculated with 10 μl of culture and incubated for 14 days at room temperature. If the absorbance at 600 nm increased from 0.00 to ≥0.15, the culture was scored as positive for growth. Difco nitrate reduction medium was inoculated and incubated similarly. It was scored according to the Difco procedure (Difco manual). Partial nitrate reduction meant that nitrite was detected after 2 weeks. Complete nitrate reduction meant that neither nitrite nor nitrate remained after that time. The solid media that were used included Difco urea agar, Difco MacConkey medium, Difco blood agar, Difco Simmon's citrate, casein decomposition agar (21), and Tween hydrolysis agar (1). The isolates were patched onto each plate and incubated at room temperature for 14 days. The media from Difco were scored according to their procedures (Difco manual). The Tween hydrolysis agar and the casein decomposition agar were scored according to standard procedures (1, 21). For antibiotic resistance, each isolate was spread onto a nutrient agar plate (Difco). Antibiotic discs (Difco) were dispensed onto the plate, and the results were scored according to the Difco procedure (Difco manual).
Statistics.
The statistical analysis of the isolates was performed with SAS (63). A chi-square test was used to determine if there was a significant difference in phenotypic properties and phylogenetic distribution between the isolate libraries from the three sources. When the P was < 0.05, scaled deviance values were analyzed using the SAS genmod procedure to determine if the differences were due to the sample source (cast versus soil versus burrow) or the sampling protocol.
Dissimilarity values of isolates.
Test results were scored as 0 or 1 (negative or positive, respectively) except for cellular morphology and growth on MacConkey's and blood agar. For morphology, cocci, small rods, medium rods, and large rods were scored as −1, 0, 1, and 2, respectively. For MacConkey's agar, no growth, growth without lactose fermentation, and growth with lactose fermentation were scored as 0, 1, and 2, respectively. For blood agar, no hemolysis, alpha-hemolysis, and beta-hemolysis were scored as 0, 1, and 2, respectively. Dissimilarity values (Di) for every possible pair of isolates were then calculated using the formula Di = |Ai − Aj| + |Bi − Bj| + |Ci − Cj|… +…|Qi − Qj| (where A through Q refer to the different tests and subscripts i and j refer to each possible pair of isolates). The maximum possible Di was 20 for the 17 phenotypic characters. Any two isolates that had a Di of 0 were considered to be of the same phenotype.
To determine if the distribution of Di was consistent with a random model, a data set was generated by randomly assigning test results to “isolates” using the same ratio of positive to negative results as obtained for the original isolates. A new matrix of Di values was then created, and the distribution of Di was determined.
To determine if the phenotypes of the isolates collected in 1996 or 1997 or the control isolates were the same as those of the isolates collected in 1999 or the test isolates, lowest-Di (LDi) analysis was performed. A matrix of Di values was constructed in Excel comparing the phenotypes of the test isolates to those of the control isolates from MMSA. The lowest Di value for each test isolate and the mean of the lowest Di value were then determined. To determine the expectation of the mean, a matrix of Di values was first constructed in Excel comparing the phenotypes within the control isolates. After removal of Di values for comparisons of the isolates to themselves, the LDi for each control isolate was determined. The means of the lowest Di values of 100 random selections equal in number to those of the test isolates were then calculated.
DNA extraction for clone libraries.
Total DNA was extracted from the soil and cast samples by a modification of the protocols of Tsai and Olson (79) and Moré et al. (52). Soil, 1 g (wet weight), from each of the five sites was placed into 10-ml centrifuge tubes. The pooled casts from five earthworms (0.2 g [wet weight]) were placed into a 1.5-ml tube. To each tube was added 2 ml (g of sample−1) of a lysis buffer (0.15 M NaCl, 0.1 M disodium EDTA [pH 8.0], and lysozyme [1.5 mg/ml]). The samples were vortexed and placed in a shaking 37°C water bath for 1 h, with additional vortexing every 15 min. The soil slurries were aliquoted into 500-μl portions in microfuge tubes. To each of the soil aliquots and the tube containing the cast material was added 400 μl of a bead beating solution (0.1 M NaCl, 0.5 M Tris Cl [pH 8.0], and 10% sodium dodecyl sulfate), and approximately 0.1 g of 0.1-mm-diameter glass beads. These tubes were then placed into a TurboMix adapter (Scientific Products) for a Vortex Genie 2 (Fisher Scientific) and treated for 3 min at maximum speed. The tubes were centrifuged for 3 min, and the supernatants were decanted into new tubes. An equal volume of phenol:chloroform:isoamyl alcohol (25:24:1) (Amresco) was added to each sample, vortexed briefly, and centrifuged for 10 min. The aqueous layer was transferred to a new tube and extracted again with chloroform:isoamyl alcohol (24:1). The aqueous layer was transferred to a new tube, and the nucleic acids were precipitated with an equal volume of isopropanol at −20°C. After the precipitation, the tubes were centrifuged for 10 min. A dark brown pellet was visible in each tube. The pellets were vacuum dried and resuspended in 500 μl of distilled water (dH2O). These preparations were treated with RNase (final concentration, 10 μg/500 μl) at 37°C for 1 h followed by passage through a Wizard DNA Clean-Up Kit (Promega). The quality and concentration of DNA were confirmed via gel electrophoresis on a 1% agarose gel.
PCR amplification of 16S rRNA genes.
16S rDNA was amplified from soil- and cast-extracted DNA for construction of bacterial and archaeal clone libraries. Each PCR consisted of 1 Ready-to-Go PCR bead (Amersham Pharmacia), 2 μl of forward primer (either 27f for bacterial libraries [5′-AGA GTT TGA TCM TGG CTC AG-3′] or 21f for archaeal libraries [5′-TTC CGG TTG ATC CYG CCG GA-3′]), 2 μl of reverse primer (1392r universal [5′-ACG GGC GGT GTG TRC-3′]), 1 or 2 μl of sample DNA (4 to 20 ng), enough dH2O to bring the reaction to 25 μl, and 30 μl of mineral oil. Each PCR was 25 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1.75 min for denaturation, annealing, and extension steps, respectively, on a Thermolyne Temp · tronic thermocycler (Dubuque, Iowa). The mineral oil was removed, and the products were visualized on a 1% agarose gel prior to cloning. PCR products were either gel purified or cloned directly into the vector.
The 16S rDNA of the 204 isolates was also PCR amplified using Ready-To-Go PCR Beads and the same bacterial primers as the clone libraries. Isolates were grown on Difco nutrient agar plates, and a single colony of each isolate was resuspended and washed two times in 100 μl of sterile dH2O. Each cell suspension was added to a reaction mixture at a concentration of 8% (vol/vol) to act as the template DNA, and each reaction mixture was heated for 5 min at 94°C. The reactions were denatured, extended, and annealed for 1 min at 94°C, 2 min at 72°C, and 1 min at 61°C for 30 cycles. Isolates which did not produce a PCR product with whole-cell cultures were treated with the same protocol used to extract DNA for the clone libraries (without the use of a Clean-Up kit), and 100 ng of the resultant DNA was used in a PCR. PCR products of the isolates were purified using Prep-A-Gene DNA Purification Systems (Bio-Rad).
Construction of clone libraries.
Clone libraries of archaeal and bacterial PCR products were constructed using a TA Cloning Kit (Invitrogen) with plasmid pCR2.1. Successful transformants were inoculated into Luria-Bertani broth containing kanomycin and allowed to grow overnight. One library was prepared for each of the five soil samples. Similarly, the cast sample was amplified twice, and a library was prepared from each PCR product. Plasmids from the libraries were extracted using a QIAprep Spin Miniprep Kit (Qiagen). Clones containing putative 16S rRNA genes were screened by EcoRI restriction endonuclease digestions. Each digestion reaction consisted of 2 μl of purified plasmid, 2 μl of buffer H (Promega), 1 μl of EcoRI (Promega), and 18 μl of dH2O and was incubated at 37°C for 1 h. Digestion products were visualized on a 1% agarose gel to ensure the presence of inserts of the expected size. Clones which exhibited no insert or inserts much larger or smaller than expected were excluded from sequencing.
Screening of archaeal clones.
Archaeal clones appeared less diverse than bacterial clones and were screened by restriction analysis prior to sequencing. Inserts in archaeal clones were reamplified using the PCR protocol described above. The restriction digestion was composed of 3 μl of PCR product, 1 μl of 10× Multicore buffer (Promega), 1 μl of PvuII (Promega), 1 μl of BglI (Promega), and 4 μl of dH2O. The reaction was incubated at 37°C for 1 h, and the entire reaction mixture was run on a 2% agarose gel.
Sequencing of 16S rRNA genes.
Partial sequencing of 16S rRNA genes was accomplished using an ABI 377 automated sequencer (Perkin-Elmer). Bacterial clones and isolates were sequenced using the primer 27f, while archaeal clones were sequenced with the primer 21f. Selected clones were also sequenced using primer 1392r. Sequences were manually examined for quality, and those containing multiple ambiguous bases or were too short for analysis were resequenced. Isolate sequences containing greater than five ambiguous bases per 500 bp were cloned into the vector pCR 2.1 (Invitrogen), and the plasmid was resequenced.
Analysis of 16S rDNA data.
The closest database relatives of all sequences were obtained using all available sequence information by FastA searches of GenBank (56). Duplicate sequences (≥99% sequence similarity) were grouped into an operational taxonomic unit (OTU), and only one sequence was used for further analysis. The low similarity of many of the clone sequences to known organisms' sequences made classification into phyla based solely on these searches difficult; therefore, all clone sequences were plotted on a phylogenetic tree with representative sequences from known phyla. Alignments of sequences and reference organisms were created using the PILEUP program of the GCG software package (26). Only sequence data corresponding to Escherichia coli bases 81 to 459 were used for the phylogenetic trees. Trees were constructed using the PHYLIP software package (22). Distances were calculated using the Jukes-Cantor algorithm of DNADIST, and the branching order was determined via a neighbor-joining algorithm using NEIGHBOR. Each tree was a consensus of 100 replicate trees (constructed by SEQBOOT) to obtain bootstrap values. Phylogenetic trees specific for phyla and candidate divisions were constructed to insure support for the positioning of clone sequences. Sequences which did not associate with any of the formally described lineages using these methods were classified as “undescribed” taxa for this analysis.
Chimera checking.
All clones exhibiting less than 95% sequence similarity to an existing GenBank sequence were examined as potential chimeras. Sequences were first analyzed with the CHIMERA_CHECK program at the Ribosomal Database Project II (RDP-II [47]) using the default settings. Potential chimeric sequences were then reanalyzed excluding Ribosomal Database Project sequences shorter than 1,000 nucleotides so that the analysis would not be biased by shorter entries that contained sequence information homologous to only a portion of the clone sequence. Clones that were not excluded as possible chimeras at this step were then examined through the use of FastA searches of the entire available sequence and of the 5′ and 3′ sequence fragments as previously suggested by CHIMERA_CHECK. Sequences where the 5′ and 3′ ends were closely related to different phylogenetic groups and did not have a close relative in this study from an independent PCR were considered chimeric and were excluded from the study.
Statistical analyses of the libraries.
Rarefaction was used to evaluate the OTU richness of the isolate libraries (35, 40). Because the sample sizes differed for the three collections, the diversity indices of the OTUs were calculated using normalized sample sizes. The sample size of soil isolates was reduced to 31 and 74 by random deletion for the comparison of diversity measurements to the burrow and cast collections, respectively. For both the isolates and clones, evenness was calculated to describe the distribution of abundance of isolates (59). Shannon diversity, a general diversity measurement, was used to evaluate OTU richness and evenness (67). The distribution of coverage (28) at various evolutionary distances (based on 16S rDNA data [D]) or dissimilarity values (based on phenotypic data [Di]) was calculated for the soil, cast, and burrow isolates. Coverage was calculated using Good's formula (28). LIBSHUFF analyses were used to compare libraries from different sources (68).
Accession numbers.
Isolate sequences were deposited in GenBank with accession numbers AY039379 to AY039509 and AY039806. Clone sequences were deposited with the accession numbers AY037556 to AY037762.
RESULTS
Construction of bacterial libraries.
Totals of 100 and 104 bacterial 16S rDNA clones were sequenced from DNA extracted from Horseshoe Bend agricultural soil and L. rubellus casts, respectively. Five soil and two cast clones were identified as chimeras and removed from further analyses, resulting in 95 and 102 clones for the libraries, respectively.
Comparison of the replica libraries suggested that the methods for their construction were reproducible because most phyla were evenly represented in the replicates (Table 1). A notable exception was the presence of relatively high numbers of Firmicutes sequences in only one of the five soil libraries and one of the two cast libraries. As all reactions were treated in a similar fashion, the explanation for the disproportionate amplification of Firmicutes sequences was not obvious. Perhaps the distribution of this taxon was uneven in the soil samples as well, although this would not explain the increased occurrence of Firmicutes in one of the cast libraries, which were both constructed from the same sample. Additionally, both the cast and soil libraries contained representatives of phyla difficult to lyse in pure culture (e.g., Actinobacteria), so biases arising from incomplete cell lysis were probably minimal (52).
TABLE 1.
Distribution of clone and isolate sequences in phylogenetic groups
| Phylogenetic group | No. of clones from each librarya
|
% Of total library (clones)
|
% Of total library (isolates) from:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Agricultural soil
|
Cast
|
Pooled clone reactions
|
Soil (1996) | Burrow (1996) | Casts (1997) | Soil (1999) | Casts (1999) | ||||||||
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | Soil | Cast | |||||||
| Acidobacteria | 3 | 2 | 3 | 4 | 1 | 3 | 7 | 13.7 | 9.8 | ||||||
| Actinobacteria | 1 | 1 | 2 | 1 | 10 | 7 | 5.3 | 16.7 | 53.5 | 51.6 | 12.2 | 57.1 | 75.0 | ||
| Candidate division TM7 | 1 | 1 | 3 | 3 | 2.1 | 5.9 | |||||||||
| Cytophagales | 3 | 2 | 3 | 1 | 1 | 8 | 5 | 10.5 | 12.7 | 1.4 | |||||
| Fibrobacter | 1 | 2 | 1 | 1 | 4.2 | 1.0 | |||||||||
| Firmicutes | 4 | 11 | 1 | 4.2 | 11.8 | 15.2 | 19.4 | 4.1 | 21.4 | 16.7 | |||||
| Chloroflexi | 2 | 1 | 3 | 2 | 2 | 8.4 | 2.0 | ||||||||
| Nitrospira | 2 | 1 | 2 | 1 | 5.3 | 1.0 | |||||||||
| Planctomycetes | 1 | 2 | 3.2 | ||||||||||||
| Proteobacteria | |||||||||||||||
| α- | 1 | 1 | 2 | 8 | 2 | 4.2 | 9.8 | 5.1 | 6.5 | 1.4 | 7.1 | ||||
| β- | 1 | 3 | 3 | 1 | 1 | 2 | 8.4 | 2.9 | 5.1 | 9.7 | 4.1 | 7.1 | 8.3 | ||
| γ- | 1 | 1 | 1 | 1 | 15 | 7 | 4.2 | 21.6 | 21.2 | 12.9 | 77.0 | 7.1 | |||
| δ- | 2 | 2 | 1 | 1 | 5.3 | 1.0 | |||||||||
| Verrucomicrobia | 2 | 1 | 2.1 | 1.0 | |||||||||||
| Undescribed groupb | |||||||||||||||
| A | 3 | 1 | 2 | 6.3 | |||||||||||
| B | 1 | 1 | 1 | 1.1 | 2.0 | ||||||||||
| C | 1 | 1 | 1 | 3.2 | |||||||||||
| D | 1 | 1 | 1.1 | 1.0 | |||||||||||
| E | 1 | 1 | 2.1 | ||||||||||||
| F | 2 | 1 | 2.1 | ||||||||||||
| G | 1 | 1.1 | |||||||||||||
| H | 1 | 1.1 | |||||||||||||
| I | 1 | 1.1 | |||||||||||||
| Total sample size (n) | 17 | 27 | 20 | 21 | 10 | 64 | 38 | 95 | 102 | 99 | 31 | 74 | 14 | 12 | |
Each column represents the number of clones from a particular PCR. Each soil PCR was derived from a separate sample, while the cast PCRs originated from the same DNA extracted from pooled cast samples. All clone sequences are from 1999.
Clones which did not associate with a known phylum. Descriptions of each group are provided in the text.
Phylogeny of clone libraries.
Sequences with ≥99% sequence similarity over the region of the 16S rRNA gene used for phylogenetic analyses were combined into OTUs. The soil library contained 86 OTUs (out of 95 clones), which were classified into 11 phyla (or candidate divisions) and 9 clades which have not been formally described (undescribed groups). No single phylum dominated the soil library, but the largest number of clones were from Acidobacteria (14%), Cytophagales (13%), Chloroflexi (8%), and β-Proteobacteria (8%). The cast library contained 81 OTUs (out of 102 clones), which were grouped into 10 phyla (or candidate divisions) and 2 undescribed clades. The library included comparable numbers of Acidobacteria (10%) and Cytophagales (13%) as the soil library, but it also contained a large number of clones from the γ-Proteobacteria (22%), Actinobacteria (17%), Firmicutes (12%), and α-Proteobacteria (10%), groups that were not well represented in the soil. All phyla and undescribed taxa found in the cast library were also present in the soil library, although the soil library contained a number of groups that were not found in the cast library.
A number of clones could not be placed into a described phylum or candidate division using our methods (Table 1) and were classified into undescribed groups A through I. Group A was the largest and was found only in soil. It possessed an average 87% sequence similarity to a clone obtained from an environment contaminated with pentachlorophenol (4). Because these clones, which possessed 85 to 93% sequence similarity to each other, were obtained from more than one sample, they were unlikely to be PCR artifacts. Group B included two cast clones and a soil clone (84 to 89% similarity) with an average sequence similarity of 91% to clones from rhizosphere (48) and arid soil (43). Group C contained three soil clones (88 to 89% similarity) which possessed an average sequence similarity of 86% to hot spring clones (34). Group D contained two clones with an average similarity of 86% to each other and clones from an arid soil (17) and a metal-contaminated soil. Group E consisted of a pair of soil clones (85% similarity) with an average similarity of 83% to Chloroflexi clones from an anaerobic bioreactor (81) and Antarctic ice (29). However, in our analyses these sequences did not possess a strong affiliation with the Chloroflexi phylum. Group F included two identical soil clones with 77% similarity to a freshwater bacterium clone (27). Groups G, H, and I each consisted of a single soil clone with sequence similarity to Acidosphaera rubrifaciens (74%), a subsurface clone (80% [10]), and a deep-sea bacterium (89% [46]), respectively.
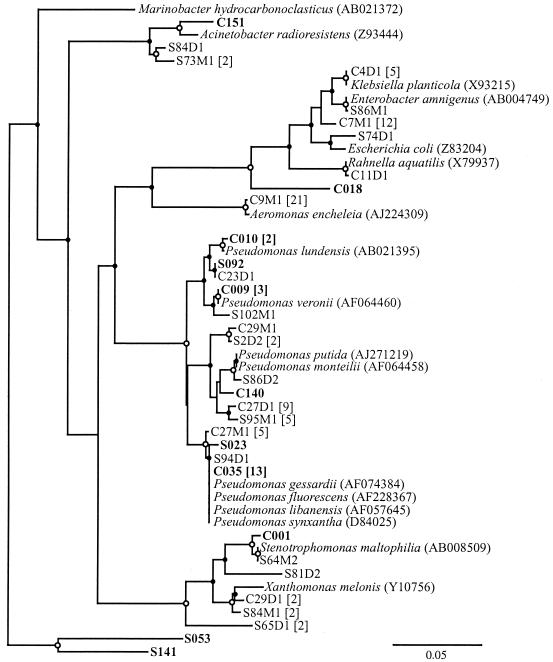
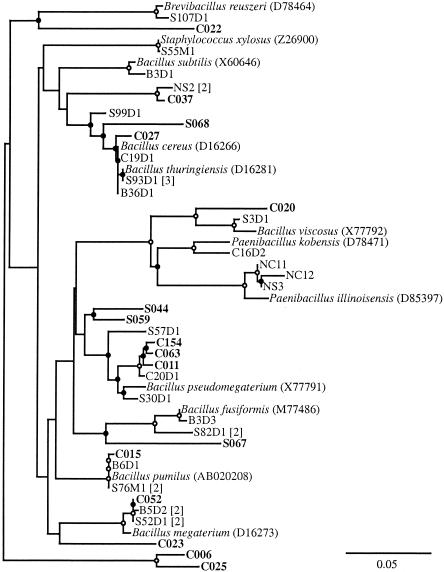
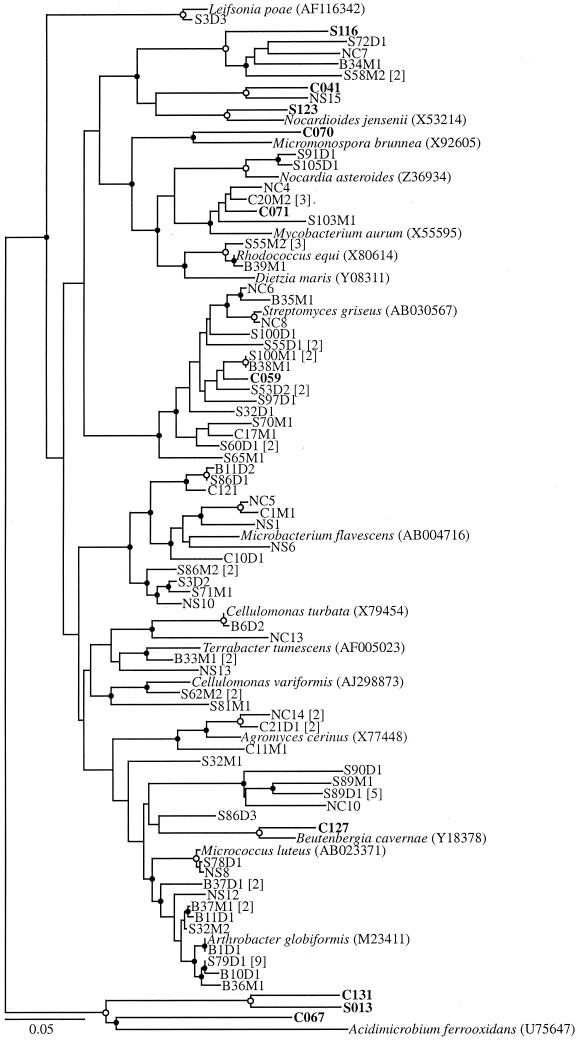
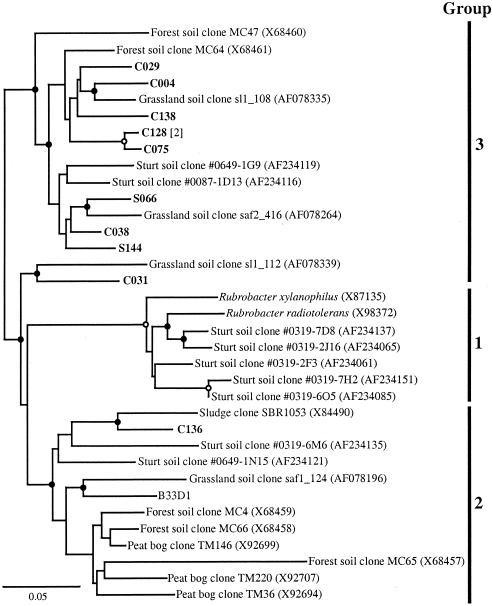
While the abundances of many of the taxa in the soil and cast libraries were similar, the abundance of other taxa changed dramatically. For instance, the γ-Proteobacteria increased from 4% of the soil library to 22% of the cast library (Table 1). The majority of cast clones in this group were closely related to a number of Pseudomonas spp. (Fig. 1). Although numerically less abundant, closely related soil clones were also observed. The Firmicutes clones also increased from 4% in the soil to 12% in the casts (Table 1),and most of the clones were phylogenetically related to Bacillus spp. and similar genera (Fig. 2). Although none of the cast clones were closely related to the soil clones, this observation may be due to the low representation of soil clones in this taxon. The Actinobacteria also increased from 5% in the soil to 17% in the cast (Table 1). While individual clones were associated with a number of different genera within the phylum (Fig. 3), the largest group of nine clones (two soil clones and seven cast clones) were related to the Rubrobacteria, which have been previously found in a number of clone libraries from soil (Fig. 4 and references 7, 33, 45, 49, and 61). A relative of Rubrobacter was also isolated from burrow soil in this study (see below).
FIG. 1.
Phylogenetic tree of the γ-Proteobacteria clones and isolates from Horseshoe Bend. Sequence names are coded. For the clones, the names are in boldface where the letter C or S before the number indicates cast or soil, respectively, e.g., C102. For the isolates from 1996 and 1997, the names include the letter C, S, or B, followed by a unique number; the letter M or D; and the number 1 or 2. The first letter indicates the habitat: cast, soil, or burrow, respectively. The first number is the sample number. The second letter indicates the isolation medium: MMSA or DNB, respectively. The last number indicates whether the isolate was the first or second isolate from the particular sample, e.g., S84D1. For the isolates from 1999, the names include the letters NC or NS for cast or soil isolates, respectively, and a unique number, e.g., NC8. Brackets after sequence names denote the total number of sequences in the OTU when that number was greater than 1. Accession numbers appear in parentheses for all reference sequences. Open circles (○) denote bootstrap support of ≥95%, and closed circles (•) denote bootstrap support of ≥50%. The scale bar represents the Jukes-Cantor evolutionary distance.
FIG. 2.
Phylogenetic tree of the Horseshoe Bend clones and isolates associated with the Firmicutes phylum. Notation is as described for Fig. 1.
FIG. 3.
Phylogenetic tree of the Horseshoe Bend clones and isolates associated with the Actinobacteria phylum except Rubrobacter. Notation is as described for Fig. 1.
FIG. 4.
Phylogenetic tree of the Horseshoe Bend clones and isolates associated with the Rubrobacteria. Groups denoted on the right side of the tree follow the nomenclature of Holmes et al. (33). Other notation is as described for Fig. 1.
Library comparisons.
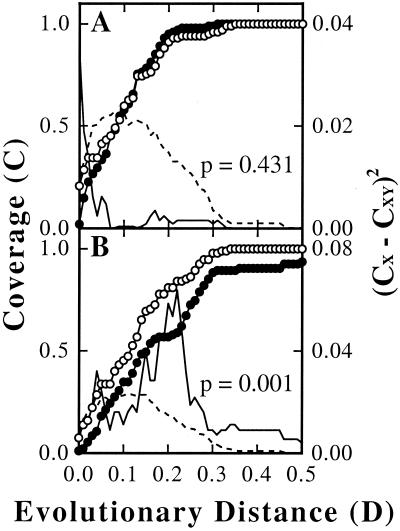
The hypothesis that the soil and cast clone libraries were not significantly different was tested by the LIBSHUFF method (68). The heterologous coverage of the cast library by the soil library was not significantly different (P = 0.431) from the homologous coverage of the cast library by itself, indicating that most sequences from the cast library had high similarity to sequences in the soil library (Fig. 5A). Additionally, a comparison of the calculated value of (CX − CXY)2 to the 95% value of (CX − CXY)2 from the random shuffles showed differences between the libraries only when D was ≤0.02. This result suggested that differences that did occur were mostly among closely related sequences. This result was also expected from the low coverage of the libraries at high levels of relatedness (see below). These conclusions were supported by the phylogenetic trees in which cast sequences often grouped near soil sequences but were rarely identical (Fig. 1 to 4). In fact, the average D (± standard deviation) from a cast clone to its closest relative in the soil libraries was only 0.09 ± 0.07, compared to a distance of 0.18 ± 0.14 for a soil clone to its nearest relative in the cast libraries. In contrast to the high similarity of the cast library to the soil library, the heterologous coverage of the soil library by the cast library was significantly different (P = 0.001) from the homologous coverage of the soil library (Fig. 5B). This result indicated that some sequences in the soil library had no close relatives in the cast library. Differences between the libraries were especially apparent when D was between 0.03 and 0.05 and well as when D ≥ 0.14 (Fig. 5B), which suggested that in addition to differences within phyla, at least one deep phylogenetic group was present in the soil library that was absent in the cast library.
FIG. 5.
LIBSHUFF comparison of HSB soil and cast clones. Homologous coverage curves are shown with open circles (○) and heterologous coverage curves with closed circles (•). Solid lines indicate the difference between the homologous and heterologous coverage curves at each value of D as determined by the Cramér-von Mises test statistic, and broken lines denote the 95% value of the random shufflings. (A) Comparison of the HSB cast clones (X) to the HSB soil clones (Y); (B) comparison of the HSB soil clones (X) to the HSB cast clones (Y).
Examination of Table 1 suggested that the increased abundance of the Actinobacteria, Firmicutes, and γ-Proteobacteria in the cast library as well as the presence of six sequences in the undescribed group A in the soil library that were absent from the cast library may have been responsible for the differences in the two libraries. When these four groups were removed from the LIBSHUFF comparison, the cast and soil libraries were no longer statistically significantly different (see comparison 2 [Table 2]). However, removal of any three of these four groups from the analysis still yielded P values that were < 0.05 (data not shown). Thus, all four taxa combined were responsible for the differences between the soil and cast bacterial libraries. In contrast, the differences in the abundance of α-Proteobacteria did not appear significant.
TABLE 2.
LIBSHUFF comparisons of isolate and clone libraries
| Comparison no. | Homologous (X) coverage data
|
Heterologous (Y) coverage data
|
|||
|---|---|---|---|---|---|
| Library | Sample yr | n | Library | P | |
| 1 | Soil clones | 1999 | 95 | Cast clones | 0.001 |
| Cast clones | 1999 | 102 | Soil clones | 0.431 | |
| 2 | Soil clone subseta | 1999 | 76 | Cast clone subseta | 0.144 |
| Cast clone subseta | 1999 | 51 | Soil clone subseta | 0.421 | |
| 3 | Soil clones | 1999 | 95 | Soil isolates | 0.001 |
| Soil isolates | 1996 | 99 | Soil clones | 0.001 | |
| 4 | Cast clones | 1999 | 102 | Cast isolates | 0.042 |
| Cast isolates | 1997 | 74 | Cast clones | 0.001 | |
| 5 | Soil isolates | 1996 | 99 | Burrow isolates | 0.192 |
| Burrow isolates | 1996 | 31 | Soil isolates | 0.551 | |
| 6 | Soil isolates | 1996 | 99 | Cast isolates | 0.001 |
| Cast isolates | 1997 | 74 | Soil isolates | 0.001 | |
| 7 | Soil isolates | 1996 | 99 | Cast isolate subsetb | 0.001 |
| Cast isolate subsetb | 1997 | 53 | Soil isolates | 0.005 | |
Does not contain clones within Actinobacteria, Firmicutes, γ-Proteobacteria, and undescribed group A.
Does not contain isolates belonging to the Aeromonas lineage.
Archaeal clone libraries.
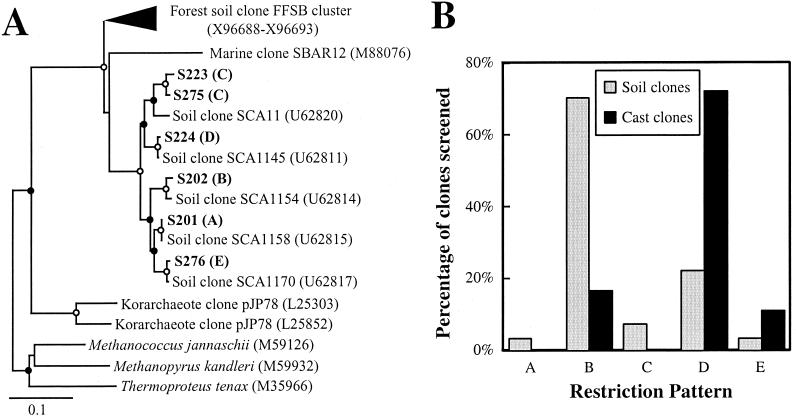
Archaeal PCR products were obtained from DNA extracted from four of the five soil samples as well as the pooled cast sample. Preliminary sequencing of some of these archaeal clones indicated that the libraries contained limited diversity. Therefore, 27 clones from the soil samples and 18 clones from the cast sample were screened by restriction fragment length polymorphism (RFLP) analysis. Five patterns (designated types A to E) were observed, and partial sequencing of representative clones indicated that they constituted a single archaeal lineage which had been detected previously in soil (Fig. 6A and reference 6). The RFLP types were not distributed evenly between the soil and cast clones (Fig. 6B). Pattern B dominated in the soil clones (70%), while pattern D accounted for the majority of the cast clones (72%). Additionally, it is possible that pattern E may have been more highly represented in the cast than the soil clones. Although only three of the five RFLP types were found in the cast library, fewer cast clones were screened, and it was not surprising that less abundant RFLP types (types A and C) were not detected in the cast sample.
FIG. 6.
(A) Phylogenetic tree of the soil archaeal clones from Horseshoe Bend. RFLP patterns are in parentheses after clone names. Other notation is as described for Fig. 1. (B) Distribution of RFLP patterns of archaeal clones in soil and cast clone libraries.
Phylogeny of the isolates.
An initial phylogenetic analysis of the 230 isolates obtained from soil and earthworm casts and burrows assigned the isolates to major bacterial groups (Table 1). Sequences belonging to the Actinobacteria were most abundant in the original soil (54%) and burrow (52%) collections, while sequences belonging to the γ-Proteobacteria group comprised 77% of the collection of original cast isolates (Table 1). A chi-square analysis of the soil and burrow collections indicated that their phylogenetic distributions were not different (P > 0.05). In contrast, the distributions of the cast and soil isolates were significantly different (P < 0.05).
Most of the Actinobacteria isolates grouped within the order Actinomyceteales and the suborders Corynebacterinaea, Streptomycinea, and Micrococcinea (Fig. 3). The soil, burrow, and cast isolates were evenly distributed throughout these divisions. It was not unexpected to find representatives of these groups, since Actinomycetales are the bacteria most commonly cultured from soil (55).
In the γ-Proteobacteria, the isolates grouped with five families: Moraxellaceae, Pseudomonadaceae, Xanthomonadaceae, Aeromonadaceae, and Enterobacteriaceae (Fig. 1). Most of the soil and burrow γ-Proteobacteria isolates (10 and 3 isolates, respectively) were associated with the family Pseudomonadaceae. The remaining γ-Proteobacteria soil isolates grouped with Moraxellaceae (three isolates), Xanthomonadaceae (six isolates), and Enterobacteriaceae (two isolates). Most of the γ-Proteobacteria cast isolates were associated with Aeromonadaceae (21 isolates). The remaining cast isolates were found in families that contained soil isolates, including Pseudomonadaceae (16 isolates), Xanthomonadaceae (2 isolates), and Enterobacteriaceae (18 isolates) groups. The large percentage (28%) of the cast isolates associated with Aeromonadaceae was unexpected. Aeromonas spp. are typically associated with fish and aquatic habitats. Although they have never been isolated from soil, they have been previously identified as a dominant isolate in the casts of the earthworm E. foetida (76) and have been associated with the gut of Pheretima sp. (77).
The Firmicutes isolates all grouped within the class Bacilli (Fig. 2). The majority of the soil, cast, and burrow isolates were associated with Bacillus spp. Since Bacillus spp. are commonly found in soil, this result was not surprising. Some of the cast and soil isolates grouped with the Paenibacillus spp., and some of the soil isolates were associated with the Brevibacillus and Staphylococcus spp.
The α- and β-Proteobacteria were also represented in the soil, cast, and burrows. The isolates in the β-Proteobacteria group were associated with the families in the order Burkholderiales, including Comamonadaceae, Burkholderiaceae, Oxalobacteraceae, and Ralstoniaceae. Soil and cast isolates grouped with the Comamonadaceae, and only one soil isolate grouped with Burkholderiaceae. The Ralstoniaceae and Oxalobacteraceae taxa each contained burrow and cast isolates. The isolates belonging to the α-Proteobacteria all grouped with the order Rhizobiales. All of these taxa are found commonly in soil.
LIBSHUFF comparisons of the isolates and clones.
The LIBSHUFF comparisons of the isolates suggested that soil and burrow isolates were derived from similar populations but that the cast isolates were from a population different from that of the soil isolates (comparisons 5 and 6 [Table 2]). Analysis of the coverage curves constructed from the comparison of the soil isolates and cast isolates indicated a significant increase in the heterologous coverage at D = 0.13 (data not shown), which appeared to be due to the presence of a large number of Aeromonas isolates from the cast but not from soil. The possibility that this taxon alone was responsible for the differences in the isolate libraries was eliminated by performing an additional LIBSHUFF analysis after removing the Aeromonas sequences from the data set (comparison 7 [Table 2]). The differences between the cast and soil isolate libraries remained significant, especially at low values of D (data not shown). More simply stated, it appeared that all the genera found in the cast isolates (except Aeromonas) were also found in the soil isolates; however, the species within those genera were different.
LIBSHUFF comparisons of the clone and isolate libraries were significantly different (comparisons 3 and 4 [Table 2]). Given that the clone libraries contained many more taxa than the isolates, it was not surprising that the isolate library provided poor heterologous coverage of the clone library. However, the clone library might have been expected to provide good heterologous coverage of the isolate libraries. Presumably, this did not occur because of the low representation of the taxa abundant among the isolates in the clone libraries and the fact that the isolates and clone libraries were collected in different years. It is worth noting that previous studies utilizing both molecular and cultivation methods have produced similar findings (5, 10, 17, 50, 58, 69, 73).
Diversity indices.
Various diversity indices were calculated for all libraries (Table 3). The diversity and evenness indices approached the maximum for both the soil and cast clone libraries, although the values for the soil library were always higher. The higher diversity of the soil clone library was reflected in the phylogenetic distribution. The soil library contained representatives of more phyla and other deep phylogenetic groups than did the cast clones (Table 1). Coverage values were low for both clone libraries, which would be expected for libraries constructed from populations of high diversity. However, the coverage of the cast library was slightly higher than the soil library's, which was consistent with the somewhat lower diversity in the cast library. Additionally, the large number of nearly identical sequences representing the genus Pseudomonas (Fig. 1) was also consistent with the higher overall coverage of the cast library.
TABLE 3.
Diversity indices for clone and isolate libraries based on 16S rDNA sequencesa
| Type of data | Clone library data
|
Isolate library data
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Soil (1999) | Castb (1999) | Maximumc | Soilb (1996) | Cast (1997) | Maximumc | Soilb (1996) | Burrow (1996) | Maximumc | |
| Shannon-Weaver | 4.40 | 4.11 | 4.61 | 3.65 | 2.47 | 4.30 | 3.21 | 3.21 | 3.43 |
| Evenness | 2.27 | 2.19 | 2.33 | 2.18 | 1.81 | 2.30 | 2.27 | 2.27 | 2.30 |
| Coverage | 0.14 | 0.20 | 1.00 | 0.57 | 0.80 | 1.00 | 0.32 | 0.32 | 1.00 |
All calculations are based on an OTU of ≥99% sequence similarity over positions 81 to 459. The sample sizes for the clone library, soil-cast isolate, and soil-burrow isolate comparisons were 95, 74, and 31, respectively.
The size of these libraries was randomly reduced to allow direct comparison to a smaller library. Values shown for the cast clone library are an average of 10 random reductions.
Maximum value for each index. The minimum values for each index were 0.
Although the diversity was much less, similar trends were also found for the isolate libraries. First, rarefaction was used to compare the OTU richness within each group of isolates at standardized sample sizes (data not shown). The original soil isolate collection contained about twice as many OTUs as the original cast isolate collection at sample sizes greater than 35 and just as many as the burrow collection at all sample sizes. The diversity and evenness indices for the soil isolates were close to the maximum possible values and higher than the values for the cast isolates (Table 3). Similarly, coverage by the soil isolates was lower. The 80% coverage of the cast isolates indicated that the majority of the cast community culturable with the methods used here was collected. At D = 0.05, the coverage was > 90% (data not shown), indicating that most of the deeper phylogenetic groups obtainable by these methods were also isolated. In contrast, the diversity and evenness indices for the soil and burrow isolates were identical (Table 3). By these criteria, those libraries appeared the same.
Phenotypic properties of isolates.
To determine the phenotypic properties of the isolates, 20 tests were designed to examine characteristics that could be relevant to the soil habitat or measured conveniently. To ensure reproducibility, each test was replicated five times using a subset of five randomly chosen isolates. In this screening, the tests for motility and cellulose and citrate utilization proved not to be reproducible and were not used to characterize the isolates.
The phenotypic properties of soil, burrow, and cast isolates collected in 1996 and 1997 were different (Table 4). A two-tailed chi-square analysis identified tests that possessed P values below 0.05 and might be statistically significantly different between sample sources. These phenotypic tests were further analyzed using scaled deviance to determine if the differences were due to the sample source or sampling procedure. In these analyses, the results of 12 of the 17 phenotypic tests varied statistically significantly with the sample source. The only tests which did not vary statistically significantly with the sample source were those for hemolysis, ampicillin, and tetracycline resistance; casein hydrolysis; and urea degradation. Pairwise phenotypic differences between cast and burrow isolates, cast and soil isolates, and burrow and soil isolates were also analyzed using a chi-square analysis. Of the tests that varied significantly with the sample source, the cast isolates were significantly different (P ≤ 0.01) from the soil and burrow isolates in 11 and 12 tests, respectively (data not shown). In contrast, only the cellular morphology and lactose utilization were different (P ≤ 0.01) between the soil and burrow isolates. Thus, the phenotypic differences between the three groups of isolates were largely due to differences between the cast isolates and the soil and burrow isolates.
TABLE 4.
Phenotypic characterization of bulk soil, burrow, and cast isolates
| Test or characteristic | % Occurrence in isolates (yr) (n)
|
chi-square Pa | Scaled deviancea | ||||
|---|---|---|---|---|---|---|---|
| Soil (1996) (99) | Soil (1999) (14) | Burrow (1996) (31) | Cast (1997) (74) | Cast (1999) (12) | |||
| Cell morphology | 0.024b | ||||||
| Cocci | 9 | 36 | 0 | 3 | 25 | 16 | |
| Small rod | 13 | 14 | 36 | 11 | 8 | 21 | |
| Rod | 46 | 36 | 36 | 58 | 25 | 20 | |
| Large rod | 33 | 14 | 29 | 28 | 42 | 38 | |
| Catalase | 0.024 | 29 | |||||
| Production | 90 | 79 | 94 | 77 | 83 | ||
| Hemolysis | 0.480b | ||||||
| Alpha- | 30 | 14 | 16 | 23 | 8 | ||
| Beta- | 7 | 7 | 10 | 12 | 8 | ||
| Hydrolysis | |||||||
| Casein | 22 | 43 | 32 | 32 | 17 | 0.286 | |
| Urea | 44 | 21 | 58 | 27 | 25 | 0.007 | 39c |
| Tween 80 | 18 | 29 | 13 | 36 | 50 | 0.006 | 14 |
| Antibiotic resistance | |||||||
| Ampicillin | 32 | 14 | 29 | 49 | 42 | 0.043 | 35c |
| Tetracycline | 22 | 0 | 0 | 0 | 0 | 0.339 | |
| Polymyxin | 29 | 7 | 23 | 7 | 25 | 0.002 | 27 |
| Streptomycin | 9 | 14 | 13 | 0 | 25 | 0.014 | 32 |
| Carbon utilization | |||||||
| Acetate | 45 | 29 | 39 | 73 | 42 | 0.001 | 22 |
| Lactose | 24 | 7 | 52 | 0 | 17 | 0.001 | 17 |
| Casamino Acids | 57 | 36 | 65 | 84 | 42 | 0.001 | 29 |
| Mannose | 35 | 21 | 42 | 7 | 25 | 0.001 | 27 |
| Cellobiose | 23 | 7 | 26 | 1 | 17 | 0.001 | 25 |
| MacConkey | 0.001b | ||||||
| Growth | 19 | 7 | 23 | 91 | 25 | 46 | |
| Fermentationd | 10 | 7 | 0 | 18 | 17 | 23 | |
| Nitrate reduction | 0.001b | ||||||
| Partial | 28 | 36 | 26 | 73 | 42 | 37 | |
| Complete | 0 | 0 | 0 | 4 | 0 | 5 | |
The null hypothesis was as follows: μbulk soil (1996) = μburrow (1996) = μcast (1997). When the chi-square analysis was statistically significant (P < 0.05), the genmod scaled deviance analysis was performed to confirm that the difference was not due to the sampling procedure.
These tests had more than one response. For the chi-square analysis, the responses were grouped together to observe overall differences in the test for the three types of isolates. For the genmod scaled deviance analysis, each response was observed separately.
These P values for these scaled deviances were not statistically significant at P = 0.05. The P values for all of the other scaled deviances were statistically significant at P < 0.05.
Fermentation of lactose.
Because cellulose is abundant in litter, many of the cast isolates were expected to degrade cellulose and cellobiose, a common intermediate in cellulose degradation. While it was not possible to test cellulose degradation in a reproducible manner, not many of the cast isolates were able to utilize cellobiose. Moreover, no other sugars tested were utilized by a large proportion of the cast isolates. Instead, they utilized amino acids and acetate. A trivial explanation for this observation is that the original media used to enrich the isolates lacked sugars and that saccharolytic organisms were selected against. Nevertheless, sugars were utilized by about one-third of the soil and burrow isolates, so this cannot be the full explanation.
Since nitrate and nitrite are more abundant in casts than in surrounding soil (44) and since N2O is emitted from the alimentary canal of L. rubellus (37), bacteria capable of reducing nitrate were expected to be abundant in casts. A high percentage of the cast isolates, 77%, were capable of reducing nitrate. The samples studied were obtained from no-till soil, which contains lower levels of nitrate (14, 16, 51, 74) and more denitrifiers (16) than does adjacent tilled soil. Likewise, no-tilled soil contains five times more earthworms than tilled soil (32). Therefore, the higher number of denitrifying bacteria in the no-till soil might come from the alimentary canal flora of the earthworms.
The phenotypic properties of the isolates from burrows and soil were very similar. Because urea is excreted through the surface of the earthworm body (18, 44), bacteria that degrade urea were expected to be more abundant in the burrow than the soil and casts. However, no differences in the proportion of urea-degrading isolates were found between the sample sources.
Phenotypic dissimilarity of the isolates.
As a measure of the diversity within each group of isolates, Dis were calculated. Isolates with a Di of 0 had the same responses on all of the phenotypic tests and were defined as possessing the same phenotype. For the isolates from 1996 and 1997, the 99 soil isolates possessed 90 different phenotypes, the 31 burrow isolates possessed 30 phenotypes, and the 74 cast isolates possessed 56 phenotypes. This corresponded to coverages of 14.4, 6.5, and 40.5% for soil, burrow, and cast phenotypes, respectively, and indicated that the sources contained about 630, 460, and 140 phenotypes of bacteria culturable by our methods, respectively. Even though not all of the culturable phenotypes were represented within the collection of isolates, the coverage of culturable phenons with a Di of ≤3 was nearly 100% by both the soil and cast isolates (data not shown). Thus, the collections of isolates provided a good representation of the phenotypic diversity of the organisms culturable by our sampling protocols at the time of isolation.
Very few phenotypes (i.e., Di = 0) were found in more than one sample source. Only four phenotypes were common to both the cast and soil isolates, three phenotypes were shared by the burrow and soil isolates, and no phenotypes were shared between the cast and the burrow isolates. Therefore, any phenotypic similarity between the burrow and soil isolates was not due to a large number of identical phenotypes found in both sources. Instead, the similarity was apparently due to different phenotypes with similar properties.
Within each group of isolates, those from the casts were phenotypically more similar to each other than were the isolates from the soil or burrows. For instance, the mean Di (± standard deviation) for the cast isolates was 5.2 ± 2.4. In contrast, the comparable values for the soil and burrow isolates were 7.2 ± 2.6 and 6.9 ± 2.5, respectively. When the same analysis was performed at standard sample sizes (n = 74 and 29 for the soil and cast, respectively) the mean Di ± standard deviation did not change greatly. The lower mean Di for the cast isolates suggested that their phenotypes were more similar to each other than were the burrow or soil phenotypes.
Isolates from the same particular cast, burrow, or soil sample had mean Di (± standard deviation) of 4.7 ± 2.5, 6.1 ± 3.6, and 5.9 ± 3.0, respectively. These values were similar to those found for all the isolates from each of the three sources. Therefore, the phenotypic properties of the isolates were not highly correlated with the individual sample.
Correlation of phenotypic and rDNA relatedness.
The analysis of both of the 16S rRNA genes and the phenotypic data showed that the burrow and soil isolates were more diverse than the cast isolates. However, the phenotypic data or Di was only weakly correlated with the rRNA evolutionary distance or D. The correlation coefficient r was only 0.49, which was statistically significant only because of the large number of comparisons (n = 2,775). Thus, the phenotypic dissimilarity (Di) was a poor predictor of phylogenetic relationships (D). This conclusion was further supported by the observation that the distribution of Di for the cast and soil isolates was very close to that expected if the test results were assigned to the isolates according to a random model (data not shown). For instance, the means and standard deviations of Di for the casts and soil isolates in a random model would be 5.3 ± 1.9 and 7.3 ± 2.0, respectively, or very close to the observed values. This result suggests that the phenotypic characters were not linked, as might be expected if the Di comparisons contained significant phylogenetic information.
Community stability.
Although the cast and soil isolates were both collected in the spring, the soil isolates were taken 1 year prior to the cast isolates. Therefore, it was possible that the observed differences between the soil and cast isolates were due to sampling time. To address this concern, 14 soil and 12 cast isolates were collected on the same day in May 1999. In this experiment, the isolation was performed on MMSA, and only a single colony type was chosen from each enrichment. However, all other aspects of the sampling protocol were identical to that used for the original isolates. These isolates were characterized by 16S rRNA gene sequencing (Table 1) and by the phenotypic tests used previously (Table 4).
Because the mean Di and chi-square analyses were sensitive to sample size (data not shown), the new collections were compared to the original isolates by the LDi mean method. First, the expected LDi mean was calculated from the data on the original isolates collected in 1996 and 1997. If the bacterial populations were the same in 1999, then the LDi mean of the new isolates should be within the range of the expected values. The LDi mean (± standard deviation) for the new soil isolates was 2.3 ± 0.4. This value was close to the expected LDi mean (± standard deviation of the mean) of 2.1 ± 0.3 and indicated that the phenotypes of the culturable soil bacteria had not changed. This conclusion was unchanged if the analysis was performed with only the original isolates from MMSA. In contrast, the expected LDi mean for the cast isolates was 1.4 ± 0.3, which was much less than 3.3 ± 0.4, the value observed. Therefore, the phenotypes of the culturable cast bacteria had changed. When compared to the original soil isolates, the LDi mean for the new cast isolates was 2.8 ± 0.4, indicating that the cast isolates were also different from the soil isolates of both 1996 and 1999. Therefore, while the phenotypes of the population of culturable bacteria from soil appeared to be stable, those of the casts changed over the period examined. This conclusion was also supported by the distribution of the ribotypes in two sets of samples. For the soil isolates, the distribution of ribotypes was essentially the same (Table 1). In contrast, the ribotypes of the cast isolates varied greatly. In the 1997 sample, 72% of the isolates were γ-Proteobacteria. In 1999, this phylum was not found and 57% of the isolates were Actinobacteria. A change in crop may have caused the change in phenotype of the cast population. In 1997, maize (Z. mays L.) was planted during the growth season, and corn litter was observed in the sample site. In 1998, kenaf (H. cannabinus) was planted and that litter was observed at the sampling site in 1999.
DISCUSSION
The comparisons of the cast and soil libraries are consistent with the model that most of the prokaryotic community in casts is derived from soil with the amplification of only a few prokaryotic groups. L. rubellus is an epigeic earthworm that typically inhabits and feeds within the litter layer of the soil. However, nutritional studies have shown that L. rubellus ingests soil when feeding and actually prefers a combination of soil and litter to litter alone (15). Indeed, the cast samples taken in this study were from earthworms found in the soil profile, well below the litter layer. Thus, it would not be unreasonable to hypothesize that many, if not all, of the prokaryotes deposited in the cast may have originated in the soil. In fact, close relatives of nearly all clone sequences from the L. rubellus cast library were found in the soil library, including those which could not be placed into a known phylum or candidate division. Moreover, some prokaryotic groups were more abundant in the cast than soil libraries, suggesting that these groups may have been specifically amplified by passage through the earthworm gut. This interpretation is consistent with lower diversity indices for the clone library and the isolate collection from the casts than from soil. The lower phenotypic diversity observed in the cast isolates is also consistent with the lower genotypic diversity.
The alternative, that all other taxa were selectively digested, seems unlikely. If degradation was the major process, taxa with the same cell wall type might be expected to fare similarly. This was not observed. For example, the γ-Proteobacteria dramatically increased in abundance while the other proteobacteria increased modestly or not at all. If the numbers of some taxa increased, the abundance of others would be expected to decrease just through dilution. This model alone would appear sufficient to explain the absence of many of the less common soil taxa in the cast library and the lower diversity of the cast library. However, it is also possible that some groups, such as undescribed group A, whose abundance was dramatically lower in casts, were either selectively degraded or avoided during feeding.
The increased numbers of γ-Proteobacteria was the most dramatic difference between the cast and soil libraries. This phenomenon has been observed previously in L. terrestris, in which members of the γ-Proteobacteria increased nearly 20-fold between the foregut and the cast (23). Of the γ-Proteobacteria sequences taken from the cast library, 91% were highly related to Pseudomonas species. Examinations of the numbers of Pseudomonas spp. associated with earthworms sometimes contradict. In one study, 1-day-old casts of L. terrestris contained up to 141 times more fluorescent pseudomonads than a corresponding untreated soil (13). In contrast, a study by Pedersen and Hendriksen (57) found that numbers of Pseudomonas putida decreased after passage through the gut of various Lumbricus earthworm species. Pseudomonas spp. utilize a wide variety of growth substrates and can grow well in the presence of other organisms (53), which could be important given that the alimentary canal of the earthworm contains higher levels of moisture, organic carbon, and total nitrogen than does soil (12, 38). Additionally, many pseudomonads are also resistant to antibiotics produced by Actinobacteria (53), a group which also constituted a significant portion of the cast clone and isolate libraries. Given these characteristics, members of the Pseudomonas genus are likely candidates for amplification by passage through the earthworm.
Members of the Actinobacteria and Firmicutes phyla also appeared in greater numbers in the cast library than in the soil library. Actinobacteria have been commonly isolated from earthworms, and the genera Streptomyces and Micromonospora have been associated with L. rubellus (11, 41, 54). While a number of isolates in this study associated with the Streptomyces genus, only two cast clones were closely related to either of these genera. While no single organism dominated the Actinobacteria cast clones, the Rubrobacteria were the most represented group. Brown (8) hypothesized that antibiotic production by Actinobacteria in the earthworm gut may inhibit the growth of other organisms, particularly other gram-positive and sensitive, gram-negative organisms. Our observations were consistent with this hypothesis, where two of the three taxa with increased abundance in the cast libraries are related to groups that are commonly antibiotic resistant. If this hypothesis is correct, the apparent increase in Firmicutes clones, which are expected to be sensitive to antibiotics, in the cast library may have resulted from endospore germination but not growth. When Fisher et al. (24) examined the passage of Bacillus megaterium endospores through L. terrestris, they found that spores germinated in the gut but found little evidence for the division of vegetative cells. Another study followed the passage of Streptomyces spores through the earthworm E. foetida and found that, while cell numbers decreased in the gut of the worm, numbers of cells and mycelium length significantly increased in the cast (60). The gut and cast conditions of L. rubellus could similarly induce germination of endospores. The loss of the spore coat could then have rendered the cells more susceptible to cellular lysis and increased their representation in the cast libraries.
Although Archaea in soils appear to have limited diversity, they include a number of deep phylogenetic groups that have never been cultivated (6, 9, 36, 39, 62, 80). The archaeal sequences recovered in this study were similar to sequences found in other agricultural soils (6, 80). The sequenced representative of the dominant soil library RFLP type (B) was a close relative of archaeal clones SCA1154 and FIE16 from other soils (6, 80). Although RFLP type B accounted for 47% of all archaeal clones screened in this study (soil and cast), soil clone SCA1154 was found only once in a screening of 34 archaeal sequences from a Wisconsin soil. Thus, it seems likely that this organism is a larger fraction of the archaeal community in HSB than in Wisconsin soil. In contrast, RFLP type D was abundant at HSB and was similar to clone SCA1145, which accounted for 47% of the archaeal sequences obtained in the Wisconsin soil (6) and was similar to a ribotype common in soils from Michigan (9). Therefore, this phylogenetic group appears to be widely distributed. Interestingly, RFLP type D also dominated the libraries constructed from cast material, and it would be worthwhile to know if this group was associated with earthworms at other sites as well.
Because most of the isolates were obtained in different years from those in which the clone libraries were developed, it is not possible to make direct comparisons between the two. However, it is somewhat remarkable that the isolates were mostly representatives of the γ-Proteobacteria, Actinobacteria, and Firmicutes or the same taxa that were amplified in the bacterial cast libraries. Presumably, some phyla of soil prokaryotes are inherently more readily culturable on standard heterotrophic media, and this property correlates with the ability to respond to passage through the earthworm gut. However, the differences in phenotypes for the cast and soil isolates support the hypothesis that the taxa amplified in the cast are not identical to the culturable soil prokaryotes, even though they may be closely related. For instance, the phenotypes of the soil and cast isolates from 1999 were different, even though they represented the same phylogenetic groups.
Acknowledgments
We thank the Institute of Ecology at the University of Georgia for allowing access to Horseshoe Bend, especially Paul Hendrix. We also acknowledge Mark Mackiewicz at the Genome Analysis Facility in the Department of Botany at the University of Georgia for his help with the sequencing of clones. We also acknowledge A. Schramm for sharing results prior to publication.
This work was supported in part by a Research Training Grant in Prokaryotic Diversity (NSF BIR9413235) and an NSF Long Term Research in Environmental Biology (LTREB) grant to study organic matter dynamics in subtropical agroecosystems.
REFERENCES
- 1.Atlas, R. M. 1995. Handbook of media for environmental microbiology. CRC Press, Boca Raton, Fla.
- 2.Barois, I. 1992. Mucus production and microbial activity in the gut of two species of Amynthas (Megascolecidae) from cold and warm tropical climates. Soil Biol. Biochem. 24:1507-1510. [Google Scholar]
- 3.Beare, M. H., P. F. Hendrix, and D. C. Coleman. 1994. Water-stable aggregates and organic matter fractions in conventional- and no-tillage soils. Soil Sci. Soc. Am. J. 58:777-786. [Google Scholar]
- 4.Beaulieu, M., V. Bécaert, L. Deschênes, and R. Villemur. 2000. Evolution of bacterial diversity during enrichment of PCP-degrading activated soils. Microb. Ecol. 40:345-355. [DOI] [PubMed] [Google Scholar]
- 5.Benlloch, S., F. Rodríguez-Valera, and A. J. Martinez-Murcia. 1995. Bacterial diversity in two coastal lagoons deduced from 16S rDNA PCR amplification and partial sequencing. FEMS Microbiol. Ecol. 18:267-280. [Google Scholar]
- 6.Bintrim, S. B., T. J. Donohue, J. Handelsman, G. P. Roberts, and R. M. Goodman. 1997. Molecular phylogeny of archaea from soil. Proc. Natl. Acad. Sci. USA 94:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bond, P. L., P. Hugenholtz, J. Keller, and L. L. Blackall. 1995. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl. Environ. Microbiol. 61:1910-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, G. G. 1995. How do earthworms affect microfloral and faunal diversity? Plant Soil 170:209-231. [Google Scholar]
- 9.Buckley, D. H., J. R. Graber, and T. M. Schmidt. 1998. Phylogenetic analysis of nonthermophilic members of the kingdom Crenarchaeota and their diversity and abundance in soils. Appl. Environ. Microbiol. 64:4333-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandler, D. P., S.-M. Li, C. M. Spadoni, G. R. Drake, D. L. Balkwill, J. K. Fredrickson, and F. J. Brockman. 1997. A molecular comparison of culturable aerobic heterotrophic bacteria and 16S rDNA clones derived from a deep subsurface sediment. FEMS Microbiol. Ecol. 23:131-144. [Google Scholar]
- 11.Contreras, E. 1980. Studies on the intestinal actinomycete flora of Eisenia lucens (Annelida, Oligochaeta). Pedobiologia 20:411-416. [Google Scholar]
- 12.Daniel, O., and J. M. Anderson. 1992. Microbial biomass and activity in contrasting soil materials after passage through the gut of the earthworm Lumbricus rubellus Hoffmeister. Soil Biol. Biochem. 24:465-470. [Google Scholar]
- 13.Devliegher, W., and W. Verstraete. 1997. Microorganisms and soil physico-chemical conditions in the drilosphere of Lumbricus terrestris. Soil. Biol. Biochem. 29:1721-1729. [Google Scholar]
- 14.Doran, J. W. 1980. Soil microbial and biochemical changes associated with reduced tillage. Soil Sci. Soc. Am. J. 44:765-771. [Google Scholar]
- 15.Doube, B. M., O. Schmidt, K. Killham, and R. Correll. 1997. Influence of mineral soil on the palatability of organic matter for lumbricid earthworms: a simple food preference study. Soil Biol. Biochem. 29:569-575. [Google Scholar]
- 16.Dowdell, R. J., and R. Q. Cannell. 1975. Effect of ploughing and direct drilling on soil nitrate content. J. Soil Sci. 26:53-61. [Google Scholar]
- 17.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. M. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards, C. A., and P. J. Bohlen. 1996. Biology of earthworms. Chapman and Hall, London, United Kingdom.
- 19.Edwards, C. A., M. J. Shipitalo, L. B. Owens, and L. D. Norton. 1990. Effects of Lumbricus terrestris L. burrows on hydrology of continuous no-till corn fields. Geoderma 46:73-84. [Google Scholar]
- 20.Edwards, C. A., and K. E. Fletcher. 1988. Interactions between earthworms and microorganisms in organic-matter breakdown. Agric. Ecosys. Environ. 24:235-247. [Google Scholar]
- 21.Farrar, W. R., and A. C. Reboli. 1981. The genus Bacillus--Medical, p. 1763. In A. Balows, H. Truper, M. Dworkin, W. Harder, and K. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, New York, N.Y.
- 22.Felenstein, J. 1989. PHYLIP--phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 23.Fisher, K., D. Hahn, R. I. Amann, O. Daniel, and J. Zeyer. 1995. In situ analysis of the bacterial community in the gut of the earthworm Lumbricus terrestris L. by whole-cell hybridization. Can. J. Microbiol. 41:666-673. [Google Scholar]
- 24.Fisher, K., D. Hahn, W. Hönerlage, and J. Zeyer. 1997. Effect of passage through the gut on the earthworm Lumbricus terrestris L. on Bacillus megaterium studied by whole cell hybridization. Soil. Biol. Biochem. 29:1149-1152. [Google Scholar]
- 25.Foster, R. C. 1988. Microenvironments of soil microorganisms. Biol. Fertil. Soils 6:189-203. [Google Scholar]
- 26.Genetics Computer Group. 2000. Wisconsin Package Version 10.1. Genetics Computer Group, Madison, Wis.
- 27.Glöckner, F. O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Good, I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237-264. [Google Scholar]
- 29.Gordon, D. A., J. C. Priscu, and S. Giovannoni. 2000. Origin and phylogeny of microbes living in permanent antarctic lake ice. Microb. Ecol. 39:197-202. [DOI] [PubMed] [Google Scholar]
- 30.Groffman, P. M., G. J. House, P. F. Hendrix, D. E. Scott, and D. A. Crossley, Jr. 1986. Nitrogen cycling as affected by interactions of components in a Georgia Piedmont agroecosystem. Ecology 67:80-87. [Google Scholar]
- 31.Hendrix, P. F. 1997. Long-term patterns of plant production and soil carbon dynamics in a Georgia Piedmont agroecosystem, p. 235-245. In E. A. Paul, J. Paustian, E. T. Elliott, and C. V. Cole (ed.), Soil organic matter in temperate agroecosystems: long-term experiments in North America. CRC Press, Boca Raton, Fla.
- 32.Hendrix, P. F., D. A. Crossley, Jr., D. C. Coleman, R. W. Parmelee, and M. H. Beare. 1987. Carbon dynamics in soil microbes and fauna in conventional and no-tillage agroecosystems. INTECOL Bull. 15:59-63. [Google Scholar]
- 33.Holmes, A. J., J. Bowyer, M. P. Holley, M. O'Donoghue, M. Montgomery, and M. R. Grillings. 2000. Diverse, yet-to-be-cultured members of the Rubrobacter subdivision of the Actinobacteria are widespread in Australian arid soils. FEMS Microbiol. Ecol. 33:111-120. [DOI] [PubMed] [Google Scholar]
- 34.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1988. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hurlburt, S. H. 1971. The nonconcept diversity: a critique and alternative parameters. Ecology 52:577-586. [DOI] [PubMed] [Google Scholar]
- 36.Jurgens, G., K. Lindström, and A. Saano. 1997. Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl. Environ. Microbiol. 63:803-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karsten, G. R., and H. L. Drake. 1997. Denitrifying bacteria in the earthworm gastrointestinal tract and in vivo emission of nitrous oxide (N2O) by earthworms. Appl. Environ. Microbiol. 63:1878-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karsten, G. R., and H. L. Drake. 1995. Comparative assessment of the aerobic and anaerobic microfloras of earthworm guts and forest soils. Appl. Environ. Microbiol. 61:1039-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, H., D. Honda, S. Hanada, N. Kanamori, S. Shibata, T. Miyaki, K. Nakamura, and H. Oyaizu. 2000. A deeply branched novel phylotype found in Japanese paddy soils. Microbiology 146:2309-2315. [DOI] [PubMed] [Google Scholar]
- 40.Krebs, C. J. 1989. Ecological methodology. Harper & Row, New York, N.Y.
- 41.Kristufek, V., K. Ravasz, and V. Pizl. 1993. Actinomycete communities in earthworm guts and surrounding soil. Pedobiologia 37:379-384. [Google Scholar]
- 42.Kristufek, V., K. Ravasz, and V. Pizl. 1992. Changes in densities of bacteria and microfungi during gut transit in Lumbricus rubellus and Aporrectodea caliginosa (Oligochaeta: Lumbricidae). Soil Biol. Biochem. 24:1499-1500. [Google Scholar]
- 43.Kuske, C. R., S. M. Barns, and J. D. Busch. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl. Environ. Microbiol. 63:3614-3621. [DOI] [PMC free article] [PubMed]
- 44.Lee, K. E. 1985. Earthworms: their ecology and relationships with soils and land use. Academic Press, Sydney, Australia.
- 45.Leisack, W., and E. Stackebrandt. 1992. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J. Bacteriol. 174:5072-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, L., C. Kato, and K. Horikoshi. 1999. Bacterial diversity in deep-sea sediments from different depths. Biodivers. Conserv. 8:659-677. [Google Scholar]
- 47.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marilley, L., and M. Aragno. 1999. Phylogenetic diversity of bacterial communities differing in degree of proximity of Lolium perenne and Trifolium repens roots. Appl. Soil Ecol. 13:127-136. [Google Scholar]
- 49.McCaig, A. E., L. A. Glover, and J. I. Prosser. 1999. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl. Environ. Microbiol. 65:1721-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCaig, A. E., S. J. Grayston, J. I. Prosser, and L. A. Glover. 2001. Impact of cultivation on characterisation of species composition of soil bacterial communities. FEMS Microbiol. Ecol. 35:37-48. [DOI] [PubMed] [Google Scholar]
- 51.Moody, J. E., J. H. Billard, and T. W. Edminster. 1952. Mulch tillage: some effects on plant and soil properties. Soil Sci. Soc. Am. Proc. 16:190-194. [Google Scholar]
- 52.Moré, M. I., J. B. Herrick, M. C. Silva, W. C. Ghiorse, and E. L. Madsen. 1994. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl. Environ. Microbiol. 60:1572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palleroni, N. J. 1992. Introduction to the family Pseudomonadaceae, p. 3071-3085. In W. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes. Springer-Verlag, New York, N.Y.
- 54.Parle, J. N. 1963. Micro-organisms in the intestines of earthworms. J. Gen. Microbiol. 31:1-11. [Google Scholar]
- 55.Paul, E. A., and F. E. Clark. 1996. Soil microbiology and biochemistry, 2nd ed. Academic Press, San Diego, Calif.
- 56.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pedersen, J. C., and N. B. Hendriksen. 1993. Effect of passage through the intestinal tract of detritovore earthworms (Lumbricus spp.) on the number of selected Gram-negative and total bacteria. Biol. Fertil. Soils 16:227-232. [Google Scholar]
- 58.Pedersen, K., J. Arlinger, S. Ekendahl, and L. Hallbeck. 1996. 16S rRNA gene diversity of attached and unattached bacteria in boreholes along the access tunnel to the Äspö hard rock laboratory, Sweden. FEMS Microbiol. Ecol. 19:249-262. [Google Scholar]
- 59.Pielou, E. C. 1977. Mathematical ecology, p. 385. Wiley Interscience, New York, N.Y.
- 60.Polyanskaya, L. M., N. I. Babkina, G. M. Zenova, and G. G. Zvyagintsev. 1996. Fate of actinomycetes in the intestinal tract of soil invertebrates fed on streptomycete spores. Microbiology 65:493-498. [Google Scholar]
- 61.Rheims, H., C. Spröer, F. A. Rainey, and E. Stackebrandt. 1996. Molecular biological evidence for the occurrence of uncultured members of the actinomycete line of descent in different environments and geographical locations. Microbiology 142:2862-2870. [DOI] [PubMed] [Google Scholar]
- 62.Sandaa, R.-A., Ø. Enger, and V. Torsvik. 1999. Abundance and diversity of Archaea in heavy-metal-contaminated soils. Appl. Environ. Microbiol. 65:3293-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.SAS. 1998. SAS Institute, Cary, N.C.
- 64.Satchell, J. E. 1955. An electrical method of sampling earthworm populations, p. 356-364. In D. K. McKevan (ed.), Soil zoology. Proceedings of the University of Nottingham Second Easter School in Agricultural Science. Academic Press, New York, N.Y.
- 65.Scheu, S. 1987. Microbial activity and nutrient dynamics in earthworm casts (Lumbricidae). Biol. Fertil. Soils 5:230-234. [Google Scholar]
- 66.Schönholzer, F., D. Hahn, and J. Zeyer. 1999. Origins and fate of fungi and bacteria in the gut of Lumbricus terrestris L. studied by image analysis. FEMS Microbiol. Ecol. 28:235-248. [Google Scholar]
- 67.Shannon, C. E., and W. Weaver. 1963. The mathematical theory of communication, p. 117. University of Illinois Press, Urbana.
- 68.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smit, E., P. Leeflang, S. Gommans, J. van den Broek, S. van Mil, and K. Wernars. 2001. Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl. Environ. Microbiol. 67:2284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smitbert, R., and N. Krieg. 1994. Phenotypic characterization, p. 614. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 71.Stackebrandt, E., and H. Prauser. 1981. The family Cellulomonadaceae, p. 1323-1345. In A. Balows, H. Trüper, M. Dworkin, W. Harder, and K. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, New York, N.Y.
- 72.Stinner, B. R., D. A. Crossley, Jr., E. P. Odum, and R. L. Todd. 1984. Nutrient budgets and internal cycling of N, P, K, Ca, and Mg in conventional tillage. Ecology 65:354-369. [Google Scholar]
- 73.Suzuki, M. T., M. S. Rappé, Z. W. Haimberger, H. Winfield, N. Adair, J. Ströbel, and S. J. Giovannoni. 1997. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl. Environ. Microbiol. 63:983-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thomas, G. W., R. L. Blevins, R. E. Phillips, and M. A. McMahon. 1973. Effect of a killed soil mulch on nitrate movement and corn yield. Agron. J. 63:736-739. [Google Scholar]
- 75.Tomlin, A. D., M. J. Shipitalo, W. M. Edwards, and R. Protz. 1995. Earthworms and their influence on soil structure and infiltration, p. 159-184. In P. F. Hendrix (ed.), Earthworm ecology and biogeography. Lewis Publishers, Chelsea, Mich.
- 76.Toyota, K., and M. Kimura. 2000. Microbial community indigenous to the earthworm Eisenia foetida. Biol. Fertil. Soils 31:187-190. [Google Scholar]
- 77.Toyota, K., and M. Kimura. 1994. Earthworms disseminate a soil-borne plant pathogen. Fusarium oxysporum f. sp. raphani. Biol. Fertil. Soils 18:32-36. [Google Scholar]
- 78.Trigo, D., and P. Lavelle. 1993. Changes in respiration rate and some physicochemical properties of soil during gut transit through Allolobophora molleri (Lumbricidae, Oligochaeta). Biol. Fertil. Soils 15:185-188. [Google Scholar]
- 79.Tsai, Y.-L., and B. H. Olson. 1991. Rapid method for direct extraction of DNA from soil and sediments. Appl. Environ. Microbiol. 57:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ueda, T., Y. Suga, and T. Matsuguchi. 1995. Molecular phylogenetic analysis of a soil microbial community in a soybean field. Eur. J. Soil Sci. 46:415-421. [Google Scholar]
- 81.von Wintzingerode, F., B. Selent, W. Hehemann, and U. B. Göbel. 1999. Phylogenetic analysis of an anaerobic, trichlorobenezene-transforming microbial consortium. Appl. Environ. Microbiol. 65:283-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Whitman, W. B., J. Shieh, S. Sohn, D. S. Caras, and U. Premachandran. 1986. Isolation and characterization of 22 mesophilic Methanococci. Syst. Appl. Microbiol. 7:235-240. [Google Scholar]
- 83.Wolter, C., and S. Scheu. 1999. Changes in bacterial numbers and hyphal lengths during the gut passage through Lumbricus terrestris (Lumbricidae, Oligochaeta). Pedobiologia 43:891-900. [Google Scholar]