Abstract
Algal-virus-specific PCR primers were used to amplify DNA polymerase (pol) gene fragments from geographically isolated natural virus communities. Natural algal virus communities were obtained from coastal sites in the Pacific Ocean in British Columbia, Canada, and the Southern Ocean near the Antarctic peninsula. Genetic fingerprints of algal virus communities were generated using denaturing gradient gel electrophoresis (DGGE). Sequencing efforts recovered 33 sequences from the gradient gel. Of the 33 sequences examined, 25 encoded a conserved amino acid motif indicating that the sequences were pol gene fragments. Furthermore, the 25 pol sequences were related to pol gene fragments from known algal viruses. In addition, similar virus sequences (>98% sequence identity) were recovered from British Columbia and Antarctica. Results from this study demonstrate that DGGE with degenerate primers can be used to qualitatively fingerprint and assess genetic diversity in specific subsets of natural virus communities and that closely related viruses occur in distant geographic locations. DGGE is a powerful tool for genetically fingerprinting natural virus communities and may be used to examine how specific components of virus communities respond to experimental manipulations.
Renewed interest in oceanic microbial processes stimulated marine virus research during the 1990s. The results of early investigations demonstrated that viruses that infect and lyse marine primary producers are abundant, active components of marine ecosystems (2, 4, 16, 24, 30). Subsequently, it was shown that viruses infecting the marine eukaryotic phytoplankton Micromonas pusilla were widespread, genetically diverse, dynamic, and caused significant mortality in M. pusilla populations (8, 9).
These findings inspired the development of the degenerate algal-virus-specific PCR primers AVS1 and AVS2, which amplify a 700-bp fragment of algal virus DNA polymerase (pol) genes (5). To permit unambiguous identification of PCR products amplified with the AVS primers, they were designed to span a short sequence encoding a highly conserved amino acid motif (YGDTDS) found in DNA polymerases (18). Phylogenetic analysis of amplified pol fragments revealed that cultured algal viruses formed a monophyletic group compared to double-stranded DNA viruses (viruses belonging to several families) (6). Recently, known algal viruses were assigned the virus family name Phycodnaviridae. Subsequently, the AVS primers were used to amplify unknown algal virus pol fragments from natural virus communities, demonstrating that molecular techniques can be used to study algal virus diversity (7).
Denaturing gradient gel electrophoresis (DGGE) is widely used to examine the diversity of PCR products. The technique separates DNA fragments based on sequence rather than length (12, 13) and has been widely used to examine the diversity of gene sequences in complex microbial communities (e.g., see references 1, 11, 21, and 34). Studies have indicated that DGGE analysis of PCR products amplified with AVS1 and AVS2 can be used to resolve genetically distinct algal viruses in artificial mixtures and to examine the diversity of natural algal virus communities (26, 27).
In general, it is agreed that viruses exert significant control on microbial communities and likely influence host community composition and succession (14, 29, 35). For example, Peduzzi and Weinbauer (23) demonstrated that virus enrichment influenced microbial community dynamics and succession, while Hennes et al. (17) used fluorescently labeled viruses to demonstrate that viruses could control the abundance of susceptible hosts. In addition, recent experiments with Pseudoalteromonas sp. revealed that the growth of virus-resistant cells was correlated to the lysis of sensitive cells (20). Furthermore, the virus-bacterium population dynamics model analyzed by Thingstad predicts that viruses ensure the coexistence of competing bacteria by infecting only the most abundant hosts or “killing the winner” (31). Similarly, the model of virioplankton control of community diversity proposed by Wommack and Colwell (36) predicts that virus community composition should be dynamic and that dominant viruses should be ephemeral. Examination of natural virus communities using pulsed-field gel electrophoresis supported these hypotheses. However, it should be noted that pulsed-field gel electrophoresis permits determination of genome sizes but not sequence identities of individual viruses. While it seems apparent that viruses influence host community composition, the effect of viruses on phytoplankton communities remains, for the most part, unexamined.
A long-term goal of our research is to understand the dynamics and effects of marine phytoplankton viruses. A critical part of this understanding requires examining the genetic composition of natural virus communities. Thus, the purpose of the present study was to use PCR and DGGE to recover and compare unknown algal virus sequences from the natural environment.
MATERIALS AND METHODS
Sample collection and preparation.
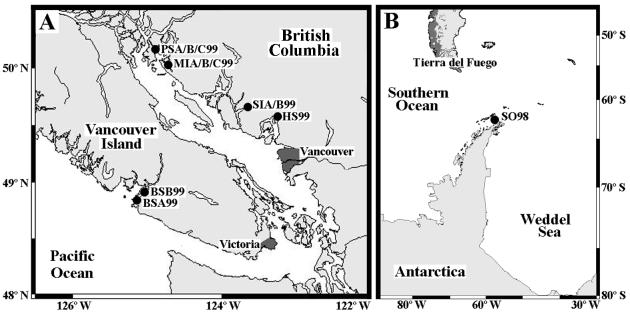
Natural virus communities were concentrated from several coastal stations in British Columbia, Canada, during the summer of 1999. A coastal Southern Ocean sample was collected near the Antarctic peninsula from the RV Lawrence M. Gould in 1998. The details of stations sampled and the labels assigned to each sample are listed in Table 1. Maps of the sample locations are shown in Fig. 1. The Southern Ocean and Barkley Sound samples SO98 and BSB99 were collected using a submersible pump, the Barkley Sound sample BSA99 was collected with a bucket, and all others were collected using GO-FLO bottles (General Oceanics, Miami, Fla.) mounted on a rosette. The virus size fraction was concentrated using previously established methods (7). Briefly, samples were pressure filtered (<17 kPa) through 142-mm-diameter glass fiber (GC50; nominal pore size, 1.2 μm; Advantec MFS, Dublin, Calif.) and polyvinylidene difluoride filters (GVWP; pore size, 0.45 μm; Millipore, Bedford, Mass.) connected in series. Remaining particulate material in the filtrates was concentrated using 30-kDa cutoff ultrafiltration; Amicon S1Y30 or S10Y30 (Millipore) cartridges were used according to the manufacturer's recommendations. After processing, concentrated virus samples were stored in the dark at 4°C until use.
TABLE 1.
Details of samples used in this study
| Sample | Date collected (mo/day/yr) | Location | Latitude | Longitude | Depth (m) | Salinity (‰) | Temp (°C) | Lane no. in gels |
|---|---|---|---|---|---|---|---|---|
| SO98 | 12/08/1998 | Southern Ocean | 62°36′S | 58° 15′ W | 0.5 | 33.9 | −1.0 | 1 |
| BSA99 | 06/25/1999 | Barkley Sound | 48°51′N | 125° 07′ W | 0 | 25.0 | NA | 2 |
| BSB99 | 07/12/1999 | Barkley Sound | 48°54′N | 125° 01′ W | 8.0 | 35.0 | NA | 3 |
| MIA99 | 08/19/1999 | Malaspina Inlet | 50°02′N | 124° 44′ W | 4.0 | 21.7 | 18.8 | 4 |
| HS99 | 08/22/1999 | Howe Sound | 49°27′N | 123° 17′ W | 6.0 | 17.8 | 15.0 | 5 |
| SIA99 | 08/18/1999 | Salmon Inlet | 49°36′N | 123° 48′ W | 15.0 | 23.4 | 13.5 | 6 |
| SIB99 | 08/18/1999 | Salmon Inlet | 49°36′N | 123° 48′ W | 4.0 | 22.6 | 15.0 | 7 |
| MIB99 | 08/20/1999 | Malaspina Inlet | 49°59′N | 124° 41′ W | 22.0 | 25.4 | 13.7 | 8 |
| MIC99 | 08/20/1999 | Malaspina Inlet | 49°59′N | 124° 41′ W | 5.0 | 22.2 | 18.3 | 9 |
| PSA99 | 08/21/1999 | Pendrell Sound | 50°18′N | 124° 44′ W | 25.0 | 28.4 | 10.1 | 10 |
| PSB99 | 08/21/1999 | Pendrell Sound | 50°18′N | 124° 44′ W | 19.0 | 27.7 | 11.4 | 11 |
| PSC99 | 08/21/1999 | Pendrell Sound | 50°18′N | 124° 44′ W | 0.5 | 14.6 | 22.6 | 12 |
FIG. 1.
Locations of sample collection are shown on two maps. (A) Positions of samples collected from coastal British Columbia, Canada, are shown. (B) The location of the sample collected in the Southern Ocean near the Antarctic peninsula is shown. Station abbreviations are listed in Table 1.
PCR and DGGE.
In order to ensure detection of rare sequences from the environment, PCR of the concentrated virus samples was conducted in two stages. Template DNA for PCR was serially diluted and extracted from virus concentrates using a previously described hot-cold treatment (7). Five microliters of each virus concentrate subsample was then added to a 45-μl first-stage PCR mixture containing Taq DNA polymerase assay buffer (50 mM KCl, 20 mM Tris-HCl [pH 8.4]), 1.5 mM MgCl2, a 0.16 mM concentration of each deoxyribonucleoside triphosphate, 60 pmol of each algal-virus-specific primer AVS1 and AVS2 (5), and 0.625 U of PLATINUM Taq DNA polymerase (Invitrogen Life Technologies, Carlsbad, Calif.). Negative controls contained all reagents except template. PCR was carried out with the following cycle parameters: denaturation at 95°C for 90 s, 32 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 45 s, extension at 72°C for 1 min, and a final extension at 72°C for 5 min. A 2-μl subsample from each first-stage reaction was added to a 73-μl second-stage PCR mixture as above except AVS1 and AVS2 were increased to 90 pmol and 1.0 U of enzyme was used. To minimize the production of PCR artifacts generated from high cycle number, second-stage PCR was limited to 20 cycles. To investigate unexpected PCR products produced in several first-stage reactions, a modified second-stage PCR (SIB99 M) was set up using 2 μl of the first-stage reaction mixture from one of the samples (SIB99) as a template. Reaction conditions were as described above for second-stage PCR, except the upstream primer AVS1 was used alone. PCR products were electrophoresed in 1.5% LE agarose (FMC BioProducts, Rockland, Maine) in 0.5× TBE buffer (45 mM Tris-borate, 1 mM EDTA [pH 8.0]) at 90 V for 75 min. The gels were stained with ethidium bromide, visualized on a UV transilluminator, and photographed with a Nikon Coolpix 950 digital camera. Digital images were manipulated using Adobe Photoshop 5.0 LE.
DGGE of second-stage PCR products was conducted using 20 to 55% (100% denaturant is defined as 7 M urea and 40% deionized formamide) linear denaturing gradient 6% polyacrylamide gels. Thirty microliters of products from SIB99 M and 50 μl from all other reactions were loaded into wells with 5 μl of 10× loading buffer (50% [vol/vol] glycerol, 0.33 M EDTA [pH 8.0], and 0.08% [wt/vol] bromophenol blue). Electrophoresis was carried out for 15 h in 1× TAE buffer (40 mM Tris-base, 20 mM sodium acetate, 1 mM EDTA [pH 8.5]) at 135 V and a constant temperature of 58°C using the D-code electrophoresis system (Bio-Rad Laboratories, Hercules, Calif.). Gels were stained in a 0.1× SYBR Green I (Molecular Probes, Eugene, Oreg.) solution overnight and were visualized and photographed as described above for agarose gels.
Sequencing.
To obtain sequence information from the denaturing gradient gels, individual bands were excised, reamplified, cloned, and sequenced. In total, 31 bands were excised and sequenced from the gradient gel. Excised bands are referred to by sample abbreviation and position in the gel from top to bottom; e.g., band BSB99-2 refers to the second band from the top of the lane loaded with sample BSB99. After excision, the gel slices were placed in sterile microcentrifuge tubes with 200 μl of 1× TAE buffer and were heated to 95°C for 5 min to elute the DNA band. One microliter of the resulting eluant was PCR amplified as described above for second-stage reactions. DNA fragments amplified from DGGE bands were cloned into the vector pGEM-5zf(+) (Promega, Madison, Wis.) by TA cloning. Subsamples from ligation mixtures were used to transform competent Escherichia coli JM109. To check if multiple sequences could be obtained from one DGGE band, cloned PCR fragments in three colonies from one transformation were sequenced (BSA99-1a, BSA99-1b, and BSA99-1c). Plasmid DNA was harvested from the cultures using a QIAprep spin miniprep kit according to the manufacturer's recommendations (Qiagen, Valencia, Calif.). Plasmid DNA (300 to 500 ng) was added to sequencing reactions using AmpliTaq FS BIGDYE Terminator cycle sequencing chemistry (Applied Biosystems, Foster City, Calif.) according to the manufacturer's recommendations. Excess Dye-Terminators were removed from the completed sequencing reactions using CENTRI-SEP spin columns (Princeton Separations, Adelphia, N.J.), and reactions were run in ABI Model 373 Stretch or ABI Prism 377 automated sequencers (Applied Biosystems) at the University of British Columbia sequencing facility.
Sequence analysis.
Sequences obtained from DGGE bands were compared to each other and known virus pol genes. The GenBank accession numbers for all sequences used in this study are listed below. All sequences were edited and translated using BioEdit (version 5.0.7) (15). Using the same program, a pairwise DNA identity matrix showing the proportion of identical residues was constructed from DGGE band sequences; only one sequence from any group of sequences with >98% identity was included in further analysis. Inferred amino acids of the unknown sequences were aligned with virus pol sequences from GenBank using the multiple sequence alignment program CLUSTAL W and the protein weight matrix BLOSUM (32). The alignment was then edited so that all sequences, with gaps included, were the same length. The alignment of the resulting 337 amino acid positions was used to construct maximum-likelihood and neighbor-joining trees using the programs TREE-PUZZLE (version 5.0) (28) and PHYLIP (version 3.57c) (10), respectively. Finally, phylogenetic trees were drawn and visualized using the program TreeView (Win32; version 1.6.1) (22).
Virus sequences used in phylogenetic analysis.
The designations and GenBank accession numbers of other virus sequences used in the phylogenetic analysis are as follows: Prasinovirus (MpV)-SP1, U32975; MpV-GM1, U32977; MpV-PB8, U32980; MpV-PL1, U32982; MpV-SG1, U32981; Prymnesiovirus (CbV)-PW1, U32983; CbV-PW3, U32984; Chlorovirus PBCV-1, M86836; Chlorovirus NY-2A, M86837; Chlorovirus CVA-1, U32985; Operational Taxonomic Unit 1 (OTU1), U36931; OTU2, U36932; OTU3, U36933; OTU4, U36934; OTU5, U36935; herpes simplex virus type 1, X04771; herpes simplex virus type 2, M16321; human cytomegalovirus, M14709; murine cytomegalovirus, M73549; Epstein-Barr virus, V01555; Autographa californica nuclear polyhedrosis virus (NPV), M20744; Bombyx mori NPV, D16231; Lymantria dispar NPV, D11476; African swine fever virus, X73330.
Nucleotide sequence accession numbers.
The 33 sequences obtained in this study were deposited in the GenBank database. The accession numbers for the sequences SO98-1, SO98-2, SO98-3, SO98-4, SO98-5, BSA99-1, BSA99-2, BSA99-3, BSA99-4, BSA99-5, BSA99-6, BSA99-7a, BSA99-7b, BSA99-7c, BSA99-8, BSB99-1, BSB99-2, BSB99-4, SIA99-1, MIB99-1, MIB99-2, PSB99-1, PSB99-2, PSB99-3, PSB99-4, PSC99-1, PSC99-2, BSA99-9, BSA99-10, BSB99-3, PSA99-1, PSC99-3, and SIB99 M-1 are AF405572 through AF405604, respectively.
RESULTS
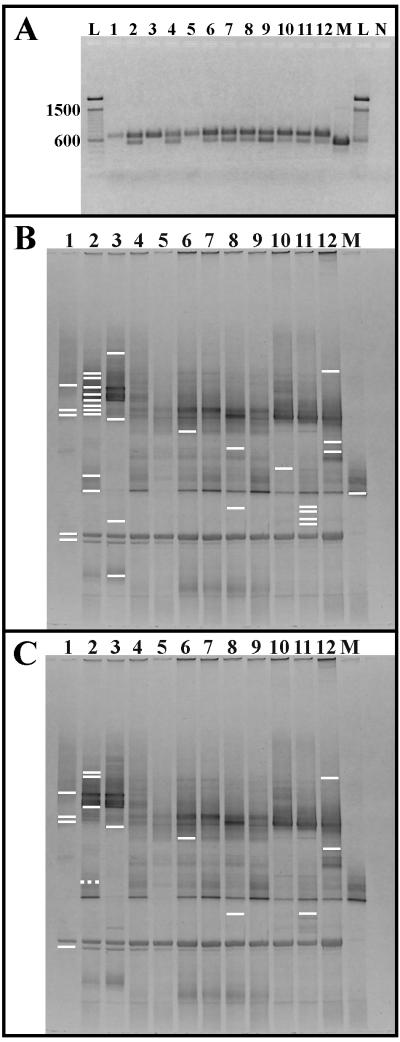
For all samples, different initial template concentrations were required for optimal PCR yield. Nonetheless, agarose gel electrophoresis revealed the presence of well-defined bands in all lanes (Fig. 2A). All reactions produced the expected 700-bp fragment, yet samples BSA99, MIA99, SIA99, SIB99, MIB99, MIC99, PSA99, PSB99, and PSC99 produced both 700- and 550-bp products (lanes 2, 4, and 6 to 12). The reaction with primer AVS1 only (SIB99 M) produced only the 550-bp product (lane M). DGGE revealed numerous bands in each lane (Fig. 2B and C). Several bands were present in all lanes, while other bands appeared in only a few. For example, the strong band near the bottom of lane 1 can be seen in every lane while the strong bands at the top of lanes 2 and 3 can only be observed in those lanes. As a final example, a single focused band can be observed in lane M, and bands at the same position are present at the same position in lanes 2, 4, and 6 to 12.
FIG. 2.
Negative images of gel electrophoresis of PCR fragments amplified in second-stage PCR. All lane numbers correspond to those listed in Table 1. Lane M was loaded with the products of the modified reaction (SIB99 M) using only the upstream primer AVS1. (A) PCR products were electrophoresed in 1.5% LE agarose in 1× TAE buffer. Numbers to the left of the gel correspond to the size of standardsloaded in lanes L, and lane N was loaded with the negative control. (B) The lines specify the position of all bands excised from the gel. (C) Lines indicate the 14 bands, SO98-1, SO98-2, SO98-3, SO98-5, BSA99-1, BSA99-2, BSA99-5, BSA99-9, BSB99-2, SIA99-1, MIB99-2, PSB99-1, PSC99-1, and PSC99-2, that produced unique pol-like sequences (solid lines) and a band, BSA99-9, that did not produce a pol-like sequence (broken line).
Sequence information was recovered from 31 excised bands (Fig. 2B). Three sequences, BSA99-7a, BSA99-7b, and BSA99-7c, were obtained from clones produced by transformation with sample BSA99-7; while these sequences were not the same, they were approximately 99% identical (range, 98.5 to 99.1%). Similarly, two groups of bands at similar positions in different lanes produced nearly identical sequences. Sequences from bands SO98-2 and BSA99-7a, -7b, and -7c were approximately 99% identical (range, 98.9 to 99.5%). Bands BSA99-10 and SIB99 M-1 produced sequences that were 99.2% identical. However, gel position and sequence similarity were not related, and many similar sequences were recovered from different positions in the gel. A pairwise identity matrix of the 33 sequences recovered from excised bands (data not shown) revealed 26 that formed 7 groups of sequences with >98% identity. Sequences with identities >98% were grouped as follows: SO98-1 and SO98-4; SO98-2, BSA99-6, BSA99-7a, BSA99-7b, BSA99-7c, BSA99-8, and BSB99-4; BSA99-1, BSA99-3, BSA99-4, and BSB99-1; BSA99-2 and MIB99-1; BSA99-9, BSA99-10, BSB99-3, PSA99-1, PSC99-3, and SIB99 M-1; BSB99-2 and PSB99-3; PSB99-1, PSB99-2, and PSB99-4. The other seven sequences (SO98-3, SO98-5, BSA99-5, SIA99-1, MIB99-2, PSC99-1, and PSC99-2) were not >98% identical to any other sequence.
The first sequences in each of the seven groups and the seven unique sequences were phylogenetically analyzed. Translation of the 14 DNA sequences revealed, in 12 instances, the presence of the conserved pol motif YGDTDS, and in one case the presence of a very similar sequence, HGDTDS (sequence BSA99-5). All of the sequences in the group starting with BSA99-9 did not contain amino acids resembling the conserved motif. As expected, this group of sequences without the pol motif included the band excised from lane M, which was loaded with products from the PCR with the primer AVS1 alone (SIB99 M). As stated previously, only 12 of the 14 analyzed sequences coded the pol motif YGDTDS, and one contained a similar sequence, HGDTDS. Not surprisingly, these sequences aligned at this motif with 24 virus pol sequences catalogued in GenBank. An amino acid alignment also revealed extensive sequence conservation among the unknown sequences and pol fragments from viruses belonging to the family Phycodnaviridae.
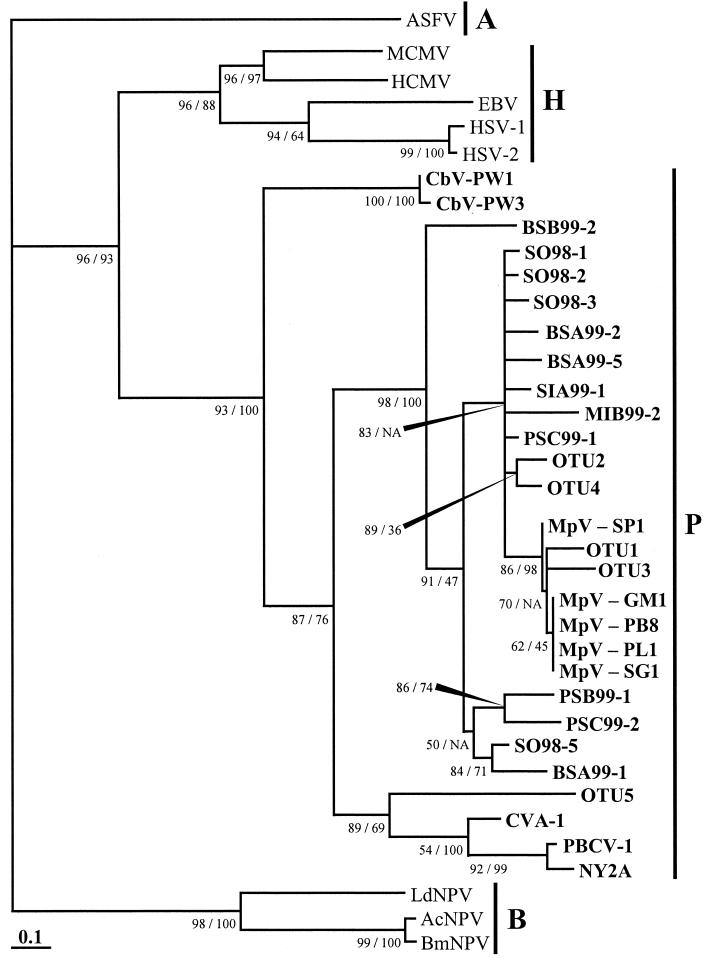
Phylogenetic analysis of aligned amino acids revealed robust relationships between sequences recovered from the gradient gel and other groups of known viruses (Fig. 3). Both algorithms (maximum-likelihood and neighbor-joining) used for phylogenetic reconstruction produced identical tree topology at major nodes. The topologies varied only at nodes near branch tips. Furthermore, the likelihood support and bootstrap values at nodes separating families of viruses were greater than 93%. All of the sequences recovered by DGGE were closely related to and clustered among pol sequences of known phycodnaviruses when compared to virus pol sequences from three other families of double-stranded DNA viruses, including Asfarviridae, Baculoviridae, and Herpesviridae. Several sequences from DGGE (SO98-1, SO98-2, SO98-3, BSA99-2, BSA99-5, SIA99-1, MIB99-2, and PSC99-1) were closely related to pol sequences from the genus Prasinovirus; prasinoviruses (abbreviated MpV) infect the prasinophyte alga M. pusilla. Four sequences (SO98-5, BSA99-1, PSB99-1, and PSC99-2) formed their own clade yet were also closely related to prasinoviruses, and one sequence (BSB99-2) formed a separate branch between prasinoviruses and viruses of the genus Prymnesiovirus (abbreviated CbV). Finally, viruses from the families Baculoviridae and Herpesviridae all clustered with viruses of the same family.
FIG. 3.
Maximum-likelihood tree of DNA pol fragments. Quartet puzzling support values for the maximum-likelihood tree and bootstrap values for a corresponding neighbor-joining tree are shown as percentages to the lower left of the appropriate node or as indicated by elongated black arrowheads; NA means the NJ tree topology differs at that node. The vertical black lines and letters to the right of the tree indicate virus families as follows: A, Asfarviridae; B, Baculoviridae; H, Herpesviridae; P, Phycodnaviridae. The Phycodnaviridae are also indicated in boldface lettering. The scale bar represents the number of amino acid substitutions per residue.
DISCUSSION
Three main results were realized during this study. First, PCR and DGGE were used to recover pol sequences from marine algal virus communities. Second, the pol sequences obtained in this study were closely related to known algal viruses. And the third, but perhaps most important, result is that nearly identical sequences were recovered from distant oceans. The following discussion considers PCR and DGGE as tools to genetically fingerprint virus communities and considers the phylogeny of sequences obtained in this study.
PCRs produced expected and unexpected products. The unexpected 550-bp products were the result of amplification with the primer AVS1 only, suggesting that they were PCR artifacts or the result of amplification of extant targets in the environment. PCR artifacts are often produced during reactions with mixed templates. For example, chimeric sequences form through recombination of similar PCR products and therefore have regions with different phylogenetic affinities. Because of this, many researchers suggest that nearest-neighbor analysis of different domains of suspect sequences can reveal PCR chimeras (19, 25, 33). To determine if any sequences recovered from DGGE were chimeric, phylogenetic trees were constructed from upstream, middle, and downstream regions of all sequences. The topology of phylogenetic trees constructed from the different regions did not differ, indicating that the sequences were likely not chimeric (data not shown). To check for the possibility of amplification of other targets, and to determine their identity, the 550-bp sequence SIB99 M-1 was compared to sequences in the public databases GenBank, DDBJ, and EMBL using the Basic Local Alignment Search Tool (BLAST) at the National Center for Biological Information web site (http://www.ncbi.nlm.nih.gov). This comparison revealed that the closest relatives of SIB99 M were phycodnavirus pol sequences; however, only the upstream primer region was related to the phycodnavirus sequences, and therefore no conclusions about the identities of the small amplicons were reached. Although the origins and identities of the small PCR products remain unknown, they did not interfere with DGGE analysis of desired PCR products.
DGGE facilitated the resolution of heterogeneous DNA fragments amplified from natural environments. However, one must be careful inferring diversity or richness from DGGE fingerprints. This is due to the fact that different sequences can migrate similarly and similar sequences can migrate differently. For example, SO98-1 and SO98-4 were 98% identical, yet SO98-1 migrated one-third of the gel length while SO98-4 migrated two-thirds of the gel. On the other hand, SO98-3 and BSB99-2 were only 36% identical, yet they migrated the same distance (Fig. 2B). Nonetheless, the community fingerprints obtained in this study were reproducible; multiple independent PCRs of the same sample analyzed by DGGE revealed the same fingerprint (data not shown). Furthermore, because DGGE resolved multiple bands from the PCR products of each reaction, qualitative comparison of PCR products was possible. For example, the banding patterns or “fingerprints” of samples from Salmon Inlet (SIA99 and SIB99) were identical and easily distinguished from other fingerprints. These samples were collected at the same time and location but different depths, suggesting that PCR templates were homogenously distributed with depth. This is reasonable considering the salinity and temperature of Salmon Inlet did not vary greatly with depth, indicating that the water and viruses were well mixed. On the other hand, the fingerprint of pol fragments amplified from surface water at Pendrell Sound (Fig. 2B, lane 12) is different than the fingerprints from greater depths (Fig. 2B, lanes 10 and 11). At this location the salinity of surface water was half of that at depth, indicating the water and viruses were stratified with depth. In general, environmental parameters agreed with qualitative lane comparison, reinforcing the idea that DGGE can be used to distinguish pol fragments amplified from natural algal virus communities.
Preliminary analysis of sequences recovered from DGGE revealed many that were more than 98% identical. On the other hand, the sequences BSA99-7a, BSA99-7b, and BSA99-7c from three different clones of PCR products reamplified from a single DGGE band were not identical. Although some of the nearly identical sequences may represent strain variation, the methods used in this study cannot distinguish the possibility that some were due to PCR and/or sequencing error. Therefore, only one sequence from each group of sequences with >98% identity was included in phylogenetic analyses.
To confirm that the sequences from DGGE were pol gene fragments, they were translated and searched for the conserved amino acid sequence corresponding to the putative pol catalytic site, YGDTDS (18). Of the 14 sequences analyzed 1 (BSA99-9) was not a recognizable pol fragment. This sequence was from the group that included the 550-bp sequence SIB99 M-1 mentioned previously. Twelve sequences encoded the pol catalytic motif, and one sequence (BSA99-5) encoded the unusual motif HGDTDS. There are two possible explanations for this sequence variation. First, HGDTDS is not an algal virus pol sequence and was the result of PCR and/or sequencing error. Second, HGDTDS may represent natural sequence variation in B-family pol genes. The amino acids histidine (H) and tyrosine (Y) are both polar, and therefore it is possible that this substitution does not affect pol function. Future studies will support the latter argument if they reveal that this unusual motif is a feature of some algal virus pol genes. On the other hand, if the HGDTDS sequence is not observed again, it is likely this pol motif was an artifact.
Phylogenetic inference revealed that the putative pol fragments recovered in this study were closely related to pol genes from the Phycodnaviridae. High likelihood support and bootstrap values indicated that the deep branches of the maximum-likelihood tree were robust (Fig. 3). For example, viruses belonging to the family Baculoviridae branched from the Asfarviridae outgroup with a likelihood support value of 98%. Similarly, viruses belonging to the closely related families Herpesviridae and Phycodnaviridae branched from the Asfarviridae and Baculoviridae with a support value of 96%. The Herpesviridae and Phycodnaviridae formed their own clusters with support values of 96 and 93%, respectively. In addition to having high support values, the phylogeny from this study resembled previously published pol phylogenies (3, 6). Therefore, it is likely that relationships obtained in this study provide accurate relationships of pol genes. Chen and Suttle (6) demonstrated that pol phylogeny reflects the phylogeny of total genomic DNA in M. pusilla viruses (MpV). Interestingly, the results of a previous phylogenetic analysis of phycodnavirus pol sequences based on nucleotide sequences (7) do not agree with the results presented in this study, yet previous phylogenies based on inferred amino acids (6) do. For example, the nucleotide-based phylogeny placed the Chlorella sp. viruses (CVA-1, PBCV-1, and NY-2A) more distant from MpV isolates than Chrysochromulina sp. viruses (CbV-PW1 and CbV-PW3) (7), whereas in our study CbV isolates were more distant from MpV isolates than Chlorella viruses. An explanation of this minor discrepancy may be that for the sake of reproducibility no eye refinement was performed on the present amino acid alignment. While the unrefined alignment presented in this work differed slightly from and may not be as accurate as previous alignments, a strong argument for unrefined alignments can be made because of the inherent irreproducibility of eye refinement. Nonetheless, these subtle differences do not affect the interpretation of our main result; all pol sequences obtained in this study clustered within the Phycodnaviridae, although outside of established genera.
Several sequences obtained in this study were not closely related to any known phycodnaviruses, yet many others (SO98-1, SO98-2, SO98-3, BSA99-2, BSA99-5, SIA99-1, MIB99-2, and PSC99-1) were most closely related to viruses in the genus Prasinovirus (MpV). However, because none of these sequences clustered among MpV, their identity remains unknown. Nonetheless, it is remarkable that sequences from the Southern Ocean were closely related to sequences from coastal British Columbia, in some cases being >98% identical, even though the environments were very different. This is the first time nearly identical viruses have been shown to occur in the Southern Ocean as well as in the temperate waters of the Northeast Pacific Ocean. It should be noted that although the samples were collected across the planet, the sample locations were all coastal and may have supported similar algal communities. In addition, the results of this study agree with previous work demonstrating the cosmopolitan nature of some algal viruses and their hosts (8). Five of the analyzed sequences obtained in this study formed two distinct groups within the Phycodnaviridae. These sequences may represent unknown genera within the Phycodnaviridae; further study will be necessary to refine the phylogeny of these unknown sequences. Previously, only one sequence (OTU5) has been recovered from the environment that was not closely related to other prasinoviruses (7). This study is the first to examine sequence diversity of algal viruses in more than one natural sample and provides further evidence that the PCR primers AVS1 and AVS2 can amplify unknown algal viruses from marine environments.
This study demonstrated that PCR and DGGE can be used to recover and identify unknown algal virus pol sequences from the natural environment and that very similar sequences can be recovered from different oceanic provinces across the planet. As more viruses are isolated and cultured from the environment and more sequences become known, we may find viruses that cluster with currently unidentified viruses. This will permit putative identification of uncultured virus sequences. In addition, we demonstrated that DGGE fingerprints can be used for qualitative comparison of virus communities. Therefore, the molecular methods described in this study can be used to relate the genetic composition of marine algal virus communities obtained from different environments. Furthermore, the ability to track changes in virus community composition may lead us closer to understanding the effects of viruses on host community structure. For example, it may be possible to relate changes in host populations to changes in the virus community. This study clearly demonstrates that the genetic composition of algal virus communities can be examined using the molecular techniques presented. Therefore, we are confident our long-term goal to understand the dynamics and effects of marine algal viruses can be obtained, in part, by examining the genetic composition of phytoplankton virus communities.
Acknowledgments
We thank W. H. Jeffrey for his role in the collection of the Southern Ocean sample, M. L. Berbee for her advice on phylogenetic analysis, and Cindy Frederickson for her assistance with digital images. We also acknowledge the crews of the CCGS Vector and RV Lawrence M. Gould for their assistance in sample collection.
This research was supported in part by an NSERC postgraduate scholarship to S. M. Short and NSERC research grants to C. A. Suttle.
REFERENCES
- 1.Bano, N., and J. T. Hollibaugh. 2000. Diversity and distribution of DNA sequences with affinity to ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in the Arctic Ocean. Appl. Environ. Microbiol. 66:1960-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergh, O., K. Y. Borsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. [DOI] [PubMed] [Google Scholar]
- 3.Braithwaite, D. K., and J. Ito. 1993. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 21:787-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bratbak, G., M. Heldal, S. Norland, and T. F. Thingstad. 1990. Viruses as partners in spring bloom microbial trophodynamics. Appl. Environ. Microbiol. 56:1400-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, F., and C. A. Suttle. 1995. Amplification of DNA-polymerase gene fragments from viruses infecting microalgae. Appl. Environ. Microbiol. 61:1274-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, F., and C. A. Suttle. 1996. Evolutionary relationships among large double-stranded DNA viruses that infect microalgae and other organisms as inferred from DNA polymerase genes. Virology 219:170-178. [DOI] [PubMed] [Google Scholar]
- 7.Chen, F., C. A. Suttle, and S. M. Short. 1996. Genetic diversity in marine algal virus communities as revealed by sequence analysis of DNA polymerase genes. Appl. Environ. Microbiol. 62:2869-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cottrell, M. T., and C. A. Suttle. 1991. Wide-spread occurrence and clonal variation in viruses which cause lysis of a cosmopolitan, eukaryotic marine phytoplankter, Micromonas pusilla. Mar. Ecol. Prog. Ser. 78:1-9. [Google Scholar]
- 9.Cottrell, M. T., and C. A. Suttle. 1995. Dynamics of a lytic virus infecting the photosynthetic marine picoflagellate Micromonas pusilla. Limnol. Oceanogr. 40:730-739. [Google Scholar]
- 10.Felsenstein, J. 1993. PHYLIP: phylogeny inference package (version 3.5). Cladistics 5:164-166. [Google Scholar]
- 11.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer, S. G., and L. S. Lerman. 1979. Length-independent separation of DNA restriction fragments in two dimensional gel electrophoresis. Cell 16:191-200. [DOI] [PubMed] [Google Scholar]
- 13.Fischer, S. G., and L. S. Lerman. 1983. DNA fragments differing by single basepair substitutions are separated in denaturing gradient gels: correspondence with melting theory. Proc. Natl. Acad. Sci. USA 80:1579-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuhrman, J. A. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541-548. [DOI] [PubMed] [Google Scholar]
- 15.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 16.Hara, S., K. Terauchi, and I. Koike. 1991. Abundance of viruses in marine waters: assessment by epifluorescence and transmission electron microscopy. Appl. Environ. Microbiol. 57:2731-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hennes, K. P., C. A. Suttle, and A. M. Chan. 1995. Fluorescently labeled virus probes show that natural virus populations can control the structure of marine microbial communities. Appl. Environ. Microbiol. 61:3623-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, J., and D. K. Braithwaite. 1991. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 19:4045-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komatsoulis, G. A., and M. S. Waterman. 1997. A new computational method for detection of chimeric 16S rRNA artifacts generated by PCR amplification from mixed bacterial populations. Appl. Environ. Microbiol. 63:2338-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Middelboe, M. 2000. Bacterial growth rate and marine virus-host dynamics. Microb. Ecol. 40:114-124. [DOI] [PubMed] [Google Scholar]
- 21.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 23.Peduzzi, P., and M. G. Weinbauer. 1993. The submicron size fraction of seawater containing high numbers of virus particles as bioactive agent in unicellular plankton community successions. J. Plankton Res. 15:1375-1386. [Google Scholar]
- 24.Proctor, L. M., and J. A. Fuhrman. 1990. Viral mortality of marine bacteria and cyanobacteria. Nature 343:60-62. [Google Scholar]
- 25.Robison-Cox, J. F., M. M. Bateson, and D. M. Ward. 1995. Evaluation of nearest-neighbor methods for detection of chimeric small-subunit rRNA sequences. Appl. Environ. Microbiol. 61:1240-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Short, S. M., and C. A. Suttle. 1999. Use of the polymerase chain reaction and denaturing gradient gel electrophoresis to study diversity in natural virus communities. Hydrobiologia 401:19-32. [Google Scholar]
- 27.Short, S. M., and C. A. Suttle. 2000. Denaturing gradient gel electrophoresis resolves virus sequences amplified with degenerate primers. BioTechniques 28:20-26. [DOI] [PubMed] [Google Scholar]
- 28.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum-likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 29.Suttle, C. A. 1999. Do viruses control the oceans? Nat. Hist. 108:48-51. [Google Scholar]
- 30.Suttle, C. A., A. M. Chan, and M. T. Cottrell. 1990. Infection of phytoplankton by viruses and reduction of primary productivity. Nature 347:467-469. [Google Scholar]
- 31.Thingstad, T. F. 2000. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45:1320-1328. [Google Scholar]
- 32.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, G. C. Y., and Y. Wang. 1997. Frequency of formation of chimeric molecules is a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl. Environ. Microbiol. 63:4645-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson, W. H., N. J. Fuller, I. R. Joint, and N. H. Mann. 2000. Analysis of cyanophage diversity in the marine environment using denaturing gradient gel electrophoresis, p. 565-571. In C. R. Bell, M. Brylinsky, and P. Johnson-Green (ed.), Microbial biosystems: new frontiers. Proceeding of the 8th International Symposium on Microbial Ecology. Atlantic Canada Society for Microbial Ecology, Halifax, Nova Scotia, Canada.
- 35.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wommack, K. E., J. Ravel, R. T. Hill, and R. R. Colwell. 1999. Hybridization analysis of Chesapeake Bay virioplankton. Appl. Environ. Microbiol. 65:241-250. [DOI] [PMC free article] [PubMed] [Google Scholar]