Abstract
Rhizobium sp. strain AC100, which is capable of degrading carbaryl (1-naphthyl-N-methylcarbamate), was isolated from soil treated with carbaryl. This bacterium hydrolyzed carbaryl to 1-naphthol and methylamine. Carbaryl hydrolase from the strain was purified to homogeneity, and its N-terminal sequence, molecular mass (82 kDa), and enzymatic properties were determined. The purified enzyme hydrolyzed 1-naphthyl acetate and 4-nitrophenyl acetate indicating that the enzyme is an esterase. We then cloned the carbaryl hydrolase gene (cehA) from the plasmid DNA of the strain and determined the nucleotide sequence of the 10-kb region containing cehA. No homologous sequences were found by a database homology search using the nucleotide and deduced amino acid sequences of the cehA gene. Six open reading frames including the cehA gene were found in the 10-kb region, and sequencing analysis shows that the cehA gene is flanked by two copies of insertion sequence-like sequence, suggesting that it makes part of a composite transposon.
Carbamate insecticides such as carbaryl (1-naphthyl N-methylcarbamate) are broad-spectrum insecticides that comprise the major proportion of agricultural pesticides used in today's agricultural industry. These compounds are considered hazardous because they potently inhibit acetylcholine esterase (9) and the N-nitrosocarbamates formed are potent mutagens (8). On the other hand, these pesticides generally do not persist in soil for a long time, and the persistence of these compounds in agricultural soil is due to repeated applications (10, 38). From these perspectives, an understanding of the degradation mechanism is needed to control the persistence of these pesticides in soil.
Soil microorganisms are thought to play a significant role in the reduction of pesticides in soil, and many soil bacteria capable of degrading carbamate pesticides have been isolated and characterized (4, 6, 16, 17, 21, 28). The biochemical characteristics of carbamate pesticide hydrolases have also been reported (5, 17, 18, 20, 25). However, little is known about the genes for these enzymes. The mcd gene, which encodes a carbofuran hydrolase, is located on a 100-kb plasmid called pPDL11, and it has been cloned from Achromobacter sp. strain WM111 (41). This gene was shown to be present in many bacteria and to be encoded on a 100-, 105-, 115-, or 124-kb plasmid found in diverse bacteria isolated from geographically distant areas (6, 29). However, the structure of the carbamate insecticide degradative gene has not as yet been reported, although the nucleotide sequence of mcd gene is available (accession no. AF160188).
The present study characterizes a carbaryl hydrolase purified from Rhizobium sp. strain AC100. From the result of a plasmid-curing experiment, it was suggested that the carbamate insecticide degradative gene is encoded on the plasmid. We then cloned the degradative gene from the plasmid DNA and determined the nucleotide sequence of the 10-kb region containing the degradative gene. The sequence analysis suggested that the genes in the sequenced region likely comprise a composite transposon.
MATERIALS AND METHODS
Isolation and culture conditions.
The minimal medium with carbaryl (MMC medium) contained the following constituents (in grams per liter of 20 mM potassium phosphate buffer [pH 7.0]): (NH4)2SO4, 1; NaCl, 0.2; MgSO4 · 7H2O, 0.1; CaCl2 · H2O, 0.05; FeSO4, 0.02; ZnSO4, 0.002; yeast extract, 0.05; and carbaryl, 0.1. Glucose (0.2%), Bacto Tryptone (0.4%), and yeast extract (0.2%) supplemented with MMC medium instead of carbaryl were used as MMGTY medium. Carbaryl-treated soil collected from agricultural fields with a history of carbaryl application was inoculated into MMC medium, and then carbaryl-degrading bacteria were enriched by repeating subcultivations. Appropriate dilutions of the enrichment culture were placed on MMC agar plates and incubated at 30°C for a few days. We selected a colony that hydrolyzed carbaryl to 1-naphthol by spraying the colony with 0.5 mM carbaryl and 0.01% fast blue B salt dissolved in sterilized distilled water (17). Colonies positive for carbaryl hydrolysis quickly turned brown. Spontaneous rifampin-resistant (Rifr) and streptomycin-resistant (Strr) mutants of strain AC100 were isolated by plating MMGTY medium-grown cells on 1/2 nutrient agar supplemented with 25 μg of rifampin or streptomycin per ml (16). Bacterial strains and plasmids used are shown in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| HB101 | supE44 hsdS20 (rB− mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | Takara, Osaka, Japan |
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) | |
| F′ [traD36 proAB+lacIqlacZΔM15] | Takara, Osaka, Japan | |
| Rhizobium sp. | ||
| AC100 | Wild type | This study |
| AC101 | Car− | This study |
| AC1012 | Car− Strr Rifr | This study |
| Plasmids | ||
| pRK2013 | Kmrtra+; triparental mating helper, ColE1 replication | 13 |
| pBI101 | Kmr; broad-host-range cloning vector | Clontech |
| pBluescript II SK(+) | Ampr; sequencing vector | Stratagene |
| pCEH-B | 23-kb BamHI fragment from pAC200 cloned into pBI101 | This study |
| pCEH-Sa | 6.2-kb SalI fragment from pAC200 cloned into pBI101 | This study |
| pCEH-Sm | 7.2-kb SmaI fragment from pAC200 cloned into pBI101 | This study |
| pCEH-EVB | 3.2-kb EcoRV-BamHI fragment from pCEH-B subcloned into pBI101 | This study |
| pCEH-EVSc | 2.7-kb EcoRV-SacII fragment from pCEH-B subcloned into pBI101 | This study |
Taxonomic identification.
The strain was identified on the basis of classification schemes published in Bergey's Manual of Systematic Bacteriology (19). The G+C content of the bacterial DNA was determined as described by Tamaoka and Komagata (39). The quinone type was detected by high-performance liquid chromatography (HPLC) using reference standards. The 16S rRNA gene was amplified by PCR, and the nucleotide sequences of the PCR products purified were determined as described (16). Computer analysis was performed using DDBJ software. The sequence of the 16S rRNA of the strain was compared with other published 16S rRNA sequences using the BLAST search option of the DDBJ database to determine the closest phylogenetic neighbors.
Carbaryl hydrolase activity.
Carbaryl hydrolase activity was assayed in a reaction mixture containing 0.5 mM carbaryl, 50 mM sodium phosphate buffer (pH 7.0), and enzyme solution in a total volume of 1.0 ml. The reaction was started by adding the enzyme and incubated at 30°C. The reaction was stopped by adding 100 μl of 2 mM HgCl2, and the amount of 1-naphthol formed was quantified as described (17). One unit of enzyme activity was defined as the amount of enzyme that produced 1 μmol of 1-naphthol per min at 30°C. In studies on the effects of pH and temperature, carbaryl hydrolase activity was assayed with metolcarb instead of carbaryl because carbaryl is unstable in alkaline solutions and at high temperatures. To examine pH stability, the enzyme was incubated in buffers at various pHs for 14 h at 4°C, and the remaining activity was determined at 30°C. For determination of the effects of metal ions and chemical reagents on the enzyme activity, the purified enzyme was preincubated with 1.0 mM of various compounds or metal ions in 1.0 ml of 50 mM sodium phosphate buffer (pH 7.0) for 20 min at 30°C, and the remaining activity was determined at 30°C.
Enzyme purification.
Shaking flasks containing 1/2 nutrient broth medium were inoculated with a late-logarithmic-stage culture in the same medium and incubated at 30°C on a reciprocal shaker. When the culture reached a late-logarithmic stage, the cells were harvested by centrifugation at 10,000 × g for 20 min at 4°C. The cell pellets were washed with 50 mM sodium phosphate buffer (pH 7.0). All operations were performed at 0 to 4°C, and sodium phosphate buffer (pH 7.0) (P buffer) was used unless otherwise mentioned. Cells resuspended in the same buffer were disrupted by sonication, and the cell debris was removed by centrifugation at 10,000 × g for 30 min. Protamine sulfate (2%) was added to the supernatant to a final concentration of 0.2 mg per mg of protein. After 15 min, the mixture was centrifuged at 10,000 × g for 30 min, and the precipitate was discarded. Solid ammonium sulfate was added to the supernatant to 25% saturation. After 30 min of stirring, the mixture was centrifuged at 10,000 × g for 15 min, and the precipitate was discarded. The supernatant was brought to 50% saturation with solid ammonium sulfate. After 30 min of stirring, the precipitate was collected by centrifugation at 10,000 × g for 30 min. The precipitate was dissolved in 50 mM P buffer and dialyzed against 20 mM Tris-HCl buffer (pH 8.0). The dialysate was applied to a column of Q Sepharose (prepacked HiTrap HP). The column was washed with 3 bed volumes of the same buffer and eluted with a linear gradient of 200 ml of NaCl (0 to 300 mM) in the same buffer. Active fractions were dialyzed against 20 mM P buffer (pH 7.0). The dialysate was applied to a column of SP Sepharose (prepacked HiTrap HP). The column was washed with 3 bed volumes of the same buffer and eluted with a linear gradient of 200 ml of NaCl (0 to 500 mM) in the same buffer. The active fractions were collected and concentrated to 6 ml using an ultrafiltration cell (model 8050; Millipore, Bedford, Mass.) with a membrane molecular weight cutoff of 10,000. The concentrated fractions were applied to a column of Sephacryl S-200 HR (26/60) with 50 mM P buffer (pH 7.0). All column chromatography was performed using ÄKTA explorer 10S (Pharmacia Biotech). The protein was eluted with the same buffer, and the active fractions were dialyzed against 50 mM sodium phosphate buffer (pH 7.0) containing 50% (vol/vol) glycerol. The enzyme solution was stored at −20°C.
Electrophoresis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described by Laemmli (22). The stacking and resolving gels contained 4.5 and 10% acrylamide, respectively. Proteins in the gel were stained with Coomassie brilliant blue R-250. Molecular mass standards for SDS-PAGE were as follows: myosin (205 kDa), β-galactosidase (116 kDa), phosphorylase b (97 kDa), fructose-6-phosphate kinase (84 kDa), bovine serum albumin (66 kDa), glutamic dehydrogenase (55 kDa), egg albumin (45 kDa), and glyceraldehyde-3-phosphate dehydrogenase (36 kDa).
Determination of substrate specificity and Km.
The reaction mixture consisted of 50 μmol of sodium phosphate buffer (pH 7.0), 0.5 μmol of substrate, and 0.01 U of the purified enzyme in a total volume of 1.0 ml. After an incubation at 30°C for 10 min, the reaction was stopped with 1.0 ml of methanol. The enzyme reaction was linear over time for at least 20 min. Substrate hydrolysis was measured by HPLC using an octyldecyl silane C18 column (ODS-UG-3 column; 4.6 by 35 mm; Nomura Kagaku Co., Aichi, Japan). The ability to hydrolyze 4-nitrophenyl acetate was measured by monitoring the A400. The Km values of carbaryl hydrolase were determined by Lineweaver-Burk plots. The reaction mixture consisted of 50 μmol of sodium phosphate buffer (pH 7.0), 10 to 200 nmol of substrate, and appropriate amounts of the purified enzyme in a total volume of 1.0 ml.
Analytical methods.
Protein concentrations were determined using the Bio-Rad DC protein assay kit with bovine serum albumin as the reference standard. Protein eluted during column chromatography was monitored by measuring the A280. Pesticides and 1-naphthyl acetate were analyzed by HPLC on a C18 column with a mobile phase of acetonitrile-distilled water (50:50 [vol/vol]) at a flow rate of 1.0 ml/min. The A220 of the column effluent was monitored. The analytical-grade pesticides carbaryl, xylylcarb (3,4-dimethylphenyl N-methylcarbamate), XMC (3,5-dimethylphenyl N-methylcarbamate), metolcarb (3-methylphenyl N-methylcarbamate), propoxur (2-isopropoxyphenyl N-methylcarbamate), fenobucarb (2-s-butylphenyl N-methylcarbamate), isoprocarb (2-isopropylphenyl N-methylcarbamate), methylparathion (O,O-dimethyl-O-4-nitrophenylthiophosphate), chloropropham (isopropyl-3-chlorocarbanilate), and carbofuran (2,3-dihydro-2,2-dimethyl-7-benzofuranyl methylcarbamate) and other chemicals were purchased from Wako Pure Chemical Industries Ltd. (Osaka, Japan). The prepacked columns—HiTrap Q Sepharose HP, HiTrap SP Sepharose HP, and HiPrep Sephacryl S-200 HR (26/60)—were obtained from Pharmacia Fine Chemicals, Ltd.
Plasmid preparation and curing.
Plasmids from strain AC100 and its derivative mutants were prepared by a modified alkaline-SDS extraction method (2, 15). For plasmid curing, single colonies of AC100 were inoculated into MMGTY medium containing mitomycin C (4 μg/ml) and shaken at 30°C overnight. A series of dilutions was made from the overnight culture and spread onto 1/2 nutrient agar plates. After several days of incubation at 30°C, mutants of AC100 unable to degrade carbaryl were selected by spraying the colony with 0.5 mM carbaryl and 0.01% fast blue B as described above.
Cloning of carbaryl degradative gene(s).
Plasmid DNA of strain AC100 was digested with BamHI, SalI, or SmaI, and the fragments were inserted into a broad-host-range plasmid pBI101 at the appropriate restriction enzyme sites, respectively. Escherichia coli HB101 transformed with the recombinant plasmids and strain AC1012 (Table 1) were used for triparental conjugation as recipient and donor, respectively. Triparental matings were performed as described by Ditta et al. (7). Donor, recipient, and helper (E. coli HB101 harboring pRK2013) were cultured for 16 h and then washed, mixed, and incubated for 16 h on 1/2 nutrient agar plates. After appropriate dilution, cells collected from the plates with sterile water were spread onto 1/2 nutrient agar plates containing rifampin and kanamycin. Carbaryl-degrading transconjugants were selected as described above.
DNA manipulation.
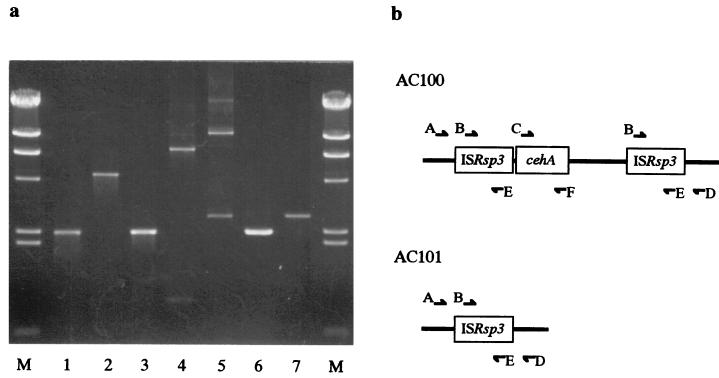
Digestion with restriction enzymes and DNA ligation proceeded basically as described by Sambrook et al. (33). DNA fragments were routinely subcloned in the plasmid vector pBluescript II SK(+). E. coli HB101 or JM109 was transformed either by the method of Hanahan (14) or by electroporation using an Electro Cell Manipulator 600 M (BTX, San Diego, Calif.). Total DNA from strain AC100 was prepared by the method of Marmur (23). Southern blot hybridization onto nylon membrane (Hybond-N+; Amersham, Buckinghamshire, United Kingdom) was performed by the capillary blot method (G capillary blotter; Taitec Co., Japan). Southern hybridization experiments were performed using a digoxigenin labeling kit (Boehringer, Mannheim, Germany). Overlapping restriction fragments of pCEH-B or pCEH-Sm were subcloned into pBluescript II SK(+) for sequencing. Nucleotides were sequenced using an automated laser fluorescence DNA sequencer (model 4000L; LI-COR). PCR amplification proceeded using Ex Taq DNA polymerase (Takara Biomedicals, Osaka, Japan) and the following primers: A, 5′-CGCGCGAATTCCAGCTTTGGCCAGACCTATTTCG-3′; B, 5′-ATGGACCTA-CTCAGTGTGATCCGCC-3′; C, 5′-CGCGGGAATTCATGGACCAACCATTCAAAC-CAGATCG-3′; D, 5′-CCCCCGAATTCAAGGCGCAGAAAATAGAAAGCGACC-3′; E, 5′-TCAAGCGGGTGATTTGTCCTTTCTG-3′; and F, 5′-GCCGCGAATTCACGTTAAGT-CGCTTTCGGCGATGATCC-3′. Plasmid DNA of strain AC100 and AC101 was used as templates, and the primer sets are indicated in Fig. 6.
FIG. 6.
PCR analyses of wild type and mutants. (a) Agarose gel electrophoresis of PCR products. Plasmid DNA from strain AC100 was template for lanes 1 to 5 and from AC101 for lanes 6 and 7. Primer sets for each lane were as follows: lanes 1 and 6, primer sets B and E; lane 2, primer sets B and F; lane 3, primer sets C and F; lane 4, primer sets C and E; lanes 5 and 7, primer sets A and D. (b) Scheme of primer annealing sites in sequenced region. Nucleotide sequences of primers are described in Materials and Methods. Arrows show primer annealing sites.
Nucleotide sequence accession numbers.
The nucleotide and amino acid sequence data reported in this study have been submitted to the nucleotide sequence databases under accession numbers AB069723 (Tnceh) and AB069724 (16S ribosomal DNA [rDNA]).
RESULTS
Isolation and identification.
The enrichment procedure generated a pure culture designated AC100 that hydrolyzed carbaryl to 1-naphthol. The following taxonomic properties of AC100 were determined: cell shape, motile straight rods or pleomorphic rods with a single polar flagellum and dimensions of 0.5 to 0.8 μm in length and 0.6 to 2.0 μm in width; gram stain negative; oxidase and catalase positive; nitrate reduction positive; urease negative; G+C content, 56.5 mol% ± 2.0 mol%; major quinone type, ubiquinone Q10. About 1,482 bases of 16S rRNA of AC100 were determined. A phylogenetic tree was constructed from evolutionary distances by the neighbor-joining method (data not shown). After phylogenetic analysis, strain AC100 was placed in a cluster making up the genus Rhizobium. The highest degree of similarity was 98%, which was obtained with the 16S rRNA gene of Rhizobium leguminosarum (DDBJ accession no. D14513) and Rhizobium mongolense (DDBJ accession no. U89820). Based on these observations, the isolate was designated Rhizobium sp. strain AC100.
Purification and properties of carbaryl hydrolase.
The purification scheme of the carbaryl hydrolase of AC100 is shown in Table 2. The enzyme was purified about 614-fold, with a yield of 27% from the cell extract of AC100. The purified enzyme preparation migrated as a single band on SDS-PAGE. The molecular mass of the purified enzyme estimated by SDS-PAGE was 82,000 Da. The estimated molecular mass of the native protein was 160,000 Da according to gel filtration on HiPrep Sephacryl S-200 HR (26/60), suggesting that the enzyme was a dimer composed of identical subunits. The optimal pH for carbaryl hydrolase activity was around 9 at 30°C, although the range was broad as it retained over 80% of its maximal activity between pH values of 6 to 11. The hydrolase was stable within a pH range of 10 to 11. The optimum temperature for the enzymatic reaction was about 45°C under the conditions described in Materials and Methods. The enzyme was stable for 10 min at temperatures up to 30°C, and 50% of the initial activity was retained at 50°C. The purified enzyme was incubated with a 1.0 mM concentration of various compounds or metal ions in 1.0 ml of 50 mM potassium phosphate buffer (pH 7.0) for 20 min at 30°C. Thereafter, the remaining activity was measured as described in Materials and Methods. The enzyme activity was completely inhibited by Hg2+, whereas iodoacetamide, diisopropylfluorophosphate, and paraoxon had no effect on the enzyme activity. The enzyme was incubated with various concentrations of EDTA (0.1 to 10 mM) for 30 min at 30°C. The enzyme activity was not affected by EDTA, suggesting the absence of a metal ion requirement. The Km value obtained for carbaryl as substrate was 62 μM. The substrate specificities of the purified carbaryl hydrolase are shown in Table 3. The specificity of the hydrolyzing activity was broad with substrates of N-methylcarbamate insecticide. The enzyme was less reactive with propoxur, fenobucarb, and isoprocarb. Hydrolase activity was undetectable with the substrates of carbofuran and chloropropham. The purified enzyme hydrolyzed 1-naphthyl acetate and 4-nitrophenyl acetate, indicating that the carbaryl hydrolase is an esterase.
TABLE 2.
Purification of the carbaryl hydrolase
| Purification step | Total activity (U) | Total protein (mg) | Sp act (U/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Crude extract | 69.3 | 1,520 | 0.05 | 1.0 | 100 |
| Protamine treatment | 69.4 | 953 | 0.07 | 1.4 | 100 |
| Ammonium sulfate (25-50%) | 72.1 | 262 | 0.27 | 5.6 | 104 |
| Q Sepharose | 114 | 12.3 | 9.30 | 185 | 165 |
| SP Sepharose | 78.8 | 3.9 | 20.2 | 404 | 114 |
| Gel filtration | 18.4 | 0.6 | 30.7 | 614 | 27 |
TABLE 3.
Substrate specificity of carbaryl hydrolase
| Substrate | Relative activitya (%) |
|---|---|
| Carbaryl | 100 |
| XMC | 115 |
| Metolcarb | 54 |
| Xylylcarb | 61 |
| Propoxur | 9 |
| Isoprocarb | 21 |
| Fenobucarb | 11 |
| Carbofuran | ND |
| Chloropropham | ND |
Rates of hydrolysis are expressed as percentages of rate observed with carbaryl. ND, not detectable.
Plasmid curing.
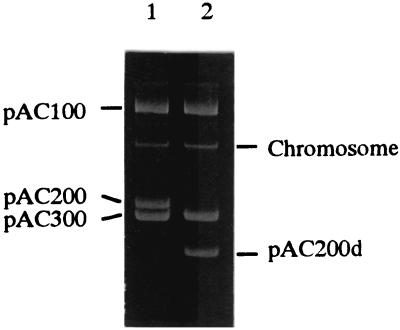
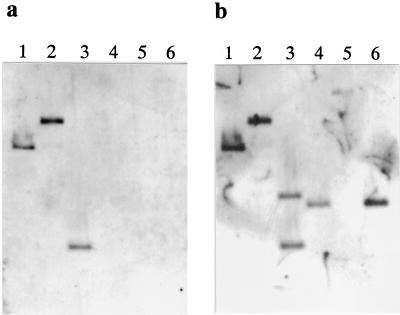
The involvement of naturally occurring plasmids in the degradation of synthetic organic compounds has been extensively documented (34). Catabolic plasmids are thought to play an important role in the evolution of pesticide-degrading ability in microorganisms (4, 34). We therefore examined the plasmid content of Rhizobium sp. strain AC100. The AC100 harbored three large plasmids, designated pAC100, pAC200 and pAC300 (Fig. 1, lane 1). To confirm that the carbaryl-degrading activity is controlled by these plasmids, we performed a curing experiment with mitomycin C. When AC100 was treated with mitomycin C, 0.4% of the cells lost carbaryl hydrolyzing activity. These mutants unable to hydrolyze carbaryl harbored a smaller plasmid of pAC200 which was termed pAC200d. The mutant was designated AC101 (Fig. 1, lane 2). These results suggested that the carbaryl hydrolase gene is encoded on pAC200.
FIG. 1.
Plasmid profiles of Rhizobium sp. strain AC100 and its mutant. Lanes: 1, strain AC100; 2, strain AC101.
Cloning and sequencing of carbaryl hydrolase gene.
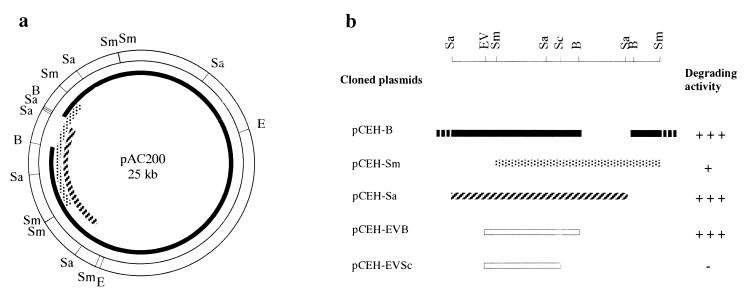
To clone the degradative gene, a library of genes in plasmid DNA from strain AC100 was constructed in E. coli HB101 by use of broad-host-range plasmid pBI101. The plasmid DNA library was mobilized from E. coli into Rhizobium sp. strain AC1012 by triparental mating. The transconjugants that complemented the carbaryl-degrading ability were selected by a fast blue solution spraying technique. Three positive clones that contained a 23-kb BamHI fragment, a 6.2-kb SalI fragment, and a 7.2-kb SmaI fragment of plasmid DNA from strain AC100 were further analyzed. The activity of the clone containing the SmaI fragment was very low. To determine the location of the degradative gene, 3.2-kb EcoRV-BamHI and 2.7-kb EcoRV-SacII fragments from pCEH-Sa were subcloned into pBI101, and the resulting plasmids were mobilized into AC1012. The activity of each transconjugant is shown in Fig. 2b. The results showed that the gene for degradation of carbaryl is located in the 3.2-kb EcoRV-BamHI fragment. On the other hand, a comparison of the restriction maps of the cloned fragments showed that the 23-kb large BamHI fragment contained most of the region of pAC200 and that the SmaI fragment included the remainder. Thus, we constructed a physical map of pAC200 (Fig. 2a).
FIG. 2.
(a) Restriction map of pAC200. Cloned regions are indicated as following: 23-kb BamHI fragment, solid line; 7.2-kb SmaI fragment, stippled line; 6.2-kb SalI fragment, striped line. (b) Scheme of a DNA region containing the cehA gene and its carbaryl-degrading activities. Abbreviations: B, BamHI; E, EcoRI; EV, EcoRV; Sa, SalI; Sc, SacII; Sm, SmaI.
The nucleotide sequence of the 3.2-kb EcoRV-BamHI fragment was determined on both strands. This region contained a large open reading frame (ORF) of 2,382-bp that encoded 794 amino acids. TTG was the estimated initiation codon of the ORF, and it was preceded by the putative ribosome-binding site AGGAAGG. The N-terminal amino acid sequence of purified carbaryl esterase (STDAIEPQPYFA) determined by automated Edman degradation was identical to that deduced from the nucleotide sequence starting from Ser-30, indicating that this gene encodes the carbaryl hydrolase. The gene was designated cehA. The amino acid sequence from the putative start codon to the N-terminal of the mature protein showed the characteristic features of a signal peptide. The calculated molecular weight of the mature protein deduced from the DNA sequence was 84,100, which was consistent with that of the purified protein estimated by SDS-PAGE. No significant homology sequences with cehA were found by a database homology search.
Nucleotide sequence of the cehA surrounding region.
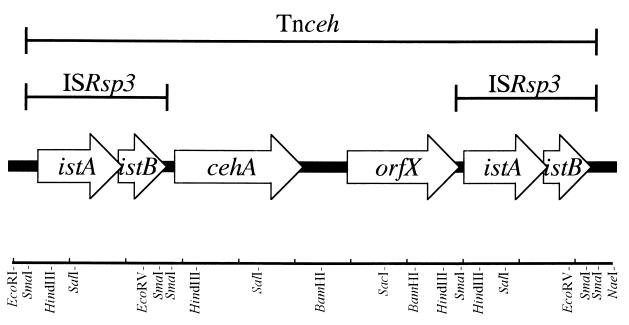
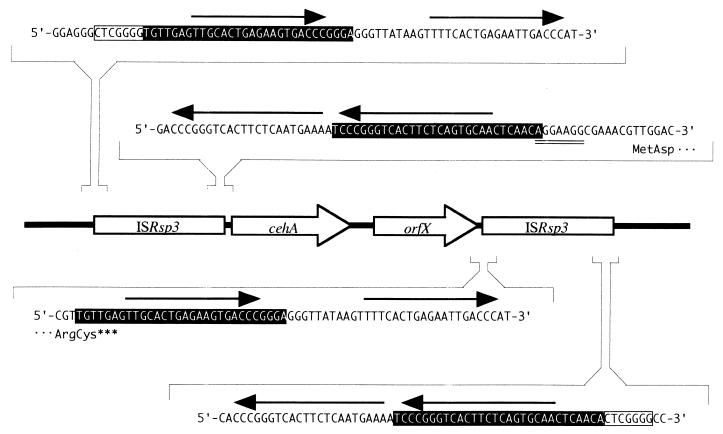
Since a part of another ORF located at upstream of cehA was highly homologous to istB of IS1600 from Alcaligenes eutrophus NH9 (26), the nucleotide sequence of the cehA surrounding region (10-kb EcoRI-NaeI fragment) was determined on both strands. It was found that the DNA fragment had six ORFs containing cehA (Fig. 3). The deduced amino acid sequence encoded by two ORFs located upstream of cehA gene showed significant homology to IstA and IstB of IS1600 from A. eutrophus NH9 and of IS1326 from Pseudomonas aeruginosa, which belong to the IS21 family (1, 26). The nucleotide sequence of the region containing IstA- and IstB-like sequences showed 67.7% homology with IS1600 and 60.5% with IS1326. In addition, a long terminal inverted repeat (IR) was found at both ends of the region (Fig. 4), indicating that the region had an insertion sequence (IS) of the IS21 family. Thus, the region and the two genes were designated ISRsp3, istA and istB, respectively. ISRsp3 was 2,490 bp long and was delimited by 30 bp of perfect IR, and parts of the terminal IR were directly repeated (arrows in Fig. 4). The two adjacent ORFs, istA and istB, overlapped by 8 bp. The IstA of ISRsp3 carried a putative helix-turn-helix motif in the N-terminal region and a motif related to the widespread integrase DDE motif but lacked the conserved K or R residue (35). IstB of ISRsp3 contained the ATP- or GTP-binding motif. ISRsp3 was adjacent to the cehA gene, and the putative ribosome-binding site and right terminal IR overlapped by 1 bp. We therefore presumed that the promoter of cehA is located in the ISRsp3 region and that the expression of cehA is affected by the insertion of ISRsp3.
FIG. 3.
Physical map of the locations of ORFs found in the sequenced region of pAC200. Open arrows indicate ORFs.
FIG. 4.
Characteristic nucleotide sequences at the ends of ISRsp3. IRs of ISRsp3 are shown on a black background. Arrows indicate direct repeats. The double-underlined region is the putative ribosome-binding site of the cehA gene. Boxed CTCGGGG sequences are target site duplicated sequences of Tnceh.
Further analysis revealed an identical sequence to ISRsp3 downstream of the cehA gene (Fig. 3). Although no target site duplication was found at either end of the ISRsp3s, a 7-bp direct repeat (CTCGGGG) was found at both ends of the 10-kb region containing ISRsp3s and cehA (Fig. 4). Therefore, the genes in the 10-kb region had a transposon-like structure that was designated Tnceh. The target site direct repeat at both ends of Tnceh suggested that Tnceh is transposed into a precursor plasmid of pAC200.
An ORF of considerable length was found in a 3-kb region between cehA and lower ISRsp3, and it was designated orfX. The deduced amino acid sequence of orfX showed homology to the putative salicin receptor SalC (27.2%) from Azospirillum irakense (36), ORF1201 (25.5%) from Sphingomonas aromaticivorans (32), and the putative TonB-dependent receptor SftP (23.5%) from P. aeruginosa (37). The amino acid sequences exhibited all the characteristics of the FepA/FhuA family outer membrane receptor that are implicated in the transport of several molecules, such as siderophores or vitamin B12. However OrfX seemed to be nonfunctional, since the result of the alignment analysis of OrfX and its homologous proteins revealed that OrfX (663 amino acid) was suddenly interrupted by lower ISRsp3 and it was 50 amino acids shorter at the end of the C-terminal region than the homologous proteins.
Southern hybridization and PCR analysis.
The genetic structure of the Tnceh region was examined by Southern blot hybridization and PCR analysis. Total DNA from strain AC100 and AC101 was used for Southern hybridization (Fig. 5). Labeled internal fragments of the cehA gene and ISRsp3 were used as probes to examine total DNA digested with EcoRI and/or BamHI. The sizes of all hybridized fragments from AC100 were consistent with sizes estimated from the nucleotide sequence (Fig. 5, lanes 1 to 3). On the other hand, AC101 did not hybridize with the cehA probe and signals from the hybridized ISRsp3 probe indicated that the EcoRI-digested DNA fragment was truncated and that the BamHI site in pAC200 had disappeared (Fig. 5, lanes 4 to 6). We analyzed the PCR products amplified using some primer sets (Fig. 6). Although a 2.8-kb fragment was coamplified with the entire transposon region (Fig. 6, lane 5), the lengths of all amplified products from strain AC100 agreed with the sizes estimated from the nucleotide sequence. Therefore, the genetic structure of Tnceh was further supported by the PCR results. On the other hand, PCR analyses of carbaryl-nondegrading mutant AC101 were carried out to disclose changes in the mutant. A 2.8-kb fragment was amplified from strain AC101 using primers that anneal to the flanking regions of Tnceh (primers A and D). The results of restriction fragment length polymorphism analysis of the fragment revealed that the fragment contained an ISRsp3. Thus, the catabolic region was deleted due to homologous recombination between the two ISRsp3s in strain AC101 during the curing procedure. This finding was consistent with those of Southern hybridization. Furthermore, restriction fragment length polymorphism analysis showed that the 2.8-kb fragment from AC100 that coamplified with the entire transposon region was identical to the ISRsp3 containing the 2.8-kb fragment amplified from AC101. This indicated that the deletion event occurred spontaneously while AC100 was in culture.
FIG. 5.
Southern blot hybridization of total DNA of strain AC100 and AC101 digested with EcoRI and/or BamHI. Hybridization proceeded overnight at 50°C. Lanes: 1 to 3, total DNA of AC100 digested with restriction enzymes (lane 1, EcoRI; lane 2, BamHI; lane 3, EcoRI and BamHI); 4 to 6, total DNA of AC101 digested with restriction enzymes (lane 4, EcoRI; lane 5, BamHI; lane 6, EcoRI and BamHI). (a) Labeled HindIII-SalI fragment containing internal sequence of the cehA gene was the probe. (b) Labeled PstI-SalI fragment containing internal sequence of upper IS Rsp3 was the probe.
DISCUSSION
We analyzed the properties of carbaryl hydrolase and its gene from Rhizobium sp. strain AC100. Some reports have revealed that genes encoding carbamate pesticide hydrolases are located in plasmids (11, 16, 28, 29, 41). The present study constructed the restriction map of a carbaryl degradative plasmid (pAC200). In addition, this is the first report to our knowledge on the nucleotide sequence of a gene encoding an enzyme that hydrolyzes N-methylcarbamate insecticides, although only one sequence was submitted to DNA databases. The submitted mcd gene from Achromobacter sp. strain WM111 encodes an enzyme that hydrolyzes carbofuran, which is a kind of carbamate insecticide (database accession no. AF160188). No homologous sequences were found by database homology search using nucleotide and deduced amino acid sequences of the mcd gene. Those of the cehA gene also had no homologous sequences, and no significant homology was apparent between cehA and mcd in either nucleotide and amino acid sequences. Therefore, the two carbamate hydrolyzing enzymes are not evolutionarily related, so cehA seems to be a novel gene. On the other hand, the enzymatic properties, except for substrate specificities, of the carbaryl hydrolase from strain AC100 were very similar to those of Blastobacter sp. strain M501 (17). Furthermore, 12 amino acid residues of the N-terminal sequences of both enzymes were identical (unpublished data).
The results of the ORF analysis of the cehA gene did not reveal a start ATG codon, and two potential start codons, both of which were TTG, were found in the frame of cehA gene. The distances between the TTG codons and the codon of the N-terminal amino acid of the mature protein were 87 and 36 bp, respectively. Since the putative amino acid sequence from the upper TTG codon to the N-terminal region of the mature protein displayed the common feature of signal peptide, the upper was predicted as a start codon. A putative ribosome-binding sequence, AGGAAGG, was then found at 7 nucleotides before the start codon. The distance between the IR of ISRsp3 and the initiation codon of the cehA gene was 13 bp, and the putative ribosome-binding site and the IR overlapped by 1 bp. These results suggested that the promoter of the cehA gene is located in the ISRsp3 region and that the transcription of cehA gene is activated by the insertion of IS. Since ISRsp3 is highly homologous to IS1600 of the IS21 family, ISRsp3 was also classified into the family. IS21 was found in the R68 plasmid of P. aeruginosa and transcriptional activations by IS21 and other ISs belonging to the IS21 family have been suggested (24, 30, 31). The nucleotide sequence and/or multiple repeated structure of terminal IRs of the IS21 family seem to be involved in the transcriptional activation. Ogawa and Miyashita suggested that IstAs of IS1600 and homologous ISs are clustered on the phylogenetic tree of the IstAs of the IS21 family (27). The cluster included IS1600 from A. eutrophus, orfSA from Pseudomonas sp. strain P51, IS1326 from P. aeruginosa, and nmoT from Chelatobacter heintzii (1, 42). C. heintzii and Rhizobium sp. strain AC100 are allied species that belong to the α-subdivision of the Proteobacteria, but ISRsp3 is most homologous to IS1600 of A. eutrophus, which belongs to the β-subdivision. The clustered ISs were identified from antibiotic resistant and xenobiotic degrading bacteria. However, no relationships were shown among the degradative genes, the substrate compounds and the bacterial species. This indicated that the ISs are involved in acquiring the effective ability to degrade various foreign organic hydrocarbons.
The carbofuran-degrading bacterium Sphingomonas sp. strain CF06 contains five plasmids, at least some of which are required for the metabolism of carbofuran (11). Observations of rearrangements, deletions, and the loss of individual plasmids resulting in the loss of carbofuran-degrading ability suggested the involvement of mobile DNA elements such as transposons or ISs. However, the relationship between the carbofuran degradative gene and the mobile elements has not been clarified. On the other hand, the 10-kb region in Rhizobium sp. strain AC100 has likely been transposed since the 7 bp of duplicate sequences of the target site were found at both ends of Tnceh. Besides, the partial sequence of the other region of pAC200 was homologous to the traA gene, which was concerned with conjugation (data not shown). Many xenobiotic degradative genes are located in transposons and plasmids (40). These genes may have repeatedly transferred and evolved to adapt to the variety of substrates and to degrade them more effectively. Associations between plasmids or transposable elements and the diversity of carbamate-pesticide degradative genes have been predicted or observed (11, 12, 29). This agrees with the present study report that the carbaryl degradative gene cehA is located in a transposon on a plasmid. Desaint et al. observed the genetic diversity of 128 carbofuran-degrading bacteria isolated from English and French soils, using amplified DNA restriction analyses of the 16S rDNA and 16S-23S rDNA spacer region (6). These were classified into six widely spread clusters, of high- and low-G+C-content gram-positive bacteria; Cytophaga; and α-, β- and γ-Proteobacteria. Although 58 of the 128 studied strains harbor the mcd gene, there was no clear relationship between the presence of the gene and the phylogenetic position of the strain. On the other hand, Chapalamadugu and Chaudhry have noted that 15 bacteria isolated from North America soils do not contain sequences homologous to the mcd gene (3). Since the N-terminal sequences of carbaryl hydrolases from strain AC100 and M501, which were isolated from different sites in Japan, were identical, diverse carbaryl-degrading bacteria in Japanese soil are likely to contain the cehA homologous gene. The worldwide genetic diversity of carbamate degradative genes should be studied in more detail.
Environments contaminated with carbaryl are regarded as hazardous because the pesticide is considered to be an endocrine-disrupting chemical. Thus, a technique for rapid decomposition of the compound is required. The effectiveness of bioremediation can be measured by factors such as the survival of the introduced microorganism, the stability of the genes encoding the appropriate catalytic functions, and the degree of contaminant removal. The catabolic region of strain AC100 was spontaneously deleted without selective pressure. Further study of the gene stability and survivability of Rhizobium sp. strain AC100 in various environments is needed before the bacteria can be applied to the process of bioremediation.
Acknowledgments
This work was supported by a program for the promotion of basic research activities for innovative biosciences of the Bio-Oriented Technology Research Advancement Institution (Minatoku, Japan).
REFERENCES
- 1.Brown, H. J., H. W. Stokes, and R. M. Hall. 1996. The integrons In0, In2, and In5 are defective transposon derivatives. J. Bacteriol. 178:4429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casse, F., C. Boucher, J. S. Julliot, M. Michel, and J. Denarie. 1979. Identification and characterization of large plasmids in Rhizobium meliloti using agarose gel electrophoresis. J. Gen. Microbiol. 113:229-242. [Google Scholar]
- 3.Chapalamadugu, S., and G. R. Chaudhry. 1992. Microbiological and biotechnological aspects of metabolism of carbamates and organophosphates. Crit. Rev. Biotechnol. 12:357-389. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhry, G. R., and A. N. Ali. 1988. Bacterial metabolism of carbofuran. Appl. Environ. Microbiol. 54:1414-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derbyshire, M. K., J. S. Karns, P. C. Kearney, and J. O. Nelson. 1987. Purification and characterization of an N-methylcarbamate pesticide hydrolyzing enzyme. J. Agric. Food Chem. 35:871-877. [Google Scholar]
- 6.Desaint, S., A. Hartmann, N. R. Parekh, and J. Fournier. 2000. Genetic diversity of carbofuran-degrading soil bacteria. FEMS Microbiol. Ecol. 34:173-180. [DOI] [PubMed] [Google Scholar]
- 7.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elespuru, R., W. Lijinsky, and J. K. Setlow. 1974. Nitrosocarbaryl as a potent mutagen of environmental significance. Nature 247:386-387. [DOI] [PubMed] [Google Scholar]
- 9.Fahmy, M. A., T. R. Fukuto, R. O. Myers, and R. B. March. 1970. The selective toxicity of new N-phosphorothioyl-carbamate esters. J. Agric. Food Chem. 18:793-796. [DOI] [PubMed] [Google Scholar]
- 10.Felsot, A., J. V. Maddox, and W. Bruce. 1981. Enhanced microbial degradation of carbofuran in soils with histories of furadan use. Bull. Environ. Contam. Toxicol. 26:781-788. [DOI] [PubMed] [Google Scholar]
- 11.Feng, X., L. T. Ou, and A. Ogram. 1997. Plasmid-mediated mineralization of carbofuran by Sphingomonas sp. strain CF06. Appl. Environ. Microbiol. 63:1332-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng, X., L. T. Ou, and A. Ogram. 1997. Cloning and sequence analysis of a novel insertion element from plasmids harbored by the carbofuran-degrading bacterium, Sphingomonas sp. CFO6. Plasmid 37:169-179. [DOI] [PubMed] [Google Scholar]
- 13.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 15.Hansen, J. B., and R. H. Olsen. 1978. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J. Bacteriol. 135:227-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayatsu, M., M. Hirano, and T. Nagata. 1999. Involvement of two plasmids in the degradation of carbaryl by Arthrobacter sp. strain RC100. Appl. Environ. Microbiol. 65:1015-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayatsu, M., and T. Nagata. 1993. Purification and characterization of carbaryl hydrolase from Blastobacter sp. strain M501. Appl. Environ. Microbiol. 59:2121-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayatsu, M., A. Mizutani., M. Hashimoto, K. Sato, and K. Hayano. 2001. Purification and characterization of carbaryl hydrolase from Arthrobacter sp. RC100. FEMS Microbiol. Lett. 201:99-103. [DOI] [PubMed] [Google Scholar]
- 19.Jordan, D. C. 1986. Gram-negative aerobic rods and cocci, genus Rhizobium, p. 235-242. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins Co., Baltimore, Md. [Google Scholar]
- 20.Karns, J. S., and P. H. Tomasek. 1991. Carbofuran hydrolase purification and properties. J. Agric. Food Chem. 39:1004-1008. [Google Scholar]
- 21.Karpouzas, D. G., J. A. Morgan, and A. Walker. 2000. Isolation and characterization of 23 carbofuran-degrading bacteria from soils from distant geographical areas. Lett. Appl. Microbiol. 31:353-358. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 277:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Marmur, J. 1963. A procedure for the isolation of deoxyribonucleic acid from microorganisms. Methods Enzymol. 6:726-738. [Google Scholar]
- 24.Menou, G., J. Mahillon, M. M. Lecadet, and D. Lereclus. 1990. Structural and genetic organization of IS232, a new insertion sequence of Bacillus thuringiensis. J. Bacteriol. 172:6689-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulbry, W. W., and R. W. Eaton. 1991. Purification and characterization of the N-methylcarbamate hydrolase from Pseudomonas strain CRL-OK. Appl. Environ. Microbiol. 57:3679-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogawa, N., and K. Miyashita. 1995. Recombination of a 3-chlorobenzoate catabolic plasmid from Alcaligenes eutrophus NH9 mediated by direct repeat elements. Appl. Environ. Microbiol. 61:3788-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogawa, N., and K. Miyashita. 1999. The chlorocatechol-catabolic transposon Tn5707 of Alcaligenes eutrophus NH9, carrying a gene cluster highly homologous to that in the 1,2,4-trichlorobenzene-degrading bacterium Pseudomonas sp. strain P51, confers the ability to grow on 3-chlorobenzoate. Appl. Environ. Microbiol. 65:724-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogram, A. V., Y. P. Duan, S. L. Trabue, X. Feng, H. Castro, and L. T. Ou. 2000. Carbofuran degradation mediated by three related plasmid systems. FEMS Microbiol. Ecol. 32:197-203. [DOI] [PubMed] [Google Scholar]
- 29.Parekh, N. R., A. Hartmann, M. P. Charney, and J. C. Fournier. 1995. Diversity of carbofuran-degrading soil bacteria and detection of plasmid-encoded sequences homologous to the mcd gene. FEMS Microbiol. Ecol. 17:149-160. [Google Scholar]
- 30.Reimmann, C., R. Moore, S. Little, A. Savioz, N. S. Willetts, and D. Haas. 1989. Genetic structure, function and regulation of the transposable element IS21. Mol. Gen. Genet. 215:416-424. [DOI] [PubMed] [Google Scholar]
- 31.Rogers, M. B., T. K. Bennett, C. M. Payne, and C. J. Smith. 1994. Insertional activation of cepA leads to high-level β-lactamase expression in Bacteroides fragilis clinical isolates. J. Bacteriol. 176:4376-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romine, M. F., L. C. Stillwell, K. K. Wong, S. J. Thurston, E. C. Sisk, C. Sensen, T. Gaasterland, J. K. Fredrickson, and J. D. Saffer. 1999. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 181:1585-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Sayler, G. S., S. W. Hooper, A. C. Layton, and J. M. Henry King. 1990. Catabolic plasmids of environmental and ecological significance. Microb. Ecol. 19:1-20. [DOI] [PubMed] [Google Scholar]
- 35.Solinas, F., A. M. Marconi, M. Ruzzi, and E. Zennaro. 1995. Characterization and sequence of a novel insertion sequence, IS1162, from Pseudomonas fluorescens. Gene 155:77-82. [DOI] [PubMed] [Google Scholar]
- 36.Somers, E., V. Keijers, D. Ptacek, M. Halvorsen Ottoy, M. Srinivasan, J. Vanderleyden, and D. Faure. 2000. The salCAB operon of Azospirillum irakense, required for growth on salicin, is repressed by SalR, a transcriptional regulator that belongs to the Lacl/GalR family. Mol. Gen. Genet. 263:1038-1046. [DOI] [PubMed] [Google Scholar]
- 37.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 38.Tal, A., B. Rubbin, and J. Katan. 1989. Accelerated degradation of thiocarbamate herbicides in Israeli soils following repeated use of vernolate. Pestic. Sci. 25:343-353. [Google Scholar]
- 39.Tamaoka, J., and K. Komagata. 1984. Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol. Lett. 25:125-128. [Google Scholar]
- 40.Tan, H. M. 1999. Bacterial catabolic transposons. Appl. Microbiol. Biotechnol. 51:1-12. [DOI] [PubMed] [Google Scholar]
- 41.Tomasek, P. H., and J. S. Karns. 1989. Cloning of a carbofuran hydrolase gene from Achromobacter sp. strain WM111 and its expression in gram-negative bacteria. J. Bacteriol. 171:4038-4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu, Y., M. W. Mortimer, T. S. Fisher, M. L. Kahn, F. J. Brockman, and L. Xun. 1997. Cloning, sequencing, and analysis of a gene cluster from Chelatobacter heintzii ATCC 29600 encoding nitrilotriacetate monooxygenase and NADH: flavin mononucleotide oxidoreductase. J. Bacteriol. 179:1112-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]