Abstract
Double Homeobox 4 (DUX4) is a potent transcription factor encoded by a retrogene mapped in D4Z4 repeated elements on chromosome 4q35. DUX4 has emerged as pivotal in the pathomechanisms of facioscapulohumeral muscular dystrophy (FSHD), a relatively common hereditary muscle wasting condition, although classified as a rare disease. DUX4 contributes to zygote genome activation before its expression is repressed in most somatic tissues through epigenetic mechanisms, including DNA methylation and chromatin modifications. In FSHD, inappropriate activation of DUX4 expression is driven by a complex interplay of genomic and epigenetic alterations. The ectopic presence of DUX4 in skeletal muscle cells activates genes, viral elements and pathways that are typical of very early embryonic development, disturbing cell function and ultimately contributing to muscle weakness and wasting. This review first traces the history of DUX4, from the FSHD genetic linkage studies in the early 1990s, through to identification and characterization of the DUX4 gene in 1999. We then discuss the seminal studies that showed how and why DUX4 is expressed in FSHD and the effects of this ectopic expression in muscle, notably cellular toxicity. Other pathological roles of DUX4, such as participation in cancer and viral infection, are also highlighted. Maintenance of DUX4 in the genome was explained by discovery of the function of DUX4 in zygotic genome activation to institute the totipotent cells of the embryo. Thus, we encompass the gradual transition of DUX4 over the past 25 years from being considered a pseudogene in “junk DNA” to becoming central to understanding the molecular pathogenesis of FSHD and the primary focus for FSHD therapeutics.
Keywords: DUX4, D4Z4, FSHD, Facioscapulohumeral muscular dystrophy, Muscle, Pathology
Introduction
DUX4 came to prominence during the search for the genetic cause of facioscapulohumeral muscular dystrophy (FSHD). This disorder is the third most common inherited muscular dystrophy after Duchenne muscular dystrophy and Myotonic dystrophy, with an estimated mean prevalence of 5/100,000 and incidence of 0.3/100,000 person-years in Western Europe [1–3]. FSHD is divided into the more common FSHD1 (OMIM 158900) and the rarer (~ 5% of cases) digenic FSHD2 (OMIM 158901). In both cases, pathology is caused by a DUX4 gain of function that requires (1) genetic predisposition: a complete DUX4 gene with a polyadenylation signal, (2) a particular epigenetic conformation: DNA hypomethylation and ‘open’ chromatin structure allowing DUX4 transcription. In most somatic cells, DUX4 is not expressed because it maps in a large array of hypermethylated repeated elements named D4Z4 that is ‘buried’ in compact chromatin. Epigenetic alterations leading to DUX4 expression result either from a decrease in D4Z4 repeat array size below a critical level (FSHD1) or a less contracted array accompanied by mutation in a gene encoding a protein involved in DNA methylation and/or chromatin modification (FSHD2) [4].
Clinically, FSHD is often described as a descending skeletal muscle weakness and wasting, in most cases with left/right asymmetry [5]. FSHD usually presents in the second decade of life in males but there is evidence of later onset in females [6]. Symptom onset and severity demonstrate inter-patient heterogeneity, which has even been observed in monozygotic twins [7]. Facial weakness is typically the earliest symptom, but weakness in the shoulder girdle and upper arms are the most common presenting symptoms [8–12]. As disease progresses, notable abdominal, lower limb and hip girdle musculature weakness are common [8, 13]. Lower limb weakness can necessitate use of ambulatory aids and wheelchairs [14, 15]. This ‘classical’ FSHD occurs in most patients, with the remainder being atypical, such as a facial sparing variant [11, 16, 17]. Extra-muscular features can include a retinal vascular pathology resembling Coat’s disease [18], and, rarer, sensorineural hearing loss [19] and asymptomatic electrocardiogram abnormalities [20]. Infantile onset (< 10 years of age) accounts for ~ 10% of cases, usually with rapid ‘classical’ progression [21] and higher prevalence of extra-muscular features [22].
Newcomers to the field of FSHD often think that DUX4 history started in 2010 with the key publication of a large multicentre genetic study led by Silvère van der Maarel [23], showing that a permissive allele (4qA) provides a polyadenylation signal (PAS) to stabilize DUX4 mRNA allowing for translation to the muscle-toxic DUX4 protein [23]. This “unifying theory of FSHD”, was built upon a body of research conducted over many years, which was instrumental to its formulation. Here, we first outline the research that shed light on many aspects of the genetics and molecular biology of FSHD, which contributed to the current model of pathogenesis. We describe the history of DUX4, starting with the FSHD genetic linkage studies of the early 1990s, and continuing through three key milestones: identification of the DUX4 gene in the D4Z4 repeat array in 1999 [24], discovery of DUX4 toxicity [25], DUX4 mRNA characterization with a PAS 3’ of D4Z4 and DUX4 protein detection in FSHD muscle cells [26]. How our understanding of DUX4 has gradually shifted from being considered a pseudogene in “junk DNA”, to acceptance as fundamental to molecular pathology in FSHD is then discussed [4, 27–29]. We finish by highlighting some of the outstanding questions about DUX4 and its role in FSHD.
The foundations of FSHD clinical symptoms and inheritance
In the mid-1800s, neurologists believed that muscle paralysis or atrophy could only result from a nerve lesion. The first account of a primary muscle disease was probably by Edward Meryon in 1852, who described 8 boys from 3 families with early onset muscle wasting. This seminal study reported muscle wasting without apparent involvement of nerves, which was both inherited and X-linked, and was likely Duchenne muscular dystrophy [30]. At around this time, Jean Cruveilhier described the autopsy of an 18-year old man who presented a severe facioscapulohumeral muscle wasting syndrome with unaffected brain, spinal cord or peripheral nerves [31], now considered the first case of FSHD [5]. Later, when Guillaume-Benjamin-Amand Duchenne (de Boulogne) published his ground breaking work on muscular dystrophies, he also included the classification of “l’atrophie musculaire graisseuse progressive de l’enfance” [32] that included facial muscle weakness and a descending progression of muscular involvement: essentially highlighting the muscular features of FSHD. A few years later, Louis Théophile Joseph Landouzy and Joseph Jules Dejerine described patients with selective facial, then shoulder/upper arm, followed by trunk/pelvic musculature involvement [33, 34]. Muscle atrophy with light sclerosis and adiposity was noted from post-mortem examination of a 24-year-old patient. Importantly, brain, spinal cord, peripheral nerve and intramuscular nerve endings were normal, indicating no neurological ‘disturbance’ [35]. This pedigree, and other cases, led Landouzy and Dejerine to term the disorder ‘facioscapulohumeral type of progressive myopathy’, extend the definition to include infant onset cases and encompass both facial muscle and/or shoulder girdle weakness [33, 34]. This explains why FSHD is also known as Landouzy-Dejerine muscular dystrophy. Wilhelm Erb independently confirmed that the muscle wasting conditions that he called ‘dystrophia muscularis progressiva’ were primarily muscle disorders and so distinct from secondary progressive muscular atrophy due to spinal cord disease [36, 37]. Based on initial muscle involvement, muscles affected and clinical symptoms, Erb classified four categories of “dystrophia muscularis progressiva” that included the facioscapulohumeral type described by Landouzy and Dejerine [38].
Landouzy and Dejerine also found that FSHD was inherited in a five-generation pedigree with the proband’s father, younger brother and sister similarly affected. Typical Mendelian inheritance with complete penetrance and highly variable expression was described in the 1950s by Frank Tyler and Fayette Stephens with a study of 1249 descendants of a man who emigrated to Utah from England in the 1850s [8]. It is of note that the Tyler and Stephens work was funded by the first NIH grant, after the Public Health Service Act of 1944 allowed the NIH to give grants to researchers [39]. In this Utah kindred, FSHD is linked to a 20-kb D4Z4 repeat array in 4q35, conserved in multiple, distantly-related branches, confirming the meiotic stability of the deletion [40]. George Padberg further explored the Mendelian inheritance and was central to the genetic search for the FSHD locus. During his PhD thesis, he toured The Netherlands to document families comprising affected and non-affected individuals and obtained numerous blood samples correlated with clinical description and family history [5].
The hunt for the genetic locus linked to FSHD
Shortly after the discovery that mutations in the DMD (dystrophin) gene caused Duchenne muscular dystrophy [41], an international consortium was established to find the ‘FSHD’ gene [42]. This initially led to exclusion of > 80% of the genome [43]. Advent of multiallelic microsatellite markers facilitated more efficient screening and one of these (Mfd22) displayed linkage at 13 centi morgan (cM) from the FSHD locus, with a LOD score above 6. This was the first genetic linkage success with such microsatellite markers. The corresponding locus D4S171, was assigned to chromosome 4 by the Peter Harper and Padberg labs in 1990 [44, 45]. Mapping was refined when cosmid 13E, comprising genomic DNA from that region, was isolated in a collaboration between the groups of Robert Williamson, Padberg and Rune Frants [46]. Different repeated sequences in this region strongly complicated the search. However, extensive subcloning finally isolated an almost single copy probe termed p13E-11 (D4F104S1, formerly D4S810), that recognised EcoRI restriction fragments (Fig. 1) of up to ~ 40 kb on Southern blot [46]. Association was confirmed by examining DNA from eight sporadic FSHD patients who had D4F104S1 EcoRI fragments that were shorter than 30 kb, while their unaffected parents had much longer fragments [46] and in a mosaic individual who passed on the rearranged D4F104S1 fragment to his affected son [47]. These seminal contributions from Frants’ group were key to the diagnosis of FSHD, anticipating that the p13E-11 probe “has immediate diagnostic value” [46]. The authors were also optimistic about identifying the gene responsible for FSHD, stating in 1992 that “the cloning of the FSHD gene should now be imminent” [46].
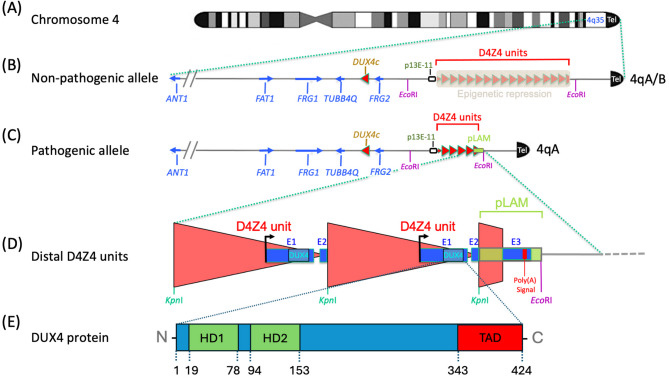
Fig. 1.
The FSHD locus at 4q35 contains a tandem D4Z4 unit array, each with a DUX4 ORF. (A) The FSHD locus is located adjacent to a telomere (Tel) of chromosome 4 at 4q35. (B) The locus usually contains a microsatellite array of > 11 D4Z4 units (red triangles) arranged head to tail, associated with epigenetic repression. This can be isolated on an EcoRI fragment identified by hybridization to the p13E-11 probe. Sequence differences telomeric to the D4Z4 repeat array define either a 4qA or 4qB haplotype. Centromeric to the locus are neighbouring genes including FRG2, DUX4c, TUBB4q, FRG1, FAT1 and ANT1. (C) In FSHD1, there is a reduced number of D4Z4 repeats to between 1 − 10 units, which leads to epigenetic derepression. FSHD1 also requires a 4qA haplotype in cis with the contracted array, containing the pLAM region (green box). Shortening of the D4Z4 repeat array generates a smaller EcoRI fragment identified using the p13E-11 probe. (D) A complete 3.3-kb D4Z4 unit is delimited by KpnI sites. Each D4Z4 unit contains the promoter, transcription start site, and entire open reading frame for the DUX4 retrotransposed gene in exon 1 (E1), together with the non-coding exon 2 (E2). The pLAM region on the 4qA haplotype provides intron 2 and exon 3 (E3) containing a polyadenylation (Poly(A)) signal required to stabilise DUX4 mRNA, allowing for its translation. (E) The main regions of DUX4 protein include the two DNA binding homeodomains (HD1 and HD2) and the transactivation domain (TAD) at the carboxyl-terminus. Locations of domains are given with respect to amino acid residues
Importantly, 13E was isolated by a screen in search for homeobox genes by hybridization to a homeobox probe [46], and each D4Z4 unit was bound [48]. Fine restriction mapping of the EcoRI fragment detected with p13E-11 hybridization by the Frants and Jane Hewitt groups revealed that it contained multiple copies of this homeobox-containing element. Digestion with KpnI delineated these as 3.2-kb repeat units, which were then termed D4Z4 [48–50]. Crucially, the proximal and distal sequences were identical on D4F104S1 EcoRI fragments from both unaffected and FSHD-affected individuals [50]. This EcoRI fragment, however, was shorter in FSHD and differed in size by multiples of 3.2-kb, suggesting rearrangements by homologous recombination resulting in loss of entire D4Z4 units [50]. Thus, the restriction map of this genomic fragment started with the 5’ EcoRI site, followed by the D4F104S1 single copy sequence detected by p13E-11, then by several homeobox-containing 3.2-kb D4Z4 units delineated by KpnI sites, and ended with the 3’ EcoRI site (Fig. 1). The region between the distal KpnI and EcoRI sites had been identified by a probe called pLAM1 and was either 1.5 or 2.9 kb long [50]. This telomeric region became known as pLAM, beginning with the Kpn1 site of a truncated D4Z4 unit, and containing a distal part composed of 68-bp tandem repeats similar to the Sau3A repeat family [50] (Fig. 1). The D4Z4 repeat array also begins with an incomplete D4Z4 unit (911-bp) located 5’ of the first KpnI site [51].
D4Z4 sequence reveals an open reading frame encoding two homeodomains
The full sequence of D4Z4 was published by a joint effort of the Hewitt, Frants and Williams labs in 1994 [52], following a partial sequence published earlier in that year [53]. A D4Z4 unit was 3,261 bp long, hence the revision in unit size to 3.3-kb. The sequence was GC-rich (71% G + C), containing GC-rich repeats designated LSau and a low copy repeat termed hhspm3, followed by two homeoboxes presenting 67% sequence identity between them and separated by 45-bp [52]. The encoded proximal homeodomain was named HD1 and the distal, HD2 (Fig. 1), sharing 52% amino acid sequence identity [52]. These two homeodomains had the greatest similarity to homeodomains of the paired and orthodenticle classes [52]. In searching for open reading frames (ORF), while none went through an entire D4Z4 unit, a long ORF encoding both homeodomains was detected. However, no evidence for expression was found from screening cDNA libraries [52]. Interestingly, the homeoboxes from D4Z4 were identical in sequence (across 405 bp) to the centromeric, inverted homeobox-containing region [52] previously identified with the homeobox probe [48]: this was later found to be part of the DUX4c gene.
Homologous D4Z4 tandem repeat arrays are on both chromosomes 4 and 10
Location of the tandem array of D4Z4 repeats on chromosome 4 was further refined to heterochromatin adjacent (within 215 kb) to the 4q telomere at 4q35 [53, 54]. The p13E-11 probe hybridized to two nonallelic EcoRI polymorphic fragments, generating a total of four fragments, of which only two were assigned to 4q35. The mystery was solved by the Frants and Luciano Felicetti groups who showed that the other two EcoRI fragments delimited tandem D4Z4 units located instead on chromosome 10q26 [55, 56]. These 4q35 and 10q26 homologous subtelomeric regions arose from duplication events and evolved independently [55, 57, 58]. Importantly, the D4Z4 repeat array at 10q26 was not associated with FSHD [56] although D4Z4 units on chromosomes 4 and 10 had 98–100% sequence identity. D4Z4 units on chromosome 10 had a BlnI restriction site absent in the repeats on chromosome 4 [59], which contained a unique XapI site. This polymorphism permitted detection of 4/10 inter-chromosomal exchanges and cases of hybrid chromosome 4 and 10-derived D4Z4 units [60]. A puzzle at the time however, was that this region had all the characteristics of so called non-functional “junk DNA” [61].
FSHD is only associated with contracted D4Z4 arrays on chromosome 4
The four bands observed on Southern blot when human genomic DNA was digested with EcoRI and hybridized with p13E-11 were thus explained: two originating from chromosome 4 and two from chromosome 10. Conversion of EcoRI fragment size (kb) to D4Z4 unit number is determined by subtracting 8.5 kb (the combined size of the 6 and 2.5 kb DNA segments flanking the D4Z4 array) and dividing by 3.3, the D4Z4 unit size in kb [62]. Most EcoRI fragments were 38 kb to > 300 kb in length, so accounting for ~10 to > 100 D4Z4 units. FSHD however, was associated with at least one D4F104S1 EcoRI fragment of 10 to 38 kb, so containing 1 − 10 D4Z4 units, as shown by Frants and Gert-Jan van Ommen's groups [50, 63].
Association was then found between D4F104S1 EcoRI fragment size and age of disease onset, with smaller fragments in the range of 10–18 kb (1–3 D4Z4 units) being severe childhood cases, 18–34 kb (3–8 units) usually associated with typical teenage onset, while ~ 30–38 kb (7–10 units) often associated with late onset [64, 65]. Also, high inter- and intra-familial variability in clinical presentation was noted, which even occurred in monozygotic twins [7, 66, 67]. Significantly, Rossella Tupler and colleagues found a healthy individual with a total loss of D4Z4 units on one 4q35 allele, indicating that FSHD was not associated with haploinsufficiency of a key gene(s) at the 4q subtelomeric region [68]. In fact, this enigmatic result underpinned the cryptic pathogenic mechanism of the disease, as it indicated either a dominant-negative or gain-of-function effect.
FSHD was only associated with a contracted D4F104S1 EcoRI fragment on chromosome 4 [69] on which the D4Z4 repeat array was located in heterochromatin and adjacent to the 4q telomere [53, 54]. Because homologous D4Z4 repeat arrays were found on both chromosomes 4 and 10 but only chromosome 4 was linked to the pathology, the mysterious FSHD-causing gene(s) was proposed to map outside of the D4Z4 repeat array and its expression to be subject to a position effect [52]. In this model, expression levels of a gene result from the degree of telomeric heterochromatin extension onto it, as initially described in yeast [70, 71]. Telomeres are composed of a 6-bp repeated sequence (TTAGGG in human) associated with heterochromatin, and repeat copy number is correlated with extension of heterochromatin outside of the telomeric region. Changes in the D4Z4 repeat array length/chromatin structure was proposed to favour or block telomere heterochromatin extension to the unknown FSHD gene(s) and so affect its expression [52]. A decade later, this model proved correct for DUX4, where telomere length alterations in isogenic FSHD myoblasts/tubes showed increased DUX4 gene expression inversely proportional to telomere length [72].
FSHD is only associated with the 4qA haplotype distal to the contracted D4Z4 array
Another important piece of the puzzle was contributed by Van der Maarel’s group, who observed that 4q35 subtelomeric DNA sequences could be segregated into 4qA and 4qB haplotypes, found with approximately equal frequency in the general population [73]. In 4qA, the telomeric-most complete 3.3-kb D4Z4 unit is followed by pLAM, which starts with the truncated D4Z4 unit and ends with only the first portion of an 8 kb of 68-bp beta-satellite repeated DNA, which itself is followed by a 1-kb divergent (TTAGGG)n array (Fig. 1). In the 4qB allele by contrast, the D4Z4 array ends with just the first 570 bp of a truncated D4Z4 unit but no satellite repeats [73]. The D4Z4 array on 10q was also followed by a 68-bp beta-satellite array, with 4qA and 10qter subtelomeres being > 98% identical, so closer in sequence than the 92% similarity between 4qA and 4qB [74]. Importantly, FSHD was only associated with contracted D4Z4 tandem arrays in cis on a 4qA haplotype [73] (Fig. 1).
The D4Z4 array is normally epigenetically repressed
The D4Z4 array contained CpG islands [48] and was located within heterochromatin adjacent to the 4q telomere [53, 54]. Sequencing had shown that each D4Z4 unit was GC-rich (71% G + C) and contained a CpG/GpC dinucleotide ratio of 0.8 and GC-rich hhspm3 [52] and LSau repetitive sequences [52, 53], associated with heterochromatin. D4Z4 was noted to bind a multiprotein complex consisting of YY1, together with HMGB2 and nucleolin, that suppressed expression of a linked reporter gene [75]. Furthermore, van der Maarel and colleagues reported that while many CpG dinucleotides in the D4Z4 array were methylated in unaffected individuals, there was reduced DNA methylation in contracted alleles, associated with an epigenetic state more conducive to gene expression [76, 77]. Intriguingly, this was also the case in FSHD2, where larger “non-contracted” D4Z4 arrays on both chromosomes 4 and 10 were hypomethylated [78]. Later, a reduction in binding of HP1γ and cohesin at D4Z4, accompanied by loss of the histone mark H3K9 trimethylation, was found to hallmark FSHD1 [79]. Davide Gabellini’s lab revealed that contraction of the D4Z4 array resulted in loss of Polycomb silencing and gain of Trithorax activation. This led to expression of a long non-coding RNA (lncRNA) termed DBE-T that could recruit Ash1L to facilitate gene derepression [80]. Thus, in FSHD, the D4Z4 array exhibits a less repressed epigenetic state, and the DUX4 gene can be stochastically activated in rare myoblasts or myonuclei by ‘available’ regulatory elements (e.g. DUX4 myogenic enhancer 1 and 2) [81] and transcription factors such as Sp1, p53, WDR5 and SIX family members [24, 82–84]. Indeed, Peter Jones and colleagues showed that the epigenetic status assessed via the DNA methylation of the distal D4Z4 unit on a 4qA haplotype correlates with disease: hypomethylation in FSHD1 but intermediate methylation in non-manifesting, and hypermethylation in healthy, individuals [85]. Moreover, the even lower methylation in FSHD2 allowed the Jones lab to develop a differential diagnosis method based on the presence of a 4qA allele, and the distal D4Z4 methylation level [86].
Discovery of the DUX4 gene fails to set the field alight
So, what made the combination of a small D4Z4 array and a 4qA haplotype pathogenic? Nine years elapsed from mapping the chromosomal location of the FSHD locus to chromosome 4 [44, 45], to discovery of the DUX4 gene by Alexandra Belayew’s group [24, 87] based on the Hewitt publication of the D4F104S1 EcoRI fragment sequence [52]. A large ORF containing the two homeoboxes was within each 3.3-kb D4Z4 unit but neither a promoter nor a cDNA clone with sequence identical to the D4Z4 ORF was found [51, 52]. Prophetically, it had been suggested in Hewitt et al. that “the overall structure of D4Z4 makes it unlikely to encode a functional protein; however, it cannot be ruled out that one copy of the repeat may produce a protein” [52].
Searching for target genes of Helicase-like Transcription Factor (HLTF/SMARCA3) via chromatin immunoprecipitation identified a 182-bp fragment containing a putative TATAA box that was named HLTF target 1 (HEFT1) [87]. Multiple potential HEFT1 promoter regions were found in 3.3-kb elements repeated throughout the genome in so called “junk DNA” including at D4Z4. This HEFT1 promoter had 87% sequence identity with a region inside the D4Z4 ORF, upstream of the two homeoboxes. The HEFT1 TATAA box corresponded to a functional variant TACAA sequence in D4Z4, generating a shorter ORF with a potential start codon 135 bp downstream from that of Hewitt et al. 1994 [52] but still in frame with the two homeoboxes. The ORF sequence known at the time encoded a putative 424-residue protein with two homeodomains [87]. Both promoter and ORF were also present in a 17.5 kb fragment of patient genomic DNA provided by the Frants group [24]. Of note, several GC quadruplex structures (G4s) have been identified in D4Z4 units and DUX4 promoter/enhancer regions [88, 89] and recent studies demonstrate that HLTF interacts with and destabilizes such G4s to facilitate error-free DNA replication [90].
This putative gene within each D4Z4 unit was named “Double Homeobox on chromosome 4” or DUX4 [24] (Fig. 1). The DUX4 promoter was active in human rhabdomyosarcoma cells and depended on the TACAA and a GC box [24]. Intriguingly, this promoter overlapped with the hhspm3 sequence identified in 1987 by Melanie Ehrlich [91] as hypo-methylated in sperm cells, an epigenetic feature that was shown 23 years later to associate with DUX4 expression in testis [92]. At the time of discovery however, many considered DUX4 a pseudogene due to the mutated TATAA box, lack of both introns and PAS, and, above all, inability to identify either a cDNA corresponding to its mRNA or an encoded protein. Attempts at amplifying DUX4 mRNA by RT-PCR picked up highly similar RNAs with no, or minimal, ORFs derived from hundreds of homologous 3.3-kb repeats dispersed throughout the genome, mostly on all acrocentric chromosomes, interspersed with ribosomal RNA gene clusters [53, 87, 93–95]. This reinforced the notion that the 4q35 D4Z4 array was devoid of a gene.
Position effect variegation and effects on potential candidate FSHD genes
The hypothesis was advanced that D4Z4 was instead involved in position effect variegation, with changes in its chromatin structure affecting expression of neighbouring genes [75, 96]. Position effect variegation was discovered in drosophila using an eye colour phenotype. When the white gene was inserted in the vicinity of heterochromatin, this inhibitory conformation could spread to different extents in each cell, resulting in a variable pattern of white gene expression causing mosaic red eye pigmentation [97].
Only a few genes have been mapped in the vicinity of the FSHD locus: the gene encoding mitochondrial channel Adenine Nucleotide Translocator (ANT1, also known as SLC24A4) in 1989 [98, 99], then FSHD Region Gene 1 (FRG1) in 1996 [100], TUBB4Q in 2000 [101] and FRG2 in 2002 [74] (Fig. 1). The Tupler group reported that the closer a gene was to the D4Z4 units, the more it was inhibited in healthy control muscle and inappropriately overexpressed in FSHD [75]. According to this study, each D4Z4 element harboured a repressor named DBE, and DBE multimerization inhibited a linked reporter gene. Consistent with this, 4q35 sequences were shown to be hypomethylated, and so epigenetically derepressed, on chromosome 4 variants associated with FSHD [76]. However early transcriptomic profiling using microarrays did not find misregulation of these genes in FSHD muscle [102]. A homologous DUX4c (centromeric, also known as DUX4L9) gene had also been mapped to a single truncated, inverted D4Z4 unit proximal to FRG2 [48, 103]. The encoded 46-kDa protein had high sequence identity to DUX4 but with a shorter carboxyl-terminal region of 32 residues (instead of the 82 residues of DUX4) due to a frameshift in the ORF that made DUX4c a significantly less potent transcriptional activator than DUX4 [52, 103–105]. FAT Atypical Cadherin 1 (FAT1) is also near the FSHD locus and because loss-of-function mutations were found in patients who had D4Z4 copy numbers close to the normal range, they were suggested to cause FSHD, despite the disease dominant transmission [106] (Fig. 1).
DUX4: the wilderness years
From its discovery in 1999 [24], there followed a period until 2010 when DUX4 was not widely considered relevant to FSHD, except by a few hardy acolytes. Indeed, the next publication to include the word DUX4 in the abstract was 4 years later in 2003 [107], while DUX4 did not make it into a title until 2005, debuting in a published meeting abstract [108] and then a peer-reviewed paper in 2006 [109].
DUX genes from 3.3-kb repeat elements located on other chromosomes were actively transcribed and DUX1 generated a protein with DNA binding activity [87, 93]. The DUX4 promoter activated a linked luciferase reporter gene (DUX4-luc) in human rhabdomyosarcoma and C2C12 myoblasts, activity that was strongly reduced by mutations in either its TACAA box or a 5’ GC box binding Sp1 [24]. However, the DBE repressor [75] overlapped this DUX4 promoter. DBE bound a protein complex including HMGB2 and Nucleolin associated with the transcription factor YY1 that can act as either an inhibitor or activator. In HeLa cells, a mutation that suppressed YY1 binding and so repression by DBE, activated a linked reporter gene [75]. Confusingly, this same mutation had no impact on DUX4-luc activity in murine C2C12 myoblasts, indicating that DBE was not a repressor in muscle [110]. Furthermore, the encoded DUX4 protein had a carboxyl-terminal domain with powerful transcriptional activity in the yeast one hybrid system and was localised to the nucleus when expressed in C2C12 myoblasts [24, 110]. Crucially, an antiserum raised against a DUX double homeodomain detected several spots on immunoblotted 2D gels, one of which was in primary myoblast extracts from an FSHD patient but not from unaffected individuals [110]. The DUX4 ORF was later found to be evolutionarily conserved by the Hewitt group, prompting the statement in 2007 that “Together with the conservation of the DUX4 ORF for > 100 million years, this strongly supports a coding function for D4Z4 and necessitates re-examination of current models of the FSHD disease mechanism” [111]. Indeed, transfecting C2C12 myoblasts with D4Z4 units had been shown to perturb myogenic differentiation, although DUX4 mRNA could not be detected via RT-PCR [107].
Another important piece of the puzzle came from the cancer field in 2006. A novel chromosomal translocation t(4;19)(q35;q13) in Ewing-like sarcomas generates a hybrid oncogene containing most of the CIC gene fused in frame to the 3’ region of the DUX4 ORF at 4q35 [109]. The encoded CIC–DUX4 chimeric protein retained the CIC DNA-binding domain and its target genes. Interestingly, while CIC-DUX4 was a better transcriptional activator than wild type CIC, the DUX4 carboxyl-terminal domain alone was even more effective [109]. This key study also implied that there may be a functional PAS somewhere downstream of DUX4 at 4q35 [109].
DUX4 expression is cytotoxic
A breakthrough was demonstration that DUX4 was cytotoxic by Alberto L. Rosa’s group, with a clear dose-response (Fig. 2A) [25]. Rabbit sera raised against synthetic DUX4 short peptides revealed its nuclear localization in transfected cells in vitro. These DUX4-expressing cells showed apoptotic features such as Annexin V staining, caspase 3 activation and emerin redistribution at the nuclear envelope [25]. Ubiquitous DUX4 expression in vivo was also found incompatible with normal Drosophila, zebrafish and Xenopus development, confirming that DUX4 was cytotoxic across species [112–114]. Such observations indicated that the DUX4 ORF was linked to FSHD pathogenesis [27] and suggested that the pathogenic mechanism was different from the prevailing position variegation effect model [75].
Fig. 2.
Milestones in the journey to DUX4 detection and function
(A) DUX4 toxicity as shown in co-transfection experiments. Hep2 cells transfected with a GFP reporter gene (pGFP) with or without empty control plasmid (pcDNA3.1) present increasing percentage of cells with GFP fluorescence over 50 h. In contrast, the proportion of GFP fluorescent cells after co-transfection with pGFP and a DUX4 expression plasmid (pDUX4 A) reached a plateau after 8 h, corresponding to the time cytotoxic DUX4 protein could be detected by immunofluorescence, as well as in cells transfected with pDUX4 A alone. From a poster presented by the Rosa group at the FSHD-IRC in Los Angeles in 2003 and published in Kowaljow et al. 2007 [25]
(B) First immunodetection of DUX4 protein with MAb 9A12 on a Western blot of proteins extracted from FSHD (FSHD10) but not healthy (NO36) myoblasts, with a positive control (C+) of TE671 cells transfected with a pCI-Neo-DUX4 expression vector (loaded protein amounts are indicated). Actin immunodetected with a rabbit polyclonal was used as a loading control. Performed by Alexandra Tassin for Supplemental Fig. 7 of Dixit et al.. 2007 [26] (Copyright (2007) National Academy of Sciences, U.S.A.)
(C) The secondary structure of DUX4 using the trRosetta algorithm and PyMOL software (https://pymol.org/2/) reveals an intrinsically disordered protein with three α-helical domains at each homeodomain (H1 and H2) and four α-helical domains in the carboxyl-terminal region (α-1, α-2, α-3, and α-4). The positions of the amino-terminal methionine (M1) and carboxyl-terminal leucine (L424) are indicated. Generated by Alberto L. Rosa for Supplemental Fig. 1 of Quintero et al. 2022 [158]
Why would high D4Z4 copy number not give rise to more DUX4 expression and so cause FSHD? A clue came from early attempts at producing “giant” transgenic mice with additional growth hormone gene copies [115] that revealed integration of high copy number concatemers was associated with less growth (although genome integration site effects could not be discounted). Similarly, mice generated with different copy numbers of a lacZ reporter gene at the same chromosome location only exhibited β-galactosidase activity if they carried small lacZ copy numbers: these were associated with low DNA methylation and decreased chromatin compaction [116]. This led Belayew and Hewitt to hypothesize that compact chromatin developed on large D4Z4 repeat arrays, preventing DUX4 gene expression and FSHD development, while short D4Z4 arrays favoured chromatin opening and DUX4 transcription [24].
Initial characterization of DUX4 mRNAs from D4Z4
A strategy to specifically detect DUX4 mRNA by RT-PCR, despite the hundreds of highly similar DUX genes/3.3-kb elements transcribed in the human genome was developed by the Belayew/Coppée group. Transfected murine C2C12 myoblasts with human genomic plasmids containing 1 or 2 D4Z4 units revealed 1.4 and 1.5 kb mRNAs on Northern blot when hybridized to a double homeobox probe, so demonstrating expression from the native DUX4 promoter. Two transcription start sites were found by rapid amplification of cDNA ends (5’RACE). 70% mapped to an initiator 3’ of the TACAA box and 5’ of the ATG start codon while 30% intriguingly mapped 90 bp upstream, in the DBE repressor element [75] further indicating its inactivity in myogenic cells. Multiple 3’ ends existed downstream from the STOP codon. mRNAs with the full DUX4 ORF could be amplified using RT-PCR from the transfected mouse cells. Refining primer sequences enabled detection of mRNA covering the full DUX4 ORF in differentiating myoblasts from FSHD patients but not unaffected controls [25], reinforcing the idea of DUX4 expression as a potential pathogenic mechanism in FSHD.
Detection of DUX4 protein
Characterization of DUX4 protein was difficult because the gene sequence was so GC rich that sequencing errors precluded clear definition of the ORF end. Initial studies of transcription/translation in vitro of a D4Z4 element indicated a 75-kDa protein in SDS-PAGE. This was predicted to be a dimer since the ORF sequence suggested a 42-kDa protein with 9 Cysteines potentially involved in dimerization [24]. Later, both the Rosa and Belayew/Coppée labs independently raised rabbit sera against synthetic peptides of the DUX4 ORF that immunodetected a 52-kDa protein on a Western blot prepared with total protein extracts of cells transfected with constructs expressing DUX4 or carrying 1 or 2 D4Z4 units [25]. However, the end of the DUX4 protein sequence could not be deduced from the ORF because of difficulties in its sequencing. The approach used by the Belayew/Coppée group was to get the DUX4 protein expressed by C2C12 cells transfected with the D4Z4 genomic fragments. Total cell proteins were separated on PAGE-SDS gels and those in the 50-kDa size range were extracted from the gel, digested with trypsin and the resulting peptides sequenced by MALDI-TOF to reveal the actual DUX4 ORF; these data were added to Rosa’s publication on DUX4 toxicity [25].
The Belayew/Coppée group also developed the first mouse monoclonal anti-DUX4 antibody - MAb 9A12, raised against the 253 carboxyl-terminal residues. This protein domain was so toxic that bacteria had to be transformed with the inducible expression plasmid for each production. In combination with a new high sensitivity western blot immunodetection, MAb 9A12 identified a ~ 55-kDa DUX4 protein in primary FSHD but not healthy myotube cultures [26]. Unfortunately, this first DUX4 immunodetection only appeared as Supplemental Information (Fig. 2B). Since MAb 9A12 was the only antibody that could detect endogenous DUX4, the FSHD Society (USA) provided funding to produce and distribute it to ~ 20 research groups. MAb 9A12 was also used to validate new monoclonal antibodies developed by the Stephen J. Tapscott lab against different DUX4 domains [117]. However, because of its rare stochastic expression and rapid protein turnover [118], it is only recently that DUX4 could be detected in FSHD muscle biopsies by a highly sensitive proximity ligation assay with two monoclonal antibodies targeting different DUX4 domains [119]. Cytoplasmic DUX4 was also immunodetected in MRI-guided FSHD muscle biopsies with MAbs 9A12 and E5-5 in a cluster of activated satellite cells and in very few abnormal muscle fibres with features of abortive regeneration [120]. Although MAb 9A12 was raised against an antigen partly common to DUX4 and DUX4c, it does not detect endogenous DUX4c in muscle cells. Post translational modifications (PTM) may mask the DUX4c epitope but not affect the longer DUX4 protein, allowing DUX4 specificity [117].
Size differences on western blots for DUX4 proteins synthesized in vitro or in vivo result from specific PTM that can target 17 different residues in DUX4, as recently shown by Scott Harper’s group [121]. Of therapeutic interest, these researchers identified residues for which a change in PTM could suppress DUX4 toxicity: (i) increased phosphorylation of Ser/Thr residues by PKA or (ii) decreased Arg methylation by PRMT1.
DUX4 RNA and protein are expressed in FSHD primary myogenic cultures
Using the optimized primers, the Belayew/Coppée group amplified the full length DUX4 mRNA from total RNA of FSHD myotubes by RT-PCR. The cDNA sequence corresponded to DUX4 transcription initiated in the distal D4Z4 unit and, unexpectedly, extended to the downstream pLAM region that provided a 3’ UTR with intron 2 and exon 3 with a PAS [26]. This PAS variant (ATTAAA) had not been previously detected by sequence analysis because it differed from the AATAAA consensus. Of note, a later analysis of multiple 3’ end sequencing data sets identified this variant in 14.5% of human mRNA 3’ ends, while the “consensus” PAS occurred in 47% [122]. Discovery of this PAS in pLAM in 2007 sparked discussion of whether DUX4 mRNA transcribed from the most distal D4Z4 unit possessed a uniquely extended half-life for translation due to polyadenylation from this PAS, as well as how differences between 4qA and 4qB haplotypes could affect both splicing and polyadenylation [26].
Detection of DUX4 mRNA in FSHD muscle cells had been so difficult up to this point that a reviewer requested a point to point comparison of conditions used for RNA extraction and RT-PCR alongside the methodology used by the 6 previous publications that could not detect DUX4 transcripts [52, 94, 102, 107, 123, 124]. The optimized procedure detailed in Supplemental Table 3 [26] was used by the Tapscott group to independently confirm detection of a full length DUX4 mRNA encoding DUX4-fl, and identify other spliced isoforms, including a shorter mRNA with the ORF limited to the homeoboxes encoding DUX4-s [112].
DUX4 is a potent transcription factor
Misregulation of gene expression was observed in FSHD muscle, indicating possible perturbed regulation of transcription [125]. Yi Wen Chen performed RNA profiling of muscle biopsies from controls and 12 different neuromuscular disorders and found that PITX1 RNA was specifically upregulated 11-fold in less affected, and 24-fold in clearly affected, muscles from FSHD patients [25]. PITX1 is a paired type homeodomain transcription factor involved in embryonic development and intriguingly, left/right asymmetry [126]. Although its expression level pointed to involvement in FSHD, PITX1 maps to chromosome 5q31. As homeobox genes often act in regulatory networks the hypothesis was that DUX4 might activate PITX1 expression. An evolutionarily conserved cis-element was identified in the murine Pitx1 promoter, with a central TAAT core typical of homeodomain targets [26] and high similarity to the DUX1 binding site [87]. A mouse Pitx1 promoter fragment carrying this homeodomain target sequence (but not a mutated version) bound DUX4 in vitro and activated expression of a linked reporter gene in C2C12 myoblasts, thus also creating the first DUX4 reporter gene [26]. Data from the Chen and Belayew/Coppée groups were pooled in a key publication showing DUX4 mRNA extension to a PAS, detection of DUX4 mRNA and protein (with MAb 9A12) in FSHD muscle extracts, and DUX4 function as a transcription factor with Pitx1/PITX proposed as the first DUX4 target gene [26]. These observations provided strong support for the relevance of DUX4 to FSHD pathology, with DUX4 and DUX4c metaphorically referred to as “pearls in the junk” [127].
Crucial evidence that DUX4 was the culprit in FSHD was generated by Michael Kyba’s group who directly compared the effects of each of the then 6 proposed FSHD candidate genes (FRG1, FRG2, TUBB4q, ANT1, DUX4 and DUX4c). Each was expressed individually from the same genetic locus in the iC2C12 myoblast model [128]. Strikingly, only DUX4 caused overt toxicity, with inhibition of the glutathione redox pathway and increased sensitivity to oxidative stress, as well as repression of the crucial muscle regulatory gene MyoD, MyoD target genes and myogenic differentiation [128]. Notably, pathways and processes perturbed by DUX4 over-expression were similar to those affected in FSHD myoblast cultures [102, 129], and altered expression of proteins linked to oxidative stress had also been found in FSHD muscle biopsies [130]. The DUX4 homeodomains had been noted to share a high degree of sequence similarity with paired box transcription factors [52], notably PAX3 and PAX7 [128]: master regulators of skeletal myogenesis [131]. Overexpression of Pax3 or Pax7 was shown to mitigate the ability of DUX4 to cause death in murine cells [128], leading to the idea that in FSHD, DUX4 also interferes with the capacity of PAX3 and PAX7 to regulate their target genes [128, 132].
Such engineered myoblast cell lines with inducible and variable transgenic DUX4 expression proved an invaluable tool for DUX4 research, first in murine C2C12 myoblasts (iC2C12-DUX4) [128], and later in human LHCN-M2 myoblasts (LHCN-M2iDUX4) [133] and with a codon-optimised inducible DUX4 transgene in human MB135 myoblasts (iDUX4) [134].
A crucial role for the 4qA permissive allele in FSHD pathology
Nearly identical D4Z4 arrays on both chromosome 4 and 10 but only those on chromosome 4 being linked to FSHD, combined with the perceived ‘lack’ of detectable DUX4 transcript, had been used to argue against a role for DUX4 in FSHD. In addition, the polymorphisms distal to D4Z4, with only the 4qA allele being associated with FSHD, remained intriguing, specifically because this sequence was > 98% identical to the homologous region in 10q26 [73, 74]. The answer however, rested on a single-nucleotide polymorphism (SNP) distal to the D4Z4 array. It had been found that DUX4 mRNAs initiated from the most telomeric D4Z4 unit, extended to the flanking pLAM region that provided a non-canonical PAS to give DUX4 mRNA a poly-A tail [26]. Later, a multicentre collaboration led by van der Maarel performed elegant genetic studies on a large population of patients with FSHD and unaffected relatives revealing that the ‘permissive’ 4qA allele carried this PAS, and that it was suppressed by a SNP in the non-permissive 4qB and 10qA alleles [23]. DUX4 mRNA stability required polyadenylation, which allowed for DUX4 protein synthesis [23].
Further evidence of the 4qA permissive role subsequently emerged in the form of a family in which FSHD was attributed to a rearranged chromosome 10 containing a contracted D4Z4 array in which the non-permissive 10qA distal sequences had been replaced with a 4qA permissive region including the PAS. These genetic configurations led to DUX4 expression from chromosome 10 causing FSHD [135]. Recently, human myoblasts were engineered to create the opposite sequence exchange i.e. replace the region distal to a contracted D4Z4 array on chromosome 4 with a non-permissive 10q distal region lacking the PAS, which improved the pathogenic cell phenotype [136].
DUX4 to the fore by unifying FSHD1 and FSHD2 pathomechanisms
Requirement for both D4Z4 epigenetic derepression with DNA hypomethylation on contracted (FSHD1) and less contracted arrays (FSHD2) [76, 78] and a permissive 4qA haplotype supplying a PAS [23, 26] ‘unified’ FSHD1 and FSHD2 by a common pathomechanism: DUX4 expression. This link was explained when the van der Maarel group demonstrated that FSHD2 was digenic [137]. In addition to an approximate 11-20 D4Z4 unit array on a 4qA allele, FSHD2 was frequently associated with mutations in structural maintenance of chromosomes flexible hinge domain containing 1 (SMCHD1) on chromosome 18 [137, 138]. SMCHD1 is involved in epigenetic suppression of transcription of repeated elements and X-linked genes [139] and so SMCHD1 loss of function mutations favour DNA hypomethylation and open chromatin structure at D4Z4 and DUX4 transcription. SMCHD1 also constitutes a disease modifier for FSHD1, some sequence variations explaining more severe clinical presentations than would be expected based on the patient’s number of D4Z4 units [140, 141]. Interestingly, Smchd1 favours DNA methylation by antagonizing ten-eleven translocation (TET) enzymes that initiate reversion of methylation, so Smchd1 suppression allows DNA hypomethylation, activation of Dux expression and establishment of a 2-cell like stage in mouse ES cells [142]. It was recently reported that SMCHD1 is required for activation of genes involved in myogenic differentiation and muscle regeneration, so SMCHD1 variations could also independently contribute to FSHD pathogenesis [143]. Furthermore, skeletal muscle may be more vulnerable to DUX4 expression in general, as SMCHD1 protein levels dramatically decrease as myoblasts undergo myogenic differentiation [144].
The vast majority of FSHD2 cases are associated with mutations in SMCHD1, but other chromatin modifiers were found mutated in rare FSHD2 pedigrees, such as the DNA methyl transferase DNMT3B [145] and LRIF1 [146]. Whole exome sequencing of clinically defined patients presenting either D4Z4 repeat array of typical short size or longer ones not usually associated to FSHD, recently identified further mutations in genes involved in chromatin structure that could contribute to epigenetic derepression at D4Z4, including DNMT1, DNMT3A, EZH2, CTCF and SUV39H1 [147]. These genes may thus constitute disease modifiers for FSHD1 and FSHD2.
Structural and functional domains of DUX4
The 424-residue DUX4 is actively transported into the nucleus [25, 148]. Harper and colleagues found that mutation of the first DUX4 homeodomain, suppressing DNA binding, prevented toxicity in zebrafish and mice [149]. Rosa’s group identified two nuclear localisation signals (NLS1 and NLS2), which along with the homeodomains and carboxyl-terminal domain, were also required for DUX4 cytotoxicity [108, 150]. Mutations affecting NLS1 and NLS2 and motifs IWF-65 in HD1 and IWF-140 in HD2 all decrease DUX4 cytotoxicity [150]. Given the similarity in sequence between the homeodomains in DUX4 and that in PAX3 or PAX7, the Kyba group reported that when DUX4 homeodomain HD1 was replaced by the mouse Pax7 homeodomain, DUX4 retained the ability to inhibit differentiation and induce cytotoxicity [151].
Chromatin immunoprecipitation combined with high throughput sequencing (ChIP-Seq) identified TAAYBBAATCA as the DUX4-binding site with two tandem homeodomain binding motifs (TAAT), separated by a single nucleotide [92]. Later studies showed that the optimal DNA sequence preferably bound by DUX4 is TAATCTAATCA, also being the most transcriptionally active sequence [152]. The crystal structure of tandem HD1 and HD2 bound to DNA revealed that they are arranged head-to-head. They also recognize different core sequences: HD1 binding TAAT (the HD1-altered target specificity unique to primates) and HD2 targeting TGAT [153]. Indeed, for transcription activity and cytotoxicity, only the two homeodomains and carboxyl-terminal region were required in a DUX4 ‘miniprotein’ [133].
The isolated carboxyl-terminal domain of DUX4 is a potent transcriptional activator [109], with most activity mapping to its last 20 residues [154]. DUX4 recruits the histone acetyltransferases p300/CBP to DUX4 target genes, allowing DUX4 to act as a ‘pioneer’ transcription factor, mediating a dramatic increase in acetylation at H3K27 and H3K18 to open chromatin at target genes [133, 155]. This powerful DUX4 transcriptional activity is also linked to interaction of a KIX motif in its carboxyl-terminal domain with a protein of the Mediator complex that could thus be recruited with RNA Polymerase II at DUX4 target promoters [156].
DUX4 can also indirectly regulate gene expression: it contains evolutionarily conserved LXXLL (NR box-like) motifs in its carboxyl-terminal domain [157, 158], which are also found in co-regulators of nuclear hormone receptors [158]. Rosa’s lab has demonstrated that DUX4 functions as a co-repressor of progesterone and glucocorticoid nuclear receptors [158], a phenomenon that may contribute to the sex differences observed in the onset and severity of FSHD [6]. Thus, in addition to its transcriptional activation role, the DUX4 carboxyl-terminal domain may also contribute to a potential endocrine function [158]. There is also a φXXφφ motif (AQPLL388–392) found in corepressors of hormone receptors (CoRNR boxes), which form a three-turn α-helical structure, similar to that observed in the predicted structure of the DUX4 carboxyl-terminal domain (Fig. 2C) [158].
In addition to its transcriptional functions in the nucleus, DUX4 protein unexpectedly interacts with cytoplasmic partners such as sarcomere Z-disk linked proteins desmin and LMCD1, as well as RNA-binding proteins C1QBP, SRSF9, RBM3, FUS/TLS and SFPQ [159, 160], which may contribute to its effects on RNA-processing [92, 161–163]. Many protein partners are shared by DUX4 and DUX4c, and so part of DUX4 toxicity could be linked to competition for partners normally associated with DUX4c [120]. DNA binding competition also occurs because of their identical homeodomains. DUX4c can compete for DUX4 target genes, for example those involved in the β-catenin pathway [164], but lacking a potent transcriptional activation domain, DUX4c would thus reduce DUX4 cytotoxicity [165].
Predictions of DUX4 3D structure in silico consistently show a largely disordered protein [154, 158], with α-helical regions in the two DNA-binding homeodomains (HD1 residues 19–78 and HD2 residues 94–153), and in the carboxyl-terminal region (residues ∼365–424), which includes the transactivation domain (Fig. 2C), missing in DUX4c.
In embryonic stem cells and mesenchymal stromal cells differentiating to osteoblasts or adipocytes [166], a DUX4 mRNA was induced that initiated further upstream, encoding a 58-kDa protein from a 60-codon extended ORF starting at the initiator ATG proposed by Hewitt [52]. An additional 70-kDa DUX4 protein was also detected in cells but the ends of the encoding mRNA could not be mapped [24]. Neither of these DUX4 protein isoforms was toxic in the cell cultures used and their functions have not been reported [166].
DUX4 transcriptional target genes and repetitive sequences
As a pioneer transcription factor, DUX4 has a cohort of target genes that could provide insight into its pathological functions. PITX1 was the first DUX4 target gene identified [26]. Microarray transcriptional profiling on the murine iC2C12-DUX4 myoblast model to identify differentially expressed genes after 4 and 12 h of induction showed that most activated genes classified via gene ontology under ‘regulation of growth/development’ and ‘signal transduction’ [128].
To determine DUX4 target genes in human, the Tapscott group undertook microarray profiling of healthy human primary myoblasts transduced with a lentivirus expressing DUX4. Up-regulated genes were clearly involved with gamete/spermatogenesis, RNA polymerase II mediator complexes, and RNA splicing and processing, while down-regulated genes associated with immune response pathways [92]. Importantly, they identified several genes now considered to be canonical DUX4 target genes, including ZSCAN4, PRAMEF1, TRIM43, MBD3L2 and KHDC1 and showed up-regulation of these genes in FSHD muscle. DUX4 also bound and activated long terminal repeat (LTR) elements from a class of endogenous primate Mammalian apparent LTR-Retrotransposon (MaLR) and the related endogenous retroviruses (ERVs) family [92]. These retroviral DNA sequences, some of which have integrated near protein coding sequences, are normally epigenetically silenced except in early embryos where they can act as promoter/enhancers to express RNAs and proteins needed for early development. This pivotal paper also generated a useful DUX4 reporter using a 1.9-kb enhancer and promoter region of ZSCAN4 that included four DUX4 binding sites, to drive luciferase expression [92]. The ‘newer’ technology of RNA-Seq was then used to measure transcription of DUX4 target genes [167, 168]. Because it binds to retrotransposon promoters, an issue in the study of DUX4 expressed in mouse cells is that these differ between human and rodents, so that DUX4 also activates a unique set of genes in either species in addition to common targets [105, 169].
Why is it so difficult to detect DUX4 protein in FSHD muscle yet the consequences are so profound? DUX4 is difficult to detect because its expression is rare and random [161], estimated to be active in 1/1000 myoblasts [170] and 1/200 myonuclei in myotubes [118] at any one time. Skeletal muscle fibres are syncytial, which likely explains how such a low abundance protein causes a myopathy. DUX4 transcripts produced in a few myonuclei are translated in the cytoplasm and the newly synthesized DUX4 proteins diffuse to adjacent nuclei, so activation of target genes is amplified [118].
DUX4 activity as a molecular biomarker
A major bottleneck in FSHD research and drug development was the lack of sensitive and specific molecular biomarkers. Initially RNA expression profiling was performed on FSHD versus healthy muscle to define such specific biomarkers [171, 172]. While DUX4 mRNA is detectable in muscle from FSHD foetuses [173, 174], it is much more difficult to detect in muscle biopsies from adult patients. Thus, indirect measures were developed to provide evidence that DUX4 is/was active in a muscle sample by quantifying mRNAs of activated DUX4 target genes as a DUX4 ‘footprint’ or ‘signature’. The first such footprint from Tapscott and colleagues consisted of 114 DUX4 target genes detected in FSHD but not control muscles [168], supporting the hypothesis that DUX4 misexpression is a causal factor for FSHD. Christopher Banerji and Peter Zammit developed further signatures. DUX4 biomarkers include the “early” DUX4 signature from a study of human myoblasts overexpressing DUX4 for 6 h from a doxycycline inducible promoter [133], consisting of 212 significantly upregulated transcripts [132]. The “late” DUX4 signature used a study of human myoblasts transduced with DUX4 lentivirus for 24 h [92], and comprises 165 significantly upregulated transcripts [132]. Given the similarity between the DUX4 and PAX7 homeodomains, a PAX7 signature was also derived from differential expression analysis consisting of 311 upregulated and 290 downregulated, PAX7 target genes [132]. Repression of the PAX7 target gene signature was found to be an equivalent biomarker for FSHD disease to activation of DUX4 target gene signatures in MRI-guided muscle biopsies [175]. Crucially, DUX4 target genes, the three validated DUX4 target gene signatures, MRI and histopathology measures failed to change in a cohort of FSHD patients with 1-year follow-up [176, 177]. In contrast, PAX7 signature repression increased over a year in paired FSHD samples, and so is a biomarker of FSHD progression over the relative short term, and so useful to monitor progress in clinical trials [176]. These observations also indicate that DUX4 interferes with PAX7 function in FSHD [4].
Consequences of ectopic DUX4 activation
DUX4 activates so many target genes/repetitive elements that it is unsurprising that aberrant DUX4 expression in muscle cells has many consequences, with the most obvious usually being rapid cell death. This was elegantly demonstrated by Dan Miller and colleagues, who used a DUX4-responsive nuclear GFP reporter gene in primary FSHD muscle cell cultures, where live imaging revealed rapid cell death after reporter gene activation by endogenous DUX4 [161]. Apoptotic pathways associated with DUX4-induced cell death were initially thought to be dependent on p53 [25, 149, 178] but this was later challenged [179, 180]. This discrepancy probably stems from the observation that p53 can activate DUX4 expression by interaction with an enhancer in a LTR 18 kb 3’ of the DUX4 gene in FSHD iPSC cells [83].
DUX4 expression in human myogenic cells also correlates with the accumulation of DNA damage [181, 182], and DUX4-expressing cells exhibit impaired DNA damage response [183]. DUX4-induced cellular toxicity is linked to accumulation of double stranded transcripts of human satellite II DNA (HSATII) [167] and induces formation of intranuclear HSATII dsRNA foci that bind and sequester nuclear proteins [180]. Interestingly, DUX4 and HSATII expression are highly correlated during early human embryonic development [182], suggesting a normal role for DUX4 in these mechanisms.
Reactive oxygen species (ROS) were shown to induce DUX4 gene expression [184] and earlier studies described known signs of oxidative stress damage in FSHD muscle cells [102, 129, 130, 185]. DUX4 increases sensitivity to oxidative stress by repression of the glutathione redox pathway [128] and disruption of mitochondrial function [130, 186]. FSHD muscle biopsies or primary myoblast cultures show altered expression of proteins linked to oxidative stress [102, 129], not only because of DUX4 but also possibly due to ANT1 misexpression [187]. ROS cause DNA breaks, which activate p53, leading to the DNA damage response (DDR). These findings combined, suggest that p53 is not only induced by DUX4/DUX4 target genes but may also directly trigger DUX4 expression in some cells or stages of development, potentially indicating a vicious cycle of ROS-p53-DUX4-ROS. Addition of antioxidants to DUX4-transfected, and FSHD, myoblasts reduced both DNA damage and morphological defects in myotube formation, suggesting that these pathological phenotypes are due to oxidative stress [181, 188]. Intriguingly PITX1 is among the DUX4-activated genes decreased by antioxidants, suggesting a role of NRF2, the transcription factor involved in cyto protection against oxidative stress [188]. A study on DUX4 binding had indicated that the homeodomain binding site conserved in the PITX1/Pitx1 gene is not optimal [152], so maybe combination of weak DUX4 and NRF2 activation explains the strong PITX1 increase previously observed [26].
FSHD myoblasts also show defective myogenic differentiation into myotubes [102], and primary FSHD myotubes were described as often having either an ‘atrophic’ or ‘disorganised’ morphology [189]. Indeed, ectopic DUX4 expression inhibits differentiation and causes such ‘atrophic’ myotubes [178] although a later time lapse study revealed that FSHD myotubes were actually hypotrophic [190]. This is likely related to the observation that in mouse, DUX4 downregulates MyoD and its target genes [128], creating a more stem-cell like transcriptome [105] and in human cells, DUX4 inhibits MYOD and MYF5, despite binding to the MYF5 enhancer [191].
Numerous other cellular activities are also perturbed by DUX4, including RNA metabolism/processing and translation [92, 161, 163] and immune responses [92], indicating that DUX4 initiates a cascade of dysregulated gene expression with multiple and interconnected processes affected [192, 193].
Modelling DUX4 function in vivo
A major limitation of animal models to investigate DUX4 function in vivo is that the D4Z4 tandem repeats and DUX4 are only strongly conserved in old world primates [111, 194] and so there is no ‘natural’ equivalent in most standard model animals [195]. Thus, the genetic and epigenetic mechanisms underlying the rare, stochastic expression of DUX4 in FSHD are difficult to recapitulate in vivo [196]. There is also the issue of the limited overlap between DUX4 target genes in human and standard models such as the mouse [105, 169], and the fact that many repetitive elements and other retrotransposons are only found in human [92]. Finally, DUX4 is highly toxic to most cells in most organisms [25].
Non-mammalian DUX4 animal models
Ubiquitous DUX4 expression was found incompatible with normal Drosophila, zebrafish and Xenopus development, thus limiting their use [112–114]. The Harper lab showed that muscle-directed transgenic DUX4 expression produced viable zebrafish with approximately half of the embryos malformed and with defective muscle structure [149]. Injecting low levels of DUX4 mRNA at the zebrafish one cell stage resulted in asymmetric muscle disorganisation and degeneration. Interestingly, the homeodomain-containing DUX4-s mRNA reduced toxicity of DUX4-fl [197]. However, recombination-controlled muscle specific DUX4-mCherry expression in zebrafish enables onset of DUX4 to be controlled and visualized [198].
Mammalian DUX4 animal models
After much effort and frustration due to cryptic Sp1-dependent promoters in GC rich sequences allowing for DUX4 expression, DUX4 toxicity and the normal limited expression window in early embryogenesis [199], murine DUX4 models finally emerged. Harper and colleagues used intramuscular injection of AAV-DUX4, leading to local myofiber degeneration, infiltrating mononuclear cells, and p53-dependent apoptosis [149]. The van der Maarel group created transgenic mice using the lambda-42 phage containing an FSHD patient-derived genomic fragment with 2.5 repeats of D4Z4 and the PAS associated with the permissive 4qA haplotype [196]. Their single D4Z4-2.5 mouse line had relaxed chromatin and hypomethylation with DUX4 mRNA in testes, embryonic cells and skeletal muscle tissues. No alterations in muscle structure or function were found in D4Z4-2.5 mice [196] but DUX4 expression was up-regulated during muscle regeneration [105].
The Kyba lab generated the iDUX4 [2.7] mouse line with ubiquitous doxycycline inducible (rtTA driver) DUX4 expression. However, the promoter was leaky, and use of a SV40 PAS linked to the DUX4 gene led to very efficient mRNA polyadenylation and stability, leading to high DUX4 protein levels and lethality. Rare surviving males had smaller and weaker, but not overtly dystrophic, muscles and a skin phenotype, before dying after ~ 2 months [200, 201]. Instead, the iDUX4pA mouse had a doxycycline-inducible genomic fragment from an FSHD 4qA161 allele that included the terminal D4Z4 repeat with the DUX4 ORF and 3ʹ UTR with the less effective native PAS, inserted into the X chromosome [202]. iDUX4pA mice had skin hyperkeratosis, alopecia and high-frequency hearing impairment. Males were less active, with atrophic, weaker muscles with extremely low levels of DUX4 mRNA and some DUX4 target gene expression, living only ~ 4 months. However, mice died after doxycycline induction necessitating use of a muscle-specific HSA rtTA driver to create the iDUX4pA-HSA strain [202]. High doxycycline doses trigger severe muscle damage with loss of ambulation, while low level chronic induction causes progressive muscle atrophy and weakness, with hallmarks of FSHD histopathology. Differential gene expression profiles of iDUX4pA-HSA mice and FSHD muscle have significant overlap [202, 203].
Mouse models quickly followed in which DUX4 expression was activated following recombination. Harper and colleagues developed the TIC-DUX4 mice that employed a DNA fragment encoding V5-tagged DUX4 and the natural DUX4 3′ UTR [204]. The Jones lab produced the FLExDUX4 mouse, with a modified DUX4 transgene encoding only DUX4-fl with both native 5′ and 3′ UTRs [205]. Both constructs were inserted into the Rosa26 locus and mice crossed to mice with muscle-specific Cre expression [206]. Older TIC-DUX4 mice exhibit low-level transgene expression but on exposure to tamoxifen, mice develop an FSHD-like muscle pathology. FLExDUX4 mice have mild muscle-wasting phenotype but recombination causes muscle pathology with FSHD characteristics. As with the iDUX4pA-HAS mice, DUX4 expression is conditional and titratable. Varying tamoxifen dose allows for control over time of onset and severity of muscle phenotypes. A simple non-transgenic mouse model was also developed with local intra-muscular injection/electroporation of naked plasmid DNA expressing DUX4 into the tibialis anterior that causes well delineated muscle lesions after a week [207].
Jones and collaborators have generated a large animal model using the ROSA locus to drive DUX4 expression after Cre-mediated recombination in Gottingen minipigs [208]. A pig model also has the advantage that both porcine DUXC and human DUX4 activate a very similar early embryonic program in porcine myogenic cells [209]. However, in all these inducible mammalian models, the underlying regulation of DUX4 expression is not by the native human locus.
Xenograft models of FSHD and DUX4
There is debate about how closely these non-primate inducible DUX4 models generating a ‘DUX4-opathy’ reproduce FSHD pathology [4]. Xenograft models allow examination of human DUX4 expressed from its native promoter and surrounding 4q35 DNA sequences in vivo. FSHD muscle tissue grafted into mouse generated various proportions of human cells in hybrid regenerated mouse muscle fibres [210]. Instead, xenografting immortalized FSHD muscle precursor cells into mouse resulted in organized and innervated human muscle fibres (with minimal contribution of murine myonuclei) and expression of DUX4 and DUX4 target genes. Satellite cells were also reported to be present [211]. An issue with these xenograft models though, is variability in the amount of FSHD muscle made in each graft.
DUX4 contributes to zygote genome activation
An outstanding question was why does DUX4 have such pleiotropic effects? Another enigma was why the DUX4 coding region has been conserved in “junk” repeat DNA through evolution for over 100 million years [111], which hinted at a functional role. This was uncovered in 2017 in back-to-back publications from the Bradley Cairns, Stephen J. Tapscott and Didier Trono groups who reported that DUX4 was involved in human zygote genome activation (ZGA): waves of embryonic gene transcription during early embryogenesis. The DUX4 functional homolog Dux plays a similar role in mouse [212–214]. During the cleavage stage following fertilisation, the human zygote undergoes cell divisions that are regulated by maternal RNAs and proteins from the oocyte. DUX4 accumulates during this period, peaking at the 4-cell stage in human and participates in the minor wave of ZGA, activating genes required for both trophectoderm and embryonic development, contributing to establishment of totipotent cells. As a pioneer transcription factor, DUX4 was found to induce genes transiently expressed during the cleavage stage, including ZSCAN4, KDM4E and PRAMEF, many of which are transcribed when DUX4 is expressed in myogenic cells [92]. DUX4 also activates expression of repetitive elements, including pericentromeric HSATII repeats, MaLRs, ERVs, and long interspersed nuclear elements (LINE1). Binding to such repetitive elements, DUX4 sometimes creates unorthodox promoters and first exons for nearby genes, or novel transcription start sites for long non-coding RNAs or antisense transcripts [167, 180, 182]. After its activity in the early embryo, DUX4 is epigenetically silenced, which remains throughout life, although DUX4 transcript and protein are detectable in some human tissues with high levels of apoptosis such as testis and thymus [170, 215]. DUX4 is also expressed in late differentiating keratinocytes [216], mesenchymal stem cells from umbilical cord and adipose tissue differentiating to osteoblasts [166]. However, there is no absolute requirement for a Dux gene in mice [217, 218] since redundancy with homeodomain protein Obox4 allows development of Dux-null embryos [219].
In mouse, rRNA synthesis and nucleolar maturation at the 2-cell stage limits the window of Dux activity [199], and Smchd1 binds the Dux gene to contribute to epigenetic repression [142, 220]. RNA of the LINE1 retrotransposon can act as a nuclear scaffold to recruit Nucleolin and Kap1 to facilitate Dux silencing for both ribosomal RNA gene transcription and exit from the 2-cell stage [221]. Such mechanisms may also be employed to control DUX4 at the 4–8 cell stage in human. In addition, a Dux inhibition loop is mediated by DuxBL a rodent homologue lacking an activation domain that is induced by Dux at the ZGA and then silences Dux-induced genes, allowing development to progress [222]. Again, a similar system may occur in human embryos with DUXA, a truncated DUX4 homologue [223], although other researchers propose that DUXA can activate DUX4 target genes in FSHD2 late muscle cell differentiation [224].
These observations also highlighted that there is a PAS that can be employed even with a 4qB haplotype, which was proposed to be in exon 7 [170]. The embryonic transcriptional program activated by the potent DUX4 pioneer transcription factor likely explains the many disparate effects described in FSHD muscle including interference with metabolism, RNA processing and myogenic differentiation.
Other pathological roles of DUX4
The novel chromosomal translocation generating hybrid CIC–DUX4 proteins in Ewing-like sarcomas signalled a potentially wider role for DUX4 in pathology [109]. Later, chimeric transcription factors involving DUX4 were found in another cancer, a subset of B cell acute lymphoblastic leukaemia (B-ALL) [225–228]. Some chromosomal rearrangements in B-ALL were characterised by insertion of D4Z4 repeats into the IGH locus. This encoded a hybrid transcription factor termed DUX-IGH incorporating the DUX4 amino-terminal region with the two homeodomains that dictate target genes selection, and a unique carboxyl-terminus [225]. It is of note that this chromosomal rearrangement also contains regulatory elements associated with the D4Z4 units. Expression of DUX-IGH in B-cells/their precursors in B-ALL is consistent with observations that DUX4 and transcripts of its target genes can also be detected in immortalised B-cell lymphoblastoid clones from FSHD patients, although their immortalisation with Epstein–Barr virus is a confounding factor [229, 230], and DUX4 expression was not detected in primary FSHD peripheral blood cells [231].
Crucially, DUX4 is reactivated in some solid cancers via cis-acting inherited genetic variation and trans-acting somatically acquired mutations in repressors. DUX4 induces a metastable early embryogenic stem cell transcription (ZGA, 8 cell-like program, markers of early embryogenic lineages) [232] and causes immune evasion of the cancer cells by reducing antigen presentation, since it prevents IFN-γ-mediated induction of MHC class I genes [233]. This occurs via the LXXLL (NR box-like) motifs in the carboxyl-terminal domain of DUX4 interacting with STAT1 to suppress IFNγ-induced genes by reducing bound STAT1 and RNA Pol-II [234]. DUX4 is expressed in ~ 10–50% of advanced bladder, breast, kidney, prostate, and skin cancers, revealing its high frequency in metastatic tumours, and is associated with shorter survival times [235].
Finally, DUX4 expression is induced by Herpes viruses, thus mimicking an early embryonic-like transcriptional program that prevents epigenetic silencing of the viral genome and facilitates viral gene expression and viral proliferation [236, 237].
Interestingly, DUX4 mRNA was only identified as deregulated by transcriptome analysis in other biological systems than FSHD muscle after its gene was considered functional [25, 26], and its sequence was excluded from the “gene-less repetitive regions” (“junk DNA”) not considered by the Repeat Masker software.
DUX4 today
Much focus is now on understanding the regulation of DUX4, its effects in healthy and FSHD cells, and targeting DUX4 therapeutically. A few clinical trials had been performed in patients with FSHD to evaluate various therapeutics used in other neuromuscular disorders. However, when DUX4 became widely suspected as the main cause of FSHD, focus shifted to strategies to prevent/reduce DUX4 effects by disrupting DUX4 gene expression, blocking translation of DUX4 transcripts, and/or interfering with protein function [238].
However, many questions remain about DUX4 function in health and FSHD, including:
If the mean prevalence of FSHD is 5/100,000 [1], yet approximately 13/1000 healthy individuals carry alleles with 4–8 D4Z4 repeats with a 4qA haplotype [62], then why are only 1/260 people with a ‘pathogenic allele’ affected by FSHD?
What is the normal physiological role of DUX4 in spermatocyte precursors, keratinocytes and thymic cells etc?
Does DUX4 play a normal role in regulating hormone receptors, and does it disrupt the endocrine physiology of muscle? What is its interplay with estrogens that were shown to inhibit its toxicity in cell cultures [239]?
What is the importance of DUX4 in non-myogenic cell types such as FAPs, macrophages, lymphocytes in FSHD pathology? What are the mechanisms by which DUX4 affects immune response and contributes to inflammation?
What are the functions of DUX4, DUX4c and other DUX proteins in rRNA synthesis, processing and ribosome assembly at the nucleolus?
Does DUX4 participate in DNA damage and/or normal DNA repair processes related to the occurrence of double strand breaks?
Why are there larger DUX4 isoforms found in ESCs and MSCs? Why are these not cytotoxic?
Can DUX4 diffuse between cells? Several homeoproteins can cross the plasma membrane for paracrine activity [240].
Are DUX4 and DUX4c expressed simultaneously in a given nucleus and what are the consequences?
Where, when and how does DUX4 interact with PAX7?
Will therapeutic suppression of DUX4 in adult patients with FSHD slow or suppress pathology and allow muscle regeneration?
Will targeting DUX4 in FSHD have deleterious effects on specific cell types in testis, skin and thymus?
Summary
Here, we have detailed the emergence of DUX4 from “junk DNA” status to its role in FSHD pathology (Fig. 3). Contraction of the D4Z4 array and/or mutation in epigenetic modifiers triggers re-expression of the DUX4 retrogene, whose RNA is stabilised by addition of a poly-A tail because of the PAS present on a 4qA haplotype. In skeletal muscle, this early embryonic transcription factor reactivates a totipotent stem cell program that perturbs the tissue-specific gene expression profile to ultimately cause muscle damage, weakness and wasting.
Fig. 3.
The Puzzle that is the involvement of DUX4 in FSHD
Schematic showing some of the evidence that contributed to completing the jigsaw that was the identification of the involvement of DUX4 in FSHD pathology
Many researchers participated in the initial findings that chromosome 4 harbours the FSHD disease locus and identifying the microsatellite array of D4Z4 units near the 4q telomere and the DUX4 ORF. This was followed by the discovery that epigenetic derepression and DUX4 expression from the distal unit was the culprit. As the mechanisms underlying the complex FSHD pathogenesis unfolded, much work then went into proving DUX4 was expressed in FSHD and defining its functions (Fig. 3). DUX4 is now not only central to FSHD research but roles in early embryogenesis, cancer and viral infection are widening interest in this enigmatic transcription factor. The steady increase in publications and citations since the discovery of the DUX4 gene in 1999 [24] testifies to the prominence DUX4 has gained. The total from 1999 to 2024 is now 783 publications and 22,254 citations (publications with ‘DUX4’ in an ‘All fields’ search via ‘Web of Science’ accessed on 31/12/2024). As with the last quarter of a century, the next 25 years of DUX4 research will be equally fascinating!
Acknowledgements
In a corruption of the proverb ‘It takes a village to raise a review’ we are grateful to colleagues who read and commented on this work including Christopher R. S. Banerji, Elise N. Engquist, Massimo Ganassi, Frédérique Coppée and Alexandra Tassin, and Daniel C. L. Zammit for help with figures. Valuable source material is in the 2004 book - FSHD Clinical Medicine and Molecular Cell Biology, edited by M. Upadhyaya and D.N. Cooper, Taylor & Francis LTD, but it is, sadly, not available online. We have tried to represent the journey through the history of FSHD and DUX4 accurately, within the inherent limitations of trying to organise chronologically and not making the manuscript even more unwieldly. We apologise for any omissions and/or failed attributions to the work of colleagues, and for only naming lab heads. We especially thank all the patient associations that support/supported research including The FSHD Society, Friends of FSH Research and Muscular Dystrophy Association (USA), Muscular Dystrophy UK, AFM-Telethon and Amis FSH (France), FSHD Global Foundation (Australia), FSHD Stichting (The Netherlands), ABMM (Belgium) and SOLVE FSHD (Canada). We specially acknowledge Daniel Perez of the FSHD Society (USA), who played a major role in getting researchers to network, including during the most difficult times of DUX4’s struggle to rise from the junk.
Author contributions
Alexandra Belayew, Alberto L. Rosa and Peter S. Zammit all contributed to writing this review.
Funding
Alexandra Belayew declares no relevant funding. Alberto L. Rosa is Principal Investigator from Argentina National Research Council (CONICET), and his lab is funded by grants from CONICET, The FSHD Society (#2022-4591150923) and Solve FSHD (GR003028-SA1). The Zammit lab is currently funded by the Medical Research Council UK (MR/X001520/1), SOLVE FSHD, Friends of FSH Research, AFM-Telethon and the European Union Horizon Europe project no. 101080690 - MAGIC (via UKRI under the UK government’s Horizon Europe funding guarantee grant no. 10080927, 10079726 and 10078461).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Alexandra Belayew is co-inventor on a patent “Agents useful in treating FSHD” owned by the University of Mons and licensed to Dyne Therapeutics. The other authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Deenen JC, Verbeek AL, Verschuuren JJ, van Engelen BG, Voermans NC. Prevalence and incidence rates of 17 neuromuscular disorders: an updated review of the literature. J Neuromuscul Dis. 2025;22143602241313118 [DOI] [PubMed]
- 2.Mah JK, Korngut L, Fiest KM, Dykeman J, Day LJ, Pringsheim T, Jette N. A systematic review and Meta-analysis on the epidemiology of the muscular dystrophies. Can J Neurol Sci. 2016;43(1):163–77. [DOI] [PubMed] [Google Scholar]
- 3.Kools J, Deenen JC, Blokhuis AM, Verbeek AL, Voermans NC, van Engelen BG. The Dutch registry for facioscapulohumeral muscular dystrophy: cohort profile and longitudinal patient reported outcomes. Neuromuscul Disord. 2023;33(12):964–71. [DOI] [PubMed] [Google Scholar]
- 4.Banerji CRS, Zammit PS. Pathomechanisms and biomarkers in facioscapulohumeral muscular dystrophy: roles of DUX4 and PAX7. EMBO Mol Med. 2021;13(8):e13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Padberg GWAM. Facioscapulohumeral disease. Doctoral Thesis, Leiden University 1982, (http://hdl.handle.net/1887/25818).
- 6.Zatz M, Marie SK, Cerqueira A, Vainzof M, Pavanello RC, Passos-Bueno MR. The facioscapulohumeral muscular dystrophy (FSHD1) gene affects males more severely and more frequently than females. Am J Med Genet. 1998;77(2):155–61. [PubMed] [Google Scholar]
- 7.Tawil R, Storvick D, Feasby TE, Weiffenbach B, Griggs RC. Extreme variability of expression in monozygotic twins with FSH muscular dystrophy. Neurology. 1993;43(2):345–8. [DOI] [PubMed] [Google Scholar]
- 8.Tyler FH, Stephens FE. Studies in disorders of muscle. II clinical manifestations and inheritance of facioscapulohumeral dystrophy in a large family. Ann Intern Med. 1950;32(4):640–60. [DOI] [PubMed] [Google Scholar]
- 9.Tawil R, van der Maarel SM, Tapscott SJ. Facioscapulohumeral dystrophy: the path to consensus on pathophysiology. Skelet Muscle. 2014;4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ricci G, Scionti I, Sera F, Govi M, D’Amico R, Frambolli I, Mele F, Filosto M, Vercelli L, Ruggiero L, et al. Large scale genotype-phenotype analyses indicate that novel prognostic tools are required for families with facioscapulohumeral muscular dystrophy. Brain. 2013;136(Pt 11):3408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banerji CRS, Cammish P, Evangelista T, Zammit PS, Straub V, Marini-Bettolo C. Facioscapulohumeral muscular dystrophy 1 patients participating in the UK FSHD registry can be subdivided into 4 patterns of self-reported symptoms. Neuromuscul Disord. 2020;30(4):315–28. [DOI] [PubMed] [Google Scholar]
- 12.Orrell RW. Facioscapulohumeral dystrophy and scapuloperoneal syndromes. Handb Clin Neurol. 2011;101:167–80. [DOI] [PubMed] [Google Scholar]
- 13.Eger K, Jordan B, Habermann S, Zierz S. Beevor’s sign in facioscapulohumeral muscular dystrophy: an old sign with new implications. J Neurol. 2010;257(3):436–8. [DOI] [PubMed] [Google Scholar]
- 14.Hilbert JE, Kissel JT, Luebbe EA, Martens WB, McDermott MP, Sanders DB, Tawil R, Thornton CA, Moxley RT 3. Registry scientific advisory C: if you build a rare disease registry, will they enroll and will they use it? Methods and data from the National registry of myotonic dystrophy (DM) and facioscapulohumeral muscular dystrophy (FSHD). Contemp Clin Trials. 2012;33(2):302–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichinger K, Heatwole C, Iyadurai S, King W, Baker L, Heininger S, Bartlett A, Dilek N, Martens WB, McDermott M, et al. Facioscapulohumeral muscular dystrophy functional composite outcome measure. Muscle Nerve. 2018. 10.1002:mus.26088. [DOI] [PMC free article] [PubMed]
- 16.Felice KJ, North WA, Moore SA, Mathews KD. FSH dystrophy 4q35 deletion in patients presenting with facial-sparing scapular myopathy. Neurology. 2000;54(10):1927–31. [DOI] [PubMed] [Google Scholar]
- 17.Ricci G, Cammish P, Siciliano G, Tupler R, Lochmuller H, Evangelista T. Phenotype May predict the clinical course of facioscapolohumeral muscular dystrophy. Muscle Nerve. 2019;59(6):711–3. [DOI] [PubMed] [Google Scholar]
- 18.Fitzsimons RB. Retinal vascular disease and the pathogenesis of facioscapulohumeral muscular dystrophy. A signalling message from wnt?? Neuromuscul Disord. 2011;21(4):263–71. [DOI] [PubMed] [Google Scholar]
- 19.Tawil R, Kissel JT, Heatwole C, Pandya S, Gronseth G, Benatar M, Guideline Development D, Implementation Subcommittee of the American Academy of N, Practice Issues Review Panel of the American Association of N, Electrodiagnostic M. Evidence-based guideline summary: evaluation, diagnosis, and management of facioscapulohumeral muscular dystrophy: report of the guideline development, dissemination, and implementation subcommittee of the American academy of neurology and the practice issues review panel of the American association of neuromuscular & electrodiagnostic medicine. Neurology. 2015;85(4):357–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Dijk GP, van der Kooi E, Behin A, Smeets J, Timmermans J, van der Maarel S, Padberg G, Voermans N, van Engelen B. High prevalence of incomplete right bundle branch block in facioscapulohumeral muscular dystrophy without cardiac symptoms. Funct Neurol. 2014;29(3):159–65. [PMC free article] [PubMed] [Google Scholar]
- 21.Goselink RJ, Schreuder TH, Mul K, Voermans NC, Pelsma M, de Groot IJ, van Alfen N, Franck B, Theelen T, Lemmers RJ, et al. Facioscapulohumeral dystrophy in children: design of a prospective, observational study on natural history, predictors and clinical impact (iFocus FSHD). BMC Neurol. 2016;16:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikolic A, Ricci G, Sera F, Bucci E, Govi M, Mele F, Rossi M, Ruggiero L, Vercelli L, Ravaglia S, et al. Clinical expression of facioscapulohumeral muscular dystrophy in carriers of 1–3 D4Z4 reduced alleles: experience of the FSHD Italian National registry. BMJ Open. 2016;6(1):e007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camano P, Dauwerse JG, Snider L, Straasheijm KR, van Ommen GJ, Padberg GW, et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329(5999):1650–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabriëls J, Beckers MC, Ding H, De Vriese A, Plaisance S, van der Maarel SM, Padberg GW, Frants RR, Hewitt JE, Collen D, et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236(1):25–32. [DOI] [PubMed] [Google Scholar]
- 25.Kowaljow V, Marcowycz A, Ansseau E, Conde CB, Sauvage S, Matteotti C, Arias C, Corona ED, Nunez NG, Leo O, et al. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul Disord. 2007;17(8):611–23. [DOI] [PubMed] [Google Scholar]
- 26.Dixit M, Ansseau E, Tassin A, Winokur S, Shi R, Qian H, Sauvage S, Matteotti C, van Acker AM, Leo O, et al. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci U S A. 2007;104(46):18157–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gatica LV, Rosa AL. A complex interplay of genetic and epigenetic events leads to abnormal expression of the DUX4 gene in facioscapulohumeral muscular dystrophy. Neuromuscul Disord. 2016;26(12):844–52. [DOI] [PubMed] [Google Scholar]
- 28.Richards M, Coppee F, Thomas N, Belayew A, Upadhyaya M. Facioscapulohumeral muscular dystrophy (FSHD): an enigma unravelled? Hum Genet. 2012;131(3):325–40. [DOI] [PubMed] [Google Scholar]
- 29.Campbell AE, Belleville AE, Resnick R, Shadle SC, Tapscott SJ. Facioscapulohumeral dystrophy: activating an early embryonic transcriptional program in human skeletal muscle. Hum Mol Genet. 2018;27(R2):R153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meryon E. On granular and fatty degeneration of the voluntary muscles. Med Chir Trans 1852, 35:73–84. [PMC free article] [PubMed]
- 31.Cruveilhiers J. Mémoire Sur La paralysie musculaire atrophique. Bulletins de l’Académie de Médecine 1852–3, 18:490–502, 546–583.
- 32.Duchenne G-BA. Recherches Sur La paralysie musculaire pseudohypertrophique, Ou paralysie myo-sclérosique. Arch Gén Méd 1868, 11:5–25,179–209, 305–321, 421–443, 552–588.
- 33.Landouzy L, Dejerine J. De La myopathie atrophique progressive (myopathie hérérlitaire débutant, Dans l’enfance, par La face, Sans altération du système nerveux). Comptes Rendus Hebdomadaires Des Séances De l’Académie Des Sciences. 1884;98:53–5. [Google Scholar]
- 34.Landouzy L, Dejerine J. De la myopathie atrophique progressive (myopathie sans neuropathie débutant d’ordinaire dans l’enfance par la face. In: Revue de médecine. vol. 5: Felix Alcan; 1885: 253–366.
- 35.Sorrel-Dejerine Y, Fardeau M. Birth and metamorphosis of Landouzy-Dejerine progressive atrophic myopathy. Rev Neurol (Paris). 1982;138(12):1041–51. [PubMed] [Google Scholar]
- 36.Erb W. Ueber die ‘‘juvenile form der progressiven muskelatrophie und Ihre beziehungen Zur sogenannten pseudohypertrophie der muskeln. Deutsch Archiv Klin Med. 1884;34:467–519. [Google Scholar]
- 37.Walton JN. Dystrophia Muscularis progressiva. In: Progressive Muskeldystrophie Myotonie · Myasthenie: Symposium vom 30 November bis 4 Dezember 1965 anläßlich der 125 Wiederkehr des Geburtstages von Wilhelm Erb. Edited by Kuhn E. Berlin, Heidelberg: Springer Berlin Heidelberg; 1966: 57–76.
- 38.Erb WH. Dystrophia muscularis progressiva. Klinische und pathologisch-anatomische studien. Dtsch Z Nervenheilkd 1891, 1:13–94.
- 39.Calder A. The remarkable origins of FSHD research in America. https://www.fshdsociety.org/2020/11/16/the-remarkable-origins-of-fshd-research-in-america/ 2020.
- 40.Flanigan KM, Coffeen CM, Sexton L, Stauffer D, Brunner S, Leppert MF. Genetic characterization of a large, historically significant Utah kindred with facioscapulohumeral dystrophy. Neuromuscul Disord. 2001;11(6–7):525–9. [DOI] [PubMed] [Google Scholar]
- 41.Hoffman EP, Brown RH Jr., Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51(6):919–28. [DOI] [PubMed] [Google Scholar]
- 42.Lunt PW. A workshop on facioscapulohumeral (Landouzy-Dejerine) disease, manchester, 16 to 17 November 1988. J Med Genet. 1989;26(8):535–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarfarazi M, Upadhyaya M, Padberg G, Pericak-Vance M, Siddique T, Lucotte G, Lunt P. An exclusion map for facioscapulohumeral (Landouzy-Dejerine) disease. J Med Genet. 1989;26(8):481–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Upadhyaya M, Lunt PW, Sarfarazi M, Broadhead W, Daniels J, Owen M, Harper PS. DNA marker applicable to presymptomatic and prenatal diagnosis of facioscapulohumeral disease. Lancet. 1990;336(8726):1320–1. [DOI] [PubMed] [Google Scholar]
- 45.Wijmenga C, Frants RR, Brouwer OF, Moerer P, Weber JL, Padberg GW. Location of facioscapulohumeral muscular dystrophy gene on chromosome 4. Lancet. 1990;336(8716):651–3. [DOI] [PubMed] [Google Scholar]
- 46.Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, Dauwerse HG, Gruter AM, Hofker MH, Moerer P, Williamson R, et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet. 1992;2(1):26–30. [DOI] [PubMed] [Google Scholar]
- 47.Wijmenga C, Brouwer OF, Padberg GW, Frants RR. Transmission of de-novo mutation associated with facioscapulohumeral muscular dystrophy. Lancet. 1992;340(8825):985–6. [DOI] [PubMed] [Google Scholar]
- 48.Wright TJ, Wijmenga C, Clark LN, Frants RR, Williamson R, Hewitt JE. Fine mapping of the FSHD gene region orientates the rearranged fragment detected by the probe p13E-11. Hum Mol Genet. 1993;2(10):1673–8. [DOI] [PubMed] [Google Scholar]
- 49.Wijmenga C, Wright TJ, Baan MJ, Padberg GW, Williamson R, van Ommen GJ, Hewitt JE, Hofker MH, Frants RR. Physical mapping and YAC-cloning connects four genetically distinct 4qter loci (D4S163, D4S139, D4F35S1 and D4F104S1) in the FSHD gene-region. Hum Mol Genet. 1993;2(10):1667–72. [DOI] [PubMed] [Google Scholar]
- 50.van Deutekom JC, Wijmenga C, van Tienhoven EA, Gruter AM, Hewitt JE, Padberg GW, van Ommen GJ, Hofker MH, Frants RR. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet. 1993;2(12):2037–42. [DOI] [PubMed] [Google Scholar]
- 51.Lee JH, Goto K, Matsuda C, Arahata K. Characterization of a tandemly repeated 3.3-kb Kpnl unit in the facioscapulohumeral muscular dystrophy (FSHD) gene region on chromosome 4q35. Muscle Nerve Supplement 1995(2):S6–13. [PubMed]
- 52.Hewitt JE, Lyle R, Clark LN, Valleley EM, Wright TJ, Wijmenga C, van Deutekom JC, Francis F, Sharpe PT, Hofker M, et al. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum Mol Genet. 1994;3(8):1287–95. [DOI] [PubMed] [Google Scholar]
- 53.Winokur ST, Bengtsson U, Feddersen J, Mathews KD, Weiffenbach B, Bailey H, Markovich RP, Murray JC, Wasmuth JJ, Altherr MR, et al. The DNA rearrangement associated with facioscapulohumeral muscular dystrophy involves a heterochromatin-associated repetitive element: implications for a role of chromatin structure in the pathogenesis of the disease. Chromosome Res. 1994;2(3):225–34. [DOI] [PubMed] [Google Scholar]
- 54.Bengtsson U, Altherr MR, Wasmuth JJ, Winokur ST. High resolution fluorescence in situ hybridization to linearly extended DNA visually maps a tandem repeat associated with facioscapulohumeral muscular dystrophy immediately adjacent to the telomere of 4q. Hum Mol Genet. 1994;3(10):1801–5. [DOI] [PubMed] [Google Scholar]
- 55.Deidda G, Cacurri S, Grisanti P, Vigneti E, Piazzo N, Felicetti L. Physical mapping evidence for a duplicated region on chromosome 10qter showing high homology with the facioscapulohumeral muscular dystrophy locus on chromosome 4qter. Eur J Hum Genetics. 1995;3(3):155–67. [DOI] [PubMed] [Google Scholar]
- 56.Bakker E, Wijmenga C, Vossen RH, Padberg GW, Hewitt J, van der Wielen M, Rasmussen K, Frants RR. The FSHD-linked locus D4F104S1 (p13E-11) on 4q35 has a homologue on 10qter. Muscle Nerve Supplement 1995(2):S39–44. [PubMed]
- 57.Lemmers RJ, van der Vliet PJ, van der Gaag KJ, Zuniga S, Frants RR, de Knijff P, van der Maarel SM. Worldwide population analysis of the 4q and 10q subtelomeres identifies only four discrete interchromosomal sequence transfers in human evolution. Am J Hum Genet. 2010;86(3):364–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rossi M, Ricci E, Colantoni L, Galluzzi G, Frusciante R, Tonali PA, Felicetti L. The facioscapulohumeral muscular dystrophy region on 4qter and the homologous locus on 10qter evolved independently under different evolutionary pressure. BMC Med Genet. 2007;8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deidda G, Cacurri S, Piazzo N, Felicetti L. Direct detection of 4q35 rearrangements implicated in facioscapulohumeral muscular dystrophy (FSHD). J Med Genet. 1996;33(5):361–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Maarel SM, Deidda G, Lemmers RJ, Bakker E, van der Wielen MJ, Sandkuijl L, Hewitt JE, Padberg GW, Frants RR. A new dosage test for subtelomeric 4;10 translocations improves conventional diagnosis of facioscapulohumeral muscular dystrophy (FSHD). J Med Genet. 1999;36(11):823–8. [PMC free article] [PubMed] [Google Scholar]
- 61.Ohno S. So much junk DNA in our genome. Brookhaven Symp Biol. 1972;23:366–70. [PubMed] [Google Scholar]
- 62.Scionti I, Greco F, Ricci G, Govi M, Arashiro P, Vercelli L, Berardinelli A, Angelini C, Antonini G, Cao M, et al. Large-scale population analysis challenges the current criteria for the molecular diagnosis of fascioscapulohumeral muscular dystrophy. Am J Hum Genet. 2012;90(4):628–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wijmenga C, Frants RR, Hewitt JE, van Deutekom JC, van Geel M, Wright TJ, Padberg GW, Hofker MH, van Ommen GJ. Molecular genetics of facioscapulohumeral muscular dystrophy. Neuromuscul Disord. 1993;3(5–6):487–91. [DOI] [PubMed] [Google Scholar]
- 64.Lunt PW, Jardine PE, Koch MC, Maynard J, Osborn M, Williams M, Harper PS, Upadhyaya M. Correlation between fragment size at D4F104S1 and age at onset or at wheelchair use, with a possible generational effect, accounts for much phenotypic variation in 4q35-facioscapulohumeral muscular dystrophy (FSHD). Hum Mol Genet. 1995;4(5):951–8. [DOI] [PubMed] [Google Scholar]
- 65.Zatz M, Marie SK, Passos-Bueno MR, Vainzof M, Campiotto S, Cerqueira A, Wijmenga C, Padberg G, Frants R. High proportion of new mutations and possible anticipation in Brazilian facioscapulohumeral muscular dystrophy families. Am J Hum Genet. 1995;56(1):99–105. [PMC free article] [PubMed] [Google Scholar]
- 66.Tupler R, Barbierato L, Memmi M, Sewry CA, De Grandis D, Maraschio P, Tiepolo L, Ferlini A. Identical de Novo mutation at the D4F104S1 locus in monozygotic male twins affected by facioscapulohumeral muscular dystrophy (FSHD) with different clinical expression. J Med Genet. 1998;35(9):778–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Griggs RC, Tawil R, McDermott M, Forrester J, Figlewicz D, Weiffenbach B. Monozygotic twins with facioscapulohumeral dystrophy (FSHD): implications for genotype/phenotype correlation. FSH-DY group. Muscle Nerve Supplement. 1995;2:S50–55. [PubMed] [Google Scholar]
- 68.Tupler R, Berardinelli A, Barbierato L, Frants R, Hewitt JE, Lanzi G, Maraschio P, Tiepolo L. Monosomy of distal 4q does not cause facioscapulohumeral muscular dystrophy. J Med Genet. 1996;33(5):366–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Deutekom JC, Bakker E, Lemmers RJ, van der Wielen MJ, Bik E, Hofker MH, Padberg GW, Frants RR. Evidence for subtelomeric exchange of 3.3 kb tandemly repeated units between chromosomes 4q35 and 10q26: implications for genetic counselling and etiology of FSHD1. Hum Mol Genet. 1996;5(12):1997–2003. [DOI] [PubMed] [Google Scholar]
- 70.Thompson JS, Johnson LM, Grunstein M. Specific repression of the yeast silent mating locus HMR by an adjacent telomere. Mol Cell Biol. 1994;14(1):446–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383(6595):92–6. [DOI] [PubMed] [Google Scholar]
- 72.Stadler G, Rahimov F, King OD, Chen JC, Robin JD, Wagner KR, Shay JW, Emerson CP Jr., Wright WE. Telomere position effect regulates DUX4 in human facioscapulohumeral muscular dystrophy. Nat Struct Mol Biol. 2013;20(6):671–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lemmers RJ, de Kievit P, Sandkuijl L, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet. 2002;32(2):235–6. [DOI] [PubMed] [Google Scholar]
- 74.van Geel M, Dickson MC, Beck AF, Bolland DJ, Frants RR, van der Maarel SM, de Jong PJ, Hewitt JE. Genomic analysis of human chromosome 10q and 4q telomeres suggests a common origin. Genomics. 2002;79(2):210–7. [DOI] [PubMed] [Google Scholar]
- 75.Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110(3):339–48. [DOI] [PubMed] [Google Scholar]
- 76.van Overveld PG, Lemmers RJ, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet. 2003;35(4):315–7. [DOI] [PubMed] [Google Scholar]
- 77.van Overveld PG, Enthoven L, Ricci E, Rossi M, Felicetti L, Jeanpierre M, Winokur ST, Frants RR, Padberg GW, van der Maarel SM. Variable hypomethylation of D4Z4 in facioscapulohumeral muscular dystrophy. Ann Neurol. 2005;58(4):569–76. [DOI] [PubMed] [Google Scholar]
- 78.de Greef JC, Lemmers RJ, van Engelen BG, Sacconi S, Venance SL, Frants RR, Tawil R, van der Maarel SM. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum Mutat. 2009;30(10):1449–59. [DOI] [PubMed] [Google Scholar]
- 79.Zeng W, de Greef JC, Chen YY, Chien R, Kong X, Gregson HC, Winokur ST, Pyle A, Robertson KD, Schmiesing JA, et al. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet. 2009;5(7):e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cabianca DS, Casa V, Bodega B, Xynos A, Ginelli E, Tanaka Y, Gabellini D. A long NcRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149(4):819–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Himeda CL, Debarnot C, Homma S, Beermann ML, Miller JB, Jones PL, Jones TI. Myogenic enhancers regulate expression of the facioscapulohumeral muscular Dystrophy-Associated DUX4 gene. Mol Cell Biol. 2014;34(11):1942–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mocciaro E, Giambruno R, Micheloni S, Cernilogar FM, Andolfo A, Consonni C, Pannese M, Ferri G, Runfola V, Schotta G, et al. WDR5 is required for DUX4 expression and its pathological effects in FSHD muscular dystrophy. Nucleic Acids Res. 2023;51(10):5144–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grow EJ, Weaver BD, Smith CM, Guo J, Stein P, Shadle SC, Hendrickson PG, Johnson NE, Butterfield RJ, Menafra R, et al. p53 convergently activates Dux/DUX4 in embryonic stem cells and in facioscapulohumeral muscular dystrophy cell models. Nat Genet. 2021;53(8):1207–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fox A, Oliva J, Vangipurapu R, Sverdrup FM. SIX transcription factors are necessary for the activation of DUX4 expression in facioscapulohumeral muscular dystrophy. Skelet Muscle. 2024;14(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jones TI, King OD, Himeda CL, Homma S, Chen JC, Beermann ML, Yan C, Emerson CP Jr., Miller JB, Wagner KR, et al. Individual epigenetic status of the pathogenic D4Z4 macrosatellite correlates with disease in facioscapulohumeral muscular dystrophy. Clin Epigenetics. 2015;7:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gould T, Jones TI, Jones PL. Precise epigenetic analysis using targeted bisulfite genomic sequencing distinguishes FSHD1, FSHD2, and healthy subjects. Diagnostics (Basel) 2021, 11(8):1469. [DOI] [PMC free article] [PubMed]
- 87.Ding H, Beckers MC, Plaisance S, Marynen P, Collen D, Belayew A. Characterization of a double homeodomain protein (DUX1) encoded by a cDNA homologous to 3.3 kb dispersed repeated elements. Hum Mol Genet. 1998;7(11):1681–94. [DOI] [PubMed] [Google Scholar]
- 88.Tsumagari K, Qi L, Jackson K, Shao C, Lacey M, Sowden J, Tawil R, Vedanarayanan V, Ehrlich M. Epigenetics of a tandem DNA repeat: chromatin DNaseI sensitivity and opposite methylation changes in cancers. Nucleic Acids Res. 2008;36(7):2196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ciszewski L, Lu-Nguyen N, Slater A, Brennan A, Williams HEL, Dickson G, Searle MS, Popplewell L. G-quadruplex ligands mediate downregulation of DUX4 expression. Nucleic Acids Res. 2020;48(8):4179–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bai G, Endres T, Kuhbacher U, Mengoli V, Greer BH, Peacock EM, Newton MD, Stanage T, Dello Stritto MR, Lungu R, et al. HLTF resolves G4s and promotes G4-induced replication fork slowing to maintain genome stability. Mol Cell. 2024;84(16):3044–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang XY, Loflin PT, Gehrke CW, Andrews PA, Ehrlich M. Hypermethylation of human DNA sequences in embryonal carcinoma cells and somatic tissues but not in sperm. Nucleic Acids Res. 1987;15(22):9429–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Geng LN, Yao Z, Snider L, Fong AP, Cech JN, Young JM, van der Maarel SM, Ruzzo WL, Gentleman RC, Tawil R, et al. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell. 2012;22(1):38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Beckers M, Gabriels J, van der Maarel S, De Vriese A, Frants RR, Collen D, Belayew A. Active genes in junk DNA? Characterization of DUX genes embedded within 3.3 kb repeated elements. Gene. 2001;264(1):51–7. [DOI] [PubMed] [Google Scholar]
- 94.Lyle R, Wright TJ, Clark LN, Hewitt JE. The FSHD-associated repeat, D4Z4, is a member of a dispersed family of homeobox-containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics. 1995;28(3):389–97. [DOI] [PubMed] [Google Scholar]
- 95.Winokur ST, Bengtsson U, Vargas JC, Wasmuth JJ, Altherr MR, Weiffenbach B, Jacobsen SJ. The evolutionary distribution and structural organization of the homeobox-containing repeat D4Z4 indicates a functional role for the ancestral copy in the FSHD region. Hum Mol Genet. 1996;5(10):1567–75. [DOI] [PubMed] [Google Scholar]
- 96.Tupler R, Gabellini D. Molecular basis of facioscapulohumeral muscular dystrophy. Cell Mol Life Sci. 2004;61(5):557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Girton JR, Johansen KM. Chromatin structure and the regulation of gene expression: the lessons of PEV in Drosophila. Adv Genet. 2008;61:1–43. [DOI] [PubMed] [Google Scholar]
- 98.Li K, Warner CK, Hodge JA, Minoshima S, Kudoh J, Fukuyama R, Maekawa M, Shimizu Y, Shimizu N, Wallace DC. A human muscle adenine nucleotide translocator gene has four exons, is located on chromosome 4, and is differentially expressed. J Biol Chem. 1989;264(24):13998–4004. [PubMed] [Google Scholar]
- 99.Wijmenga C, Winokur ST, Padberg GW, Skraastad MI, Altherr MR, Wasmuth JJ, Murray JC, Hofker MH, Frants RR. The human skeletal muscle adenine nucleotide translocator gene maps to chromosome 4q35 in the region of the facioscapulohumeral muscular dystrophy locus. Hum Genet. 1993;92(2):198–203. [DOI] [PubMed] [Google Scholar]
- 100.van Deutekom JC, Lemmers RJ, Grewal PK, van Geel M, Romberg S, Dauwerse HG, Wright TJ, Padberg GW, Hofker MH, Hewitt JE, et al. Identification of the first gene (FRG1) from the FSHD region on human chromosome 4q35. Hum Mol Genet. 1996;5(5):581–90. [DOI] [PubMed] [Google Scholar]
- 101.van Geel M, van Deutekom JC, van Staalduinen A, Lemmers RJ, Dickson MC, Hofker MH, Padberg GW, Hewitt JE, de Jong PJ, Frants RR. Identification of a novel beta-tubulin subfamily with one member (TUBB4Q) located near the telomere of chromosome region 4q35. Cytogenet Cell Genet. 2000;88(3–4):316–21. [DOI] [PubMed] [Google Scholar]
- 102.Winokur ST, Chen YW, Masny PS, Martin JH, Ehmsen JT, Tapscott SJ, van der Maarel SM, Hayashi Y, Flanigan KM. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum Mol Genet. 2003;12(22):2895–907. [DOI] [PubMed] [Google Scholar]
- 103.Bosnakovski D, Lamb S, Simsek T, Xu Z, Belayew A, Perlingeiro R, Kyba M. DUX4c, an FSHD candidate gene, interferes with myogenic regulators and abolishes myoblast differentiation. Exp Neurol. 2008;214(1):87–96. [DOI] [PubMed] [Google Scholar]
- 104.Ansseau E, Laoudj-Chenivesse D, Marcowycz A, Tassin A, Vanderplanck C, Sauvage S, Barro M, Mahieu I, Leroy A, Leclercq I, et al. DUX4c is up-regulated in FSHD. It induces the MYF5 protein and human myoblast proliferation. PLoS ONE. 2009;4(10):e7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Knopp P, Krom YD, Banerji CR, Panamarova M, Moyle LA, den Hamer B, van der Maarel SM, Zammit PS. DUX4 induces a transcriptome more characteristic of a less-differentiated cell state and inhibits myogenesis. J Cell Sci. 2016;129(20):3816–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Caruso N, Herberth B, Bartoli M, Puppo F, Dumonceaux J, Zimmermann A, Denadai S, Lebosse M, Roche S, Geng L, et al. Deregulation of the Protocadherin gene FAT1 alters muscle shapes: implications for the pathogenesis of facioscapulohumeral dystrophy. PLoS Genet. 2013;9(6):e1003550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yip DJ, Picketts DJ. Increasing D4Z4 repeat copy number compromises C2C12 myoblast differentiation. FEBS Lett. 2003;537(1–3):133–8. [DOI] [PubMed] [Google Scholar]
- 108.Arias C, Conde CB, Rosa AL. Cellular toxicity of DUX4, a protein encoded at the fascioscapulo- humeral muscular dystrophy locus FSHD- 1A, depends on nuclear localization. Neuromuscul Disord. 2005;15(9):709. [Google Scholar]
- 109.Kawamura-Saito M, Yamazaki Y, Kaneko K, Kawaguchi N, Kanda H, Mukai H, Gotoh T, Motoi T, Fukayama M, Aburatani H, et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet. 2006;15(13):2125–37. [DOI] [PubMed] [Google Scholar]
- 110.Coppee F, Matteotti C, Ansseau E, Sauvage S, Leclercq I, Leroy A, Marcowycz A, gerbaux D, Figlewicz D, Ding H et al. The DUX gene family and FSHD. In: FSHD Facioscapulohumeral Muscular Dystrophy: Clinical Medicine and Molecular Cell Biology. Edited by Upadhyaya M, Cooper DN: Taylor & Francis LTD; 2004.
- 111.Clapp J, Mitchell LM, Bolland DJ, Fantes J, Corcoran AE, Scotting PJ, Armour JA, Hewitt JE. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2007;81(2):264–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Snider L, Asawachaicharn A, Tyler AE, Geng LN, Petek LM, Maves L, Miller DG, Lemmers RJ, Winokur ST, Tawil R, et al. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum Mol Genet. 2009;18(13):2414–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wuebbles RD, Long SW, Hanel ML, Jones PL. Testing the effects of FSHD candidate gene expression in vertebrate muscle development. Int J Clin Exp Pathol. 2010;3(4):386–400. [PMC free article] [PubMed] [Google Scholar]
- 114.Jones TI, Parilla M, Jones PL. Transgenic Drosophila for investigating DUX4 and FRG1, two genes associated with facioscapulohumeral muscular dystrophy (FSHD). PLoS ONE. 2016;11(3):e0150938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Palmiter RD, Brinster RL, Hammer RE, Trumbauer ME, Rosenfeld MG, Birnberg NC, Evans RM. Dramatic growth of mice that develop from eggs microinjected with metallothionein-growth hormone fusion genes. Nature. 1982;300(5893):611–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene Silencing in mammals. Nat Genet. 1998;18(1):56–9. [DOI] [PubMed] [Google Scholar]
- 117.Geng LN, Tyler AE, Tapscott SJ. Immunodetection of human double homeobox 4. Hybrid (Larchmt). 2011;30(2):125–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tassin A, Laoudj-Chenivesse D, Vanderplanck C, Barro M, Charron S, Ansseau E, Chen YW, Mercier J, Coppee F, Belayew A. DUX4 expression in FSHD muscle cells: how could such a rare protein cause a myopathy? J Cell Mol Med. 2013;17(1):76–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Beermann ML, Homma S, Miller JB. Proximity ligation assay to detect DUX4 protein in FSHD1 muscle: a pilot study. BMC Res Notes. 2022;15(1):163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Claus C, Slavin M, Ansseau E, Lancelot C, Bah K, Lassche S, Fievet M, Greco A, Tomaiuolo S, Tassin A, et al. The double homeodomain protein DUX4c is associated with regenerating muscle fibers and RNA-binding proteins. Skelet Muscle. 2023;13(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Knox RN, Eidahl JO, Wallace LM, Choudury SG, Rashnonejad A, Daman K, Guggenbiller MJ, Saad NY, Hoover ME, Zhang L, et al. Post-Translational modifications of the DUX4 protein impact toxic function in FSHD cell models. Ann Neurol. 2023;94(2):398–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gruber AJ, Schmidt R, Gruber AR, Martin G, Ghosh S, Belmadani M, Keller W, Zavolan M. A comprehensive analysis of 3’ end sequencing data sets reveals novel polyadenylation signals and the repressive role of heterogeneous ribonucleoprotein C on cleavage and polyadenylation. Genome Res. 2016;26(8):1145–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Osborne RJ, Welle S, Venance SL, Thornton CA, Tawil R. Expression profile of FSHD supports a link between retinal vasculopathy and muscular dystrophy. Neurology. 2007;68(8):569–77. [DOI] [PubMed] [Google Scholar]
- 124.Alexiadis V, Ballestas ME, Sanchez C, Winokur S, Vedanarayanan V, Warren M, Ehrlich M. RNAPol-ChIP analysis of transcription from FSHD-linked tandem repeats and satellite DNA. Biochim Biophys Acta. 2007;1769(1):29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tupler R, Perini G, Pellegrino MA, Green MR. Profound misregulation of muscle-specific gene expression in facioscapulohumeral muscular dystrophy. Proc Natl Acad Sci U S A. 1999;96(22):12650–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tran TQ, Kioussi C. Pitx genes in development and disease. Cell Mol Life Sci. 2021;78(11):4921–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dmitriev P, Lipinski M, Vassetzky YS. Pearls in the junk: dissecting the molecular pathogenesis of facioscapulohumeral muscular dystrophy. Neuromuscul Disord. 2009;19(1):17–20. [DOI] [PubMed] [Google Scholar]
- 128.Bosnakovski D, Xu Z, Gang EJ, Galindo CL, Liu M, Simsek T, Garner HR, Agha-Mohammadi S, Tassin A, Coppee F, et al. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. Embo J. 2008;27(20):2766–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Winokur ST, Barrett K, Martin JH, Forrester JR, Simon M, Tawil R, Chung SA, Masny PS, Figlewicz DA. Facioscapulohumeral muscular dystrophy (FSHD) myoblasts demonstrate increased susceptibility to oxidative stress. Neuromuscul Disord. 2003;13(4):322–33. [DOI] [PubMed] [Google Scholar]
- 130.Laoudj-Chenivesse D, Carnac G, Bisbal C, Hugon G, Bouillot S, Desnuelle C, Vassetzky Y, Fernandez A. Increased levels of adenine nucleotide translocator 1 protein and response to oxidative stress are early events in facioscapulohumeral muscular dystrophy muscle. J Mol Med (Berl). 2005;83(3):216–24. [DOI] [PubMed] [Google Scholar]
- 131.Relaix F, Zammit PS. Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage. Development. 2012;139(16):2845–56. [DOI] [PubMed] [Google Scholar]
- 132.Banerji CRS, Panamarova M, Hebaishi H, White RB, Relaix F, Severini S, Zammit PS. PAX7 target genes are globally repressed in facioscapulohumeral muscular dystrophy skeletal muscle. Nat Commun. 2017;8(1):2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Choi SH, Gearhart MD, Cui Z, Bosnakovski D, Kim M, Schennum N, Kyba M. DUX4 recruits p300/CBP through its C-terminus and induces global H3K27 acetylation changes. Nucleic Acids Res. 2016;44(11):5161–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jagannathan S, Shadle SC, Resnick R, Snider L, Tawil RN, van der Maarel SM, Bradley RK, Tapscott SJ. Model systems of DUX4 expression recapitulate the transcriptional profile of FSHD cells. Hum Mol Genet. 2016;25(20):4419–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lemmers R, van der Vliet PJ, Blatnik A, Balog J, Zidar J, Henderson D, Goselink R, Tapscott SJ, Voermans NC, Tawil R et al. Chromosome 10q-linked FSHD identifies DUX4 as principal disease gene. J Med Genet 2022;59(2):180–8. [DOI] [PMC free article] [PubMed]
- 136.Ma Y, Schwager Karpukhina A, Dib C, Gautier C, Hermine O, Allemand E, Vassetzky YS. Exchange of subtelomeric regions between chromosomes 4q and 10q reverts the FSHD genotype and phenotype. Sci Adv. 2024;10(18):eadl1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lemmers RJ, Tawil R, Petek LM, Balog J, Block GJ, Santen GW, Amell AM, van der Vliet PJ, Almomani R, Straasheijm KR, et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet. 2012;44(12):1370–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mitsuhashi S, Boyden SE, Estrella EA, Jones TI, Rahimov F, Yu TW, Darras BT, Amato AA, Folkerth RD, Jones PL, et al. Exome sequencing identifies a novel SMCHD1 mutation in facioscapulohumeral muscular dystrophy 2. Neuromuscul Disord. 2013;23(12):975–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Blewitt ME, Vickaryous NK, Hemley SJ, Ashe A, Bruxner TJ, Preis JI, Arkell R, Whitelaw E. An N-ethyl-N-nitrosourea screen for genes involved in variegation in the mouse. Proc Natl Acad Sci U S A. 2005;102(21):7629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sacconi S, Lemmers RJ, Balog J, van der Vliet PJ, Lahaut P, van Nieuwenhuizen MP, Straasheijm KR, Debipersad RD, Vos-Versteeg M, Salviati L, et al. The FSHD2 gene SMCHD1 is a modifier of disease severity in families affected by FSHD1. Am J Hum Genet. 2013;93(4):744–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cascella R, Strafella C, Caputo V, Galota RM, Errichiello V, Scutifero M, Petillo R, Marella GL, Arcangeli M, Colantoni L, et al. Digenic inheritance of shortened repeat units of the D4Z4 region and a Loss-of-Function variant in SMCHD1 in a family with FSHD. Front Neurol. 2018;9:1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Huang Z, Yu J, Cui W, Johnson BK, Kim K, Pfeifer GP. The chromosomal protein SMCHD1 regulates DNA methylation and the 2c-like state of embryonic stem cells by antagonizing TET proteins. Sci Adv 2021, 7(4):eabb9149. [DOI] [PMC free article] [PubMed]
- 143.Wong MM, Hachmer S, Gardner E, Runfola V, Arezza E, Megeney LA, Emerson CP Jr., Gabellini D, Dilworth FJ. SMCHD1 activates the expression of genes required for the expansion of human myoblasts. Nucleic Acids Res. 2024;52(16):9450–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Balog J, Thijssen PE, Shadle S, Straasheijm KR, van der Vliet PJ, Krom YD, van den Boogaard ML, de Jong A, RJ FL, Tawil R et al. Increased DUX4 expression during muscle differentiation correlates with decreased SMCHD1 protein levels at D4Z4. Epigenetics 2015, 10(12):1133–1142. [DOI] [PMC free article] [PubMed]
- 145.van den Boogaard ML, Lemmers RJ, Balog J, Wohlgemuth M, Auranen M, Mitsuhashi S, van der Vliet PJ, Straasheijm KR, van den Akker RF, Kriek M, et al. Mutations in DNMT3B modify epigenetic repression of the D4Z4 repeat and the penetrance of facioscapulohumeral dystrophy. Am J Hum Genet. 2016;98(5):1020–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hamanaka K, Sikrova D, Mitsuhashi S, Masuda H, Sekiguchi Y, Sugiyama A, Shibuya K, Lemmers R, Goossens R, Ogawa M, et al. Homozygous nonsense variant in LRIF1 associated with facioscapulohumeral muscular dystrophy. Neurology. 2020;94(23):e2441–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Strafella C, Caputo V, Bortolani S, Torchia E, Megalizzi D, Trastulli G, Monforte M, Colantoni L, Caltagirone C, Ricci E, et al. Whole exome sequencing highlights rare variants in CTCF, DNMT1, DNMT3A, EZH2 and SUV39H1 as associated with FSHD. Front Genet. 2023;14:1235589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ostlund C, Garcia-Carrasquillo RM, Belayew A, Worman HJ. Intracellular trafficking and dynamics of double homeodomain proteins. Biochemistry. 2005;44(7):2378–84. [DOI] [PubMed] [Google Scholar]
- 149.Wallace LM, Garwick SE, Mei W, Belayew A, Coppee F, Ladner KJ, Guttridge D, Yang J, Harper SQ. DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann Neurol. 2010;69(3):540–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Corona ED, Jacquelin D, Gatica L, Rosa AL. Multiple protein domains contribute to nuclear import and cell toxicity of DUX4, a candidate pathogenic protein for facioscapulohumeral muscular dystrophy. PLoS ONE. 2013;8(10):e75614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bosnakovski D, Toso EA, Hartweck LM, Magli A, Lee HA, Thompson ER, Dandapat A, Perlingeiro RCR, Kyba M. The DUX4 homeodomains mediate Inhibition of myogenesis and are functionally exchangeable with the Pax7 homeodomain. J Cell Sci. 2017;130(21):3685–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang Y, Lee JK, Toso EA, Lee JS, Choi SH, Slattery M, Aihara H, Kyba M. DNA-binding sequence specificity of DUX4. Skelet Muscle. 2016;6:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lee JK, Bosnakovski D, Toso EA, Dinh T, Banerjee S, Bohl TE, Shi K, Orellana K, Kyba M, Aihara H. Crystal structure of the double homeodomain of DUX4 in complex with DNA. Cell Rep. 2018;25(11):2955–62.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mitsuhashi H, Ishimaru S, Homma S, Yu B, Honma Y, Beermann ML, Miller JB. Functional domains of the FSHD-associated DUX4 protein. Biol Open. 2018;7(4):bio033977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bosnakovski D, da Silva MT, Sunny ST, Ener ET, Toso EA, Yuan C, Cui Z, Walters MA, Jadhav A, Kyba M. A novel P300 inhibitor reverses DUX4-mediated global histone H3 hyperacetylation, target gene expression, and cell death. Sci Adv. 2019;5(9):eaaw7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Vuoristo S, Bhagat S, Hyden-Granskog C, Yoshihara M, Gawriyski L, Jouhilahti EM, Ranga V, Tamirat M, Huhtala M, Kirjanov I, et al. DUX4 is a multifunctional factor priming human embryonic genome activation. iScience. 2022;25(4):104137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Leidenroth A, Hewitt JE. A family history of DUX4: phylogenetic analysis of DUXA, B, C and duxbl reveals the ancestral DUX gene. BMC Evol Biol. 2010;10:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Quintero J, Saad NY, Pagnoni SM, Jacquelin DK, Gatica LV, Harper SQ, Rosa AL. The DUX4 protein is a co-repressor of the progesterone and glucocorticoid nuclear receptors. FEBS Lett. 2022;596(20):2644–58. [DOI] [PubMed] [Google Scholar]
- 159.Ansseau E, Eidahl JO, Lancelot C, Tassin A, Matteotti C, Yip C, Liu J, Leroy B, Hubeau C, Gerbaux C, et al. Homologous transcription factors DUX4 and DUX4c associate with cytoplasmic proteins during muscle differentiation. PLoS ONE. 2016;11(1):e0146893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.DeSimone AM, Leszyk J, Wagner K, Emerson CP Jr. Identification of the hyaluronic acid pathway as a therapeutic target for facioscapulohumeral muscular dystrophy. Sci Adv. 2019;5(12):eaaw7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rickard AM, Petek LM, Miller DG. Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Hum Mol Genet. 2015;24(20):5901–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jagannathan S, Ogata Y, Gafken PR, Tapscott SJ, Bradley RK. Quantitative proteomics reveals key roles for post-transcriptional gene regulation in the molecular pathology of facioscapulohumeral muscular dystrophy. eLife 2019, 8:e41740. [DOI] [PMC free article] [PubMed]
- 163.Feng Q, Snider L, Jagannathan S, Tawil R, van der Maarel SM, Tapscott SJ, Bradley RK. A feedback loop between nonsense-mediated decay and the retrogene DUX4 in facioscapulohumeral muscular dystrophy. eLife. 2015;4:e04996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Banerji CR, Knopp P, Moyle LA, Severini S, Orrell RW, Teschendorff AE, Zammit PS. β-Catenin is central to DUX4-driven network rewiring in facioscapulohumeral muscular dystrophy. J R Soc Interface. 2015;12(102):20140797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ganassi M, Figeac N, Reynaud M, Ortuste Quiroga HP, Zammit PS. Antagonism between DUX4 and DUX4c highlights a pathomechanism operating through beta-Catenin in facioscapulohumeral muscular dystrophy. Front Cell Dev Biology. 2022;10:802573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ansseau E, Nachtegael C, Pieters K, Vanderplanck C, Geens M, Sermon K, Wilton SD, Coppee F, Lagneaux L et al. The Role of D4Z4-Encoded Proteins in the Osteogenic Differentiation of Mesenchymal Stromal Cells Isolated from Bone Marrow. Stem Cells Dev 2015, 24(22):2674–86. [DOI] [PubMed]
- 167.Young JM, Whiddon JL, Yao Z, Kasinathan B, Snider L, Geng LN, Balog J, Tawil R, van der Maarel SM, Tapscott SJ. DUX4 binding to retroelements creates promoters that are active in FSHD muscle and testis. PLoS Genet. 2013;9(11):e1003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yao Z, Snider L, Balog J, Lemmers RJ, Van Der Maarel SM, Tawil R, Tapscott SJ. DUX4-induced gene expression is the major molecular signature in FSHD skeletal muscle. Hum Mol Genet. 2014;23(20):5342–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sharma V, Harafuji N, Belayew A, Chen YW. DUX4 differentially regulates transcriptomes of human rhabdomyosarcoma and mouse C2C12 cells. PLoS ONE. 2013;8(5):e64691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Snider L, Geng LN, Lemmers RJ, Kyba M, Ware CB, Nelson AM, Tawil R, Filippova GN, van der Maarel SM, Tapscott SJ, et al. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6(10):e1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rahimov F, King OD, Leung DG, Bibat GM, Emerson CP Jr., Kunkel LM, Wagner KR. Transcriptional profiling in facioscapulohumeral muscular dystrophy to identify candidate biomarkers. Proc Natl Acad Sci U S A. 2012;109(40):16234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tasca G, Pescatori M, Monforte M, Mirabella M, Iannaccone E, Frusciante R, Cubeddu T, Laschena F, Ottaviani P, Ricci E. Different molecular signatures in magnetic resonance imaging-staged facioscapulohumeral muscular dystrophy muscles. PLoS ONE. 2012;7(6):e38779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Broucqsault N, Morere J, Gaillard MC, Dumonceaux J, Torrents J, Salort-Campana E, Maues De Paula A, Bartoli M, Fernandez C, Chesnais AL, et al. Dysregulation of 4q35- and muscle-specific genes in fetuses with a short D4Z4 array linked to facio-scapulo-humeral dystrophy. Hum Mol Genet. 2013;22(20):4206–14. [DOI] [PubMed] [Google Scholar]
- 174.Ferreboeuf M, Mariot V, Bessieres B, Vasiljevic A, Attie-Bitach T, Collardeau S, Morere J, Roche S, Magdinier F, Robin-Ducellier J, et al. DUX4 and DUX4 downstream target genes are expressed in fetal FSHD muscles. Hum Mol Genet. 2014;23(1):171–81. [DOI] [PubMed] [Google Scholar]
- 175.Banerji CRS, Zammit PS. PAX7 target gene repression is a superior FSHD biomarker than DUX4 target gene activation, associating with pathological severity and identifying FSHD at the single-cell level. Hum Mol Genet. 2019;28(13):2224–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Banerji CRS. PAX7 target gene repression associates with FSHD progression and pathology over 1 year. Hum Mol Genet. 2020;29(13):2124–33. [DOI] [PubMed] [Google Scholar]
- 177.Wong CJ, Wang LH, Friedman SD, Shaw D, Campbell AE, Budech CB, Lewis LM, Lemmers R, Statland JM, van der Maarel SM, et al. Longitudinal measures of RNA expression and disease activity in FSHD muscle biopsies. Hum Mol Genet. 2020;29(6):1030–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Vanderplanck C, Ansseau E, Charron S, Stricwant N, Tassin A, Laoudj-Chenivesse D, Wilton SD, Coppee F, Belayew A. The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLoS ONE. 2011;6(10):e26820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bosnakovski D, Gearhart MD, Toso EA, Recht OO, Cucak A, Jain AK, Barton MC, Kyba M. p53-independent DUX4 pathology in cell and animal models of facioscapulohumeral muscular dystrophy. Dis Models Mech. 2017;10(10):1211–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shadle SC, Zhong JW, Campbell AE, Conerly ML, Jagannathan S, Wong CJ, Morello TD, van der Maarel SM, Tapscott SJ. DUX4-induced DsRNA and MYC mRNA stabilization activate apoptotic pathways in human cell models of facioscapulohumeral dystrophy. PLoS Genet. 2017;13(3):e1006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dmitriev P, Bou Saada Y, Dib C, Ansseau E, Barat A, Hamade A, Dessen P, Robert T, Lazar V, Louzada RA, et al. DUX4-induced constitutive DNA damage and oxidative stress contribute to aberrant differentiation of myoblasts from FSHD patients. Free Radic Biol Med. 2016;99:244–58. [DOI] [PubMed] [Google Scholar]
- 182.Shadle SC, Bennett SR, Wong CJ, Karreman NA, Campbell AE, van der Maarel SM, Bass BL, Tapscott SJ. DUX4-induced bidirectional HSATII satellite repeat transcripts form intranuclear double-stranded RNA foci in human cell models of FSHD. Hum Mol Genet. 2019;28(23):3997–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Arends T, Tsuchida H, Adeyemi RO, Tapscott SJ. DUX4-induced HSATII transcription causes KDM2A/B-PRC1 nuclear foci and impairs DNA damage response. J Cell Biol 2024, 223(5):e202303141. [DOI] [PMC free article] [PubMed]
- 184.Sasaki-Honda M, Jonouchi T, Arai M, Hotta A, Mitsuhashi S, Nishino I, Matsuda R, Sakurai H. A patient-derived iPSC model revealed oxidative stress increases facioscapulohumeral muscular dystrophy-causative DUX4. Hum Mol Genet. 2018;27(23):4024–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Turki A, Hayot M, Carnac G, Pillard F, Passerieux E, Bommart S, Raynaud de Mauverger E, Hugon G, Pincemail J, Pietri S, et al. Functional muscle impairment in facioscapulohumeral muscular dystrophy is correlated with oxidative stress and mitochondrial dysfunction. Free Radic Biol Med. 2012;53(5):1068–79. [DOI] [PubMed] [Google Scholar]
- 186.Heher P, Ganassi M, Weidinger A, Engquist EN, Pruller J, Nguyen TH, Tassin A, Decleves AE, Mamchaoui K, Banerji CRS, et al. Interplay between mitochondrial reactive oxygen species, oxidative stress and hypoxic adaptation in facioscapulohumeral muscular dystrophy: metabolic stress as potential therapeutic target. Redox Biol. 2022;51:102251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Arbogast S, Kotzur H, Frank C, Compagnone N, Sutra T, Pillard F, Pietri S, Hmada N, Moussa DMA, Bride J, et al. ANT1 overexpression models: some similarities with facioscapulohumeral muscular dystrophy. Redox Biol. 2022;56:102450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Karpukhina A, Galkin I, Ma Y, Dib C, Zinovkin R, Pletjushkina O, Chernyak B, Popova E, Vassetzky Y. Analysis of genes regulated by DUX4 via oxidative stress reveals potential therapeutic targets for treatment of facioscapulohumeral dystrophy. Redox Biol. 2021;43:102008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Barro M, Carnac G, Flavier S, Mercier J, Vassetzky Y, Laoudj-Chenivesse D. Myoblasts from affected and non-affected FSHD muscles exhibit morphological differentiation defects. J Cell Mol Med. 2010;14(1–2):275–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Banerji CRS, Panamarova M, Pruller J, Figeac N, Hebaishi H, Fidanis E, Saxena A, Contet J, Sacconi S, Severini S, et al. Dynamic transcriptomic analysis reveals suppression of PGC1alpha/ERRalpha drives perturbed myogenesis in facioscapulohumeral muscular dystrophy. Hum Mol Genet. 2019;28(8):1244–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bosnakovski D, Gearhart MD, Toso EA, Ener ET, Choi SH, Kyba M. Low level DUX4 expression disrupts myogenesis through deregulation of myogenic gene expression. Sci Rep. 2018;8(1):16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lim KRQ, Nguyen Q, Yokota T. DUX4 signalling in the pathogenesis of facioscapulohumeral muscular dystrophy. Int J Mol Sci. 2020;21(3):729. [DOI] [PMC free article] [PubMed]
- 193.Mocciaro E, Runfola V, Ghezzi P, Pannese M, Gabellini D. DUX4 role in normal physiology and in FSHD muscular dystrophy. Cells. 2021;10(12):3322. [DOI] [PMC free article] [PubMed]
- 194.Leidenroth A, Clapp L, Mitchell LM, Coneyworth D, Dearden FL, Iannuzzi L, Hewitt JE. Evolution of DUX gene macrosatellites in placental mammals. Chromosoma. 2012;121(5):489–97. [DOI] [PubMed] [Google Scholar]
- 195.DeSimone AM, Cohen J, Lek M, Lek A. Cellular and animal models for facioscapulohumeral muscular dystrophy. Dis Models Mech. 2020;13(10):dmm046904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Krom YD, Thijssen PE, Young JM, den Hamer B, Balog J, Yao Z, Maves L, Snider L, Knopp P, Zammit PS, et al. Intrinsic epigenetic regulation of the D4Z4 macrosatellite repeat in a Transgenic mouse model for FSHD. PLoS Genet. 2013;9(4):e1003415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Mitsuhashi H, Mitsuhashi S, Lynn-Jones T, Kawahara G, Kunkel LM. Expression of DUX4 in zebrafish development recapitulates facioscapulohumeral muscular dystrophy. Hum Mol Genet. 2013;22(3):568–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Pakula A, Lek A, Widrick J, Mitsuhashi H, Bugda Gwilt KM, Gupta VA, Rahimov F, Criscione J, Zhang Y, Gibbs D, et al. Transgenic zebrafish model of DUX4 misexpression reveals a developmental role in FSHD pathogenesis. Hum Mol Genet. 2019;28(2):320–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Xie SQ, Leeke BJ, Whilding C, Wagner RT, Garcia-Llagostera F, Low Y, Chammas P, Cheung NT, Dormann D, McManus MT, et al. Nucleolar-based dux repression is essential for embryonic two-cell stage exit. Genes Dev. 2022;36(5–6):331–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Dandapat A, Bosnakovski D, Hartweck LM, Arpke RW, Baltgalvis KA, Vang D, Baik J, Darabi R, Perlingeiro RC, Hamra FK, et al. Dominant lethal pathologies in male mice engineered to contain an X-linked DUX4 transgene. Cell Rep. 2014;8(5):1484–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Dandapat A, Perrin BJ, Cabelka C, Razzoli M, Ervasti JM, Bartolomucci A, Lowe DA, Kyba M. High frequency hearing loss and hyperactivity in DUX4 Transgenic mice. PLoS ONE. 2016;11(3):e0151467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bosnakovski D, Chan SSK, Recht OO, Hartweck LM, Gustafson CJ, Athman LL, Lowe DA, Kyba M. Muscle pathology from stochastic low level DUX4 expression in an FSHD mouse model. Nat Commun. 2017;8(1):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bosnakovski D, Shams AS, Yuan C, da Silva MT, Ener ET, Baumann CW, Lindsay AJ, Verma M, Asakura A, Lowe DA, et al. Transcriptional and cytopathological hallmarks of FSHD in chronic DUX4-expressing mice. J Clin Invest. 2020;130(5):2465–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Giesige CR, Wallace LM, Heller KN, Eidahl JO, Saad NY, Fowler AM, Pyne NK, Al-Kharsan M, Rashnonejad A, Chermahini GA, et al. AAV-mediated follistatin gene therapy improves functional outcomes in the TIC-DUX4 mouse model of FSHD. JCI Insight. 2018;3(22):e123538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Jones T, Jones PL. A cre-inducible DUX4 Transgenic mouse model for investigating facioscapulohumeral muscular dystrophy. PLoS ONE. 2018;13(2):e0192657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.McCarthy JJ, Srikuea R, Kirby TJ, Peterson CA, Esser KA. Inducible Cre Transgenic mouse strain for skeletal muscle-specific gene targeting. Skelet Muscle. 2012;2(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Derenne A, Tassin A, Nguyen TH, De Roeck E, Jenart V, Ansseau E, Belayew A, Coppee F, Decleves AE, Legrand A. Induction of a local muscular dystrophy using electroporation in vivo: an easy tool for screening therapeutics. Sci Rep. 2020;10(1):11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Jones PA. https://myfshd.org/wp-content/uploads/2021/04/myfshd-animal-model-pig1.pdf. 2021.
- 209.Nip Y, Bennett SR, Smith AA, Jones TI, Jones PL, Tapscott SJ. Human DUX4 and Porcine DUXC activate similar early embryonic programs in pig muscle cells: implications for preclinical models of FSHD. Hum Mol Genet. 2023;32(11):1864–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zhang Y, King OD, Rahimov F, Jones TI, Ward CW, Kerr JP, Liu N, Emerson CP Jr., Kunkel LM, Partridge TA, et al. Human skeletal muscle xenograft as a new preclinical model for muscle disorders. Hum Mol Genet. 2014;23(12):3180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Mueller AL, O’Neill A, Jones TI, Llach A, Rojas LA, Sakellariou P, Stadler G, Wright WE, Eyerman D, Jones PL, et al. Muscle xenografts reproduce key molecular features of facioscapulohumeral muscular dystrophy. Exp Neurol. 2019;320:113011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hendrickson PG, Dorais JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet. 2017;49(6):925–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Whiddon JL, Langford AT, Wong CJ, Zhong JW, Tapscott SJ. Conservation and innovation in the DUX4-family gene network. Nat Genet. 2017;49(6):935–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.De Iaco A, Planet E, Coluccio A, Verp S, Duc J, Trono D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet. 2017;49(6):941–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Das S, Chadwick BP. Influence of repressive histone and DNA methylation upon D4Z4 transcription in Non-Myogenic cells. PLoS ONE. 2016;11(7):e0160022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Gannon OM, Merida de Long L, Saunders NA. DUX4 is derepressed in Late-Differentiating keratinocytes in conjunction with loss of H3K9me3 epigenetic repression. J Invest Dermatol. 2016;136(6):1299–302. [DOI] [PubMed] [Google Scholar]
- 217.Bosnakovski D, Gearhart MD, Choi SH, Kyba M. Dux facilitates post-implantation development, but is not essential for zygotic genome activation. Biol Reprod. 2021;104:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Chen Z, Zhang Y. Loss of DUX causes minor defects in zygotic genome activation and is compatible with mouse development. Nat Genet. 2019;51(6):947–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Guo Y, Kitano T, Inoue K, Murano K, Hirose M, Li TD, Sakashita A, Ishizu H, Ogonuki N, Matoba S et al. Obox4 promotes zygotic genome activation upon loss of Dux. eLife 2024, 13:e95856. [DOI] [PMC free article] [PubMed]
- 220.Ruebel ML, Vincent KA, Schall PZ, Wang K, Latham KE. SMCHD1 terminates the first embryonic genome activation event in mouse two-cell embryos and contributes to a transcriptionally repressive state. Am J Physiol Cell Physiol. 2019;317(4):C655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Percharde M, Lin CJ, Yin Y, Guan J, Peixoto GA, Bulut-Karslioglu A, Biechele S, Huang B, Shen X, Ramalho-Santos M. A LINE1-Nucleolin partnership regulates early development and ESC identity. Cell. 2018;174(2):391–405. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Vega-Sendino M, Luttmann FF, Olbrich T, Chen Y, Kuenne C, Stein P, Tillo D, Carey GI, Zhong J, Savy V, et al. The homeobox transcription factor DUXBL controls exit from totipotency. Nat Genet. 2024;56(4):697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Bosnakovski D, Toso EA, Ener ET, Gearhart MD, Yin L, Luttmann FF, Magli A, Shi K, Kim J, Aihara H, et al. Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein. iScience. 2023;26(10):107823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Jiang S, Williams K, Kong X, Zeng W, Nguyen NV, Ma X, Tawil R, Yokomori K, Mortazavi A. Single-nucleus RNA-seq identifies divergent populations of FSHD2 myotube nuclei. PLoS Genet. 2020;16(5):e1008754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Yasuda T, Tsuzuki S, Kawazu M, Hayakawa F, Kojima S, Ueno T, Imoto N, Kohsaka S, Kunita A, Doi K, et al. Recurrent DUX4 fusions in B cell acute lymphoblastic leukemia of adolescents and young adults. Nat Genet. 2016;48(5):569–74. [DOI] [PubMed] [Google Scholar]
- 226.Liu YF, Wang BY, Zhang WN, Huang JY, Li BS, Zhang M, Jiang L, Li JF, Wang MJ, Dai YJ, et al. Genomic profiling of adult and pediatric B-cell acute lymphoblastic leukemia. EBioMedicine. 2016;8:173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Lilljebjorn H, Henningsson R, Hyrenius-Wittsten A, Olsson L, Orsmark-Pietras C, von Palffy S, Askmyr M, Rissler M, Schrappe M, Cario G, et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun. 2016;7:11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zhang J, McCastlain K, Yoshihara H, Xu B, Chang Y, Churchman ML, Wu G, Li Y, Wei L, Iacobucci I, et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet. 2016;48(12):1481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Jones TI, Himeda CL, Perez DP, Jones PL. Large family cohorts of lymphoblastoid cells provide a new cellular model for investigating facioscapulohumeral muscular dystrophy. Neuromuscul Disord. 2017;27(3):221–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Banerji CRS, Panamarova M, Zammit PS. DUX4-expressing immortalised FSHD lymphoblastoid cells express genes elevated in FSHD muscle biopsies, correlating with the early stages of inflammation. Hum Mol Genet. 2020;29:2285–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Signorelli M, Mason AG, Mul K, Evangelista T, Mei H, Voermans N, Tapscott SJ, Tsonaka R, van Engelen BGM, van der Maarel SM, et al. Evaluation of blood gene expression levels in facioscapulohumeral muscular dystrophy patients. Sci Rep. 2020;10(1):17547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Smith AA, Nip Y, Bennett SR, Hamm DC, Lemmers R, van der Vliet PJ, Setty M, van der Maarel SM, Tapscott SJ. DUX4 expression in cancer induces a metastable early embryonic totipotent program. Cell Rep. 2023;42(9):113114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Chew GL, Campbell AE, De Neef E, Sutliff NA, Shadle SC, Tapscott SJ, Bradley RK. DUX4 suppresses MHC class I to promote cancer immune evasion and resistance to checkpoint blockade. Dev Cell. 2019;50(5):658–71.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Spens AE, Sutliff NA, Bennett SR, Campbell AE, Tapscott SJ. Human DUX4 and mouse dux interact with STAT1 and broadly inhibit interferon-stimulated gene induction. eLife 2023, 12:e82057. [DOI] [PMC free article] [PubMed]
- 235.Pineda JMB, Bradley RK. DUX4 is a common driver of immune evasion and immunotherapy failure in metastatic cancers. eLife 2024, 12:RP89017. [DOI] [PMC free article] [PubMed]
- 236.Friedel CC, Whisnant AW, Djakovic L, Rutkowski AJ, Friedl MS, Kluge M, Williamson JC, Sai S, Vidal RO, Sauer S et al. Dissecting herpes simplex virus 1-Induced host shutoff at the RNA level. J Virol 2021, 95(3):e01399-20. [DOI] [PMC free article] [PubMed]
- 237.Neugebauer E, Bastidas-Quintero AM, Weidl D, Full F. Pioneer factors in viral infection. Front Immunol. 2023;14:1286617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Tihaya MS, Mul K, Balog J, de Greef JC, Tapscott SJ, Tawil R, Statland JM, van der Maarel SM. Facioscapulohumeral muscular dystrophy: the road to targeted therapies. Nat Rev Neurol. 2023;19(2):91–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Teveroni E, Pellegrino M, Sacconi S, Calandra P, Cascino I, Farioli-Vecchioli S, et al. Estrogens enhance myoblast differentiation in facioscapulohumeral muscular dystrophy by antagonizing DUX4 activity. J Clin Invest. 2017;127(4):1531–45. [DOI] [PMC free article] [PubMed]
- 240.Joliot A, Prochiantz A. Unconventional secretion, gate to Homeoprotein intercellular transfer. Front Cell Dev Biology. 2022;10:926421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.