Abstract
The active population of low-affinity methanotrophs in a peat soil microcosm was characterized by stable-isotope probing. “Heavy” 13C-labeled DNA, produced after microbial growth on 13CH4, was separated from naturally abundant 12C-DNA by cesium chloride density gradient centrifugation and used as a template for the PCR. Amplification products of 16S rRNA genes and pmoA, mxaF, and mmoX, which encode key enzymes in the CH4 oxidation pathway, were analyzed. Sequences related to extant type I and type II methanotrophs were identified, indicating that these methanotrophs were active in peat exposed to 8% (vol/vol) CH4. The 13C-DNA libraries also contained clones that were related to β-subclass Proteobacteria, suggesting that novel groups of bacteria may also be involved in CH4 cycling in this soil.
It is estimated that 400 to 640 Tg of methane (CH4) is produced annually in anoxic environments (9), much of which is oxidized at the anaerobic-aerobic interface, thereby mitigating the global emissions of this greenhouse gas into the atmosphere. There are two distinct populations of organisms responsible for aerobic CH4 oxidation. One population exhibits high-affinity CH4 oxidation kinetics and acts as a sink for atmospheric concentrations of CH4 in many soils (2, 8, 10, 15, 17, 20, 24). The second population (methanotrophs) can grow on CH4 as a sole source of carbon and exhibits low-affinity CH4 oxidation kinetics (2, 17), and extant strains can be divided into type I (γ-Proteobacteria) or type II (α-Proteobacteria) methanotrophs (4, 5, 7, 40).
Methane monooxygenase (MMO) and methanol dehydrogenase (MDH) are key enzymes involved in CH4 oxidation. There are two forms of MMO, both of which oxidize CH4 to methanol. A membrane-bound, particulate enzyme (pMMO) (32) has been reported in nearly all methanotrophs (12, 32). A soluble, cytoplasmic enzyme (sMMO) is only found in certain methanotrophs (32). The pMMO genes exhibit high similarity to genes encoding ammonia monooxygenase (23) from ammonia-oxidizing nitrifying bacteria (nitrifiers). Nitrifiers also oxidize CH4 but probably do not use it as a carbon source (1). MDH oxidizes methanol to formaldehyde, and it is present in all known methylotrophic bacteria within the Proteobacteria (30). Genes encoding MMO and MDH (functional genes) and 16S rRNA genes have been used to assess the diversity of methanotrophs in the environment (11, 14, 18, 24, 31, 33, 44). Phylogenetic analysis of 16S rRNA, sMMO, pMMO, and MDH gene sequences obtained from these environments has suggested that a greater diversity of methanotrophs exists than we have seen in culture, a fact demonstrated by the continued isolation of novel methanotroph taxa (12, 25, 45).
A major goal of microbial ecology is to identify the microorganisms that are actively involved in specific processes in the environment. This has recently been addressed by using the natural abundance of stable isotopes in biomarkers (22, 34) and techniques including fluorescent in situ hybridization (FISH) coupled with microautoradiography (27, 35), analysis of phospholipid fatty acids by gas chromatography-isotope ratio mass spectrometry (3, 36), FISH coupled with mass spectrometry, and stable-isotope probing (SIP) (37, 41), which use substrates enriched with stable isotopes or radioisotopes. SIP exploits the fact that DNA of an organism growing on a 13C-enriched carbon source becomes 13C labeled (“heavier”), enabling it to be resolved from the total community DNA by density gradient centrifugation (37). The ability to isolate DNA from microorganisms involved in the metabolism of 13CH4 allowed us to characterize the microbial population actively involved in low-affinity CH4 oxidation in a peat soil microcosm without any prior knowledge of the organisms involved or the need to isolate them into culture.
Sample site and microcosms
The study site was drained fenland peat soil (49 to 51% carbon; pH 6.8) from Suffolk, United Kingdom [42]). The peat sample (0 to 5 cm in depth) was collected in May 1999 from an area not cultivated for 4 years. The in situ soil water content was 65% water-holding capacity, which was optimal for low-affinity (5% [vol/vol]) CH4 oxidation (S. A. Morris, unpublished data).
Two microcosms were established for the SIP experiment. Each consisted of freshly collected peat soil (10 g at 65% water-holding capacity) in a 125-ml crimp-top serum vial, sealed with a butyl rubber stopper, and injected with 0.4 mmol (10 ml) of CH4. One microcosm was exposed to 13CH4 (99% pure, 99% 13C atom enriched; Linde Gas), and the second microcosm was exposed to an identical amount of 12CH4 (98% pure; Linde Gas). The 13CH4 microcosm was used for extraction of 13C-labeled DNA and characterization of the microbial population actively involved in low-affinity CH4 oxidation.
Microcosms were incubated in the dark at 25°C, and headspace CH4 concentrations (200-μl samples) were determined every 2 to 5 days by gas chromatography. After >90% of the CH4 was consumed (7 to 12 days), the vials were flushed with air to remove 13CO2 and ensure that the microcosms remained aerobic. A further 0.4 mmol of 13CH4 or 12CH4 was added, and the vials were incubated until 1.6 mmol of CH4 (four injections of 10 ml of CH4) had been consumed by each microcosm (40 days).
DNA extraction. DNA was extracted from 10 g of soil exposed to 13CH4 and 3 g of soil exposed to 12CH4 by a bead-beating method (46) that was scaled up to process 3-g soil samples in 12-ml glass bead beater tubes (Braun). Soil samples were shaken for 5 min in a CO2 cooled bead beater (Braun). Extracts from the 13CH4 microcosm were pooled, and large particles were removed by centrifugation for 5 min at 120 × g. Proteins in the supernatant were precipitated with potassium acetate (7.5 M; one-sixth of the sample volume) and removed by centrifugation for 5 min at 15,000 × g. To the supernatant, an equal volume of binding matrix (Bio 101) diluted 1:5 with 6 M guanidine isothiocyanate was added. The tube was inverted regularly for 5 min and centrifuged for 5 min at 15,000 × g, and then the supernatant was discarded. The binding matrix was washed twice by resuspension in an equal volume of wash buffer (70% ethanol, 100 mM sodium acetate) and centrifugation at 15,000 × g for 1 min. DNA was eluted in 3 ml of Tris-EDTA (TE) buffer, and 1 g of CsCl was added per ml of DNA solution.
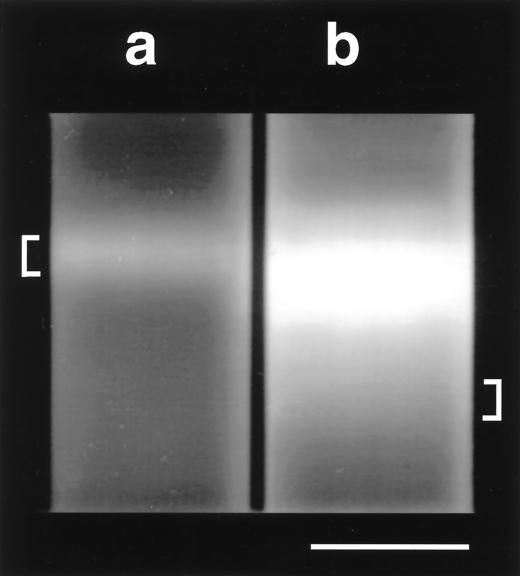
DNA fractions were resolved by equilibrium centrifugation (265,000 × g, 16 h, 20°C) in CsCl-ethidium bromide density gradients (13-by-51-mm polyallomer tubes; Beckman). A single DNA band was observed from the soil exposed to 12CH4, whereas an additional faint “smear” of more dense DNA was observed up to 1 cm below the bright band from the soil exposed to 13CH4 (Fig. 1). Assimilation of both 13C- and 12C-labeled substrates would result in intermediate-density DNA, as observed in the 13C-DNA gradient. The most dense DNA fraction (lower 0.25 cm; ca. 0.35 ml) from the 13C gradient (13C-DNA) and all DNA from the 12C gradient (12C-DNA) were collected with a syringe and needle (19 gauge). The 13C-DNA fraction was subjected to a second ultracentrifugation to remove any small amounts of coextracted 12C-DNA (not visible), providing highly enriched 13C-DNA for PCR. DNA fractions were extracted three times with an equal volume of 1-butanol, dialyzed against TE buffer, ethanol precipitated, and dissolved in 100 μl of TE buffer.
FIG. 1.
Equilibrium centrifugation of DNA extracted from soil that oxidized 12CH4 (a) or 13CH4 (b). CsCl-ethidium bromide (100 μl at 10 mg ml−1) density gradients were centrifuged at 265,000 × g for 16 h, and DNA was visualized with long-wavelength (365-nm) UV light. The positions of the 12C-DNA (indicated by “[”) and 13C-DNA (indicated by “]”) fractions collected after primary ultracentrifugation are indicated. Bar, 1 cm.
PCR amplification, cloning, sequencing, and analysis
The 13C-DNA and 12C-DNA fractions were used as a template for PCR. Primers (Table 1) specific for bacterial (Eubac27F and 1492R), archaeal (Arch21F and 1492R), and eucaryal small-subunit rRNA genes (EukF and EukR) (13) were used to determine the active methanotroph population at the domain level. With 12C-DNA as the template, PCR products of the expected size were obtained with primers that amplified Bacteria, Archaea, and Eucarya small-subunit rRNA genes. However, with 13C-DNA, PCR products were only obtained with bacterial 16S ribosomal DNA (rDNA) primers, indicating that 13CH4 was incorporated into a restricted bacterial community. Specific products of the expected size were amplified from both the 13C- and the 12C-DNA fractions with primers encoding key enzymes in CH4 metabolism (Table 1).
TABLE 1.
PCR amplification primers used to characterize the 13C- and 12C-DNA fractions
| Amplified gene(s) | Primers | Product (bp) | Library constructeda | Reference |
|---|---|---|---|---|
| 16S rRNA of Bacteria | Eubac27F | 1,450 | 13C-DNA (100) | 13 |
| 1492R | 12C-DNA (50) | |||
| 16S rRNA of Archaea | Arch21F | 1,450 | NAb | 13 |
| 1492R | ||||
| 18S rRNA of Eucarya | EukF | 1,500 | NA | 13 |
| EukR | ||||
| pmoA and amoA (putative active site subunit of pMMO and AMO) | A189 | 525 | 13C-DNA (50) | 23 |
| A682 | 12C-DNA (50) | |||
| pmoA (pMMO active site subunit) | A189 | 500 | 13C-DNA (50) | 11 |
| mb661 | ||||
| mxaF (MDH active site subunit) | mxa f1003 | 550 | 13C-DNA (50) | 30 |
| mxa r1561 | ||||
| mmoX (sMMO active site subunit) | mmoX F356 | 350 | 13C-DNA (10) | 16 |
| mmoX R898 |
DNA template used for PCR amplification and number of clones in the library (in parentheses).
NA, not applicable.
Amplification products were cloned with the TOPO-TA cloning kit (Invitrogen). Libraries were constructed from the 13C-DNA fraction for the bacterial 16S rDNA (100 clones), pmoA-A189/A682 (50 clones), pmoA-A189/mb661 (50 clones), mxaF (50 clones), and mmoX (10 clones) PCR products. Libraries were also constructed by using the 16S rDNA (50 clones) and pmoA-A189/A682 (50 clones) PCR products from the 12C-DNA fraction. Clones were grouped into operational taxonomic units (OTUs) by restriction fragment length polymorphism (RFLP) analysis by using restriction endonucleases as follows: EcoRI/RsaI and EcoRI/Sau3A for 16S rDNA clones, EcoRI/RsaI and EcoRI/PvuII/HincII for pmoA clones, and EcoRI/HincII and EcoRI/RsaI for mxaF clones.
For the 16S rDNA library, full sequence data (ca. 1,450 bp) between Escherichia coli positions 8 and 1511 (6) were obtained for one clone from each OTU containing five or more clones. A partial sequence (positions 375 to 890) was determined for each OTU that contained two to five clones and for selected OTUs with single clones. Complete sequence information (between the PCR primers) was obtained for each OTU in the pmoA libraries, eight mxaF clones, and ten mmoX clones. To verify that each unique restriction pattern represented a single clone type, partial sequence data were obtained for at least 10% of the clones within each OTU.
The ARB package (http://www.mikro.biologie.tu-muenchen.de) was used for sequence alignment and phylogenetic analysis. 16S rDNA sequences were aligned by using the ARB automatic alignment tool (Aligner v2.0) and corrected according to secondary structural constraints. Functional gene sequences were aligned manually to sequences from GenBank, and deduced amino acid sequences were used for the analyses. To evaluate tree topology, phylogenies were reconstructed by using evolutionary distance (DNA, Jukes and Cantor model; amino acids, Dayhoff PAM model), maximum parsimony (default parameters for ARB and DNAPARS or ARB and PROTPARS), and maximum likelihood (default parameters for ARB and fastDNAml or ARB and Protein_ML) analyses in conjunction with various filters and sequence subsets (28). Similarities between 16S rDNA sequences were determined by using the similarity matrix option within ARB. No chimeric 16S rDNA sequences were indicated with the Chimera Check program version 2.7 (http://rdp.cme.msu.edu). Inspection of the DNA and amino acid alignments of functional gene sequences did not identify any potential chimeric sequences. The sequences were deposited in GenBank (AF357990 to AF358055).
16S rDNA libraries
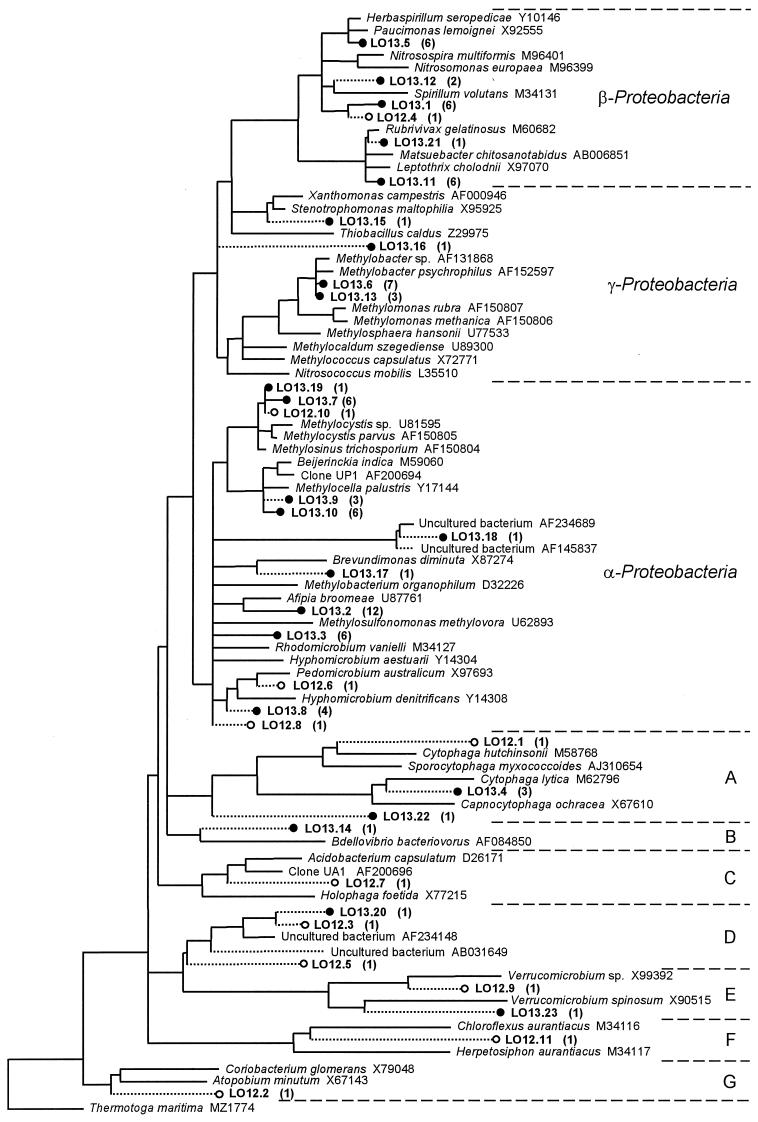
RFLP analysis of 100 16S rDNA clones from the 13C-DNA fraction (Fig. 2) assigned most of the library (70 clones) to 13 OTUs. The remaining 30 clones had unique restriction profiles (30 OTUs), and 10 were partially sequenced. Phylogenetic analysis revealed that, with the exception of five OTUs (containing seven clones), all 13C-DNA clones clustered in the α, β, or γ subclass of the Proteobacteria (40, 21, and 12 clones, respectively) (Fig. 2). In stark contrast to the 13C-DNA library, the 50 16S rDNA clones from the 12C-DNA contained 49 unique RFLPs (indicating wide diversity), of which only two were identical to an RFLP from the 13C-DNA. This difference was highlighted by sequencing 10 16S rRNA clones from the 12C-DNA; these were related to a wide variety of Bacteria (Fig. 2). One additional clone (LO12.10), with an RFLP identical to LO13.19, was sequenced and identified as an α-proteobacterial methanotroph.
FIG. 2.
Maximum-likelihood tree of 16S rDNA sequences amplified from 13C-DNA (•) or 12C-DNA (○) fractions. From the 13C-DNA library (100 clones), 23 OTUs (containing 80 clones) were sequenced, whereas the 12C-DNA library (50 clones) contained 49 OTUs (11 sequenced). The number of clones assigned to each sequenced OTU by RFLP analysis is shown in parentheses. OTU sequences were added to the tree of almost full-length sequences (>1,300 bp) in ARB by using maximum-parsimony analysis. Sequences most closely affiliated to each OTU, as well as sequences (>1,300 bp) of known methanotrophs and nitrifiers, were selected. Partial sequences (<1,300 bp) were chosen from the ARB or GenBank databases when no closely related full-length sequences were available. The tree was constructed by using Thermotoga maritima as an outgroup and a 50% conservation filter (962 nucleotides; this excludes positions of ambiguity, missing data, and positions where the frequency of a nucleotide occurring was <50% [28]). Multifurcations indicate branch points where the topology between maximum-likelihood, evolutionary-distance, and maximum-parsimony analyses of the data set were not supported by a strict consensus rule (28). With the exception of LO12.5, LO12.3, and LO13.20, the various treeing analyses consistently recovered all sequences in the groups depicted. Sequences of <1,300 bp (dashed line) were added to the tree by using a maximum-parsimony option within ARB. (A) Cytophagales; (B) δ-Proteobacteria; (C) Acidobacterium-Holophaga group; (D) uncultivated group; (E) Verrucomicrobia; (F) green nonsulfur group; (G) Actinobacteria.
Twenty-six clones in the 13C-DNA library were related to extant methanotrophs (96 to 99% 16S rDNA identity). These OTUs were most similar to the α-proteobacterial methanotrophs Methylocystis and Methylosinus (LO13.7 and LO13.19) and Methylocella palustris (LO13.9 and LO13.10) or to the γ-proteobacterial methanotroph Methylobacter (LO13.6 and LO13.13). A further 24 clones were related (92 to 95% identity) to genera in the α subclass of the Proteobacteria. Known methylotrophs (Hyphomicrobium and Methylobacterium) clustered among some (LO13.3, LO13.17, and LO13.8) but not all (LO13.2 and LO13.18) of these OTUs. The third major group of sequences (21 clones; LO13.1, LO13.5, LO13.11, LO13.12, and LO13.21) was closely related to genera within the β-Proteobacteria. Recovery of clones related to Bdellovibrio (LO13.14) and Cytophaga (LO13.4 and LO13.22) may have resulted from turnover of 13C due to predation (29, 39). The remaining sequenced clones clustered with the γ-Proteobacteria (LO13.15 and LO13.16), a group of uncultivated bacteria (LO13.20), and the Verrucomicrobia (LO13.23).
Functional gene libraries
Libraries of 50 pmoA clones were constructed with the A189-A682 primer set for both the 13C-DNA and the 12C-DNA fractions. RFLP and sequence analysis of the 13C-DNA library identified three OTUs (LOPA13.2, LOPA13.3, and LOPA13.5) that were similar to PmoA sequences of Methylocystis and Methylosinus (Fig. 3). Other clones formed a distinct group related to PmoA of type II methanotrophs (LOPA13.1), and four clones were similar to AmoA of Nitrosomonas europaea (LOPA13.4).
FIG. 3.
Maximum-likelihood tree of deduced amino acid sequences of pmoA and amoA genes amplified from 13C-DNA or 12C-DNA fractions. Clones obtained from the 13C-DNA fraction are prefixed with “LOPA13” when amplified with the A189-A682 (•) primer set or with “LOPB13” when amplified with the A189/mb661 (⧫) primer set. Clones obtained from the 12C-DNA fraction and amplified with the A189-A682 (○) primer set are prefixed with “LOPA12.” The tree was constructed with a filter (163 aligned amino acid positions; this excludes ambiguities and missing data), and the PmoA sequence of the environmental clone RA21 as an outgroup. Multifurcations indicate branch points where the topology between maximum-likelihood, evolutionary-distance, and maximum-parsimony analyses of the data set were not supported by a strict consensus rule. The number of clones (50 in each library) assigned to each OTU by RFLP analysis is shown in parentheses.
Analysis of the corresponding 12C-DNA library further demonstrated the different population within the 13C-DNA fraction. Thirty-six clones were assigned to OTUs closely related to AmoA of Nitrosomonas (LOPA12.1, LOPA12.2, LOPA12.5, and LOPA12.7) or Nitrosospira (LOPA12.3). OTU LOPA12.4 contained a distinct AmoA-like sequence. The remaining OTUs in the 12C-DNA library (LOPA12.6 and LOPA12.8) formed distinct groups that were related to the sequence of the environmental clones RA14 (24) and LOPA13.1, respectively (Fig. 3).
Since type I methanotroph sequences were detected in the 16S rDNA analysis of the 13C-DNA fraction, but not in the corresponding pmoA library with the A189-A682 primer set, we also used the methanotroph-specific primer set A189-mb661. Thirty-one clones (LOPB13.1 and LOPB13.3) contained a PmoA that was similar to that of type II methanotrophs (Fig. 3). A further 16 clones (LOPB13.2 and LOPB13.5) clustered with the PmoA of type I methanotrophs. The remaining OTU (LOPB13.4) was identical to LOPA12.6 and most closely related to the environmental clone RA14.
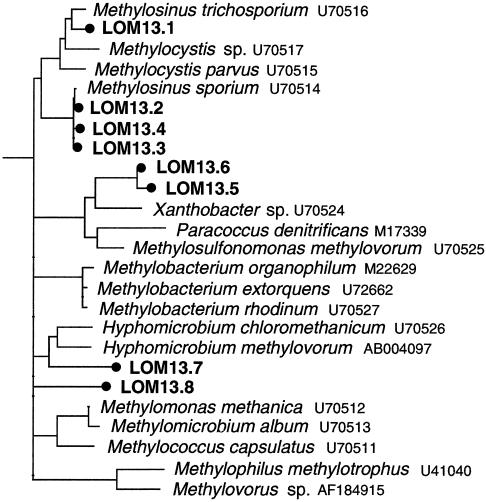
The 50 mxaF clones from the 13C-DNA grouped into 32 OTUs and, of the 20 clones sequenced, only 8 showed identity to MxaF. Phylogenetic analysis indicated that four clones (LOM13.1 to LOM13.4) clustered with the MxaF of type II methanotrophs and three (LOM13.5 to LOM13.7) clustered with other α-proteobacterial methylotrophs (Fig. 4). The different analyses of the pmoA and mxaF data sets consistently recovered the clades depicted in Fig. 3 and 4. Of the 10 mmoX clones from the 13C-DNA fraction, six different sequences were identified, all of which were similar to MmoX of the type II methanotroph Methylosinus (data not shown).
FIG. 4.
Maximum-likelihood tree of deduced amino acid sequences of mxaF genes amplified from the 13C-DNA fraction (•). The tree was constructed with a filter (171 aligned amino acid positions; this excludes ambiguities and missing data), with the amino acid sequence of the structurally related ethanol dehydrogenase of Pseudomonas aeruginosa (26) as an outgroup. Multifurcations indicate branch points where the topology between maximum-likelihood, evolutionary-distance, and maximum-parsimony analyses of the data set were not supported by a strict consensus rule.
Characterization of the population that assimilated 13C
In our study, 13C was incorporated into the DNA of metabolically active bacteria that used 13CH4 as a sole source of carbon (as shown by the extra, more dense, DNA fraction in the 13C-DNA gradient). Interestingly, the yield of 13C-DNA was very low relative to that of a methanotroph grown on 1.6 mmol of CH4 (not shown), suggesting cooxidation of CH4 in the peat, rapid turnover of methanotroph biomass, or CH4 production (not observed). Thus, it is probable that a variety of 13C-labeled compounds were produced during the 40-day incubation, including intermediates or products of methanotroph metabolism. Such 13C-labeled compounds could subsequently be assimilated by organisms other than methanotrophs. However, production of the corresponding 12C-labeled compounds (e.g., 12CO2) and other trophic interactions would considerably dilute many non-primary substrates within the complex environment of a soil microcosm. Furthermore, since identical centrifugation conditions separated the 13C-DNA and 12C-DNA fractions of a pure methanotroph culture grown on 13CH4 or 12CH4 by 1 cm (data not shown), we estimate that the 13C-DNA fraction collected (Fig. 1) contained between 75 and 100% 13C. For DNA to possess such a high 13C content and buoyant density, any non-methanotrophs must have been in very close association with the primary 13CH4 oxidizers. Therefore, it is most likely that the DNA collected in the “heavy” 13C-DNA fraction originated from microorganisms that assimilated 13C-labeled compounds as a primary carbon source.
Analysis of 16S rRNA and functional genes amplified from the 13C-DNA identified a high proportion of clones that were closely related to extant methanotrophs. These results indicate that a variety of methanotrophs had actively assimilated CH4 in the peat soil microcosm. The 16S rDNA similarity between LO13.10 and the nearest extant methanotroph, Methylocella palustris (12), was <97%, which suggested that a novel methanotroph species was involved in CH4 oxidation in the microcosm. Since PmoA/AmoA phylogeny of extant strains reflects that obtained with 16S rDNA sequences (23), the pmoA libraries support the activity of both type I and type II methano-trophs in the microcosm. The PmoA-like clones in the 13C-DNA fraction (LOPA.13.1 and LOPB13.4) also suggest that an unusual methanotroph within the α subclass of Proteobacteria was active in the peat soil. Recent studies of aerobic 13CH4 or 14CH4 oxidation have detected small amounts of uncharacteristic lipid fractions, which also suggests that unusual methanotrophs are active in soils and sediments (3, 8, 24, 38).
The distribution of clones in the libraries from the 13C-DNA fraction must be interpreted with great caution due to biases inherent in DNA extraction and PCR (43). However, one striking and unexpected feature of the 16S rDNA library is the large number of clones in OTUs that are not closely related to extant methanotrophs (Fig. 2). It is likely that these organisms had assimilated a high proportion of 13C into their DNA, even though the identity of the 13C substrate is unclear. Bacteria capable of growth on methanol and other excreted organic compounds copurify during attempts to isolate methanotrophs into culture (17). Indeed, 22 clones (LO13.2, LO13.3, and LO13.8) in the 16S rDNA library and 3 clones (LOM13.5 to LOM13.7) in the mxaF library were sufficiently related to sequences of Hyphomicrobium, Pedomicrobium, Rhodomicrobium, and Xanthobacter spp. to suggest that some methylotrophs within the microcosm may have assimilated 13CH3OH excreted by CH4-oxidizing bacteria.
The other major group of 16S rDNA sequences (LO13.1, LO13.5, LO13.11, LO13.12, and LO13.21) identified in the 13C-DNA library clustered within the β subclass of Proteobacteria. No functional genes indicative of β-proteobacterial methylotrophs were identified, a result that is similar to those of other environmental surveys done with mxaF PCR primers (18, 19, 21, 30, 37) and may be due to a primer bias. Unfortunately, therefore, it remains unclear which 13C substrate these organisms had assimilated in the peat soil microcosm. Isolates of β-proteobacterial methylotrophs are relatively rare (5, 40), and therefore their diversity might be underdescribed. Our data suggest that some β-Proteobacteria are actively involved in the cycling of carbon after CH4 oxidation in soil. These may represent novel methylotrophs, distinct from most extant strains within the α and γ subclasses of Proteobacteria. This ability of SIP to characterize the community in the 13C-DNA fraction with phylogenetic and functional gene PCR primers makes it a powerful technique for resolving microbial structure-function relationships in complex environments such as soil.
Acknowledgments
We thank Norman Sills from the Royal Society for the Protection of Birds for access to the Lakenheath Site.
This work was funded by the Natural Environment Research Council (United Kingdom) Ecological Dynamics and Genes Programme (GST/02/1864) and the European Union (BIO 4CT 960419; QLRT-1999-31528).
REFERENCES
- 1.Bédard, C., and R. Knowles. 1989. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol. Rev. 53:68-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender, M., and R. Conrad. 1992. Kinetics of CH4 oxidation in oxic soils exposed to ambient air or high CH4 mixing ratios. FEMS Microbiol. Ecol. 101:261-270. [Google Scholar]
- 3.Boschker, H. T. S., S. C. Nold, P. Wellsbury, D. Bos, W. de Graaf, R. Pel, R. J. Parkes, and T. E. Cappenberg. 1998. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 392:801-805. [Google Scholar]
- 4.Bowman, J. P., L. I. Sly, P. D. Nichols, and A. C. Hayward. 1993. Revised taxonomy of the methanotrophs: description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceace includes only the group I methanotrophs. Int. J. Syst. Bacteriol. 43:735-753. [Google Scholar]
- 5.Bratina, B. J., G. A. Brusseau, and R. S. Hanson. 1992. Use of 16S rRNA analysis to investigate phylogeny of methylotrophic bacteria. Int. J. Syst. Bacteriol. 42:645-648. [DOI] [PubMed] [Google Scholar]
- 6.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Nat. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brusseau, G. A., E. S. Bulygina, and R. S. Hanson. 1994. Phylogenetic analysis and development of probes for differentiating methylotrophic bacteria. Appl. Environ. Microbiol. 60:626-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bull, I. D., N. R. Parekh, G. H. Hall, P. Ineson, and R. P. Evershed. 2000. Detection and classification of atmospheric methane oxidizing bacteria in soil. Nature 405:175-178. [DOI] [PubMed] [Google Scholar]
- 9.Cicerone, R. J., and R. S. Oremland. 1988. Biogeochemical aspects of atmospheric methane. Global Biogeochemical Cycles 2:299-327. [Google Scholar]
- 10.Conrad, R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costello, A. M., and M. E. Lidstrom. 1999. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl. Environ. Microbiol. 65:5066-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dedysh, S. N., W. Liesack, V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, J. D. Semrau, A. M. Bares, N. S. Panikov, and J. M. Tiedje. 2000. Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int. J. Syst. Evol. Microbiol. 50:955-969. [DOI] [PubMed] [Google Scholar]
- 13.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Distel, D. L., and C. M. Cavanaugh. 1994. Independent phylogenetic origins of methanotrophic and chemoautotrophic bacterial endosymbioses in marine bivalves. J. Bacteriol. 176:1932-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunfield, P. F., W. Liesack, T. Henckel, R. Knowles, and R. Conrad. 1999. High-affinity methane oxidation by a soil enrichment culture containing a type II methanotroph. Appl. Environ. Microbiol. 65:1009-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuse, H., M. Ohta, O. Takimura, K. Murakami, H. Inoue, Y. Yamaoka, J. M. Oclarit, and T. Omori. 1998. Oxidation of trichloroethylene and dimethyl sulfide by a marine Methylomicrobium strain containing soluble methane monooxygenase. Biosci. Biotechnol. Biochem. 62:1925-1931. [DOI] [PubMed] [Google Scholar]
- 17.Hanson, R. S., and T. E. Hanson. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henckel, T., M. Friedrich, and R. Conrad. 1999. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl. Environ. Microbiol. 65:1980-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henckel, T., U. Jäckel, and R. Conrad. 2001. Vertical distribution of methanotrophic community after drainage of rice field soil. FEMS Microbiol. Ecol. 34:279-291. [DOI] [PubMed] [Google Scholar]
- 20.Henckel, T., U. Jäckel, S. Schnell, and R. Conrad. 2000. Molecular analyses of novel methanotrophic communities in forest soil that oxidize atmospheric methane. Appl. Environ. Microbiol. 66:1801-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henckel, T., P. Roslev, and R. Conrad. 2000. Effects of O2 and CH4 on presence and activity of the indigenous methanotrophic community in rice field soil. Environ. Microbiol. 2:666-679. [DOI] [PubMed] [Google Scholar]
- 22.Hinrichs, K. U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. DeLong. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 23.Holmes, A. J., A. M. Costello, M. E. Lidstrom, and J. C. Murrell. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132:203-208. [DOI] [PubMed] [Google Scholar]
- 24.Holmes, A. J., P. Roslev, I. R. McDonald, N. Iversen, K. Henriksen, and J. C. Murrell. 1999. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl. Environ. Microbiol. 65:3312-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaluzhnaya, M., V. N. Khmelenina, B. T. Eshinimaev, N. E. Suzina, D. I. Nikitin, A. Solonin, J. L. Lin, I. R. McDonald, J. C. Murrell, and Y. A. Trotsenko. 2001. Taxonomic characterization of new alkaliphilic and alkalitolerant methanotrophs from soda lakes of the Southeastern Transbaikal Region and description of Methylomicrobium buryatense sp. nov. Syst. Appl. Microbiol. 24:166-176. [DOI] [PubMed] [Google Scholar]
- 26.Keitel, T., A. Diehl, T. Knaute, J. J. Stezowski, W. Hohne, and H. Gorisch. 2000. X-ray structure of the quinoprotein ethanol dehydrogenase from Pseudomonas aeruginosa: basis of substrate specificity. J. Mol. Biol. 297:961-974. [DOI] [PubMed] [Google Scholar]
- 27.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K.-H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography: a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K.-H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 29.Mancinelli, R. L. 1995. The regulation of methane oxidation in soil. Annu. Rev. Microbiol. 49:581-605. [DOI] [PubMed] [Google Scholar]
- 30.McDonald, I. R., and J. C. Murrell. 1997. The methanol dehydrogenase structural gene mxaF and its use as a functional gene probe for methanotrophs and methylotrophs. Appl. Environ. Microbiol. 63:3218-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonald, I. R., and J. C. Murrell. 1997. The particulate methane monooxygenase gene pmoA and its use as a functional gene probe for methanotrophs. FEMS Microbiol. Lett. 156:205-210. [DOI] [PubMed] [Google Scholar]
- 32.Murrell, J. C., B. Gilbert, and I. R. McDonald. 2000. Molecular biology and regulation of methane monooxygenase. Arch. Microbiol. 173:325-332. [DOI] [PubMed] [Google Scholar]
- 33.Murrell, J. C., and S. Radajewski. 2000. Cultivation-independent techniques for studying methanotroph ecology. Res. Microbiol. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 34.Orphan, V. J., K. U. Hinrichs, W. Ussler, C. K. Paull, L. T. Taylor, S. P. Sylva, J. M. Hayes, and E. F. DeLong. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67:1922-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelz, O., C. Hesse, M. Tesar, R. B. Coffin, and W. R. Abraham. 1997. Development of methods to measure carbon isotope ratios of bacterial biomarkers in the environment. Isotopes Environ. Health Stud. 33:131-144. [Google Scholar]
- 37.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 38.Roslev, P., and N. Iversen. 1999. Radioactive fingerprinting of microorganisms that oxidize atmospheric methane in different soils. Appl. Environ. Microbiol. 65:4064-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toncheva-Panova, T., and J. Ivanova. 2000. Influence of physiological factors on the lysis effect of Cytophaga on the red microalga Rhodella reticulata. J. Appl. Microbiol. 88:358-363. [DOI] [PubMed] [Google Scholar]
- 40.Tsuji, K., H. C. Tsien, R. S. Hanson, S. R. DePalma, R. Scholtz, and S. LaRoche. 1990. 16S ribosomal RNA sequence analysis for determination of phylogenetic relationship among methylotrophs. J. Gen. Microbiol. 136:1-10. [DOI] [PubMed] [Google Scholar]
- 41.Whitby, C. B., G. Hall, R. Pickup, J. R. Saunders, P. Ineson, N. R. Parekh, and A. J. McCarthy. 2001. 13C incorporation into DNA as a means of identifying the active components of ammonia-oxidizer populations. Lett. Appl. Microbiol. 32:398-401. [DOI] [PubMed] [Google Scholar]
- 42.Willison, T. W., J. C. Baker, and D. V. Murphy. 1998. Methane fluxes and nitrogen dynamics from a drained fenland peat. Biol. Fertil. Soils 27:279-283. [Google Scholar]
- 43.Wintzingerode, F. V., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 44.Wise, M. G., J. V. McArthur, and L. J. Shimkets. 1999. Methanotroph diversity in landfill soil: isolation of novel type I and type II methanotrophs whose presence was suggested by culture-independent 16S ribosomal DNA analysis. Appl. Environ. Microbiol. 65:4887-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wise, M. G., J. V. McArthur, and L. J. Shimkets. 2001. Methylosarcina fibrata gen. nov., sp. nov. and Methylosarcina quisquiliarum sp. nov., novel type I methanotrophs. Int. J. Syst. Evol. Microbiol. 51:611-621. [DOI] [PubMed] [Google Scholar]
- 46.Yeates, C., and M. R. Gillings. 1998. Rapid purification of DNA from soil for molecular biodiversity analysis. Lett. Appl. Microbiol. 27:49-53. [Google Scholar]